Abstract
This review article explores the potential of metformin, a medication commonly used for type 2 diabetes, as an antiviral and anti-inflammatory agent in the context of coronavirus disease 2019 (COVID-19). Metformin has demonstrated inhibitory effects on the growth of SARS-CoV-2 in cell culture models and has shown promising results in reducing viral load and achieving undetectable viral levels in clinical trials. Additionally, metformin exhibits anti-inflammatory properties by reducing the production of pro-inflammatory cytokines and modulating immune cell function, which may help prevent cytokine storms associated with severe COVID-19. The drug’s ability to regulate the balance between pro-inflammatory Th17 cells and anti-inflammatory Treg cells suggests its potential in mitigating inflammation and restoring T cell functionality. Furthermore, metformin’s modulation of the gut microbiota, particularly changes in bacterial taxa and the production of short-chain fatty acids, may contribute to its therapeutic effects. The interplay between metformin, bile acids, the gut microbiome, glucagon-like peptide-1 secretion, and glycemic control has implications for the management of diabetes and potential interventions in COVID-19. By refreshing the current evidence, this review highlights the potential of metformin as a therapeutic option in the management of COVID-19, while also exploring its effects on the gut microbiome and immunometabolism.
Keywords: gut microbiota, coronavirus disease 2019, dysbiosis, metformin, diabetes
Introduction
Coronavirus disease 2019 (COVID-19), a global pandemic caused by the novel coronavirus Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has impacted millions of individuals across the globe (Atzrodt et al., 2020). People with pre-existing conditions like type 2 diabetes (T2D) are at a higher risk of experiencing severe outcomes from COVID-19 (Al-Kuraishy et al., 2021; Kamyshnyi et al., 2021). T2D is a chronic metabolic disorder characterized by insulin resistance and elevated blood glucose levels, which can lead to various complications and impaired immune function (Hameed et al., 2015).
Metformin, a commonly prescribed medication for T2D, has garnered attention for its potential benefits beyond glycemic control (Kamyshnyi et al., 2021). Recent research indicates that metformin may possess antiviral and anti-inflammatory properties, making it a promising candidate for combating SARS-CoV-2 infection and reducing the adverse effects of COVID-19 (Petakh et al., 2022a; Petakh et al., 2022b; Petakh et al., 2022c; Petakh et al., 2023a; Petakh et al., 2023b; Petakh et al., 2023c; Petakh et al., 2023d; Erickson et al., 2023).
This review article aims to present an overview of the current evidence related to metformin’s potential anti-SARS-CoV-2 effects, its anti-inflammatory properties, and its role in modulating the Th17/Treg balance. Additionally, it delves into the interactions between metformin, the gut microbiota, bile acids, and glycemic control concerning COVID-19 and T2D.
Anti-SARS-CoV-2 effect of metformin
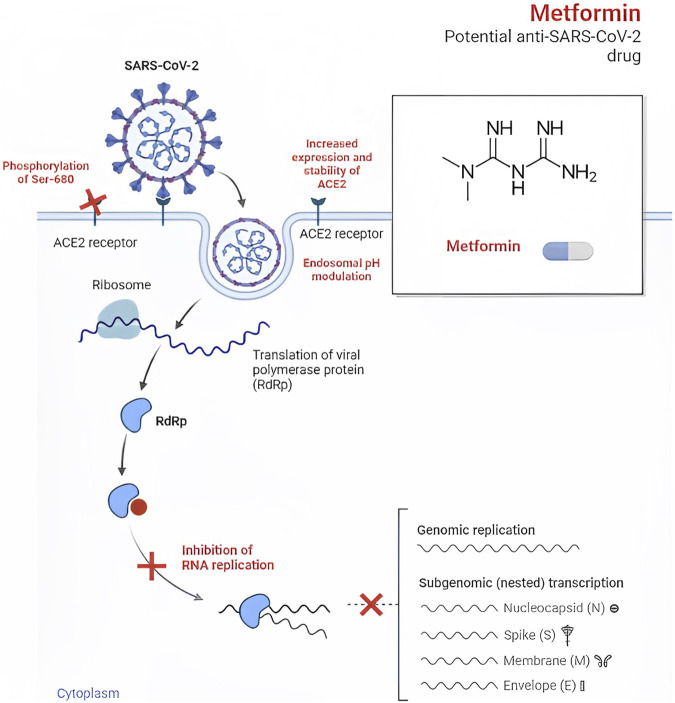
Metformin, an established medication for T2D, has recently gained attention for its potential antiviral properties against SARS-CoV-2, as well as other RNA viruses such as Zika, dengue, hepatitis B, hepatitis C, influenza, and human immunodeficiency viruses (HIV) (Farfan-Morales et al., 2021; Justice et al., 2021; Wiernsperger et al., 2022). Early studies in the 1940s even demonstrated the beneficial effects of metformin in treating patients with influenza (Ibrahim et al., 2021). In the context of SARS-CoV-2, metformin may exert its antiviral effects by preventing viral entry into cells through conformational changes in the angiotensin I converting enzyme 2 (ACE2) receptor, which is thought to be facilitated by AMP-activated protein kinase (AMPK)-mediated phosphorylation at S680 of the ACE2 protein (Sharma et al., 2020).
Additionally, metformin’s impact on intracellular pH regulation within endosomes is noteworthy. Key components involved in pH regulation within endosomes, such as Vacuolar ATPase (V-ATPase) and endosomal Na+/H+ exchangers (eNHE), play critical roles in this process. Research has indicated that metformin can directly impact eNHE and/or V-ATPase, leading to elevated pH levels within endosomes. This elevation in pH levels may suppress viral infection (Esam, 2020; Eaton et al., 2021). Moreover, metformin might also have the potential to prevent the development of pulmonary fibrosis, a complication associated with COVID-19 (Esam, 2020).
In a study conducted by Haripriya Parthasarathy et al., metformin’s significant inhibitory effect on the growth of SARS-CoV-2 was demonstrated in cell culture models (Parthasarathy et al., 2022). As the viral infection progressed, AMPK phosphorylation showed a steady increase, indicating its crucial role during the infection process. When AMPK was activated in Calu3 and Caco2 cell lines using metformin, there was a remarkable suppression of SARS-CoV-2 infectious titers, with up to 99% reduction observed in infected cells. Dose-variation studies revealed half maximal inhibitory concentration (IC50) values of 0.4 mM and 1.43 mM in Calu3 and Caco2 cells, respectively. The involvement of AMPK in metformin’s antiviral effect was further confirmed by using other pharmacological compounds such as 5-Aminoimidazole-4-carboxamide ribonucleoside (AICAR) and Compound C (Table 1; Figure 1).
TABLE 1.
Examination of the impact of metformin on SARS-CoV-2 infection models.
| Author, year | Study design | Substrate | Key findings |
|---|---|---|---|
| Bramante et al. (2023) | In vivo (randomized trial) | COVID-19 patients | Metformin demonstrated a possible benefit in preventing more severe outcomes such as emergency department visits, hospitalization, or death |
| Carolyn et al. (2023) | In vivo (randomized trial) | COVID-19 patients | Metformin showed a 42% reduction in ER visits/hospitalizations/death through 14 days and a 58% reduction in hospitalizations/death through 28 days. Furthermore, metformin demonstrated a 42% reduction in Long COVID through 10 months. Viral load analysis revealed a 3.6-fold reduction with metformin compared to placebo |
| Sun et al. (2022b) | In vitro experimental study | Human airway epithelial cell lines (BEAS2B, A549, and 16HBE) | Metformin exhibited multifaceted effects, including restoration of autophagy, suppression of pyroptosis, and attenuation of inflammatory response |
| Parthasarathy et al. (2022) | In vitro experimental study | Calu3 (respiratory epithelial cell line) and Caco2 (gut epithelial cell line) | Metformin pretreatment effectively suppressed viral replication and protein expression in both respiratory and gut epithelial cell lines |
| Ventura-López et al. (2022) | In vitro experimental study | H1299 and Vero E6 cell lines | Metformin glycinate demonstrated significant reduction in viral load and enhanced cell viability against different SARS-CoV-2 variants |
| Mercado-Gómez et al. (2022) | In vitro experimental study | Human primary hepatocytes, human upcyte second-generation hepatocytes, humanized ACE2 (hACE2) mice, and wild-type mice | Metformin exhibited hepatoprotective effects by suppressing ACE2 expression, reducing viral infection rates, and modulating inflammatory markers in hepatocytes |
| Cory et al. (2021) | In vitro experimental study | Purified classical monocytes from healthy human subjects | Metformin pretreatment resulted in the suppression of glycolytic response and downregulation of pro-inflammatory cytokines upon viral exposure |
| Chen et al. (2021) | In vitro and in vivo assays | In vivo: Midbrain dopaminergic neurons derived from H9 human embryonic stem cells injected into mice. In vitro: Midbrain dopaminergic neuron cell line derived from human pluripotent stem cells | Metformin exhibited antiviral effects by reducing viral RNA levels and preventing cellular senescence in midbrain dopaminergic neurons |
| Schaller et al. (2021) | Ex vivo and in vitro assays | Cryopreserved bank of human lung tissue and Vero E6 cell line | Metformin exhibited variable efficacy, with significant reduction in SARS-CoV-2 titers observed in lung tissues but not in Vero E6 cell line |
| Xian et al. (2021) | In vitro and in vivo assays | Bone marrow–derived macrophages from nondiabetic mice | Metformin displayed immunomodulatory effects by inhibiting NLRP3 inflammasome activation, cytokine production, and mitochondrial dysfunction |
| Gordon et al. (2020) | In vitro experimental study | Vero E6 cell line | Metformin demonstrated potent antiviral activity by inhibiting viral replication and promoting cellular viability |
FIGURE 1.
Effects against SARS-CoV-2.
Additionally, promising results were observed in the viral load analysis from a clinical trial (Carolyn et al., 2023). The mean change in viral load from baseline to follow-up was notably lower with metformin compared to the placebo group (−0.64 log10 copies/mL), indicating a 4.4-fold greater decrease in viral load. On day 5, the rate of undetectable SARS-CoV-2 viral load was 49.9% in the metformin group and 54.6% in the placebo group, with a modest odds ratio of 1.235. Similarly, on day 10, the undetectable rate was 14.3% in the metformin group and 22.6% in the placebo group, with an odds ratio of 1.663, demonstrating a statistically significant difference between the groups.
Ventura-López et al. conducted a trial where metformin treatment demonstrated substantial advantages compared to the placebo group. The metformin-treated participants experienced a significant decrease in the need for supplemental oxygen, a more pronounced reduction in the percentage of viral load, and a quicker attainment of an undetectable viral load. However, there were no significant differences in the duration of hospitalization between the metformin and placebo groups (Ventura-López et al., 2022).
In the TOGETHER Trial, the use of metformin did not yield a significant decrease in hospitalizations due to COVID-19. Hospitalization was defined as either remaining in a COVID-19 emergency setting for more than 6 h or being transferred to a tertiary care hospital within 28 days after randomization. The statistical analysis showed that there was no significant difference between the metformin and placebo group, with a relative risk of 1.14 and a 95% credible interval of 0.73–1.81. Additionally, metformin did not significantly affect viral clearance at day 7, time to hospitalization, or clinical improvement 28 days after randomization. However, when considering the per-protocol sample, which accounted for 83% of the participants, there was a reduced likelihood of emergency department visits and hospitalizations of COVID-19 patients, resulting in an absolute risk reduction of 1.4% and 3.1%, respectively (Reis et al., 2022).
Interferon responses to SARS-CoV-2 and SAMHD1-cGAS-STING-metformin interactions
Sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) negatively regulates the interferon −1 signaling pathway: the elevated innate immune response and IFN activation upon genetic loss of SAMHD1 effectively suppress SARS-CoV-2 replication (Oo et al., 2022).
The cGAS-STING pathway is central to detecting viral DNA and initiating immune responses. Activation of cGAS-STING triggers IRF3-mediated type I IFN production and autophagy-mediated antiviral activity. cGAS produces cyclic GMP-AMP (cGAMP), activating STING (Su et al., 2023). This pathway responds to viral and bacterial DNA, as well as self-DNA from senescent or dying cells.
SARS-CoV-2 infection induces micronuclei formation, activating the cGAS-STING pathway, which can damage cells. Viral proteins like ORF3a and ORF10 can interfere with this pathway to evade immune responses (Han et al., 2022).
Metformin, a versatile drug, has significant effects on cGAS-STING signaling. For example, in gastric cancer, metformin promotes cGAS/STING activation by inhibiting AKT phosphorylation (Shen et al., 2023). It also activates type I IFN signaling against hepatitis C virus through AMPK (Tsai et al., 2017).
However, it is important to note that metformin can have inhibitory effects on the type I IFN response in specific immune cells, like human CD4+ T cells (Titov et al., 2019). This complex interplay between metformin, the cGAS-STING pathway, and type I IFN signaling underscores the intricate mechanisms involved in the immune response against viral infections, including SARS-CoV-2.
Anti-inflammatory effect of metformin: a protective mechanism against cytokine storm
Metformin exerts its effects through various mechanisms, including the activation of liver kinase B1, which leads to the activation of AMPK, as well as the activation of NAD-dependent deacetylase sirtuin-1 (SIRT1) and peroxisome proliferator–activated receptor γ coactivator-1 α (PGC-1α) (Kulkarni et al., 2020; Triggle et al., 2022). Furthermore, metformin inhibits mitochondrial complex 1, nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), and mammalian target of rapamycin complex 1 (mTORC1) (Kulkarni et al., 2020).
Metformin possesses anti-inflammatory properties, beneficial for individuals both with and without diabetes (Cameron et al., 2016). In the context of COVID-19, its anti-inflammatory actions involve reducing the levels of various pro-inflammatory factors, including tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), interleukin-6 (IL-6), C-X-C motif chemokine ligand 5 (CXCL5), C-X-C motif chemokine ligand 10 (CXCL10), and monocyte chemoattractant protein-1 (MCP-1) (Justice et al., 2021). Moreover, metformin inhibits the signal transducer and activator of transcription 3 (STAT3) and has been shown to decrease the formation of neutrophil extracellular traps (Bailey and Gwilt, 2022). Another potential effect of metformin is preventing the activation of the NLR family pyrin domain-containing 3 (NLRP3) inflammasome (Wiernsperger et al., 2022).
Observational data suggest that prior usage of metformin before COVID-19 infection is associated with lower levels of peak C-reactive protein (CRP), as well as lower rates of admission and peak ferritin in a subgroup analysis of patients in the intensive care unit (Ma et al., 2022).
Metformin modulation of Th17/Treg balance: anti-inflammatory effects. Targeting immunometabolism to prevent cytokine storm in COVID-19
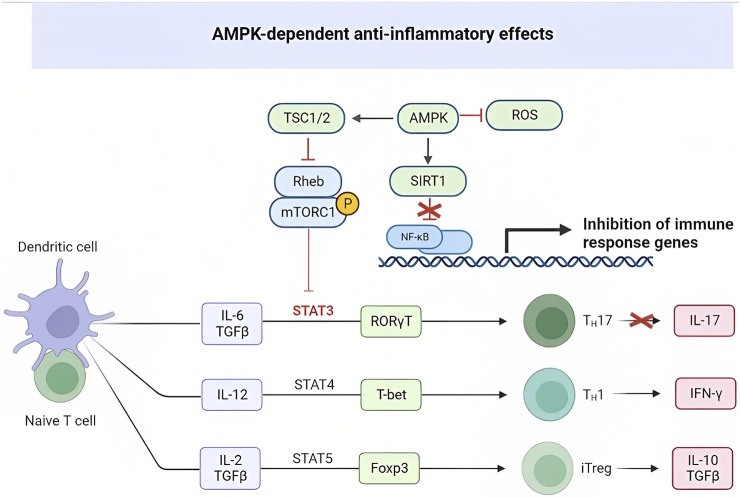
Metformin has been shown to affect Th17 cell differentiation through the AMPK/mTOR/STAT3 pathway (Tan et al., 2019). In diseases like COVID-19 and systemic lupus erythematosus (SLE), there is an increase in Th17 cells (Duan et al., 2019). Metformin’s influence on CD4+ T cell glucose metabolism, achieved by inhibiting mitochondrial complex I and oxidative phosphorylation, helps normalize cellular processes crucial for CD4+ T cell activation, proliferation, and differentiation. By targeting overactive effector T cells, including Th1 and Th17 cells, as well as proinflammatory cytokines such as interferon interferon-gamma (IFN-γ) and IL-17, metformin shows potential in reducing inflammation in Systemic Lupus Erythematosus (SLE) (Tan et al., 2019). In viral infections such as COVID-19, CD8+ T cells play a vital role in eliminating the virus by releasing cytotoxic molecules like perforin, granzyme, and IFN-γ (Omarjee et al., 2020). The use of metformin at the doses prescribed for diabetes treatment could potentially help restore T cell functionality and alleviate the cytokine storm commonly observed in COVID-19 (Salvatore et al., 2020; Scheen, 2020; Sharma et al., 2020) (Figure 2).
FIGURE 2.
AMPK-dependent anti-inflammatory effects.
AMPK and mTOR serve as metabolic detectors that influence the balance between pro-inflammatory and anti-inflammatory cells. Th17 cells rely on glycolysis for their energy needs, while regulatory T cells (Tregs) rely on fatty acid oxidation. AMPK’s ability to regulate metabolism affects the differentiation of these cell types. On the other hand, mTOR activation, leading to the induction of HIF1α, promotes glucose import and glycolysis at the transcriptional and translational level. Lack of HIF1α induction significantly reduces Th17 cells. In a rat model of collagen-induced arthritis (CIA), metformin was found to activate AMPK, inhibit mTOR, and regulate the Th17/Treg ratio. Metformin also reduced the levels of proinflammatory cytokines TNFα, IL-1β, IL-6, and IL-17 in the serum of rats with CIA, while decreasing the number of splenic CD4+/RORγt+/IL-17 + T cells (Th17s) in a dose-dependent manner. Furthermore, metformin positively correlated with the increase of regulatory T cells (CD4+/CD25+/FOXP3+) in the study (Yang et al., 2017). Likewise, metformin played a role in alleviating autoimmune insulitis in a model of Type 1 diabetes (T1D) using NOD mice. Female NOD mice were administered metformin or a control substance starting at 4 weeks of age. By the time the mice reached 12 weeks of age, they showed signs of insulitis. However, the mice treated with metformin displayed a higher number of functional β cells compared to those on the control treatment. Additionally, metformin significantly reduced the number of pro-inflammatory IFN-γ+ and IL17+ CD4 T cells in the spleen of NOD mice, while concurrently increasing the presence of regulatory IL-10+ and Foxp3+ CD4-T cells. This effect resulted in the mitigation of autoimmune insulitis (Duan et al., 2019). The imbalance in Teff/Treg ratios observed in collagen-induced arthritis (CIA) and Type 1 diabetes (T1D) is also evident in various other diseases, including T2D, obesity, aging, and rheumatoid arthritis.
Relationship between the gut microbiota and metformin
Metformin is a widely used medication for the treatment of T2D that has been shown to improve insulin sensitivity, reduce blood glucose levels, and decrease the risk of cardiovascular disease (Kamyshnyi et al., 2021; Mohammed et al., 2021). Nevertheless, the precise mechanism of how metformin works remains incompletely understood. Lately, there has been increasing attention given to the connection between metformin and the gut microbiome. Several studies have indicated that metformin could potentially influence the gut microbiome, and these changes in the gut microbiome might be linked to metformin’s therapeutic effects (Table 2; Figure 3) (Forslund et al., 2015; Wu et al., 2017a; De La Cuesta-Zuluaga et al., 2017; Barengolts et al., 2018; Sun et al., 2018; Bryrup et al., 2019; Zhang et al., 2019; Chávez-Carbajal et al., 2020).
TABLE 2.
Major findings from the research of patients with T2D with/without metformin treatment.
| Sample size | Year of publication | Age | Techique | Associated microbiota changes | References |
|---|---|---|---|---|---|
| 6 T2D with obesity | 2020 | 47.0 ± 4.5 years | Whole-genome metagenomics shotgun | ↓Firmicutes, Oribacterium, and Paenibacillus | Wang et al. (2020) |
| 6 Controls | |||||
| 183 T2D | 2012 | 13– years | Metagenomic sequencing | ↑Akkermansia muciniphila, Bacteroides caccae, Clostridium hathewayi, Clostridium ramosum, Clostridium symbiosum, Desulfovibrio spp., Eggerthella lenta, and Escherichia coli | Qin et al. (2012) |
| 185 Controls | ↓Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia intestinalis, and Roseburia inulinivorans | ||||
| Healthy controls had a ↑ abundance of butyrate-producing bacteria | |||||
| 53 TD | 2013 | 69–72 years | Metagenomic sequencing | ↑ Clostridium clostridioforme, Lactobacillus spp., and Streptococcus mutans | Karlsson et al. (2013) |
| 49 Impaired glucose tolerance | ↓ Roseburia and Clostridium spp., Eubacterium eligens, Bacteroides intestinalis | ||||
| 43 Controls | |||||
| T2D with good glycemic control (52 patients) | 2020 | T2D with good glycemic control (66.38 ± 1.314 years) | 16S rRNA sequencing | RT2D ↑ Bacteroides vulgatus and Veillonella denticariosi | Shih et al. (2020) |
| Refractory T2D (RT2D) (27 patients) | RT2D (64.37 ± 2.194 years) | RT2D ↓ Akkermansia muciniphila and Fusobacterium spp. | |||
| Newly diagnosed T2D (50 patients) 50 Controls | 2018 | 51 ± 12 years | 16S rRNA sequencing | ↑ Lactobacillus spp. | Chen et al. (2019) |
| ↓ Clostridium leptum and Clostridium coccoides | |||||
| 134 Prediabetic | 2018 | 57–68 years | 16S rRNA sequencing | ↓ Akkermansia muciniphila and Clostridium spp. | Allin et al. (2018) |
| 134 Controls | |||||
| Treatment-naïve T2D (77 patients) | 2019 | 61.75 ± 9.09 years | Whole-genome metagenomics shotgun | ↑ Escherichia coli, Eggerthella spp., Streptococcus salivarius | Zhong et al. (2019) |
| 80 Prediabetic | ↓ Clostridium spp., Faecalibacterium prausnitzii | ||||
| 97 Controls | |||||
| 18 T2D | 2010 | 31–73 | 16S rRNA sequencing | ↑ Bacteroidetes, Clostridium coccoides, Firmicutes | Larsen et al. (2010) |
| 18 Controls | ↓ Proportions of phylum Firmicutes and class Clostridia | ||||
| 40 T2D | 2016 | 40–77 | 16S rRNA sequencing | ↑ Collinsella spp., Streptococcus spp., Lactobacillus spp. | Candela et al. (2016) |
| 13 Controls | ↓ Bacteroides spp., Prevotella spp., Roseburia spp., Faecalibacterium spp. | ||||
| 25 T2D | 2021 | T2D (62.52 ± 7.58) | 16S rRNA sequencing | ↑ Bifidobacterium spp. and Lactobacillus spp. | Huang et al. (2021) |
| 25 TD2 with retinopathy | Controls (57.8 ± 10.06) | ↓Escherichia-Shigella, Faecalibacterium, Eubacterium_hallii_group and Clostridium genera | |||
| 25 Controls | TD2 with retinopathy (60.28 ± 10.5) | ||||
| Metformin-treated (MT) 93 | 2015 | 35–75 years | Metagenomic | ↑Escherichia spp., ↓Intestinibacter spp. | Forslund et al. (2015) |
| Metformin-untreated (MUT) 106 | |||||
| MT 14 | 2017 | 18–62 years | 16S rRNA sequencing | ↑ Prevotella spp., Megasphaera spp., Butyrivibrio spp | De La Cuesta-Zuluaga et al. (2017) |
| MUT 14 | Akkermansia muciniphila | ||||
| ↓ Oscillospira spp., Barnesiellaceae | |||||
| 27 healthy young men without T2D | 2019 | 18–35 years | 16S rRNA sequencing | ↑ Escherichia/Shigella spp., Bilophila wadsworthia | Bryrup et al. (2019) |
| ↓Intestinibacter spp., Clostridium spp. | |||||
| Treatment-naïve T2D (22 patients) | 2017 | NA | Metagenomic | ↑Pectobacterium spp., Pantoea spp., Serratia spp., Dickeya spp., Helicobacter spp., Cronobacter spp., Erwinia spp., Yersinia spp., Enterobacter spp., Citrobacter spp., Escherichia spp., Bacillus spp | Wu et al. (2017a) |
| Treatment-naïve T2D with metformin treatment 4 months (22 patients) | ↓ Dethiosulfovibrio spp., Deferribacter spp., Subdoligranulum spp., Intestinibacter spp. | ||||
| Treatment-naïve T2D (22 patients) | 2018 | NA | Metagenomic | ↓Bacteroides fragilis | Sun et al. (2018) |
| Treatment-naïve T2D with metformin treatment 3 days (22 patients) | |||||
| MT 21 | 2018 | 35–70 | 16S rRNA sequencing | ↑Bifidobacterium spp., Catenibacterium spp., Parabacteroides spp. | Barengolts et al. (2018) |
| MUT 11 | |||||
| T2D patients before MT 26 | 2019 | NA | 16S rRNA sequencing | ↑ Spirochaete spp., Turicibacter spp., and Fusobacterium spp. | Zhang et al. (2019) |
| T2D patients after 3 months of MT 51 | |||||
| Treatment-naïve T2D (14 patients) | 2020 | 48.1 ± 4.7 | 16S rRNA sequencing | ↑Pelomonas spp. | Chávez-Carbajal et al. (2020) |
| Treatment-naïve T2D with metformin treatment 3 days (14 patients) |
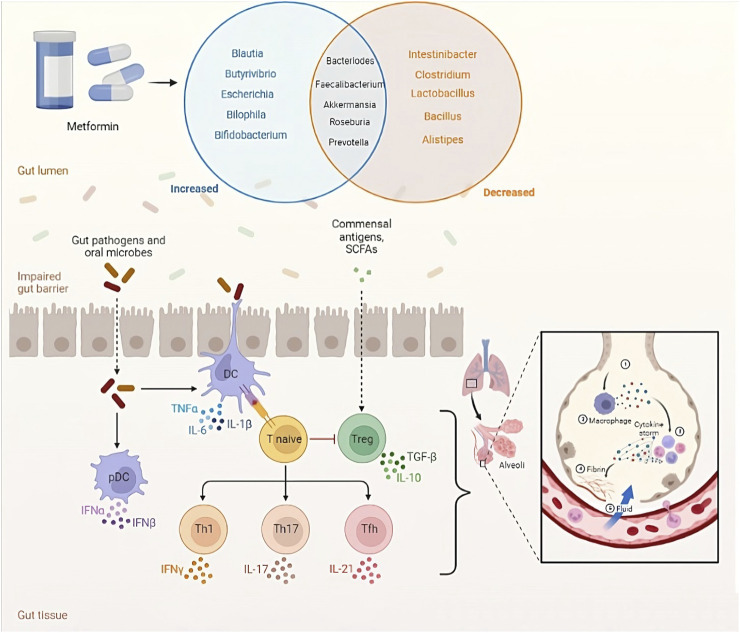
FIGURE 3.
Microbiota-mediated effects of metformin.
Research has revealed that the use of metformin is linked to alterations in the relative abundance and diversity of specific gut bacterial taxa (Petakh et al., 2023e). For instance, certain studies have found that metformin use is associated with an increased relative abundance of the genus Lactobacillus and a decreased relative abundance of the genus Bacteroides (Lee et al., 2021). Additionally, other studies have indicated that metformin use is linked to an increased relative abundance of the genus Akkermansia, a genus known for its potential to enhance insulin sensitivity (Ke et al., 2021).
The exact mechanisms by which metformin modulates the gut microbiome and how changes in the gut microbiome contribute to metformin’s therapeutic effects are not fully understood. Several potential mechanisms have been proposed, including the reduction of pro-inflammatory cytokines and the increase of short-chain fatty acids (SCFAs) (den Besten et al., 2013). In particular, SCFAs can bind to G-protein-coupled receptors (GPCRs) such as free fatty acid receptor 3 (FFAR3) and free fatty acid receptor 2 (FFAR2), which are expressed on enteroendocrine L cells, causing the release of glucagon-like peptide-1 (GLP-1) and Peptide YY. These hormones regulate glucose metabolism and insulin secretion (Holz et al., 1993). In rodent studies, an increase in acetate production caused by changes in gut microbiota activates the parasympathetic nervous system, leading to an increase in insulin secretion in response to glucose and increased secretion of ghrelin. This creates a positive feedback loop, causing excessive eating (hyperphagia), increased fat storage, and ultimately obesity (Perry et al., 2016). Long-term delivery of propionate to the colon has been found to significantly reduce weight gain and accumulation of fat in the abdomen, and also prevents a decline in insulin sensitivity in overweight adults (Chambers et al., 2015).
Human studies have demonstrated the beneficial effects of short-chain fatty acids (SCFAs) on glucose homeostasis and insulin sensitivity. For example, one study investigated the impact of delivering propionate to the colon in overweight and obese individuals using inulin-propionate ester (IPE). The supplementation improved insulin sensitivity compared to a low-fermentable fiber control (cellulose) and led to increased secretion of GLP-1 and Peptide YY. Additionally, changes in gut bacterial composition and markers of systemic inflammation were observed, highlighting the significant physiological impact of raising colonic propionate delivery in humans (Ch et al., 2019).
Another pilot-and-feasibility trial utilized high-amylose maize-resistant starch modified with acetate and butyrate (HAMSAB), resulting in increased SCFA concentrations in stools and plasma, along with alterations in gut microbiota composition and function. Subjects with the highest SCFA concentrations exhibited better glycemic control, although glucose control and insulin requirements remained unchanged (Bell et al., 2022). These findings suggest that targeting the gut microbiota and its metabolites, particularly SCFAs, may hold therapeutic potential for treating metabolic disorders like obesity and diabetes.
Metformin has been extensively researched in both animal and human subjects, and the findings suggest that it can influence the secretion of gut hormones and increase glucose uptake and utilization in the human intestine (Pénicaud et al., 1989; Bailey et al., 1992; Wilcock and Bailey, 1994; Ma et al., 2004; Duca et al., 2015; Koffert et al., 2017). However, the precise mechanism by which metformin enhances gut glucose uptake and utilization remains unclear.
Some studies have reported that metformin reduces the activity of sodium-glucose transporter 1 (SGLT1) while increasing the recruitment of glucose transporter 2 (GLUT2) to the apical membrane of rat jejunum (Sakar et al., 2010). Meanwhile, other research indicates that metformin increases SGLT1 gene expression in the duodenum and jejunum, but has no significant effect on GLUT2 gene expression in the intestine (Lenzen et al., 1996). Tongzhi Wu et al. also investigated that metformin inhibits small intestinal glucose absorption, which may contribute to augmented GLP-1 secretion (Wu et al., 2017b).
One of the mechanisms that triggers GLP-1 release is glucose absorption in the small intestine (Kuhre et al., 2015). Glucose-induced GLP-1 release is triggered by various underlying mechanisms, but it seems that SGLT1 plays a dominant role (Par et al., 2012). SGLT1 mediates the uptake of glucose with Na+, which depolarizes the membrane and stimulates Ca2+ entry. This, in turn, leads to the secretion of GLP-1 (Gorboulev et al., 2011; Kuhre et al., 2015). SGLT1’s dominant role in glucose-stimulated GLP-1 secretion is further supported by the fact that SGLT1−/− mice not only show impaired glucose absorption but also impaired GLP-1 release (Gorboulev et al., 2012).
In studies using germ-free mice as a “microbial knockout” model, the transplantation of gut microbiota from healthy mice led to modifications in genes related to glucose metabolism in the gut (El Aidy et al., 2013). Prebiotics and probiotics have also been found to influence the gut microbiome, affecting GLP-1 secretion (Ejtahed et al., 2012; Balakumar et al., 2018). Bauer et al. demonstrated that metformin can alter the gut microbiome in the upper small intestine, resulting in an increase in SGLT-1 expression. In rodents fed a high-fat diet, SGLT-1 expression was reduced but could be restored with metformin administration (Bauer et al., 2018).
Research has shown that the presence of Lactobacillus significantly increases after metformin treatment, suggesting a possible link between Lactobacillus and the modification of SGLT-1 following metformin administration. This increase in SGLT-1 mediated metabolites produced by Lactobacillus has been found to enhance glucose uptake in Caco-2 cells, supporting the idea that Lactobacillus may be involved in the regulation of glucose metabolism influenced by metformin (Rooj et al., 2010). Thus, these findings indicate that Lactobacillus might play a role in regulating glucose metabolism and may be associated with improvements in glucose levels in both rodents and humans taking probiotic supplements containing Lactobacillus (Yadav et al., 2007). However, the specific mechanism by which metformin alters the amount of Lactobacillus in the gut remains unknown.
In conclusion, the relationship between the gut microbiome and metformin is an emerging field of research. While some studies have shown that metformin use is associated with changes in the gut microbiome, the exact mechanisms by which metformin modulates the gut microbiome and how changes in the gut microbiome affect metformin’s therapeutic effects are not yet fully understood and require further investigation.
Interplay between metformin, bile acids, gut microbiome, GLP-1, and glycemic control in diabetes
Bile acids, traditionally known for their role in fat digestion and absorption, have been found to act as signaling molecules, influencing blood glucose regulation. When administered to different parts of the gastrointestinal tract, bile acids increase plasma GLP-1 concentrations and attenuate the blood glucose response to small intestinal glucose infusion in both healthy individuals and those with T2D (Wu et al., 2013a; Wu et al., 2013b; Brønden et al., 2017). The glucose-lowering effect of bile acids is believed to be mediated by the GLP-1 receptor, as evidenced by the inhibition of this effect in T2D patients treated with a GLP-1 receptor antagonist (Sansome et al., 2020).
Roux-en-Y gastric bypass surgery, a type of bariatric surgery, enhances GLP-1 secretion and metabolic improvements by diverting bile from the duodenum to the distal small intestine (Larraufie et al., 2019; Madsen et al., 2019). Researchers are exploring bile acid-based therapies that could mimic the effects of bariatric surgery by delivering bile acids to the distal gut, which holds promise for managing T2D (81).
Bile acids can interact with different receptors, including Takeda G-protein-coupled receptor 5 (TGR5) and farnesoid X receptor (FXR), both expressed in L-cells (Makishima et al., 1999; Ding et al., 2015). TGR5 activation by bile acids increases GLP-1 secretion, while FXR activation has a more variable effect (Thomas et al., 2009; Li et al., 2013; Trabelsi et al., 2015; Kuhre et al., 2018). Additionally, bile acids can activate bitter taste receptors throughout the gastrointestinal tract, leading to GLP-1 secretion and weight loss in rodents (Dotson et al., 2008).
Bile acid sequestrants have been investigated as a therapy for T2D (Hansen et al., 2017). Although they moderately reduce blood glucose, they also decrease GLP-1 secretion when combined with exogenous or endogenous bile (Adrian et al., 2012; Hansen et al., 2016; Brønden et al., 2018). The exact role of this reduction in GLP-1 in the benefits of bile acid sequestrants remains unknown, necessitating further research on their chronic administration.
Metformin inhibits bile acid resorption, resulting in increased fecal bile salt excretion (Scarpello et al., 1998). This mechanism may explain the gastrointestinal adverse effects, such as diarrhea, associated with metformin (Watson et al., 2019). Moreover, by reducing proximal bile acid reabsorption, metformin increases the exposure of the distal gut to bile acids, potentially enhancing bile acid-induced GLP-1 secretion and glucose-lowering effects (Sansome et al., 2020).
The gut microbiota, a group of bacteria in the gastrointestinal tract, can be disrupted, leading to dysbiosis and contributing to various diseases like obesity, T2D, and allergies (Gomaa, 2020). Both metformin administration and T2D have been associated with changes in gut microbial composition (Qin et al., 2012; Forslund et al., 2015). Clinical trials have shown that metformin administration can lead to changes in several bacterial strains in healthy individuals and those with T2D (Wu et al., 2017a). These changes may improve glucose tolerance and insulin sensitivity. Notably, metformin use is linked to the reduction of Bacteroides fragilis and alterations in bile acid composition, which can enhance GLP-1 secretion and inhibit FXR activity (Sun et al., 2018). Although the specific bacterial strains affected may vary between studies, it is evident that metformin independently influences the gut microbiota regardless of T2D presence (Sansome et al., 2020).
The role of gut microbiota in SARS-CoV-2 infection
SARS-CoV-2, the causative agent of COVID-19, has been shown to infect the gastrointestinal tract, and alterations in gut microbiota composition have been linked to an increased susceptibility to viral infections (Li et al., 2022; Xiang and Liu, 2022). Studies have reported a decrease in beneficial gut bacteria, such as Faecalibacterium prausnitzii and Bifidobacterium, and an increase in pathogenic bacteria, such as Enterococcus faecalis and Streptococcus, in COVID-19 patients (Yeoh et al., 2021; Ancona et al., 2023; Petakh et al., 2023a). These changes in gut microbiota composition were associated with increased levels of inflammatory markers, such as IL-6 and CRP (Petakh et al., 2023a; Gradisteanu et al., 2023). Moreover, COVID-19 patients with gastrointestinal symptoms had a higher abundance of opportunistic pathogens, suggesting that gut microbiota dysbiosis may contribute to the severity of gastrointestinal symptoms in COVID-19 patients (Wang et al., 2022).
The gut microbiota can influence the host’s immune response to viral infections by regulating the production of antiviral cytokines and modulating the activity of immune cells. For example, gut bacteria can produce SCFAs, which have been shown to enhance the antiviral immune response by increasing the production of IFN-γ and natural killer (NK) cells (Carreca et al., 2022; Govers et al., 2022). Moreover, the gut microbiota can influence the development and function of Tregs, which play a crucial role in maintaining immune homeostasis and preventing excessive inflammation (Smigiel et al., 2014).
Recent research indicates that the gut microbiota may play a role in influencing the immunogenicity of COVID-19 vaccines (Ng et al., 2023). In one study, the use of antibiotics before vaccination was associated with lower seroconversion rates and median antibody levels after receiving one dose of the BNT162b2 vaccine, although this effect was not observed after receiving two doses (Che et al., 2022). Although the study did not directly analyze fecal microbiota, it suggests that dysbiosis in the gut microbiota might have influenced the immune response to the COVID-19 vaccine.
A study involving patients with inflammatory bowel disease found that those with below-average concentrations of SARS-CoV-2-specific antibodies had lower gut microbiota beta diversity and exhibited different bacterial abundances compared to those with above-average antibody concentrations (Alexander et al., 2023). Additionally, differential abundance of fecal metabolites was observed in above- and below-average responders. Specific gut microbial species, such as Bilophila, were associated with above-average response, while others like Streptococcus were associated with below-average response. These findings imply that certain gut microbial species and metabolites, including trimethylamine, short-chain fatty acids (SCFAs), and bile acids, could be linked to COVID-19 vaccine immunogenicity. However, it is essential to consider that these studies have limitations, such as small sample sizes and the lack of analysis of other factors that might influence the gut microbiota and metabolome.
In addition to the gut microbiota’s role in modulating the immune response to viral infections and influencing COVID-19 vaccine immunogenicity, emerging evidence suggests the existence of a gut-lung axis that may contribute to the pathogenesis of COVID-19. The gut-lung axis represents a bidirectional communication pathway between the gut and the lungs, where alterations in the gut microbiota can affect lung health and vice versa (Synodinou et al., 2022).
Studies have demonstrated that gut dysbiosis, characterized by an imbalance in the gut microbial composition, can lead to systemic inflammation and immune dysregulation, which may have implications for lung diseases. In the context of COVID-19, it has been proposed that the gut-lung axis could influence the severity of respiratory symptoms and the risk of developing complications (Petakh et al., 2023a; Petakh et al., 2023c; Petakh et al., 2023d).
Metabolites produced by the gut microbiota, such as SCFAs and bile acids, have the ability to enter the bloodstream and exert effects on distant organs, including the lungs. SCFAs, generated by certain gut bacteria through the fermentation of dietary fiber, possess immunomodulatory properties and can influence immune responses in the lungs (Dang et al., 2019). They are known to regulate the production of inflammatory cytokines and promote the generation of regulatory T cells, which help maintain immune balance and reduce excessive inflammation in the lungs (de Oliveira et al., 2021).
Moreover, alterations in the composition of the gut microbiota can lead to increased gut permeability, enabling the translocation of bacterial components, such as lipopolysaccharides (LPS), into the bloodstream. This process, referred to as bacterial translocation, can trigger systemic inflammation and contribute to the development of lung injury (Sun Z. et al., 2022). In COVID-19 patients, the presence of circulating LPS has been associated with disease severity and poorer clinical outcomes (Giron et al., 2021).
Conclusion
In conclusion, metformin has shown potential as an antiviral agent against SARS-CoV-2, as well as other RNA viruses. It may inhibit viral entry into cells and suppress viral growth in cell culture models. Clinical trials have demonstrated promising results, with metformin leading to a decrease in viral load and a higher rate of undetectable viral load in COVID-19 patients. Furthermore, metformin’s anti-inflammatory effects may help prevent cytokine storms by reducing the production of pro-inflammatory cytokines and modulating immune cell function. The drug’s ability to regulate Th17/Treg balance and influence immunometabolism suggests its potential in mitigating inflammation and restoring T cell functionality in COVID-19. Additionally, metformin’s modulation of the gut microbiota, particularly changes in bacterial taxa and the production of short-chain fatty acids, may contribute to its therapeutic effects. Bile acids, gut microbiota, and their interplay with metformin and GLP-1 have implications for glycemic control and the management of diabetes. Understanding the relationship between metformin, the gut microbiome, and SARS-CoV-2 infection opens new avenues for research and potential therapeutic interventions in COVID-19.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Grant Agreement #2025-VSE “Virtual Scientist Engagement Fellowship Program for Vulnerable Ukrainian Scientists.”
Author contributions
PP: Visualization, Writing–original draft. IK: Writing–review and editing. AK: Conceptualization, Formal Analysis, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adrian T. E., Gariballa S., Parekh K. A., Thomas S. A., Saadi H., Al Kaabi J., et al. (2012). Rectal taurocholate increases L cell and insulin secretion, and decreases blood glucose and food intake in obese type 2 diabetic volunteers. Diabetologia 55 (9), 2343–2347. 10.1007/s00125-012-2593-2 [DOI] [PubMed] [Google Scholar]
- Al-Kuraishy H. M., Al-Gareeb A. I., Alblihed M., Guerreiro S. G., Cruz-Martins N., Batiha G. E.-S. (2021). COVID-19 in relation to hyperglycemia and diabetes mellitus. J Front. Cardiovasc. Med. 8, 644095. 10.3389/fcvm.2021.644095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. L., Mullish B. H., Danckert N. P., Liu Z., Olbei M. L., Saifuddin A., et al. (2023). The gut microbiota and metabolome are associated with diminished COVID-19 vaccine-induced antibody responses in immunosuppressed inflammatory bowel disease patients. EBioMedicine 88, 104430. 10.1016/j.ebiom.2022.104430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allin K. H., Tremaroli V., Caesar R., Jensen B. A., Damgaard M. T., Bahl M. I., et al. (2018). Aberrant intestinal microbiota in individuals with prediabetes. J. Diabetol. 61 (4), 810–820. 10.1007/s00125-018-4550-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancona G., Alagna L., Alteri C., Palomba E., Tonizzo A., Pastena A., et al. (2023). Gut and airway microbiota dysbiosis and their role in COVID-19 and long-COVID. Front. Immunol. 14, 1080043. 10.3389/fimmu.2023.1080043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzrodt C. L., Maknojia I., McCarthy R. D. P., Oldfield T. M., Po J., Ta K. T. L., et al. (2020). A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 287 (17), 3633–3650. 10.1111/febs.15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C. J., Gwilt M. (2022). Diabetes, Metformin and the Clinical Course of Covid-19: outcomes, Mechanisms and Suggestions on the Therapeutic Use of Metformin. Front. Pharmacol. 13, 784459. 10.3389/fphar.2022.784459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C. J., Wilcock C., Day C. (1992). Effect of metformin on glucose metabolism in the splanchnic bed. Br. J. Pharmacol. 105 (4), 1009–1013. 10.1111/j.1476-5381.1992.tb09093.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakumar M., Prabhu D., Sathishkumar C., Prabu P., Rokana N., Kumar R., et al. (2018). Improvement in glucose tolerance and insulin sensitivity by probiotic strains of Indian gut origin in high-fat diet-fed C57BL/6J mice. Eur. J. Nutr. 57 (1), 279–295. 10.1007/s00394-016-1317-7 [DOI] [PubMed] [Google Scholar]
- Barengolts E., Green S. J., Eisenberg Y., Akbar A., Reddivari B., Layden B. T., et al. (2018). Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS One 13 (3), e0194171. 10.1371/journal.pone.0194171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P. V., Duca F. A., Waise T. M. Z., Rasmussen B. A., Abraham M. A., Dranse H. J., et al. (2018). Metformin Alters Upper Small Intestinal Microbiota that Impact a Glucose-SGLT1-Sensing Glucoregulatory Pathway. Cell Metab. 27 (1), 101–117. 10.1016/j.cmet.2017.09.019 [DOI] [PubMed] [Google Scholar]
- Bell K. J., Saad S., Tillett B. J., McGuire H. M., Bordbar S., Yap Y. A., et al. (2022). Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome 10 (1), 9. 10.1186/s40168-021-01193-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramante C. T., Beckman K. B., Mehta T., Karger A. B., Odde D. J., Tignanelli C. J., et al. (2023). Metformin reduces SARS-CoV-2 in a Phase 3 Randomized Placebo Controlled Clinical Trial. medRxiv. 2023, 23290989. medRxiv. 10.1101/2023.06.06.23290989 [DOI] [Google Scholar]
- Brønden A., Albér A., Rohde U., Gasbjerg L. S., Rehfeld J. F., Holst J. J., et al. (2018). The bile acid-sequestering resin sevelamer eliminates the acute GLP-1 stimulatory effect of endogenously released bile acids in patients with type 2 diabetes. Diabetes, Obes. metabolism 20 (2), 362–369. 10.1111/dom.13080 [DOI] [PubMed] [Google Scholar]
- Brønden A., Albér A., Rohde U., Rehfeld J. F., Holst J. J., Vilsbøll T., et al. (2017). Single-Dose Metformin Enhances Bile Acid-Induced Glucagon-Like Peptide-1 Secretion in Patients With Type 2 Diabetes. J. Clin. Endocrinol. metabolism 102 (11), 4153–4162. 10.1210/jc.2017-01091 [DOI] [PubMed] [Google Scholar]
- Bryrup T., Thomsen C. W., Kern T., Allin K. H., Brandslund I., Jørgensen N. R., et al. (2019). Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia 62 (6), 1024–1035. 10.1007/s00125-019-4848-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A. R., Morrison V. L., Levin D., Mohan M., Forteath C., Beall C., et al. (2016). Anti-Inflammatory Effects of Metformin Irrespective of Diabetes Status. Circ. Res. 119 (5), 652–665. 10.1161/CIRCRESAHA.116.308445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M., Biagi E., Soverini M., Consolandi C., Quercia S., Severgnini M., et al. (2016). Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br. J. Nutr. 116 (1), 80–93. 10.1017/S0007114516001045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolyn T. B., Kenneth B. B., Tanvi M., Amy B. K., David J. O., Christopher J. T., et al. Metformin reduces SARS-CoV-2 in a Phase 3 Randomized Placebo Controlled Clinical Trial. medRxiv. 2023:2023.23290989. 10.1101/2023.06.06.23290989 [DOI] [Google Scholar]
- Carreca A. P., Gaetani M., Busà R., Francipane M. G., Gulotta M. R., Perricone U., et al. (2022). Galectin-9 and Interferon-Gamma Are Released by Natural Killer Cells upon Activation with Interferon-Alpha and Orchestrate the Suppression of Hepatitis C Virus Infection. Viruses 14 (7), 1538. 10.3390/v14071538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers E. S., Byrne C. S., Morrison D. J., Murphy K. G., Preston T., Tedford C., et al. (2019). Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut 68 (8), 1430–1438. 10.1136/gutjnl-2019-318424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers E. S., Viardot A., Psichas A., Morrison D. J., Murphy K. G., Zac-Varghese S. E., et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64 (11), 1744–1754. 10.1136/gutjnl-2014-307913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Carbajal A., Pizano-Zárate M. L., Hernández-Quiroz F., Ortiz-Luna G. F., Morales-Hernández R. M., Sales-Millán D., et al. (2020). Characterization of the gut microbiota of individuals at different T2D stages reveals a complex relationship with the host. Microorganisms 8 (1), 94. 10.3390/microorganisms8010094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K. S., Lam L. K., Zhang R., Ooi P. H., Tan J. T., To W. P., et al. (2022). Association between Recent Usage of Antibiotics and Immunogenicity within Six Months after COVID-19 Vaccination. Vaccines 10 (7), 1122. 10.3390/vaccines10071122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.-C., Chien Y.-W., Yang S.-C. (2019). The alteration of gut microbiota in newly diagnosed type 2 diabetic patients. Nutrition 63-64, 51–56. 10.1016/j.nut.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Chen S., Han Y., Yang L., Kim T., Nair M., Harschnitz O., et al. (2021). SARS-CoV-2 Infection Causes Dopaminergic Neuron Senescence. Res. square, 513461. 10.21203/rs.3.rs-513461/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory T. J., Emmons R. S., Yarbro J. R., Davis K. L., Pence B. D. (2021). Metformin Suppresses Monocyte Immunometabolic Activation by SARS-CoV-2 Spike Protein Subunit 1. Front. Immunol. 12, 733921. 10.3389/fimmu.2021.733921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang A. T., Marsland B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12 (4), 843–850. 10.1038/s41385-019-0160-6 [DOI] [PubMed] [Google Scholar]
- De La Cuesta-Zuluaga J., Mueller N. T., Corrales-Agudelo V., Velásquez-Mejía E. P., Carmona J. A., Abad J. M., et al. (2017). Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid–producing microbiota in the gut. Diabetes care 40 (1), 54–62. 10.2337/dc16-1324 [DOI] [PubMed] [Google Scholar]
- de Oliveira G. L. V., Oliveira C. N. S., Pinzan C. F., de Salis L. V. V., Cardoso C. R. B. (2021). Microbiota Modulation of the Gut-Lung Axis in COVID-19. Front. Immunol. 12, 635471. 10.3389/fimmu.2021.635471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., van Eunen K., Groen A. K., Venema K., Reijngoud D. J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. lipid Res. 54 (9), 2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Yang L., Wang Z., Huang W. (2015). Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 5 (2), 135–144. 10.1016/j.apsb.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson C. D., Zhang L., Xu H., Shin Y. K., Vigues S., Ott S. H., et al. (2008). Bitter taste receptors influence glucose homeostasis. PLoS One 3 (12), e3974. 10.1371/journal.pone.0003974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., Ding Y., Yu X., Ma D., Yang B., Li Y., et al. (2019). Metformin mitigates autoimmune insulitis by inhibiting Th1 and Th17 responses while promoting Treg production. Am. J. Transl. Res. 11 (4), 2393–2402. [PMC free article] [PubMed] [Google Scholar]
- Duca F. A., Côté C. D., Rasmussen B. A., Zadeh-Tahmasebi M., Rutter G. A., Filippi B. M., et al. (2015). Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 21 (5), 506–511. 10.1038/nm.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton A. F., Merkulova M., Brown D. (2021). The H(+)-ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am. J. physiology Cell physiology 320 (3), C392–c414. 10.1152/ajpcell.00442.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejtahed H. S., Mohtadi-Nia J., Homayouni-Rad A., Niafar M., Asghari-Jafarabadi M., Mofid V. (2012). Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition 28 (5), 539–543. 10.1016/j.nut.2011.08.013 [DOI] [PubMed] [Google Scholar]
- El Aidy S., Merrifield C. A., Derrien M., van Baarlen P., Hooiveld G., Levenez F., et al. (2013). The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 62 (9), 1306–1314. 10.1136/gutjnl-2011-301955 [DOI] [PubMed] [Google Scholar]
- Erickson S. M., Fenno S. L., Barzilai N., Kuchel G., Bartley J. M., Justice J. N., et al. (2023). Metformin for Treatment of Acute COVID-19: systematic Review of Clinical Trial Data Against SARS-CoV-2. Diabetes Care 46 (7), 1432–1442. 10.2337/dc22-2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esam Z. (2020). A proposed mechanism for the possible therapeutic potential of Metformin in COVID-19. Diabetes Res. Clin. Pract. 167, 108282. 10.1016/j.diabres.2020.108282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan-Morales C. N., Cordero-Rivera C. D., Osuna-Ramos J. F., Monroy-Muñoz I. E., De Jesús-González L. A., Muñoz-Medina J. E., et al. (2021). The antiviral effect of metformin on zika and dengue virus infection. Sci. Rep. 11 (1), 8743. 10.1038/s41598-021-87707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528 (7581), 262–266. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron L. B., Dweep H., Yin X., Wang H., Damra M., Goldman A. R., et al. (2021). Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front. Immunol. 12, 686240. 10.3389/fimmu.2021.686240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa E. Z. (2020). Human gut microbiota/microbiome in health and diseases: a review. Antonie Leeuwenhoek 113 (12), 2019–2040. 10.1007/s10482-020-01474-7 [DOI] [PubMed] [Google Scholar]
- Gorboulev V., Schürmann A., Vallon V., Kipp H., Jaschke A., Klessen D., et al. (2011). Na+-d-glucose Cotransporter SGLT1 is Pivotal for Intestinal Glucose Absorption and Glucose-Dependent Incretin Secretion. Diabetes 61 (1), 187–196. 10.2337/db11-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorboulev V., Schürmann A., Vallon V., Kipp H., Jaschke A., Klessen D., et al. (2012). Na(+)-D-glucose cotransporter SGLT1 is pivotal for intestinal glucose absorption and glucose-dependent incretin secretion. Diabetes 61 (1), 187–196. 10.2337/db11-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 (7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers C., Calder P. C., Savelkoul H. F. J., Albers R., van Neerven R. J. J. (2022). Ingestion, Immunity, and Infection: nutrition and Viral Respiratory Tract Infections. Front. Immunol. 13, 841532. 10.3389/fimmu.2022.841532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradisteanu Pircalabioru G., Grigore G. A., Czobor Barbu I., Chifiriuc M.-C., Savu O. (2023). Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines 11 (1), 179. 10.3390/biomedicines11010179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed I., Masoodi S. R., Mir S. A., Nabi M., Ghazanfar K., Ganai B. A. (2015). Type 2 diabetes mellitus: from a metabolic disorder to an inflammatory condition. World J. diabetes 6 (4), 598–612. 10.4239/wjd.v6.i4.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zheng Y., Deng J., Nan M.-L., Xiao Y., Zhuang M.-W., et al. (2022). SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. J. Med. Virol. 94 (11), 5174–5188. 10.1002/jmv.27965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Scheltema M. J., Sonne D. P., Hansen J. S., Sperling M., Rehfeld J. F., et al. (2016). Effect of chenodeoxycholic acid and the bile acid sequestrant colesevelam on glucagon-like peptide-1 secretion. Diabetes, Obes. metabolism 18 (6), 571–580. 10.1111/dom.12648 [DOI] [PubMed] [Google Scholar]
- Hansen M., Sonne D. P., Mikkelsen K. H., Gluud L. L., Vilsbøll T., Knop F. K. (2017). Bile acid sequestrants for glycemic control in patients with type 2 diabetes: A systematic review with meta-analysis of randomized controlled trials. J. diabetes its Complicat. 31 (5), 918–927. 10.1016/j.jdiacomp.2017.01.011 [DOI] [PubMed] [Google Scholar]
- Holz G. G., Kühtreiber W. M., Habener J. F. (1993). Pancreatic beta-cells are rendered glucose-competent by the insulinotropic hormone glucagon-like peptide-1(7-37). Nature 361 (6410), 362–365. 10.1038/361362a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang Z., Ma H., Ji S., Chen Z., Cui Z., et al. (2021). Dysbiosis and Implication of the Gut Microbiota in Diabetic Retinopathy, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S., Lowe J. R., Bramante C. T., Shah S., Klatt N. R., Sherwood N., et al. (2021). Metformin and Covid-19: focused Review of Mechanisms and Current Literature Suggesting Benefit. Front. Endocrinol. (Lausanne). 12, 587801. 10.3389/fendo.2021.587801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice J. N., Gubbi S., Kulkarni A. S., Bartley J. M., Kuchel G. A., Barzilai N. (2021). A geroscience perspective on immune resilience and infectious diseases: a potential case for metformin. GeroScience 43 (3), 1093–1112. 10.1007/s11357-020-00261-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamyshnyi O., Matskevych V., Lenchuk T., Strilbytska O., Storey K., Lushchak O. (2021). Metformin to decrease COVID-19 severity and mortality: molecular mechanisms and therapeutic potential. Biomed. Pharmacother. 144, 112230. 10.1016/j.biopha.2021.112230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F. H., Tremaroli V., Nookaew I., Bergström G., Behre C. J., Fagerberg B., et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498 (7452), 99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- Ke H., Li F., Deng W., Li Z., Wang S., Lv P., et al. (2021). Metformin Exerts Anti-inflammatory and Mucus Barrier Protective Effects by Enriching Akkermansia muciniphila in Mice With Ulcerative Colitis. Front. Pharmacol. 12, 726707. 10.3389/fphar.2021.726707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffert J. P., Mikkola K., Virtanen K. A., Andersson A. D., Faxius L., Hällsten K., et al. (2017). Metformin treatment significantly enhances intestinal glucose uptake in patients with type 2 diabetes: results from a randomized clinical trial. Diabetes Res. Clin. Pract. 131, 208–216. 10.1016/j.diabres.2017.07.015 [DOI] [PubMed] [Google Scholar]
- Kuhre R. E., Frost C. R., Svendsen B., Holst J. J. (2015). Molecular mechanisms of glucose-stimulated GLP-1 secretion from perfused rat small intestine. Diabetes 64 (2), 370–382. 10.2337/db14-0807 [DOI] [PubMed] [Google Scholar]
- Kuhre R. E., Wewer Albrechtsen N. J., Larsen O., Jepsen S. L., Balk-Møller E., Andersen D. B., et al. (2018). Bile acids are important direct and indirect regulators of the secretion of appetite- and metabolism-regulating hormones from the gut and pancreas. Mol. Metab. 11, 84–95. 10.1016/j.molmet.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A. S., Gubbi S., Barzilai N. (2020). Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab. 32 (1), 15–30. 10.1016/j.cmet.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larraufie P., Roberts G. P., McGavigan A. K., Kay R. G., Li J., Leiter A., et al. (2019). Important Role of the GLP-1 Axis for Glucose Homeostasis after Bariatric Surgery. Cell Rep. 26 (6), 1399–1408. 10.1016/j.celrep.2019.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Vogensen F. K., Van Den Berg F. W., Nielsen D. S., Andreasen A. S., Pedersen B. K., et al. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one 5 (2), e9085. 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. B., Chae S. U., Jo S. J., Jerng U. M., Bae S. K. (2021). The Relationship between the Gut Microbiome and Metformin as a Key for Treating Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 22 (7), 3566. 10.3390/ijms22073566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzen S., Lortz S., Tiedge M. (1996). Effect of metformin on SGLT1, GLUT2, and GLUT5 hexose transporter gene expression in small intestine from rats. Biochem. Pharmacol. 51 (7), 893–896. 10.1016/0006-2952(95)02243-0 [DOI] [PubMed] [Google Scholar]
- Li F., Jiang C., Krausz K. W., Li Y., Albert I., Hao H., et al. (2013). Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4, 2384. 10.1038/ncomms3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhou Y., Yan D., Wan Y. (2022). An Update on the Mutual Impact between SARS-CoV-2 Infection and Gut Microbiota. Viruses 14 (8), 1774. 10.3390/v14081774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannucci E., Tesi F., Bardini G., Ognibene A., Petracca M. G., Ciani S., et al. (2004). Effects of metformin on glucagon-like peptide-1 levels in obese patients with and without Type 2 diabetes. Diabetes, Nutr. metabolism 17 (6), 336–342. [PubMed] [Google Scholar]
- Ma Z., Patel N., Vemparala P., Krishnamurthy M. (2022). Metformin is associated with favorable outcomes in patients with COVID-19 and Type 2 diabetes mellitus. J. Sci. Rep. 12 (1), 5553–5557. 10.1038/s41598-022-09639-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L. R., Baggesen L. M., Richelsen B., Thomsen R. W. (2019). Effect of Roux-en-Y gastric bypass surgery on diabetes remission and complications in individuals with type 2 diabetes: a Danish population-based matched cohort study. Diabetologia 62 (4), 611–620. 10.1007/s00125-019-4816-2 [DOI] [PubMed] [Google Scholar]
- Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., et al. (1999). Identification of a nuclear receptor for bile acids. Science 284 (5418), 1362–1365. 10.1126/science.284.5418.1362 [DOI] [PubMed] [Google Scholar]
- Mercado-Gómez M., Prieto-Fernández E., Goikoetxea-Usandizaga N., Vila-Vecilla L., Azkargorta M., Bravo M., et al. (2022). The spike of SARS-CoV-2 promotes metabolic rewiring in hepatocytes. Commun. Biol. 5 (1), 827. 10.1038/s42003-022-03789-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed I., Hollenberg M. D., Ding H., Triggle C. R. (2021). A Critical Review of the Evidence That Metformin Is a Putative Anti-Aging Drug That Enhances Healthspan and Extends Lifespan. Front. Endocrinol. (Lausanne) 12, 718942. 10.3389/fendo.2021.718942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. Y., Leung W. K., Cheung K. S. (2023). Association between Gut Microbiota and SARS-CoV-2 Infection and Vaccine Immunogenicity. Microorganisms 11 (2), 452. 10.3390/microorganisms11020452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omarjee L., Janin A., Perrot F., Laviolle B., Meilhac O., Mahe G. (2020). Targeting T-cell senescence and cytokine storm with rapamycin to prevent severe progression in COVID-19. Clin. Immunol. Orl. Fla) 216, 108464. 10.1016/j.clim.2020.108464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo A., Zandi K., Shepard C., Bassit L. C., Musall K., Goh S. L., et al. (2022). Elimination of Aicardi–Goutières syndrome protein SAMHD1 activates cellular innate immunity and suppresses SARS-CoV-2 replication. J. Biol. Chem. 298 (3), 101635. 10.1016/j.jbc.2022.101635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H. E., Adriaenssens A., Rogers G., Richards P., Koepsell H., Reimann F., et al. (2012). Predominant role of active versus facilitative glucose transport for glucagon-like peptide-1 secretion. Diabetologia 55 (9), 2445–2455. 10.1007/s00125-012-2585-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy H., Tandel D., Siddiqui A. H., Harshan K. H. (2022). Metformin suppresses SARS-CoV-2 in cell culture. Virus Res. 323, 199010. 10.1016/j.virusres.2022.199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pénicaud L., Hitier Y., Ferré P., Girard J. (1989). Hypoglycaemic effect of metformin in genetically obese (fa/fa) rats results from an increased utilization of blood glucose by intestine. Biochem. J. 262 (3), 881–885. 10.1042/bj2620881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. J., Peng L., Barry N. A., Cline G. W., Zhang D., Cardone R. L., et al. (2016). Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 534 (7606), 213–217. 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Griga V., Mohammed I. B., Loshak K., Poliak I., Kamyshnyiy A. (2022a). Effects of Metformin, Insulin on Hematological Parameters of COVID-19 Patients with Type 2 Diabetes. Med. archives (Sarajevo, Bosnia Herzegovina) 76 (5), 329–332. 10.5455/medarh.2022.76.329-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Isevych V., Mohammed I., Loshak K., Poliak I., Kamyshnyiy A. (2022b). Association between Use of Metformin and Insulin with Hematological Parameters in COVID-19 Patients with Type 2 Diabetes: A Single Center, Cross-Sectional Study. Clin. Diabetol. 11 (6), 432–433. 10.5603/dk.a2022.0055 [DOI] [Google Scholar]
- Petakh P., Kamyshna I., Kamyshnyi A. (2023e). Effects of metformin on the gut microbiota: A systematic review. Mol. Metab. 77, 101805. 10.1016/j.molmet.2023.101805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Kamyshna I., Nykyforuk A., Yao R., Imbery J. F., Oksenych V., et al. (2022c). Immunoregulatory Intestinal Microbiota and COVID-19 in Patients with Type Two Diabetes: A Double-Edged Sword. J. Viruses 14 (3), 477. 10.3390/v14030477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Kamyshna I., Oksenych V., Kainov D., Kamyshnyi A. (2023c). Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: the Role of SARS-CoV-2 Variants and Antibiotic Treatment. Pharm. (Basel, Switz. 16 (6), 904. 10.3390/ph16060904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Kobyliak N., Kamyshnyi A. (2023a). Gut microbiota in patients with COVID-19 and type 2 diabetes: A culture-based method. Front. Cell. Infect. Microbiol. 13, 1142578. 10.3389/fcimb.2023.1142578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petakh P., Loshak K., Kamyshnyi A. (2023b). Hematological features of patients with type 2 diabetes depending on the variant of SARS-COV-2. Fiziolohichnyĭ Zhurnal 69 (1), 35–42. 10.15407/fz69.01.035 [DOI] [Google Scholar]
- Petakh P., Oksenych V., Kamyshnyi A. (2023d). The F/B ratio as a biomarker for inflammation in COVID-19 and T2D: impact of metformin. Biomed. Pharmacother. 163, 114892. 10.1016/j.biopha.2023.114892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490 (7418), 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Reis G., Dos Santos Moreira Silva E. A., Medeiros Silva D. C., Thabane L., Cruz Milagres A., Ferreira T. S., et al. (2022). Effect of early treatment with metformin on risk of emergency care and hospitalization among patients with COVID-19: the TOGETHER randomized platform clinical trial. Lancet reg. health Am. 6, 100142. 10.1016/j.lana.2021.100142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooj A. K., Kimura Y., Buddington R. K. (2010). Metabolites produced by probiotic Lactobacilli rapidly increase glucose uptake by Caco-2 cells. BMC Microbiol. 10, 16. 10.1186/1471-2180-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakar Y., Meddah B., Faouzi M. A., Cherrah Y., Bado A., Ducroc R. (2010). Metformin-induced regulation of the intestinal D-glucose transporters. J. physiology Pharmacol. 61 (3), 301–307. [PubMed] [Google Scholar]
- Salvatore T., Pafundi P. C., Galiero R., Gjeloshi K., Masini F., Acierno C., et al. (2020). Metformin: A Potential Therapeutic Tool for Rheumatologists. Pharm. (Basel, Switz. 13 (9), 234. 10.3390/ph13090234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome D. J., Xie C., Veedfald S., Horowitz M., Rayner C. K., Wu T. (2020). Mechanism of glucose-lowering by metformin in type 2 diabetes: role of bile acids. Diabetes, Obes. metabolism 22 (2), 141–148. 10.1111/dom.13869 [DOI] [PubMed] [Google Scholar]
- Scarpello J. H., Hodgson E., Howlett H. C. (1998). Effect of metformin on bile salt circulation and intestinal motility in type 2 diabetes mellitus. Diabet. Med. a J. Br. Diabet. Assoc. 15 (8), 651–656. [DOI] [PubMed] [Google Scholar]
- Schaller M. A., Sharma Y., Dupee Z., Nguyen D., Urueña J., Smolchek R., et al. (2021). Ex vivo SARS-CoV-2 infection of human lung reveals heterogeneous host defense and therapeutic responses. JCI insight 6 (18), e148003. 10.1172/jci.insight.148003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheen A. (2020). Metformin and COVID-19: from cellular mechanisms to reduced mortality. J. Diabetes Metab. 46 (6), 423–426. 10.1016/j.diabet.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Ray A., Sadasivam B. (2020). Metformin in COVID-19: a possible role beyond diabetes. Diabetes Res. Clin. Pract. 164, 108183. 10.1016/j.diabres.2020.108183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Yang L., Li C., Wang T., Lv J., Liu W., et al. (2023). Metformin promotes cGAS/STING signaling pathway activation by blocking AKT phosphorylation in gastric cancer. Heliyon 9 (8), e18954. 10.1016/j.heliyon.2023.e18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C.-T., Yeh Y.-T., Lin C.-C., Yang L.-Y., Chiang C.-P. (2020). Akkermansia muciniphila is Negatively Correlated with Hemoglobin A1c in Refractory Diabetes. Microorganisms 8 (9), 1360. 10.3390/microorganisms8091360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel K. S., Srivastava S., Stolley J. M., Campbell D. J. (2014). Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol. Rev. 259 (1), 40–59. 10.1111/imr.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Shen S., Hu Y., Chen S., Cheng L., Cai Y., et al. (2023). SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function. J. Med. Virol. 95 (1), e28175. 10.1002/jmv.28175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xie C., Wang G., Wu Y., Wu Q., Wang X., et al. (2018). Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 24 (12), 1919–1929. 10.1038/s41591-018-0222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Liu Y., Huang Z., Xu W., Hu W., Yi L., et al. (2022b). SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell death Differ. 29 (6), 1240–1254. 10.1038/s41418-021-00916-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Song Z.-G., Liu C., Tan S., Lin S., Zhu J., et al. (2022a). Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 20 (1), 24. 10.1186/s12916-021-02212-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synodinou K. D., Nikolaki M. D., Triantafyllou K., Kasti A. N. (2022). Immunomodulatory Effects of Probiotics on COVID-19 Infection by Targeting the Gut-Lung Axis Microbial Cross-Talk. Microorganisms 10 (9), 1764. 10.3390/microorganisms10091764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. K. X., Heng T. Y. J., Mak A. (2019). The Potential Use of Metformin, Dipyridamole, N-Acetylcysteine and Statins as Adjunctive Therapy for Systemic Lupus Erythematosus. Syst. Lupus Erythematosus. Cells. 8 (4), 323. 10.3390/cells8040323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., et al. (2009). TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10 (3), 167–177. 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov A. A., Baker H. V., Brusko T. M., Sobel E. S., Morel L. (2019). Metformin Inhibits the Type 1 IFN Response in Human CD4(+) T Cells. J. Immunol. 203 (2), 338–348. 10.4049/jimmunol.1801651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi M. S., Daoudi M., Prawitt J., Ducastel S., Touche V., Sayin S. I., et al. (2015). Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat. Commun. 6, 7629. 10.1038/ncomms8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggle C. R., Mohammed I., Bshesh K., Marei I., Ye K., Ding H., et al. (2022). Metformin: is it a drug for all reasons and diseases? Metabolism Clin. Exp. 133, 155223. 10.1016/j.metabol.2022.155223 [DOI] [PubMed] [Google Scholar]
- Tsai W.-L., Chang T.-H., Sun W.-C., Chan H.-H., Wu C.-C., Hsu P.-I., et al. (2017). Metformin activates type I interferon signaling against HCV via activation of adenosine monophosphate-activated protein kinase. Oncotarget 8 (54), 91928–91937. 10.18632/oncotarget.20248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura-López C., Cervantes-Luevano K., Aguirre-Sánchez J. S., Flores-Caballero J. C., Alvarez-Delgado C., Bernaldez-Sarabia J., et al. (2022). Treatment with metformin glycinate reduces SARS-CoV-2 viral load: an in vitro model and randomized, double-blind, Phase IIb clinical trial. Biomed. Pharmacother. = Biomedecine Pharmacother. 152, 113223. 10.1016/j.biopha.2022.113223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhang L., Wang Y., Dai T., Qin Z., Zhou F., et al. (2022). Alterations in microbiota of patients with COVID-19: potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 7 (1), 143. 10.1038/s41392-022-00986-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.-Y., Zhang X.-Q., Chen A.-L., Zhang J., Lv B.-H., Ma M.-H., et al. (2020). A comparative study of microbial community and functions of type 2 diabetes mellitus patients with obesity and healthy people. Appl. Microbiol. Biotechnol. 104 (16), 7143–7153. 10.1007/s00253-020-10689-7 [DOI] [PubMed] [Google Scholar]
- Watson L. E., Xie C., Wang X., Li Z., Phillips L. K., Sun Z., et al. (2019). Gastric Emptying in Patients With Well-Controlled Type 2 Diabetes Compared With Young and Older Control Subjects Without Diabetes. J. Clin. Endocrinol. metabolism 104 (8), 3311–3319. 10.1210/jc.2018-02736 [DOI] [PubMed] [Google Scholar]
- Wiernsperger N., Al-Salameh A., Cariou B., Lalau J. D. (2022). Protection by metformin against severe Covid-19: an in-depth mechanistic analysis. Diabetes & metabolism 48 (4), 101359. 10.1016/j.diabet.2022.101359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock C., Bailey C. J. (1994). Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica; fate foreign Compd. Biol. Syst. 24 (1), 49–57. 10.3109/00498259409043220 [DOI] [PubMed] [Google Scholar]
- Wu H., Esteve E., Tremaroli V., Khan M. T., Caesar R., Mannerås-Holm L., et al. (2017a). Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23 (7), 850–858. 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- Wu T., Bound M. J., Standfield S. D., Gedulin B., Jones K. L., Horowitz M., et al. (2013b). Effects of rectal administration of taurocholic acid on glucagon-like peptide-1 and peptide YY secretion in healthy humans. Diabetes, Obes. metabolism 15 (5), 474–477. 10.1111/dom.12043 [DOI] [PubMed] [Google Scholar]
- Wu T., Bound M. J., Standfield S. D., Jones K. L., Horowitz M., Rayner C. K. (2013a). Effects of taurocholic acid on glycemic, glucagon-like peptide-1, and insulin responses to small intestinal glucose infusion in healthy humans. J. Clin. Endocrinol. metabolism 98 (4), E718–E722. 10.1210/jc.2012-3961 [DOI] [PubMed] [Google Scholar]
- Wu T., Xie C., Wu H., Jones K. L., Horowitz M., Rayner C. K. (2017b). Metformin reduces the rate of small intestinal glucose absorption in type 2 diabetes. Diabetes, Obes. metabolism 19 (2), 290–293. 10.1111/dom.12812 [DOI] [PubMed] [Google Scholar]
- Xian H., Liu Y., Rundberg Nilsson A., Gatchalian R., Crother T. R., Tourtellotte W. G., et al. (2021). Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation. Immunity 54 (7), 1463–1477.e11. 10.1016/j.immuni.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H., Liu Q. P. (2022). Alterations of the gut microbiota in coronavirus disease 2019 and its therapeutic potential. World J. gastroenterology 28 (47), 6689–6701. 10.3748/wjg.v28.i47.6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav H., Jain S., Sinha P. R. (2007). Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition 23 (1), 62–68. 10.1016/j.nut.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Yang M., Ding Y., Wang Y., Gu J., Zhang B., Wang H. (2017). Metformin regulates of Th17/treg cell balance and reduces hyperplastic synovium via activating AMPK and inhibiting mTOR in a collagen-induced arthritis rat model. J. Int. J. Clin. Exp. Med. 10 (8), 11479–11487. [Google Scholar]
- Yeoh Y. K., Zuo T., Lui G. C.-Y., Zhang F., Liu Q., Li A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70 (4), 698–706. 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang M., Yang J., Xu Q., Liang C., Chen B., et al. (2019). Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 66 (3), 485–493. 10.1007/s12020-019-02041-5 [DOI] [PubMed] [Google Scholar]
- Zhong H., Ren H., Lu Y., Fang C., Hou G., Yang Z., et al. (2019). Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. eBioMedicine 47, 373–383. 10.1016/j.ebiom.2019.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]