On-demand computational infrastructure, such as that provided by Amazon Web Services (AWS), is ideal for at-scale parallelizable workflows (especially workflows for which demand is not constant but comes in occasional spikes), as neither computing power nor data storage are limited by local availability and costs are limited to actual resource usage. However, cloud infrastructure configuration is time-consuming and confusing, and cloud-native services that automatically monitor and scale resources can increase the workflow price. Distributed-Something (DS) is a collection of easy-to-use Python scripts that leverage the power of the former while minimizing the problems of the latter.
DS makes it possible for a developer with moderate computational comfort to design a way to deploy a new tool or program to the cloud, and for a user with relatively low computational comfort to then deploy this tool at will. It simplifies the process of using AWS by scripting the majority of the setup, triggering and monitoring of jobs, requiring only minimal human-readable config files to be edited before the run and simple, single-line commands to trigger each step. This increases the ability of novice computational users to be able to execute workflows and lowers the barrier to creating a new workflow for developers. Unlike most existing tools for running hosted containerized analyses, it does not require learning new workflow languages (either for new tool addition or end-user deployment) and minimizes the understanding requirement of the AWS components that are used (see Supplementary Table 1).
We originally sought to simplify large-scale scientific image analysis using our CellProfiler software, creating Distributed-CellProfiler1. Recognizing the utility of the framework, we herein provide Distributed-Something as a fully customizable template for the distribution of any Dockerized2 workflow. We show its extensibility with two example implementations of DS in the open-source bioimage ecosystem (though DS is in no way limited to bioimage analysis).
ImageJ is the most widely used open-source software for bioimage analysis3. Fiji is an open-source distribution of ImageJ that comes bundled with libraries and plug-ins that extend ImageJ’s functionality4. Fiji scripts can be run at scale using Distributed-Fiji, allowing the user to take advantage of its plug-in ecosystem and its ability to run user-written scripts in many coding languages. As with all DS implementations, the computational environment can be tailored to each task — for example, many small machines can be used to individually process thousands of files or a large machine can be use to perform a single task (such as stitching) on many files.
To increase shareability of especially large bioimage data, the Open Microscopy Environment5 team is creating next-generation file formats, including .ome.zarr6, to make bioimaging data more findable, accessible, interoperable and reusable (FAIR)7. We created Distributed-OMEZarrCreator to simplify the conversion of large bioimage datasets to .ome.zarr’s and thus encourage the adoption of this format and simplify sharing of bioimaging data via resources such as the Image Data Resource8.
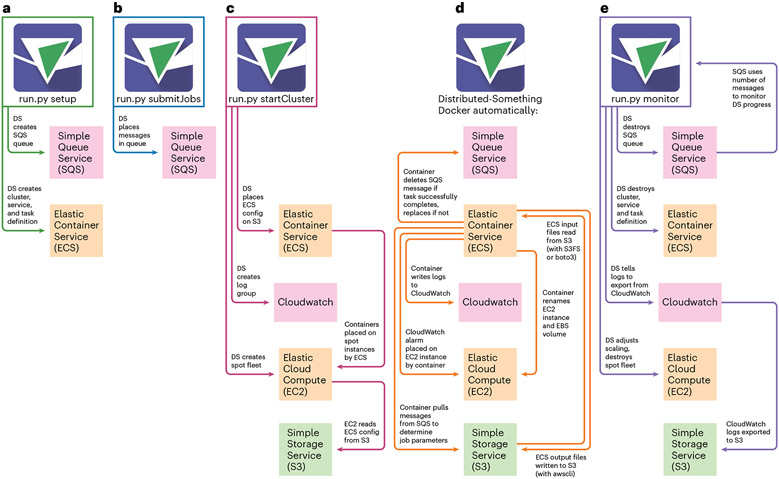
DS coordinates five separate AWS resources. Data are stored on AWS in its Simple Storage Service (S3), and ‘spot fleets’ of Elastic Compute Cloud (EC2) instances (or virtual computers) access that data, run the ‘Something’ on that data, and upload the end product back to S3. Elastic Container Services (ECS) places your customized Docker containers on the EC2 machines while Simple Queue Service (SQS) tracks the list of jobs, and Cloudwatch provides logs and metrics on the services you are using, allowing configuration, optimization and troubleshooting. A developer can easily customize DS code to download data from or upload data to the cloud and/or on-premises storage outside the AWS account used for processing.
DS stands out in the simplicity of user execution. Only two human-readable files must be edited to configure individual DS runs: the Config file and the Job file. The Config file contains information about naming, the number and size of machines to use, and the maximum price you are willing to pay for the machines, minimizing computational costs. The Job file lists all of the individual tasks to run in parallel by setting both the metadata shared between tasks and the metadata to parse individual tasks. An additional Fleet file contains information about AWS-account-specific information but does not need to be edited after initial creation.
Three single-line Python commands initiate all of the AWS architecture creation and coordination, and an optional fourth command provides additional monitoring and automated clean-up of resources (Fig. 1). Implementing a new version of DS can be done in a matter of hours by a developer with moderate Python abilities. Creating a Distributed version of software that itself takes input scripts (for example, Distributed-Fiji) makes workflow customization possibilities near limitless with no extra overhead. Over 1,000 containers are already registered on BioContainers9, and, conceptually, any could be put in the DS framework.
Fig. 1 ∣. Distributed-Something.
Distributed-Something uses four single-line commands to coordinate five separate AWS services for the parallel processing of jobs by any Dockerized software. a–c, Three execution commands prepare various aspects of AWS infrastructure: ‘setup’ (a) prepares SQS and ECS, ‘submitJobs’ (b) sends jobs to SQS, and ‘startCluster’ (c) initiates and coordinates the spot fleet request. d, After these commands, the Distributed-Something Docker automatically completes setup and jobs run. e, A fourth, optional command, ‘monitor’, assists in downscaling and cleaning up resources as they are no longer required. See documentation at https://distributedscience.github.io/Distributed-Something for a deeper discussion of each step.
We believe DS will enable the scientific community to quickly, easily and cost-effectively scale their parallelizable workflows using AWS. As this is an open-source tool, we look forward to contributions and implementations from within and outside the bioimage analysis community.
Supplementary Material
Acknowledgements
We thank Juan Caicedo and Shantanu Singh for creating the original Distributed-CellProfiler, Callum Tromans-Coia, Josh Moore and Sebastien Besson for their assistance with DOZC, and other members of the Cimini and Carpenter-Singh labs for their feedback on this project and manuscript. This study was supported by Calico Life Sciences LLC, NIH P41 GM135019, and grant 2020-225720 from the Chan Zuckerberg Initiative DAF. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Code availability
Distributed-Something is available at https://github.com/DistributedScience/Distributed-Something and https://doi.org/10.5281/zenodo.7949283. Distributed-CellProfiler is available at https://github.com/DistributedScience/Distributed-CellProfiler and https://doi.org/10.5281/zenodo.7949380. Distributed-Fiji is available at https://github.com/DistributedScience/Distributed-Fiji and https://doi.org/10.5281/zenodo.7949387. Distributed-OMEZarrCreator is available at https://github.com/DistributedScience/Distributed-OMEZarrCreator and https://doi.org/10.5281/zenodo.7949394.
Competing interests
The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-023-01918-8.
References
- 1.McQuin C. et al. PLoS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merkel D. Linux J. 2, 2014 (2014). [Google Scholar]
- 3.Rueden CT et al. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindelin J. et al. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swedlow JR, Goldberg I, Brauner E & Sorger PK Science 300, 100–102 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore J. et al. Nat. Methods 18, 1496–1498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson MD et al. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams E. et al. Nat. Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Veiga Leprevost F. et al. Bioinformatics 33, 2580–2582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.