ABSTRACT
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has revolutionized diagnostics in culture-based microbiology. Commonly used MALDI-TOF MS systems in clinical microbiology laboratories are MALDI Biotyper (Bruker Daltonics) and Vitek MS (bioMérieux), but recently the new EXS2600 (Zybio) has been launched. This study aimed to evaluate the performance of the three devices by comparing the results to 16S rRNA gene sequencing. A set of 356 previously collected difficult-to-identify bacteria was tested in parallel with the three systems. Only the direct smear method and simple formic acid extraction were applied. Valid results were achieved for 98.6%, 94.4%, and 93.3% of all isolates by MALDI Biotyper, EXS2600, and Vitek MS, respectively. Of all valid results, agreement with sequencing data was achieved in 98.9%, 98.5%, and 99.7% by MALDI Biotyper, EXS2600, and Vitek MS, respectively. Considering only the isolates with valid measurements at the single-species level, misidentification rates were 0%, 2.6%, and 1.1% for MALDI Biotyper, EXS2600, and Vitek MS, respectively. Apart from minor performance differences, our data demonstrate that the three systems provide comparable results and are suitable for use in medical diagnostic laboratories.
KEYWORDS: MALDI-TOF, mass spectrometry, identification, Biotyper, Vitek MS, EXS2600
INTRODUCTION
Early and accurate identification of pathogens from clinical specimens is pivotal to guide the therapy of infectious diseases and to ensure a favorable outcome (1). Despite the increasing significance of molecular techniques, which are characterized by short turnaround times, cultivation still represents the most common method for the detection of bacteria and fungi. Significant benefits of culture-based diagnostics are comparably low costs and the fact that it is not necessary to define the pathogen to be investigated in advance, which is in contrast to pathogen-specific methods such as PCR or serology. All colonies grown on agar plates can be subjected to phenotypic and biochemical analyses. However, even with automated systems, identification of isolates via those methods requires a significant amount of time, which typically delays the result for a minimum of 1 day (2). Identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has overcome the previous limitations since it offers timely and cost-effective results (2, 3). To date, MALDI-TOF MS has become an essential technique and the diagnostic gold standard for virtually all medical microbiology laboratories in developed healthcare systems.
As great as the importance of MALDI-TOF MS is in diagnostics, the choice of instruments and databases is particularly small (4). This puts laboratories in a weak position as customers and limits the possibilities for customization to meet specific needs. Currently, there are only two manufacturers offering FDA-approved and IVD-certified MALDI-TOF MS systems: Bruker Daltonics (Bremen, Germany) and bioMérieux (Marcy-l’Étoile, France). Recently, two novel competitors have entered the market, namely, ASTA (Suwon, South Korea) and Zybio (Chongqing, People’s Republic of China). However, until very recently, the availability of their respective MALDI-TOF MS systems has been limited to local markets. Hence, data about the performance of these devices are scarce and comprehensive comparisons with the more established MALDI-TOF MS systems are lacking (5, 6). Conducting such comparative studies is challenging since laboratories typically have access to only one device and database. Furthermore, the measurements, particularly those with discordant results, must be compared to a reference standard of identification.
In this study, we aimed to evaluate the performance of three different MALDI-TOF MS systems in a head-to-head comparison, with 16S rRNA gene sequences as reference.
MATERIALS AND METHODS
Bacterial strains
This study relies on a collection of isolates from human specimens sampled for diagnostic analysis at the Diagnostic and Research Institute for Hygiene, Microbiology and Environmental Medicine of the Medical University of Graz. This academic institution hosts the central microbiology laboratory of the University Hospital Graz, a 1,600-bed tertiary care center in Graz, Austria. In 2010, 1,145 consecutive isolates were analyzed in parallel using the MALDI Biotyper instrument with related software (Bruker Daltonics) and the AXIMA Assurance instrument (Shimadzu Company, Kyoto, Japan) with the Saramis software (AnagonsTec GmbH, Potsdam, Germany) applying the respective then-current databases. All isolates with discordant or invalid MALDI-TOF MS results were identified by 16S rRNA gene sequencing [database: GenBank (National Center for Biotechnology Information)] with sequence identities ≥98% being considered to represent a valid result. Notably, this allowed for situations, in which differing results of two MALDI-TOF MS systems were still both categorized to be correct since the two identified species demonstrate sequence identities ≥98%. In this study, 356 precharacterized strains were included. The distribution of orders and genera is summarized in Table 1.
TABLE 1.
Distribution of orders and genera of the isolates included in the study
| Order | Genus | n | % |
|---|---|---|---|
| All isolates | 356 | 100.0 | |
| Lactobacillales | 71 | 19.9 | |
| Streptococcus | 39 | 11.0 | |
| Enterococcus | 20 | 5.6 | |
| Lactobacillus | 9 | 2.5 | |
| Others | 3 | 0.8 | |
| Bacteroidales | 63 | 17.7 | |
| Prevotella | 42 | 11.8 | |
| Bacteroides | 21 | 5.9 | |
| Pseudomonadales | 45 | 12.6 | |
| Acinetobacter | 18 | 5.1 | |
| Pseudomonas | 18 | 5.1 | |
| Moraxella | 9 | 2.5 | |
| Enterobacterales | 40 | 11.2 | |
| Bacillales | 36 | 10.1 | |
| Staphylococcus | 32 | 9.0 | |
| Others | 4 | 1.1 | |
| Pasteurellales | 17 | 4.8 | |
| Micrococcales | 10 | 2.8 | |
| Burkholderiales | 12 | 3.4 | |
| Actinomycetales | 10 | 2.8 | |
| Others | 52 | 14.6 | |
MALDI-TOF MS systems, scoring scheme, and statistical analysis
The isolates were used for a comparison of three MALDI-TOF MS systems of different manufacturers and their respective current databases (as of October 2022): (i) Bruker’s MALDI Biotyper (Biotyper) using the “MBT IVD Library Revision G (Claim 6)” database, (ii) bioMérieux’s VITEK MS (Vitek MS) using the “Knowledge Base V3.2” database, and (iii) Zybio’s EXS2600 (EXS) using the “V.1.0.0.0” database.
All MALDI-TOF MS systems indicate the reliability of identification results by a color-coded categorization. For Biotyper and EXS, green results (identification score: ≥2.00) represent measurements valid at the species level and yellow results (1.70–1.99) represent measurements valid at the genus level. However, in certain cases, it is possible to get green results that do not feature a decisive species, for example, “Fusobacterium sp.” (Bruker). With red scores (<1.70), no valid identification results were obtained. Although the traffic light coding is also applied by Vitek MS, the classification is based on a considerably different algorithm: green results (confidence levels ≥ 60%) include results with one organism at the species level as well as results with a group of organisms at the species level, for example, “99.9% Lactobacillus casei/paracasei/rhamnosus.” Yellow results consist of a group of results that add up to a confidence level of 100% caused by isolates that are not distinguishable by Vitek MS, for example, “33.3% Moraxella lacunata/33.3% Neisseria avis/33.3% Moraxella catarrhalis.” Identification results with more than four organisms or no concordant spectra in the database are categorized as red. Therefore, the Vitek MS performance cannot be easily compared to the performance of Biotyper and EXS. To facilitate a quantitative comparison, this study applies different identification levels: (i) single species level, which includes all green results featuring only one distinct species, (ii) level of green results, and (iii) level of valid results, that is, all green and yellow results. Descriptive statistics were calculated with IBM SPSS Statistics for Windows, Version 28.0 (IBM, Armonk, USA).
Sample processing
Each measurement was performed from fresh colonies following overnight culture on Columbia sheep blood agar (BD; ref. no. 254071) or chocolate agar (bioMérieux; ref. no. 43109) at 5% CO2 for fastidious organisms, or on Schaedler agar in anaerobic conditions for anaerobes (BD, Franklin Lakes, USA; ref. no. 254084). The processing was carried out in parallel, whereby one trained laboratory technician applied the strain onto the three targets (one spot each), according to the respective manufacturers’ instructions using the corresponding reagents.
In a first attempt, all isolates were applied as smear directly from the agar plate to the respective MALDI-TOF MS targets. All measurements that did not yield a green result were repeated under the same conditions. When this approach also failed to generate a green result, a third measurement was performed, for which the cells were pretreated with formic acid, according to the manufacturer’s protocols.
RESULTS
Validity of results
With the routinely used standard measurement procedure, that is, direct smear application of colonies to the target, green results were obtained for 85.4%, 77.2%, and 78.7% of isolates by the Biotyper, the EXS, and the Vitek MS MALDI-TOF MS systems, respectively. Upon failure to generate green results, re-testing and (if necessary) re-testing including formic acid treatment was performed, which increased the rates for measurement results at a green level to 92.1%, 87.4%, and 86.8% for Biotyper, EXS, and Vitek MS, respectively (Table 2).
TABLE 2.
Rates of valid measurement results of the three tested MALDI-TOF MS systems a
| Biotyper | EXS | Vitek MS | |
|---|---|---|---|
| Single-species results | |||
| Direct smear | 84.3 (80.1–87.9) | 75.6 (70.8–79.9) | 74.4 (69.4–78.9) |
| Re-testing | 88.2 (84.4–91.4) | 82.0 (77.6–85.9) | 79.5 (74.9–83-6) |
| Formic acid treatment | 91.0 (87.5–93.8) | 85.7 (81.6–89.1) | 81.7 (82.8–90.1) |
| Green results | |||
| Direct smear | 85.4 (81.3–88.9) | 77.2 (72.5–81.5) | 78.7 (69.6–78.9) |
| Re-testing | 89.3 (85.6–92.3) | 83.7 (79.5–87.4) | 84.3 (80.1–87.9) |
| Formic acid treatment | 92.1 (88.8–94.7) | 87.4 (83.5–90.6) | 86.8 (82.8–90.1) |
| All valid results | |||
| Direct smear | 94.9 (92.1–97.0) | 91.9 (88.5–94.5) | 85.4 (81.3–88.9) |
| Re-testing | 97.8 (95.6–99.0) | 93.8 (90.8–96.1) | 91.6 (88.2–94.2) |
| Formic acid treatment | 98.6 (96.8–99.5) | 94.4 (91.5–96.5) | 93.3 (90.1–95.6) |
Measurements of all 356 isolates were performed with Bruker MALDI Biotyper (Biotyper), Zybio EXS2600 (EXS), and bioMérieux VITEK MS (Vitek MS). Validity is displayed at the single-species level (Biotyper and EXS: green results featuring only one distinct species; Vitek: green and yellow results featuring only one distinct species), the level of green results, and the level of all valid results (green or yellow). When the first analysis failed to yield results at the desired level, the isolate was re-tested. When the re-test also failed, the isolates were re-tested using the formic acid extraction protocol. All rates are presented as percentages with 95% confidence intervals in parentheses.
When all (green and yellow) results were accepted, the respective rates increased by up to 16% by the first measurement (Table 2). Following the protocol of re-tests with and without formic acid, a maximum success rate of 98.6% was achieved by the Biotyper system.
Notably, a direct comparison of the categories “green” and “yellow” is restricted by the different definitions by the respective manufacturers, that is, Bruker and Zybio vs bioMérieux. For instance, Streptococcus dysgalactiae was identified by all MALDI-TOF MS systems at the species level. While Biotyper and EXS categorized this finding as a green result, the Vitek MS result was only yellow, since decisive discrimination between S. dysgalactiae subspecies dysgalactiae and S. dysgalactiae subspecies equisimilis failed—a differentiation level that is not even targeted by Biotyper or EXS. Consequently, an evaluation that considers only the “green result” vs “yellow result” categories would incorrectly understate the performance of the Vitek MS in this case. In other cases, however, the structure of the Vitek MS database results in differentiation results being presented as green that would be deemed invalid by the other MALDI-TOF MS systems: the Vitek MS database includes entries, in which for one spectrum results of “organism groups” are deposited. These groups can consist of up to four species, which can even be members of different genera and still are defined as valid measurements. To facilitate a quantitative evaluation, we included another comparison, in which only those results were accepted, which featured one single species (Table 2). After up to three measurements, valid results at the single-species level were obtained for 91.0%, 85.7%, and 81.7% of isolates by the Biotyper, the EXS, and the Vitek MS system, respectively (Table S1).
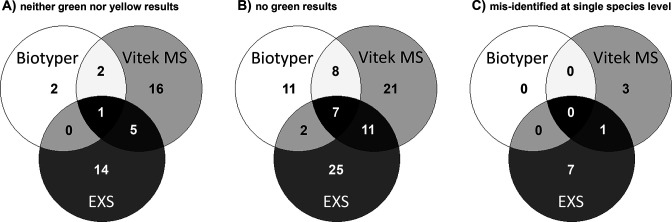
Some isolates did not yield valid results since no accurate spectra could be generated. For a certain group, however, not the quality of spectra but the database is the reason for identification failure: two of the five (40.0%) invalid Biotyper, seven of the 20 (35.0%) invalid EXS, and 12 of the 24 (50.0%) invalid Vitek MS results were attributable to missing database entries. The overlap between all measurement failures of the different MALDI-TOF MS systems is depicted in Fig. 1A (failure to generate green or yellow results) and Fig. 1B (failure to generate green results). In only seven isolates (2.0%), it was observed that all devices failed to generate a result at the species level (Biotyper and EXS) or organism group level (Vitek MS): one isolate each of Achromobacter xylosoxidans, Bifidobacterium longum, Prevotella melaninogenica, Propionimicrobium lymphophilum, Pseudomonas aeruginosa, Pseudomonas pseudoalcaligenes, and Prevotella multiformis. For the latter one, no valid measurement results at all were obtained (red results with all MALDI-TOF MS systems).
Fig 1.
Venn diagrams visualizing the overlap of isolates with neither green nor yellow results (A), no green results (B), or incorrectly identified isolates (C). The latter group included all cases, in which the respective MALDI-TOF system (Bruker MALDI Biotyper: Biotyper; bioMérieux VITEK MS: Vitek; and Zybio EXS2600: EXS) yielded a valid result at single-species level, that is, green with only one distinct species, but false species identification.
For EXS and Vitek MS, trends were observable that specific groups of organisms were particularly challenging: 10 of 20 (50.0%) invalid EXS results were obtained analyzing Prevotella spp. Aerobe gram-negative rods such as Pseudomonas spp. and Acinetobacter spp. were very prevalent [13/24 (54.1 %)] in the group of invalid Vitek MS measurements.
Accuracy of results
All MALDI-TOF MS results were compared to the results of 16S rRNA gene sequencing. When only results featuring one distinct species were considered, agreement with sequencing results was 91.0% for the Biotyper, 83.4% for the EXS, and 80.6% for the Vitek MS after up to three measurements. However, those numbers do not allow us to calculate the rate of misidentifications since the aforementioned rates refer to the overall number of isolates—including those without valid identification results. Considering only the isolates with valid measurements at the single-species level, misidentification rates were 0% (0/324) for Bruker, 2.6% for EXS (8/305), and 1.4% for Vitek MS (4/291).
The performance for correct identification at the other validity levels is summarized in Table 3. Except for single-species-level analysis, MALDI-TOF MS results featuring more than one species were accepted to be correct, when at least one of the species was in concordance with 16S rRNA gene sequencing results.
TABLE 3.
Concordance of 16S rRNA gene sequencing and MALDI-TOF MS measurements a
| Valid results | True results | |||
|---|---|---|---|---|
| n | n | Of valid tests | Of all tests | |
| Biotyper | ||||
| Single-species results | 324 | 324 | 100.0 (98.9–100) | 91.0 (87.5–93.8) |
| Green results | 328 | 324 | 98.8 (96.9–99.7) | 91.0 (87.5–93.8) |
| All valid results | 351 | 347 | 98.9 (97.1–99.7) | 97.5 (95.3–98.8) |
| EXS | ||||
| Single-species results | 305 | 297 | 97.4 (94.9–98.9) | 83.4 (79.1–87.1) |
| Green results | 311 | 300 | 96.5 (93.8–98.2) | 84.3 (80.1–87.9) |
| All valid results | 336 | 331 | 98.5 (96.6–99.5) | 93.0 (89.8–95.4) |
| Vitek MS | ||||
| Single-species results | 291 | 287 | 98.6 (96.5–99.6) | 80.6 (76.1–84.6) |
| Green results | 309 | 303 | 98.1 (95.8–99.3) | 85.1 (81.0–88.6) |
| All valid results | 332 | 331 | 99.7 (98.3–100) | 93.0 (89.8–95.4) |
The rate of MALDI-TOF MS tests, which yielded results in agreement with sequencing results (true), refers to the total measurements that yielded results at the respective validity level, that is, single-species level, level of green results, and level of valid results (green or yellow). Except for single-species-level analysis, MALDI-TOF MS results featuring more than one species were accepted to be correct, when at least one of the species was in concordance with 16S rRNA gene sequencing results. For the category “all valid results,” results featuring the correct genus according to the reference results were classified as true. Total number of isolates included in the analysis: 356; all rates are presented as percentages with 95% confidence intervals in parentheses.
Characterization of misidentified isolates
There was a notable overlap of the misidentification results observable: most isolates with results at the single-species level that were misidentified by any MALDI-TOF MS system also yielded incorrect or invalid results for at least one other MALDI-TOF MS device (Table 4; Fig. 1C). For three Vitek MS misidentifications, it must be considered that the current database does not include the analyzed species. No isolate was misidentified by all three systems. All misidentifications (EXS: eight; Vitek MS: four) represented minor errors with results at least correct at the genus level. In one case, the misidentification is particularly noteworthy since the respective false results might have triggered a misdiagnosis that also could have had therapeutic consequences. A Streptococcus mitis strain, a commensal causing rare but sometimes severe infections, was misidentified by EXS and Vitek MS as the closely related Streptococcus pneumoniae, which is much more often related to the disease. Considerably, it is good and common laboratory practice to verify the MALDI-TOF MS finding of S. pneumoniae with traditional methods, for example, bile solubility or optochin sensitivity, since the limitations of MALDI-TOF MS to differentiate these germs are well known.
TABLE 4.
Summary of isolates that yielded valid but incorrect identification results at single-species level by any MALDI-TOF MS system a
| Reference result | Biotyper result | Score | EXS result | Score | BMX result | Conf. level |
|---|---|---|---|---|---|---|
| Bacteroides dorei | Bacteroides vulgatus b , c | 2.22 | B. vulgatus | 2.43 | B. dorei | 99.9 |
| Brevibacterium massiliense | No green result | Invalid result | Brevibacterium casei | 99.9 | ||
| Brevundimonas nasdae | No green result | Brevundimonas aurantiaca | 2.17 | Invalid result | ||
| Fusobacterium gonidiaformans | No green result | Fusobacterium necrophorum | 2.16 | Invalid result | ||
| Moraxella canis | M. canis | 2.43 | No green result | M. catarrhalis | 99.9 | |
| Neisseria flavescens | Neisseria subflava | 2.16 | Neisseria lactamica | 2.25 | No single-species result | |
| Neisseria flavescens | Neisseria subflava | 2.14 | N. lactamica | 2.16 | No single-species result | |
| Prevotella timonensis | P. timonensis | 2.13 | Prevotella buccae | 2.32 | Prevotella timonensis | 99.9 |
| P. timonensis | P. timonensis | 2,06 | P. buccae | 2.05 | P. timonensis | 99.9 |
| Staphylococcus croceolyticus | Staphylococcus petrasii | 2.08 | Invalid result | Staphylococcus haemolyticus b | 99.9 | |
| S. mitis | S. mitis | 2.13 | Streptococcus pneumoniae | 2.04 | S. pneumoniae | 99.1 |
Misidentified isolates are marked by underlining. For all isolates with results at the single-species level, the respective score (Biotyper and EXS) or confidence level (Vitek MS) is indicated.
Cases of misidentification, in which the analyzed species is not included in the respective MALDI-TOF system’s database.
Classified as correct since the Biotyper provides the hint that upon the result Bacteroides vulgatus a differentiation between B. vulgatus and B. dorei is not possible.
DISCUSSION
Already in 2014, McElvania TeKippe and Burnham evaluated the performance of Biotyper and Vitek MS for analyzing “unusual and/or difficult-to-identify microorganisms” and reported identification rates comparable to our findings despite relying on much less comprehensive databases (7). However, their collection of 172 isolates representing 50 genera featured a very specific focus on organisms rarely encountered in routine diagnostics. In this setting, MALDI-TOF MS became the primary tool for rapid, accurate, and cheap identification of cultivated bacteria and yeasts. Accordingly, there is plenty of data analyzing Biotyper and Vitek MS performance (8 – 15). Contrarily, studies evaluating the EXS system are very scarce: to date, only three studies have been published. However, focusing on the detection of Shewanella (16), on antifungal susceptibility testing in Talaromyces marneffei (17), and classification of Listeria species (18), those data do not feature an insight into the capability of EXS to support routine diagnostics in a medical microbiology laboratory.
This study attempted to present the results not only as a narrative enumeration of individual results but to make these results evaluable with the help of quantitative indicators. These indicators aim to give an overview of the MALDI-TOF MS performance in terms of providing (i) valid and (ii) correct identification results. With valid results ranging from 93.3% to 98.6% and correct results of 98.5% to 99.7% in this cohort, all three MALDI-TOF MS systems proved to be valuable tools for the microbiology laboratory. Inevitably, details suffer in this attempt to aggregate and summarize data. As an example, 17 isolates (4.8%) with a Biotyper score greater than 1.90 but just not the target score of 2.00 (=green result) had to be measured repeatedly by the Biotyper system. In a practical and pragmatic approach, which might be the daily routine in many diagnostic laboratories that aim to provide results for the attending physicians as soon as possible, such close results would probably not have triggered retesting— even when this practice must be validated and the results should be interpreted with caution. Consequently, it can be assumed that the critical evaluation of this study even tends to underestimate the performance of all three MALDIs.
Previously, anaerobe bacteria represented a particular challenge for MALDI-TOF MS analysis with notably lower identification rates compared to other groups of microorganisms (11, 12). Due to the continuous expansion and improvement of the spectra databases, identification rates for anaerobes today exceed 80% at the species level and 90% at the genus level, with a slight advantage reported favoring Vitek MS (13). Contrarily, the identification of Enterobacterales and non-fermenting Gram-negative bacilli was reported to be a specific strength of Biotyper (14, 15). The methodic limitations to differentiate between closely related Gram-negative species are reflected by the peculiarity of the Vitek MS not to commit to a single species ID for certain organism groups, for example, “Citrobacter braakii/freundii” or “Enterobacter cloacae/asburiae” (19). Also in our study, invalid Vitek MS results were observed more frequently with those organisms. A comparable finding occurred for EXS and anaerobe bacteria. However, the strain collection of this investigation had no particular focus on those groups and therefore lacks an adequate number of respective organisms to substantiate this finding, which should be addressed in more specific comparisons. Contrarily, the Biotyper system has some sort of intrinsic limitations regarding some organism groups: Bruker is the only manufacturer to provide a modular database with add-ons that are not available for all users but must be purchased separately. Depending on whether the respective databases are installed, identification of specific organism groups, for example, filamentous fungi or Mycobacteria, is not possible. Notably, the sole mycobacterial isolate included in this study (Mycobacterium abscessus) was not identified by the Bruker Biotyper using the IVD reference library but only after the optional Mycobacteria IVD Module was applied. Furthermore, the required database must be selected as a reference before analyzing the isolate, which has certain implications for laboratories and their workflows.
To our knowledge, this is the first study presenting a parallel comparison of three different MALDI-TOF MS devices and databases. Furthermore, all MALDI-TOF MS results were compared to the results of 16S rRNA gene sequencing representing the gold standard of identification. However, this study also has some limitations, that is, (i) the low sample number, (ii) the specific species spectrum, and (iii) the applied test systems. In the case of the latter, it should be considered that neither the bioMérieux nor the Bruker device used in this work represent the latest models. On the other hand, the respective differences between the bioMérieux VITEK MS PRIME (market launch: 2022) and the Bruker MALDI Biotyper sirius (market launch: 2019) are less to be found in the measurement technology but rather in the handling and the capability for high-throughput measurement. Both the most recent and the precursor models use the same spectrum database of the respective manufacturer. Hence, this study still allows a comparison of the performance in terms of measurement validity and database quality. With regard to the overall low sample size, it is evident that a study based on 356 isolates cannot provide a definitive comparison of the MALDI-TOF MS systems. However, the identity of all analyzed isolates was specified by 16S rRNA DNA gene sequencing representing a high-quality reference method. This gives this study an edge over other investigations just relying on a method comparison between two devices, which would not allow the detection of concordantly incorrect results. Notably, such results were observed in our analysis. Finally, the species included in this analysis are not representative of the spectrum of microorganisms that we are encountering in the daily routine of medical microbiology. However, with respect to the excellent performance reported for the identification of the most common pathogens, for example, Staphylococcus aureus or Klebsiella pneumoniae, it would not have strengthened the significance of this study to expand the sample collection with strains that do not represent any challenge for MALDI-TOF MS (8 – 10). Instead, this study focused on species that neither Biotyper nor Vitek MS has been successful in identifying in the past, which makes that organism collection an interesting study object.
Altogether, our data suggest that all three devices are suitable for the use in a diagnostic laboratory for medical microbiology, although specific weaknesses for each MALDI-TOF MS system were observed. Further studies are necessary to provide a more profound comparison.
ACKNOWLEDGMENTS
We thank bioMérieux, Bruker Daltonic, and Zybio for technical support. We thank Zybio for providing the instrument and reagents.
The study was funded by the research budget of the Diagnostic and Research Institute of Hygiene, Microbiology and Environmental Medicine, Medical University of Graz.
Conceptualization: K.D., I.S., and E.L. Methodology: K.D., and E.L. Formal analysis: K.D., I.K., H.G., and E.L. Investigation: K.D., I.K., H.G., J.L., J.K., S.F., and E.L. Resources: K.D., I.S., and E.L. Writing—original draft: K.D. Writing—review and editing: K.D., I.K., I.S., and E.L. Visualization: K.D., and I.K. Supervision: K.D. and E.L. Project administration: K.D., S.F., and E.L.
Authors declare no conflict of interest.
None of the three manufacturers, whose products were compared in this work, were involved in the design or evaluation of this study.
Contributor Information
Karl Dichtl, Email: karl.h.dichtl@gmail.com.
Erin McElvania, NorthShore University HealthSystem, Evanston, Illinois, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.01913-22.
MALDI-TOF measurement results for each included isolate.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Vallicelli C, Santandrea G, Sartelli M, Coccolini F, Ansaloni L, Agnoletti V, Bravi F, Catena F. 2022. Sepsis team organizational model to decrease mortality for intra-abdominal infections: is antibiotic stewardship enough? Antibiotics (Basel) 11:1460. doi: 10.3390/antibiotics11111460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torres-Sangiao E, Leal Rodriguez C, García-Riestra C. 2021. Application and perspectives of MALDI-TOF mass spectrometry in clinical microbiology laboratories. Microorganisms 9:1539. doi: 10.3390/microorganisms9071539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen XF, Hou X, Xiao M, Zhang L, Cheng JW, Zhou ML, Huang JJ, Zhang JJ, Xu YC, Hsueh PR. 2021. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) analysis for the identification of pathogenic microorganisms: a review. Microorganisms 9:1536. doi: 10.3390/microorganisms9071536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee H, Park JH, Oh J, Cho S, Koo J, Park IC, Kim J, Park S, Choi JS, Shin SY, Sung GH, Kim J. 2019. Evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for the identification of yeast isolation. J Clin Lab Anal 33:e22685. doi: 10.1002/jcla.22685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi Y, Kim D, Choe KW, Lee H, Kim J-S, Ahn J-Y, Lee M-K. 2022. Performance evaluation of Bruker Biotyper, ASTA microidsys, and Vitek-MS and three extraction methods for filamentous fungal identification in clinical laboratories. J Clin Microbiol 60:e0081222. doi: 10.1128/jcm.00812-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ko YJ, Lee OJ, Lee SB, Kim CM, Lee J, Kook JK, Park SN, Shin JH, Kim SH, Won EJ, Park G, Kang SH, Jang SJ. 2022. Accuracy of ASTA microidsys, a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system, for the identification of Korean reference and clinical bacterial and yeast strains. Diagn Microbiol Infect Dis 103:115658. doi: 10.1016/j.diagmicrobio.2022.115658 [DOI] [PubMed] [Google Scholar]
- 7. McElvania TeKippe E, Burnham C-AD. 2014. Evaluation of the Bruker Biotyper and Vitek MS MALDI-TOF MS systems for the identification of unusual and/or difficult-to-identify microorganisms isolated from clinical specimens. Eur J Clin Microbiol Infect Dis 33:2163–2171. doi: 10.1007/s10096-014-2183-y [DOI] [PubMed] [Google Scholar]
- 8. Bilecen K, Yaman G, Ciftci U, Laleli YR. 2015. Performances and reliability of Bruker microflex LT and Vitek MS MALDI-TOF mass spectrometry systems for the identification of clinical microorganisms. Biomed Res Int 2015:516410. doi: 10.1155/2015/516410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Porte L, García P, Braun S, Ulloa MT, Lafourcade M, Montaña A, Miranda C, Acosta-Jamett G, Weitzel T. 2017. Head-to-head comparison of microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI-TOF mass spectrometry in Chile. PLoS One 12:e0177929. doi: 10.1371/journal.pone.0177929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lévesque S, Dufresne PJ, Soualhine H, Domingo M-C, Bekal S, Lefebvre B, Tremblay C. 2015. A side by side comparison of Bruker Biotyper and Vitek MS: utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One 10:e0144878. doi: 10.1371/journal.pone.0144878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagy E, Becker S, Kostrzewa M, Barta N, Urbán E. 2012. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J Med Microbiol 61:1393–1400. doi: 10.1099/jmm.0.043927-0 [DOI] [PubMed] [Google Scholar]
- 12. La Scola B, Fournier P-E, Raoult D. 2011. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe 17:106–112. doi: 10.1016/j.anaerobe.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 13. Li Y, Shan M, Zhu Z, Mao X, Yan M, Chen Y, Zhu Q, Li H, Gu B. 2019. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect Dis 19:941. doi: 10.1186/s12879-019-4584-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, Howard W, Pruessner J, Safwat N, Cockerill FR, Bossler AD, Patel R, Richter SS. 2012. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J Clin Microbiol 50:2034–2039. doi: 10.1128/JCM.00330-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alby K, Gilligan PH, Miller MB. 2013. Comparison of matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry platforms for the identification of gram-negative rods from patients with cystic fibrosis. J Clin Microbiol 51:3852–3854. doi: 10.1128/JCM.01618-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu K, Huang Z, Li Y, Fu Q, Lin L, Wu S, Dai H, Cai H, Xiao Y, Lan R, Wang D. 2021. Establishment and application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for detection of Shewanella genus. Front Microbiol 12:625821. doi: 10.3389/fmicb.2021.625821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang L, Liu M, Huang C, Ma X, Zheng Y, Wu W, Guo J, Huang J, Xu H. 2022. MALDI-TOF MS-based clustering and antifungal susceptibility tests of Talaromyces marneffei isolates from Fujian and Guangxi (China). Infect Drug Resist 15:3449–3457. doi: 10.2147/IDR.S364439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Gan Z, Zhou X, Chen Z. 2022. Accurate classification of listeria species by MALDI-TOF mass spectrometry incorporating denoising autoencoder and machine learning. J Microbiol Methods 192:106378. doi: 10.1016/j.mimet.2021.106378 [DOI] [PubMed] [Google Scholar]
- 19. Lee Y, Sung JY, Kim H, Yong D, Lee K. 2017. Comparison of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform, ASTA MicroIDSys, with Bruker Biotyper for species identification. Ann Lab Med 37:531–535. doi: 10.3343/alm.2017.37.6.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MALDI-TOF measurement results for each included isolate.