Abstract
We examined the response of Streptococcus pneumoniae 7785 to clinafloxacin, a novel C-8-substituted fluoroquinolone which is being developed as an antipneumococcal agent. Clinafloxacin was highly active against S. pneumoniae 7785 (MIC, 0.125 μg/ml), and neither gyrA nor parC quinolone resistance mutations alone had much effect on this activity. A combination of both mutations was needed to register resistance, suggesting that both gyrase and topoisomerase IV are clinafloxacin targets in vivo. The sparfloxacin and ciprofloxacin MICs for the parC-gyrA mutants were 16 to 32 and 32 to 64 μg/ml, respectively, but the clinafloxacin MIC was 1 μg/ml, i.e., within clinafloxacin levels achievable in human serum. S. pneumoniae 7785 mutants could be selected stepwise with clinafloxacin at a low frequency, yielding first-, second-, third-, and fourth-step mutants for which clinafloxacin MICs were 0.25, 1, 6, and 32 to 64 μg/ml, respectively. Thus, high-level resistance to clinafloxacin required four steps. Characterization of the quinolone resistance-determining regions of the gyrA, parC, gyrB, and parE genes by PCR, HinfI restriction fragment length polymorphism, and DNA sequence analysis revealed an invariant resistance pathway involving sequential mutations in gyrA or gyrB, in parC, in gyrA, and finally in parC or parE. No evidence was found for other resistance mechanisms. The gyrA mutations in first- and third-step mutants altered GyrA hot spots Ser-83 to Phe or Tyr (Escherichia coli coordinates) and Glu-87 to Gln or Lys; second- and fourth-step parC mutations changed equivalent hot spots Ser-79 to Phe or Tyr and Asp-83 to Ala. gyrB and parE changes produced novel alterations of GyrB Glu-474 to Lys and of Pro-454 to Ser in the ParE PLRGK motif. Difficulty in selecting first-step gyrase mutants (isolated with 0.125 [but not 0.25] μg of clinafloxacin per ml at a frequency of 5.0 × 10−10 to 8.5 × 10−10) accompanied by the small (twofold) MIC increase suggested only a modest drug preference for gyrase. Given the susceptibility of defined gyrA or parC mutants, the results suggested that clinafloxacin displays comparable if unequal targeting of gyrase and topoisomerase IV. Dual targeting and the intrinsic potency of clinafloxacin against S. pneumoniae and its first- and second-step mutants are desirable features in limiting the emergence of bacterial resistance.
Streptococcus pneumoniae is an important human pathogen. It is the main cause of community-acquired pneumonia and is frequently involved in exacerbations of chronic bronchitis and in meningitis, acute otitis media, and sinusitis (3). Treatment of S. pneumoniae infections relies heavily on antimicrobial therapy with penicillin or other beta-lactams. Over the past two decades, the emergence and, in some areas, the prevalence of pneumococci with decreased susceptibility to penicillins have emphasized the need for new therapeutic agents and have focused attention on the fluoroquinolones (14, 25, 34). However, ciprofloxacin, the main quinolone in current clinical use, has modest activity against gram-positive bacteria such as S. pneumoniae. Consequently, it has had relatively little impact on the treatment of respiratory tract infections.
Clinafloxacin (AM-1091, CI-960, and PD127391) is a novel fluoroquinolone with potent broad-spectrum in vitro activity against gram-positive, gram-negative, and anaerobic pathogens (reviewed in reference 18). The drug has a structure somewhat different from that of ciprofloxacin—notably, the presence of a chlorine C-8 substituent—and is much more active than ciprofloxacin against gram-positive species, including S. pneumoniae. Clinafloxacin has been identified as the most active fluoroquinolone against S. pneumoniae when compared with ofloxacin, levofloxacin, sparfloxacin, grepafloxacin, and trovafloxacin (22a) and is currently being evaluated as an antipneumococcal agent. However, its mechanism of action has yet to be examined in detail.
Previous studies showed that quinolones inhibit DNA gyrase and DNA topoisomerase IV (reviewed in reference 6). Both enzymes act by a double-strand DNA break mechanism and are essential for bacterial growth (19, 35). They cooperate in DNA replication to facilitate DNA unlinking and chromosome segregation (41). Gyrase, an A2B2 tetramer encoded by the gyrA and gyrB genes, catalyzes negative DNA supercoiling (10, 35) and is thought to act ahead of the replication fork, neutralizing positive supercoils arising from DNA unwinding and thereby promoting fork movement (41). Topoisomerase IV, a C2E2 complex specified by the parC and parE genes, allows the segregation of daughter chromosomes at cell division (1, 17, 41). Point mutations in discrete regions of the gyrase and topoisomerase IV genes—the quinolone resistance-determining regions (QRDRs)—are responsible for the development of resistance (22, 39, 40). Ciprofloxacin resistance in Staphylococcus aureus and S. pneumoniae arises through mutation of the parC or parE genes before changes in the gyrase genes take place, suggesting that topoisomerase IV is the primary ciprofloxacin target and that gyrase is the secondary target in these gram-positive species (7, 15, 21, 24, 26, 27, 33). Interestingly, in Escherichia coli and other gram-negative bacteria, gyrase is the primary target (5, 6, 9, 13, 22), initially suggesting that there could be fundamental differences in drug responses within the bacterial kingdom. However, in recent work, we have shown that whereas ciprofloxacin targets topoisomerase IV in S. pneumoniae, sparfloxacin targets gyrase, indicating that the molecular structure of the quinolone determines the target preference (28). We and others have proposed that a quinolone acting equally through gyrase and topoisomerase IV would be desirable, as the onset of resistance would require selection for two mutations, which would be a rare event (24, 26). It is not known which if any of the new antipneumococcal fluoroquinolones exhibits this property.
Given the potency of clinafloxacin against S. pneumoniae, we have sought to determine its mechanism of action in this pathogen. We have studied the activity of clinafloxacin against representative S. pneumoniae mutants with characterized mutations in topoisomerase genes. We also report a detailed analysis of the gyrA, parC, gyrB, and parE QRDRs of S. pneumoniae mutants selected stepwise for resistance to clinafloxacin.
MATERIALS AND METHODS
Bacterial strains.
S. pneumoniae 7785, a quinolone-susceptible clinical isolate, has been described previously (27). Characterization of mutants of strain 7785 selected in vitro for resistance to ciprofloxacin (1C1, 2C6, 2C7, and 3C4) or to sparfloxacin (1S1, 1S4, 2S1, and 2S4) also has been reported (26, 28).
Drug susceptibilities.
Clinafloxacin hydrochloride was from Parke-Davis, Ann Arbor, Mich. A stock solution was made up in water and stored at −70°C. Ciprofloxacin was provided by Bayer U.K., Newbury, United Kingdom. Sparfloxacin was obtained from Dainippon Pharmaceutical Co., Suita, Japan. MICs were determined by the twofold dilution method with brain heart infusion medium supplemented with 10% horse blood. Approximately 105 CFU of bacteria was spotted on plates, which were examined after overnight aerobic incubation at 37°C.
Bacterial growth rates.
Strains 7785 and 4CLN9 were taken from plates, diluted to 50 to 100 CFU/ml in T broth (Sanofi Diagnostics Pasteur), and grown aerobically on an orbital shaker at 37°C. At time zero and at 1-h intervals over a 7-h period, 100-μl samples of the culture were removed and, after appropriate dilution, spread on brain heart infusion plates containing 10% horse blood. Plates were incubated overnight at 37°C for the determination of viable counts. Experiments were done in duplicate, and doubling times were obtained from semilog plots of CFU versus time.
Stepwise selection of clinafloxacin-resistant S. pneumoniae strains.
Mutants were selected by plating approximately 1011 CFU of strain 7785 or its drug-resistant derivatives on brain heart infusion plates containing 10% horse blood and clinafloxacin. Plates were incubated aerobically at 37°C for 24 to 48 h. Mutant frequencies were determined by comparing the number of colonies that grew on plates containing drug with the number of colonies obtained in the absence of drug. All procedures were as described previously (26, 28).
PCR and RFLP analysis.
Conditions for bacterial growth and the protocol for the isolation of genomic DNA were as described previously (27). PCR was used to amplify DNA from the QRDRs of the gyrase and topoisomerase IV genes of clinafloxacin-selected S. pneumoniae mutants. PCR conditions have been reported previously, as have the primers: VGA3 and VGA4 for gyrA, M0363 and M4721 for parC, H4025 and H4026 for gyrB, and S6398 and S6399 for parE (28). Restriction fragment length polymorphism (RFLP) analysis by HinfI digestion of the PCR products was carried out as reported previously (27).
AsPCR and DNA sequence analysis.
Asymmetric PCR (AsPCR) was used to generate single-stranded DNA for direct DNA sequencing by the chain termination method (32). AsPCR conditions, purification of AsPCR products, and DNA sequencing were as described previously (28).
RESULTS
Activity of clinafloxacin against S. pneumoniae gyrA and parC mutants.
As an initial step in defining the target specificity of clinafloxacin, we examined the susceptibilities of S. pneumoniae 7785 and its mutants bearing characterized quinolone resistance mutations in parC, in gyrA, and in both parC and gyrA (Table 1). These mutants were obtained previously by stepwise selection for resistance to ciprofloxacin (strains 1C1, 2C6, 2C7, and 3C4) or sparfloxacin (strains 1S1, 1S4, 2S1, and 2S4) (28). The mutations in these strains affect hot spots for quinolone resistance (Ser-79 and Asp-83 in ParC and the residue in S. pneumoniae GyrA equivalent to Ser-83 in E. coli GyrA). From Table 1, it can be seen that the parC mutants were about threefold more resistant to ciprofloxacin (compared to parent 1C1) and that the gyrA mutants were eightfold more resistant to sparfloxacin. Mutations in gyrA did not affect the response to ciprofloxacin, and parC changes had no effect on susceptibility to sparfloxacin (Table 1). Strains expressing both parC and gyrA mutations were extremely resistant to both ciprofloxacin and sparfloxacin; MICs were 16 to 64 μg/ml, i.e., ∼64 to 128-fold higher than those for the wild type.
TABLE 1.
Quinolone susceptibilities of S. pneumoniae mutants
| Strain | MIC (μg/ml) ofa:
|
Mutation identified in the QRDRs of the gene encoding
|
|||
|---|---|---|---|---|---|
| CIP | SPAR | CLN | GyrAb | ParC | |
| 7785 | 1 | 0.25 | 0.125 | ||
| 1C1 | 3 | 0.25 | 0.125 | None | None |
| 2C6 | 8 | 0.25 | 0.25 | None | Ser-79→Tyr |
| 2C7 | 8 | 0.25 | 0.25 | None | Ser-79→Phe |
| 1S1 | 1 | 2 | 0.25 | Ser-83→Phe | None |
| 1S4 | 1 | 2 | 0.25 | Ser-83→Tyr | None |
| 3C4 | 64 | 32 | 1 | Ser-83→Tyr | Ser-79→Tyr |
| 2S1 | 64 | 32 | 1 | Ser-83→Phe | Ser-79→Tyr |
| 2S4 | 32 | 16 | 1 | Ser-83→Phe | Asp-83→Asn |
CIP, ciprofloxacin; SPAR, sparfloxacin; CLN, clinafloxacin.
GyrA residue identified by analogy with the equivalent residue in E. coli GyrA.
The pattern of clinafloxacin susceptibility was strikingly different. First, wild-type strain 7785 was highly susceptible to clinafloxacin (MIC, 0.125 μg/ml, twofold lower than that of sparfloxacin and eightfold lower than that of ciprofloxacin). Second, in contrast to the results obtained with ciprofloxacin and sparfloxacin, neither parC mutations (in strains 2C6 and 2C7) nor gyrA mutations (in strains 1S1 and 1S4) had much effect on clinafloxacin activity; in each case, the clinafloxacin MICs were twofold higher than those for the parent, within experimental error (Table 1). Third, for strains 3C4, 2S1, and 2S4, bearing both gyrA and parC mutations, the clinafloxacin MIC was 1 μg/ml, an eightfold increase over that for the wild type (compared with 16- to 64-fold increases in the ciprofloxacin and sparfloxacin MICs). These observations indicate that gyrase and topoisomerase IV are each targeted appreciably by clinafloxacin and that mutations in both enzymes are needed to register a significant increase in resistance. Moreover, clinafloxacin retained useful activity even against the double mutants (Table 1).
Stepwise selection of clinafloxacin-resistant S. pneumoniae mutants.
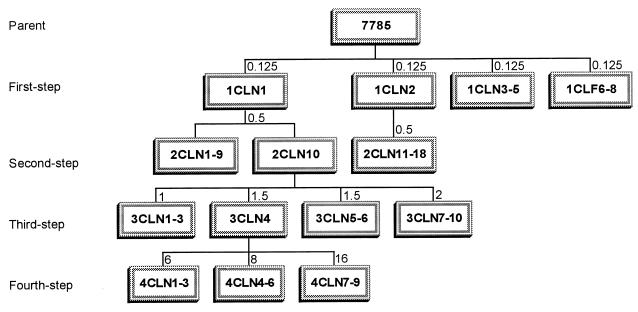
Studies with characterized gyrA and parC mutants have provided evidence that clinafloxacin targets gyrase and topoisomerase IV with approximate parity in vivo but are insufficiently sensitive to distinguish whether one or the other target is favored. Were clinafloxacin to exert a target preference, then in a series of mutants selected in a stepwise manner, we should expect resistance mutations to appear first and invariably in that target. To examine this question, we generated S. pneumoniae 7785 mutants by stepwise challenge using increasing concentrations of clinafloxacin, an approach similar to that adopted in studies of other quinolones (26, 28) (Fig. 1).
FIG. 1.
Relationships among S. pneumoniae 7785 and its resistant mutants selected by stepwise exposure to clinafloxacin. First-, second-, third-, and fourth-step mutants are designated by the prefixes 1, 2, 3, and 4, respectively. Numbers above the boxes indicate the concentrations of clinafloxacin (in micrograms per milliliter) used for each selection step.
From the results shown in Table 1, we realized that it might be difficult to select mutants from strain 7785 using clinafloxacin at >0.25 μg/ml, as two mutations (in gyrA and parC) would be needed for growth. Therefore, approximately 1011 CFU of strain 7785 was plated on brain heart infusion agar plates containing 10% horse blood and clinafloxacin at 0.125 and 0.25 μg/ml, i.e., one and two times the MIC. Resistant mutants appeared on the plates containing drug at 0.125 μg/ml after 48 h of aerobic incubation at 37°C. (No mutants were obtained with clinafloxacin at 0.25 μg/ml, even when 5 × 1011 CFU was screened). Colonies were restreaked on plates containing the same level of clinafloxacin. From two independent experiments, eight of the first-step clinafloxacin-resistant mutants—1CLN1 to 1CLN5 and 1CLN6 to 1CLN8—were chosen for analysis (Fig. 1). Strains 1CLN1 and 1CLN2 were challenged independently with clinafloxacin at 0.5 μg/ml, yielding second-step mutants 2CLN1 to 2CLN10 and 2CLN11 to 2CLN18, respectively (Fig. 1). Third-step mutants 3CLN1 to 3CLN3, 3CLN4 to 3CLN6, and 3CLN7 to 3CLN10 were generated from strain 2CLN10 by exposure to clinafloxacin at 1, 1.5, and 2 μg/ml, respectively. Finally, fourth-step mutants were selected from strain 3CLN4 with clinafloxacin at 6, 8, and 16 μg/ml (Fig. 1). The mutant frequencies were similar for all steps of selection and were in the range of 5 × 10−10 to 1.2 × 10−9 (Table 2). These frequencies were very low, despite selection with clinafloxacin concentrations that were only one to three times the MICs for the parent strains. By comparison, the frequency of mutants selected with ciprofloxacin at similar multiples of the ciprofloxacin MIC was higher by one to several orders of magnitude (12, 26).
TABLE 2.
Frequencies of clinafloxacin (CLN)-selected mutants generated from S. pneumoniae 7785 and its derivatives
| Strain | CLN MIC (μg/ml) | CLN concn (μg/ml) used for selection | Mutant frequency |
|---|---|---|---|
| 7785 | 0.125 | 0.125 | 5 × 10−10–8.5 × 10−10 |
| 7785 | 0.125 | 0.25 | <10−11 |
| 1CLN1 | 0.25 | 0.5 | 1 × 10−9 |
| 1CLN2 | 0.25 | 0.5 | 5 × 10−10 |
| 2CLN10 | 1.0 | 1.0 | 1.2 × 10−9 |
| 2CLN10 | 1.0 | 1.5 | 5.5 × 10−10 |
| 2CLN10 | 1.0 | 2.0 | 5.5 × 10−10 |
| 3CLN4 | 6 | 6 | 7 × 10−10–8 × 10−10 |
| 3CLN4 | 6 | 8 | 7 × 10−10–8 × 10−10 |
| 3CLN4 | 6 | 16 | 7 × 10−10–8 × 10−10 |
Clinafloxacin has a target preference for DNA gyrase in S. pneumoniae.
Resistant strains were characterized for their drug susceptibilities, and the status of their gyrA, parC, gyrB, and parE QRDRs was determined by DNA sequence analysis of PCR products generated with genomic DNA as a template (see reference 26 for details). A rapid RFLP method was particularly useful in the initial screening of S. pneumoniae gyrA and parC PCR products for resistance mutations affecting codons for Ser-83 and Ser-79 (26).
For each of the first-step mutants 1CLN1 to 1CLN8, there was a twofold increase in the clinafloxacin MIC (Table 3). All eight mutants produced a 366-bp parC PCR product which retained the wild-type HinfI digestion pattern, producing 183-, 127-, and 56-bp fragments; this result indicated the likely absence of a mutation affecting ParC Ser-79. All of the mutants (except for 1CLN2 and 1CLN8) yielded a 382-bp gyrA PCR product which did not undergo cleavage at an internal HinfI site overlapping codon 83, suggesting that this codon carried a mutation. PCR products from 1CLN2 and 1CLN8 yielded the 110- and 272-bp fragments seen for the wild-type gyrA gene. By DNA sequence analysis of AsPCR products from the gyrA, parC, gyrB, and parE QRDRs, it could be shown that selected strains 1CLN1, 1CLN3, 1CLN6, and 1CLN7 carried an acquired mutation in gyrA resulting in a change of Ser-83 to Phe or Tyr at the protein level. Strain 1CLN2 carried a GyrA change of Asp-87 to Lys, and 1CLN8 carried a GyrB change of Glu-474 to Lys (Table 3). None of the strains carried parC mutations. These sequencing results are consistent with the RFLP analysis. As 1CLN1 to 1CLN8 were produced in two independent drug challenges, it would seem that the gyrA and gyrB mutations are consistently selected in the first step.
TABLE 3.
Mutations identified in the QRDRs of the genes encoding the gyrase and topoisomerase IV proteins in clinafloxacin-selected S. pneumoniae mutants
| Strain | MIC (μg/ml) ofa:
|
Mutation(s)b in the QRDR of the gene encoding
|
|||||
|---|---|---|---|---|---|---|---|
| CLN | SPAR | CIP | GyrAc | ParC | GyrB | ParE | |
| 7785 | 0.125 | 0.25 | 1 | ||||
| 1CLN1d | 0.25 | 2 | 1–2 | Ser-83→Phe | None | None | None |
| 1CLN2d | 0.25 | 2 | 1–2 | Glu-87→Lys | None | ||
| 1CLN3 | 0.25 | 2 | 1–2 | Ser-83→Tyr | None | ||
| 1CLN6 | 0.25 | 2 | 1–2 | Ser-83→Tyr | None | ||
| 1CLN7 | 0.25 | 2 | 1–2 | Ser-83→Phe | None | ||
| 1CLN8 | 0.25 | 1 | 1–2 | None | None | Glu-474→Lys | None |
| 2CLN1 | 1 | 32 | 64 | Ser-83→Phe | Ser-79→Tyr | ||
| 2CLN6 | 1 | 32 | 64 | Ser-83→Phe | Ser-79→Tyr | ||
| 2CLN10d | 1 | 32 | 64 | Ser-83→Phe | Ser-79→Tyr | None | None |
| 2CLN11 | 1 | 16 | 32 | Glu-87→Lys | Ser-79→Tyr | ||
| 2CLN14 | 1 | 16 | 32 | Glu-87→Lys | Ser-79→Phe | ||
| 3CLN1 | 6 | 64 | 64 | Ser-83→Phe | Ser-79→Tyr | None | None |
| Glu-87→Gln | |||||||
| 3CLN4d | 6 | 64 | 64 | Ser-83→Phe | Ser-79→Tyr | None | None |
| Glu-87→Lys | |||||||
| 3CLN10 | 6 | 64 | 64 | Ser-83→Phe | Ser-79→Tyr | None | None |
| Glu-87→Gln | |||||||
| 4CLN3 | 32–64 | 64 | 128 | Ser-83→Phe | Ser-79→Tyr | None | Pro-454→Ser |
| Glu-87→Lys | |||||||
| 4CLN9 | 64 | 64 | 128 | Ser-83→Phe | Ser-79→Tyr | None | None |
| Glu-87→Lys | Asp-83→Ala | ||||||
CLN, clinafloxacin; SPAR, sparfloxacin; CIP, ciprofloxacin.
The indicated GyrA mutations resulted from the following nucleotide changes: Ser-83→Phe, TCC→TTC; Ser-83→Tyr, TCC→TAC; Glu-87→Lys, GAA→AAA; Glu-87→Gln, GAA→CAA. The indicated ParC mutations resulted from the following changes: Ser-79→Tyr, TCT→TAT; Ser-79→Phe, TCT→TTT; Asp-83→Ala, GAT→GCT. The GyrB mutation Glu-474→Lys resulted from a GAA→AAA change. The ParE mutatioin Pro-454→Ser resulted from a CCT→TCT change.
S. pneumoniae GyrA residues are identified by analogy with E. coli GyrA.
Mutant strain used as a parent for the subsequent step of selection.
All 18 second-step mutants generated from two different parent strains, 1CLN1 (GyrA Phe-83) and 1CLN2 (GyrA Lys-87), produced parC PCR products that had lost the HinfI site overlapping codon 79, yielding a 183-bp doublet on HinfI digestion; this result indicated a substitution at ParC Ser-79. The parC and gyrA PCR products of five selected mutants (clinafloxacin MIC, 1 μg/ml) were sequenced. Strains 2CLN1, 2CLN6, 2CLN10, 2CLN11, and 2CLN14, in addition to the expected GyrA mutation, had all acquired a mutation altering Ser-79 to Tyr or Phe in ParC, consistent with the RFLP analysis (Table 3). There were no additional mutations in GyrA. For strain 2CLN10, there were no alterations in GyrB or ParE. Thus, it appears that the increased clinafloxacin resistance of second-step mutants is associated with mutation of the ParC protein.
For 3 of the 10 third-step mutants obtained from parent 2CLN10 (Fig. 1), the QRDRs of the four topoisomerase genes were sequenced. No mutations were detected in the gyrB or parE QRDRs of strains 3CLN1, 3CLN4, and 3CLN10, and no additional mutations were detected in the parC QRDR (Table 3). However, all three strains had acquired a second mutation in GyrA (Glu-87 to Gln or Lys) associated with a clinafloxacin MIC of 6 μg/ml. Finally, for the nine fourth-step mutants, the four topoisomerase gene QRDRs were examined for strains 4CLN3 and 4CLN9 derived from strain 3CLN4 by independent challenge with clinafloxacin at 6 and 16 μg/ml (Table 3). These strains were highly resistant to clinafloxacin (MICs, 32 to 64 μg/ml). In addition to the ParC and two GyrA mutations present in the parent, both mutants had acquired a second mutation affecting topoisomerase IV: a ParE Pro-454 alteration to Ser in 4CLN3 and a ParC Asp-83 change to Ala in 4CLN9. In summary, the development of high-level resistance occurred in four steps, each associated with a mutation in a topoisomerase gene and involving alternate changes in gyrase and topoisomerase IV.
Growth properties of quinolone-resistant mutants.
Given that gyrase and topoisomerase IV are essential bacterial enzymes, it was of interest to examine whether the growth of resistant mutants was impaired. We found that in the absence of drug, the growth on plates and morphology of fourth-step mutants 4CLN3 and 4CLN9 were indistinguishable from those of the wild type (data not shown). In a more detailed analysis, the generation times for 7785 and 4CLN9 in liquid cultures, determined from duplicate experiments, were 26.6 ± 2.8 and 28.6 ± 3.2 min, respectively (data not shown). Thus, although we cannot rule out the possibility that fourth-step mutants exhibit other phenotypic changes, e.g., altered pathogenicity, it is clear that the altered topoisomerases in these mutants do not have a marked effect on bacterial growth in the laboratory.
DISCUSSION
We have determined the target specificity of clinafloxacin in S. pneumoniae by using defined gyrA and parC strains and through analysis of mutants selected in a stepwise manner. A key finding was that representative gyrA or parC mutations did not affect the activity of clinafloxacin (Table 1). The combined presence of the same mutations led to a small but significant increase in the clinafloxacin MIC. These results are consistent with the idea that clinafloxacin targets both gyrase and topoisomerase IV in S. pneumoniae. In further support of the dual-target hypothesis, it proved very difficult to isolate clinafloxacin-resistant mutants by stepwise challenge (Tables 2 and 3). First-step mutants could be obtained at a frequency of 5 × 10−10 only by challenge with clinafloxacin at 0.125 μg/ml, the MIC for the parent strain. These mutants exhibited an increase in resistance of only less than or equal to twofold (clinafloxacin MIC, 0.25 μg/ml); this finding was associated with the acquisition of a gyrA or gyrB mutation, suggesting a modest drug preference for gyrase. The subsequent selection of second-, third-, and fourth-step mutants, again at low frequencies and involving small incremental changes in clinafloxacin MICs, was associated with alternate mutations in the topoisomerase IV and gyrase genes (Table 3). The drug retained potency against first- and second-step mutants; indeed, four topoisomerase IV mutations were necessary to generate high-level resistance. These results indicate that clinafloxacin is intrinsically highly active against S. pneumoniae and exhibits approximate parity in targeting gyrase and topoisomerase IV in vivo.
Access to a panel of S. pneumoniae mutants expressing ParC and/or GyrA proteins with characterized quinolone resistance mutations proved extremely useful in assessing the activities of clinafloxacin and its target preferences. The responses of mutant strains to clinafloxacin could be rapidly examined and compared with parallel data obtained for sparfloxacin and ciprofloxacin, agents that target gyrase and topoisomerase IV, respectively (Table 1). Clinafloxacin was the most potent of the three agents tested against wild-type S. pneumoniae, parC, or gyrA mutants, and particularly the parC-gyrA double mutants (Table 1). For the parent strain and single mutants, the clinafloxacin MICs were 0.125 and 0.25 μg/ml, respectively. For the double mutants, the clinafloxacin MIC was increased fourfold to 1 μg/ml. However, the sparfloxacin and ciprofloxacin MICs for the double mutants each climbed steeply to 16 to 64 μg/ml, i.e., beyond levels attainable in serum or tissue (Table 1). Although we did not examine trovafloxacin, recent studies with a similar approach have measured trovafloxacin MICs of 0.5 and 6 μg/ml for representative parC and parC-gyrA S. pneumoniae mutants isolated as first-step and second-step mutants by challenge with ciprofloxacin, respectively (12). The fourfold increase in the trovafloxacin MIC for parC mutants indicates that trovafloxacin targets topoisomerase IV, and from human pharmacokinetic data, it appears that the drug retains clinically relevant activity against the first-step mutants (12). For clinafloxacin, pharmacokinetic studies have shown that for twice-daily oral doses of 200 and 400 mg, respective peak concentrations in human serum reached 2.75 and 5.22 μg/ml, with half-lives for elimination of 5.7 and 7.6 h (18, 31). Although further pharmacokinetic studies may be needed, the data presented in Table 1 indicate that clinafloxacin should retain potent clinical activity against gyrA, parC, and gyrA-parC mutants.
One limitation in using mutant strains to test drug activities is that small preferences in drug targeting are not apparent. Thus, for both gyrA and parC mutants of S. pneumoniae there was a twofold increase in the clinafloxacin MIC (Table 1). These small changes are usually considered to be within experimental error for the twofold dilution method used for MIC determinations. The stepwise selection of mutants is a more sensitive approach for determining target preferences. The development of high-level resistance to clinafloxacin required multiple steps with a defined and invariant sequence of mutations of single residues in gyrase or topoisomerase IV (Table 3). That first-step mutants displaying an increase in the clinafloxacin MIC of less than or equal to twofold in association with a mutation in either gyrA or gyrB could be selected is consistent with dual targeting with a modest drug preference for gyrase. Recent work with trovafloxacin has shown that this agent selects resistant S. pneumoniae mutants at a frequency of ∼10−8 when used at the MIC. The MICs and QRDRs of the trovafloxacin-resistant mutants were not examined (12).
We found no evidence for clinafloxacin resistance mechanisms other than topoisomerase gene mutations. In contrast, selection with ciprofloxacin led to first-step mutants, such as 1C1, whose resistance was not attributable to topoisomerase changes (Table 1) (26). Interestingly, 1C1 was completely susceptible to clinafloxacin. We have speculated that 1C1 may be a permeation mutant whose resistance accrues from the upregulation of an efflux pump, akin to the NorA protein of S. aureus, for which ciprofloxacin is a substrate. Basal-level expression of NorA in S. aureus is involved in setting wild-type susceptibility to ciprofloxacin (38), and several ciprofloxacin-resistant S. aureus strains in which NorA is upregulated have been identified (16, 23). S. pneumoniae is known to express one or more efflux pumps (2), and our failure to isolate such mutants could suggest that clinafloxacin is not a substrate, a property that would contribute to its intrinsic activity. These issues require further study.
The mutations selected by clinafloxacin challenge were predominantly those described previously for other quinolones (Tables 1 and 3) (22). Thus, the gyrA mutations altered GyrA Ser-83 (to Phe or Tyr) or Glu-87 (to Gln or Lys) (Table 3). The parC mutations resulted in ParC Ser-79 (to Phe or Tyr) and Asp-83 (to Ala) changes. These changes occurred at the expected GyrA and ParC hot spots. In the crystal structure of an N-terminal 59-kDa fragment of the E. coli GyrA protein, the equivalent residues lie in an α helix that is adjacent to the catalytic Tyr-122 residues involved in DNA breakage and reunion (20). This helix likely functions in DNA recognition and binding. It is not known how the resistance mutations interfere with quinolone action, although this interference may arise from steric inhibition of drug binding (5, 37).
In contrast to the GyrA and ParC changes, the GyrB and ParE mutations acquired in first-step strain 1CLN8 and fourth-step strain 4CLN3 are novel. First, the Glu-474→Lys GyrB mutation does not occur in the EGDSA and P(I/L)RGK motifs of GyrB, commonly implicated in resistance and identified as the GyrB QRDR (40). Instead, the mutation lies C terminal to the PLRGK motif. Previous studies of quinolone-resistant Salmonella typhimurium identified a Ser-463→Lys alteration in GyrB located at a position similar to that which is altered in GyrB of strain 1CLN8 (11). Moreover, an Asn-470→Asp mutation lying C terminal to the PLRGK motif has been described for S. aureus ParE (8). Together, the data suggest that the GyrB QRDR may occupy a more extensive region of the protein than was previously defined by studies with E. coli (40). Second, the Pro-454→Ser ParE mutation lies within the PLRGK motif but is a novel change, as the Arg residue of this sequence usually is mutated, at least in GyrB. Other studies with S. pneumoniae have shown that first-step mutants resistant to ciprofloxacin carry ParE mutations of Asp-435 to Asn in the EGDSA motif (30). The functional role of the GyrB or ParE QRDR region is unknown. The equivalent region in the crystal structure of a fragment of Saccharomyces cerevisiae topoisomerase II lies distant from the DNA binding domain, and it is not clear how mutations in this region may affect drug susceptibility (4). Mutations in the conserved PLRGK motif and DNA recognition helix A′α4 of yeast topoisomerase II confer resistance to the anticancer agents amsacrine and doxorubicin, respectively (29, 36). Thus, there appear to be close similarities in the mechanisms of action of these topoisomerase-targeting antitumor agents and the antibacterial fluoroquinolones.
Interest in drugs that act on two or more targets stems in part from the possibility of limiting the emergence of resistance. Were a drug to act with equal potency through two targets, then simultaneous acquisition of two mutations would be required for the development of resistance. Given a typical frequency of 10−8 for single mutations, the frequency of double mutations would be 10−16, an extremely rare occurrence. It may be difficult to obtain agents that act exactly equally through two topoisomerase targets. However, comparable if unequal targeting is also advantageous. In this case, the level of resistance arising from mutation of the preferred target will be severely limited by the appreciable contribution to susceptibility of the second target. The results of the stepwise selection (Table 3) indicate that clinafloxacin acts in this manner. To our knowledge, clinafloxacin is the first reported example of a dual-targeting quinolone for S. pneumoniae and extends our previous work indicating that quinolone structure determines target preference in S. pneumoniae (28). Interestingly, recent studies with defined quinolone-resistant S. aureus mutants suggest that sparfloxacin and DU6895a also act against both gyrase and topoisomerase IV in S. aureus (8). In the absence of data from mutants selected in a stepwise manner, it is an open question whether these fluoroquinolones act equally or with approximate parity on their two S. aureus targets. However, the results suggest that dual-targeting quinolones may not be as rare as might be imagined. It is notable that sparfloxacin targets gyrase in S. pneumoniae but acts against both gyrase and topoisomerase IV in S. aureus. Evidently, targeting depends not only on drug structure but also on relative structural differences in topoisomerases. It remains to be seen whether dual targeting for one bacterial species can be retained for other pathogens.
Finally, the target preferences for clinafloxacin described in this paper indicate the relative importance of cell-killing pathways initiated through drug inhibition of gyrase or topoisomerase IV in S. pneumoniae. The molecular determinants favoring one or the other of these pathways are poorly understood. A C-8 substituent on the fluoroquinolone appears to favor cell killing through gyrase in S. pneumoniae. The gyrase-targeting agents sparfloxacin and clinafloxacin have fluorine and chlorine substituents at C-8, respectively. In contrast, neither ciprofloxacin nor trovafloxacin carries a C-8 substituent, and it is significant that both target topoisomerase IV in S. pneumoniae. We do not know whether these preferences arise from enhanced affinity for the target or enhanced lethality, as recently described for the 8-methoxy derivative of ciprofloxacin against E. coli gyrA mutants (42). Our working hypothesis is that the high potency of clinafloxacin against S. pneumoniae and its gyrA, parC, and gyrA-parC mutants arises from intrinsic tight binding of the drug to the target enzyme-DNA complexes, perhaps aided by poor efflux. Biochemical studies on clinafloxacin in progress in our laboratory should provide further information on the factors underlying its potent antipneumococcal activity.
ACKNOWLEDGMENTS
We thank Stephen J. Gracheck, Jing Li, and Michael A. Cohen for helpful comments.
This work was supported by a grant from Parke-Davis Co.
REFERENCES
- 1.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning DNA replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 2.Baranova N N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett J G, Grundy L M. Community-acquired pneumonia. N Engl J Med. 1995;333:1618–1624. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 4.Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure and mechanism of DNA topoisomerase IV. Nature (London) 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 5.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrero L, Cameron B, Manse B, Lagneux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target for quinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 8.Fournier B, Hooper D C. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother. 1998;42:121–128. doi: 10.1128/aac.42.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gellert M, Mizuuchi K, O’Dea M H, Itoh T, Tomizawa J-I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gellert M, Mizuuchi K, O’Dea M H, Nash H A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gensberg K, Jin Y F, Piddock L J V. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol Lett. 1995;132:57–60. doi: 10.1111/j.1574-6968.1995.tb07810.x. [DOI] [PubMed] [Google Scholar]
- 12.Gootz T D, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Polzer R J. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann J, Cetron M S, Farley M M, Baughman W S, Facklam R R, Elliott J A, Deaver K A, Breiman R F. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med. 1995;333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 15.Janoir C, Keller V, Kitzis M-D, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaatz G W, Seo S M. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:2650–2655. doi: 10.1128/aac.39.12.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato J, Nishimura Y, Imamura R, Niki H, Higara S, Suzuki H. New topoisomerase essential for chromosomal segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 18.Kucers A, Crowe S M, Grayson M L, Hoy J F, editors. The use of antibiotics. A clinical review of antibacterial, antifungal and antiviral drugs. 5th ed. Oxford, England: Butterworth-Heinemann; 1997. pp. 1169–1171. [Google Scholar]
- 19.Mizuuchi K, Fisher L M, O’Dea M H, Gellert M. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morais-Cabral J H, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 21.Munoz R, de la Campa A. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of quinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 22a.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial testing; eighth informational supplement. NCCLS document M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 23.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallares R, Linares J, Vadillo M, Carbellos C, Manresa F, Viladrich P F, Martin R, Gudiol F. Resistance to penicillin and cephalosporin and mortality from severe pneumococcal pneumonia in Barcelona, Spain. N Engl J Med. 1995;333:474–480. doi: 10.1056/NEJM199508243330802. [DOI] [PubMed] [Google Scholar]
- 26.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X-S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel S, Sprung A U, Keller B A, Heaton V J, Fisher L M. Identification of yeast DNA topoisomerase II mutants resistant to the antitumor drug doxorubicin: implications for the mechanisms of doxorubicin action and cytotoxicity. Mol Pharmacol. 1997;52:658–666. doi: 10.1124/mol.52.4.658. [DOI] [PubMed] [Google Scholar]
- 30.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randinitis, E. J. 1998. Personal communication.
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in the gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2502–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasz A. The pneumococcus at the gates. N Engl J Med. 1995;333:314–315. doi: 10.1056/NEJM199508243330810. [DOI] [PubMed] [Google Scholar]
- 35.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 36.Wasserman R A, Wang J C. Analysis of yeast DNA topoisomerase II mutants resistant to the antitumor drug amsacrine. Cancer Res. 1994;54:1795–1800. [PubMed] [Google Scholar]
- 37.Willmott C J, Maxwell A. A point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada H, Kurose-Hamada S, Fukuda Y, Mitsuyama J, Takahata M, Minami S, Watanabe Y, Narita H. Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2308–2309. doi: 10.1128/aac.41.10.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]