Abstract
Rationale
Lung disease is the major cause of morbidity and mortality in persons with cystic fibrosis (pwCF). Variability in CF lung disease has substantial non-CFTR (CF transmembrane conductance regulator) genetic influence. Identification of genetic modifiers has prognostic and therapeutic importance.
Objectives
Identify genetic modifier loci and genes/pathways associated with pulmonary disease severity.
Methods
Whole-genome sequencing data on 4,248 unique pwCF with pancreatic insufficiency and lung function measures were combined with imputed genotypes from an additional 3,592 patients with pancreatic insufficiency from the United States, Canada, and France. This report describes association of approximately 15.9 million SNPs using the quantitative Kulich normal residual mortality-adjusted (KNoRMA) lung disease phenotype in 7,840 pwCF using premodulator lung function data.
Measurements and Main Results
Testing included common and rare SNPs, transcriptome-wide association, gene-level, and pathway analyses. Pathway analyses identified novel associations with genes that have key roles in organ development, and we hypothesize that these genes may relate to dysanapsis and/or variability in lung repair. Results confirmed and extended previous genome-wide association study findings. These whole-genome sequencing data provide finely mapped genetic information to support mechanistic studies. No novel primary associations with common single variants or rare variants were found. Multilocus effects at chr5p13 (SLC9A3/CEP72) and chr11p13 (EHF/APIP) were identified. Variant effect size estimates at associated loci were consistently ordered across the cohorts, indicating possible age or birth cohort effects.
Conclusions
This premodulator genomic, transcriptomic, and pathway association study of 7,840 pwCF will facilitate mechanistic and postmodulator genetic studies and the development of novel therapeutics for CF lung disease.
Keywords: cystic fibrosis, whole-genome sequencing, lung disease severity, GWAS/TWAS, pathway analyses
At a Glance Commentary
Scientific Knowledge on the Subject
Genetic modifiers affect lung disease severity.
What This Study Adds to the Field
This is the largest premodulator genome-wide association study to date for lung function in cystic fibrosis, using recently generated whole-genome sequencing. Genetic variation controlling cystic fibrosis lung disease severity spans the biological spectrum, from innate immunity to lung development, and defines key genes and pathways for future exploration.
Lung disease is the major cause of morbidity and mortality in cystic fibrosis (CF) (1), but the severity can vary widely among individuals. In part, this variation reflects genetic variants in CFTR (CF transmembrane conductance regulator) (2) that span a spectrum of severity from complete loss-of-function mutations that are associated with exocrine pancreatic insufficiency (PI) to CFTR variants with residual function (2). Additionally, although environmental influences contribute to lung disease variability, non-CFTR modifier genes also play a role (heritability, 0.54) (3, 4). Although recent advances in CFTR modulator therapies have improved outcomes for many persons with CF (pwCF), some do not benefit as a result of nonresponsive genetic variants in CFTR. Continued exploration of non-CFTR genetic modifiers is expected to provide new therapeutic targets (5).
A previously published genome-wide association study (GWAS) for lung disease severity in pwCF with PI reported modifier variants at five loci (6), with an additional significant GWAS locus identified using improved imputation of SNP genotypes from the primary paper (7). These previous studies used whole-genome SNP arrays and a validated lung disease phenotype, Kulich normal residual mortality-adjusted (KNoRMA), which is based on multiple measurements of FEV1 corrected for sex, age, and survival, enabling analysis across different ages and cohorts (6).
Whole-genome sequencing (WGS) has made it possible to study genotype–phenotype associations at high resolution. The Cystic Fibrosis Genome Project (CFGP) is a multisite consortium to dissect molecular sources of the variability of phenotypes in pwCF (8). We reasoned that combining data from WGS samples with samples and data from prior GWASs would provide a highly resolved picture of CF lung disease phenotype–genotype associations and a more detailed biological understanding of CF lung disease. We report extensive analyses using KNoRMA, calculated from lung function data before modulator therapy, to identify genetic modifiers of pulmonary disease in 7,840 pwCF, the largest such study to date. These rigorous premodulator data will inform ongoing therapeutic development studies and serve as a basis for future postmodulator genome studies.
Some of the results of these studies have been previously reported in the form of an abstract (9).
Methods
The online supplement includes numerous details, with brief descriptions provided here.
A total of 5,199 CFGP samples were sequenced (8). Of those, 4,248 were patients with PI with sufficient premodulator lung function measures for inclusion from three studies/sites: the Gene Modifier Study at the University of North Carolina (UNC), the Twin and Sibling Study and CF-related Diabetes Studies at Johns Hopkins University, and the Early Pseudomonas Infection Control Study at the University of Washington (UW) (Table 1). These WGS data were combined with an independent set of 3,592 patients with genome-wide genotypes imputed from TOPMed data (10) from array-based genotypes (6) (total N = 7,840).
Table 1.
Characteristics of Patients with CF (All with Pancreatic Insufficiency) in the Present Study
| Cohort | Total Pts. | CFGP WGS Pts. | Imputed Pts. | KNoRMA [Mean (SD)] | Age, yr* |
Male sex [n (%)] | European† [n (%)] | F508del/F508del [n (%)] | |
|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median | ||||||||
| JHU | 1,683 | 1,466 | 217 | 0.54 (0.86) | 20.6 (9.8) | 19.0 | 893 (53.1) | 1,565 (93.0) | 947 (56.3) |
| UNC | 2,159 | 1,605 | 554 | 0.60 (0.92) | 26.8 (11.2) | 24.9 | 1,170 (54.1) | 2,057 (95.3) | 1,606 (74.4) |
| UW | 1,177 | 1,177 | 0 | 0.51 (0.73) | 13.1 (3.5) | 12.9 | 592 (50.3) | 1,088 (92.4) | 710 (60.3) |
| FrGMS | 1,207 | 0 | 1,207 | 0.32 (0.77) | 21.1 (9.2) | 20.1 | 619 (51.3) | 1,196 (99.1) | 707 (58.6) |
| CGS | 1,614 | 0 | 1,614 | 0.38 (0.82) | 17.3 (9.2) | 14.9 | 865 (53.6) | 1,531 (94.9) | 1,015 (62.9) |
| Overall | 7,840 | 4,248 | 3,592 | 0.48 (0.84) | 20.6 (10.4) | 18.4 | 4,139 (52.8) | 7,437 (94.9) | 4,985 (63.6) |
Definition of abbreviations: CFGP = Cystic Fibrosis Genome Project; CGS = Canadian CF Gene Modifier Study (population-based); FrGMS = French CF Gene Modifier Consortium (population-based); JHU = Johns Hopkins University (twin-siblings design); KNoRMA = Kulich normal residual mortality-adjusted; UNC = University of North Carolina (extremes of phenotype); UW = University of Washington (longitudinal study for effect of Pseudomonas aeruginosa acquisition on lung disease); WGS = whole-genome sequencing.
Age for lung function phenotyping for KNoRMA calculation.
Based on self-reported ancestry, confirmed by ancestry by genotyping.
A validated quantitative lung function trait was calculated using the KNoRMA phenotype (6), which allows analyses across age, sex, and cohort. KNoRMA is based on multiple measures of FEV1 over 3 years using data from the CF Foundation Patient Registry (2017) (11) and is corrected for age, sex, and survival. A disease progression– and mortality-adjusted phenotype such as KNoRMA increases statistical power while reducing the need for stratification or additional covariates. For this study, to avoid the confounding effects of recently approved modulators, KNoRMA was calculated from FEV1 before modulator therapy (see online supplement for details).
CFGP samples were sequenced to approximately 30× coverage with careful quality/identity checks (see online supplement for details). The GWAS array-based data and cohorts were described previously (6). Genetic imputation for the non-CFGP samples was performed as described previously (10).
Analyses used a quantitative trait of lung disease severity (KNoRMA) (6). The primary analyses included nonrare single-variant SNP testing (minor allele count ⩾20). Association was tested using KNoRMA as a response in an additive effect mixed model using ancestry, sex, and terms for site/platform combinations as covariates. A genetic relatedness matrix was used to account for the small proportion of families and cryptic relatedness. Results were combined across site × platform as a fixed-effect meta-analysis. P value thresholds were applied at the genome-wide significance level (P < 5 × 10−8) (12), and we considered SNPs with P < 5 × 10−7 to be suggestive.
For the significant GWAS loci, we ran Causal Variants Identification in Associated Regions to assess evidence of SNP causality (13). The Ensembl Variant Effect Predictor was used to determine putative effects of variants on genes, transcripts, protein sequences, and regulatory regions (14).
Transcriptome-wide association (TWAS) evidence was determined from 50 tissues using a summary association z-statistic (15). This approach uses SNP-level gene-expression weights from 48 tissues from the Genotype-Tissue Expression project v8 (16), peripheral blood from the Netherlands Twin Registry (17), and whole blood from the Young Finns Study (18). For these and all gene-based approaches, including gene association summaries and rare-variant methods, we used a false-discovery q value of less than 0.1 to declare significance.
Although the KNoRMA phenotype is corrected for age-dependent effects on survival, we additionally devised a reverse regression approach to investigate potential age interactions for genotype associations (see online supplement). In addition, a method was devised to assess concordant effect size ordering across site cohorts for different loci using a summary of pairwise correlations of estimated effect sizes across cohorts, with statistical significance assessed by permutation. For our meta-analysis statistic, we show that this permutation approach remains valid under selection for genome-wide significance (see online supplement). Gene-level summary analyses were performed using VEGAS2 (19) for intragenic SNPs and SNPs within a flanking region of 20 kb around each gene. Gene-based pathway analyses were performed using the Gene Set Enrichment Analysis (GSEA) method (20), available in the clusterProfiler R package (21). Rare variant methods (minor allele count <20) were performed at the gene level using the GENESIS R package for the burden test, SMMAT, and SKAT-O.
Results
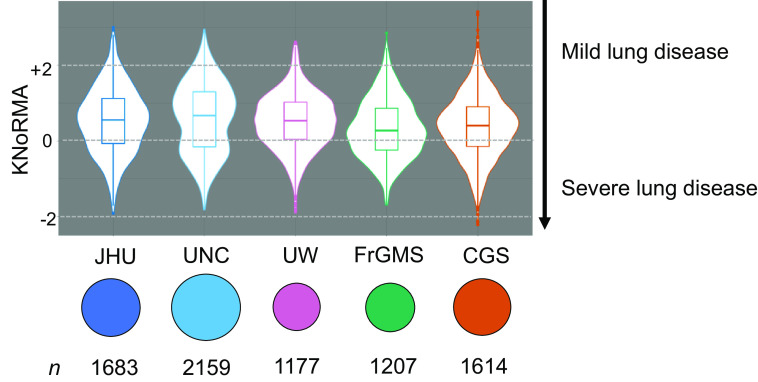
Table 1 describes key features of the five cohorts, including the country of enrollment. The majority (n = 4,248) of these pwCF with PI had WGS, and the remainder (n = 3,592) had genotypes imputed from WGS (10). The means and standard deviations for lung disease severity (KNoRMA) were comparable across cohorts despite considerable differences in mean (and median) age. On average, the UW cohort includes the youngest patients (mean, 13.1 yr) and the UNC cohort the oldest (mean, 26.8 yr). The vast majority of these pwCF are of European ancestry (∼95%), and most (∼64%) are c.1521_1523del (p.Phe508del; legacy: F508del) homozygotes. Effective ancestry control could be achieved by four genotype principal components (22), and we used six principal components to be conservative. The violin plot in Figure 1 illustrates the distribution of KNoRMA by cohort, and the UNC plot shows two distinct modes, reflecting an extremes-of-phenotype design (6).
Figure 1.
Distributions of the Kulich normal residual mortality-adjusted (KNoRMA) age-adjusted lung function phenotype by site cohort. The line inside each box is the median KNoRMA, and the box represents the interquartile range, or distance between the first and third quartiles (the 25th and 75th percentiles). Violin plots show the overall population distribution. Sample sizes are shown, with symbol areas proportional to sample size. CGS = Canadian CF Gene Modifier Study (population-based); FrGMS = French CF Gene Modifier Consortium (population-based); JHU = Johns Hopkins University (twin-siblings design); UNC = University of North Carolina (extremes of phenotype); UW = University of Washington (longitudinal study for effect of Pseudomonas aeruginosa acquisition on lung disease).
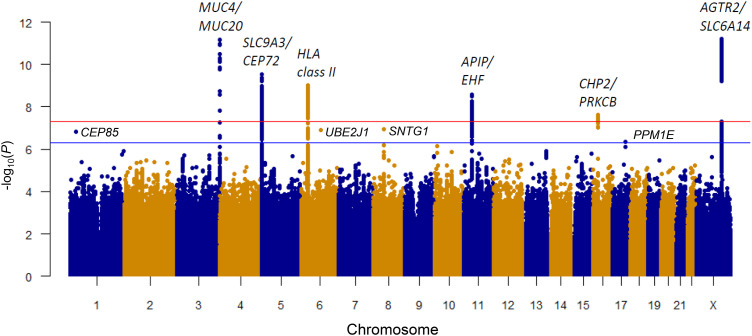
The GWAS analysis for KNoRMA using nonrare variants (minor allele count >20, hereafter termed “common”) identified six genome-wide significant (P < 5 × 10−8) (12) loci (Figure 2, Table 2, and Figures E1 and E2 in the online supplement). The present analyses increased the significance of four of five loci reported in our previous GWAS (Table 2) (6). The sixth locus at 16p12.2 near CHP2 and PRKCB (P = 2.5 × 10−8) (Table 2 and Figure E2) was not reported in our previous GWAS (6) but was identified in a separate analysis using updated and improved imputation of SNP genotypes (7). Each of these six loci contains genes of high biological relevance to the pathophysiology of CF lung disease (4, 7). Four suggestive loci (P < 5 × 10−7; all with low minor allele frequency; range, 0.005–0.009) were also identified, including chr1p36 (CEP85), chr6q15 (UBE2J1), chr8q11.2 (SNTG1), and chr17q22 (PPM1E) (Table 2 and Figure E3). Finally, we identified many associations (P < 10−5) with KNoRMA in all pwCF (N = 7,840) and 4,985 F508del homozygotes (Table E1 in the online supplement).
Figure 2.
Genetic loci significantly associated with Kulich normal residual mortality-adjusted (KNoRMA) lung phenotype. Genome-wide Manhattan plot of associations with KNoRMA in all 7,840 patients. Red line shows genome-wide significance of P < 5 × 10−8. Blue line shows a suggestive significance of P < 5 × 10−7.
Table 2.
Genome-Wide Significant (P < 5 × 10−8) and Suggestive (P < 5 × 10−7) Association Results
| Chromosome Band |
Gene(s) | Base Pair Position | SNP | Risk/Protective Allele |
PAF | β* | P Value | Prior GWAS Regional P Value† |
|---|---|---|---|---|---|---|---|---|
| Significant findings | ||||||||
| 3q29 | MUC20/MUC4 | 195,760,866 | rs2246771 | G/A | 0.29 | 0.1 | 6.7 × 10−12‡ | 3.3 × 10−11‡ |
| 5p15.33 | SLC9A3/CEP72 | 537,775 | rs56108664 | T/C | 0.83 | 0.11 | 2.8 × 10−10‡ | 6.8 × 10−12‡ |
| 6p21 | HLA class II | 32,462,048 | rs9268860 | T/C | 0.68 | 0.08 | 9.9 × 10−10‡ | 1.2 × 10−8‡ |
| 11p13 | EHF/APIP | 34,808,842 | rs485845 | A/C | 0.64 | 0.09 | 2.6 × 10−9‡ | 4.8 × 10−9‡ |
| 16p12.2 | CHP2/PRKCB | 23,779,017 | rs194788 | A/T | 0.44 | 0.07 | 2.5 × 10−8‡ | 7.7 × 10−7 |
| Xq23 | AGTR2/SLC6A14 | 116,230,240 | rs12009976 | G/A | 0.49 | 0.08 | 6.1 × 10−12‡ | 1.8 × 10−9‡ |
| Suggestive findings | ||||||||
| 1p36 | CEP85 | 26,257,354 | rs41284341 | A/C | 0.009 | 0.39 | 1.6 × 10−7 | 9.1 × 10−3 |
| 6q15 | UBE2J1 | 89,330,626 | rs9294434 | T/C | 0.009 | 0.41 | 1.3 × 10−7 | 8.0 × 10−3 |
| 8q11.2 | SNTG1 | 50,730,869 | rs140650336 | C/T | 0.005 | 0.65 | 1.2 × 10−7 | 7.2 × 10−4 |
| 17q22 | PPM1E | 58,950,377 | rs72828739 | C/T | 0.991 | 0.36 | 4.7 × 10−7 | 7.1 × 10−2 |
Gene listed if intergenic; flanking genes are listed otherwise.
Definition of abbreviations: GWAS = genome-wide association study; PAF = frequency of protective allele.
β-Coefficient refers to increased average Kulich normal residual mortality-adjusted for each copy of the protective allele.
From Corvol and colleagues, 2015 (6).
P values with genome-wide significant association, P < 5 × 10−8; others listed are suggestive association, P < 5 × 10−7.
Conditioning on the top-ranked SNP in six regions with genome-wide significance eliminated significant secondary signals in four regions, but two loci (chr5p15.33; SLC9A3/CEP72 and chr11p13; APIP/EHF) displayed regional significance for secondary SNPs (Figure E2). By fitting all regional two-SNP models for chr5p15 and chr11p13 (see Methods), we determined the best-fitting SNP pair for each region. For chr5p15, conditioning on the primary SNP (rs56108664) revealed a significant secondary SNP (rs111275646) and other SNPs in linkage disequilibrium (LD) (Figure E4). Conditioning on the secondary SNP at chr5p15 recapitulated the original signal (conditional P ≈ 3 × 10−8 for the original SNP rs56108664 after conditioning on rs111275646). For the chr11p13 locus, the use of the best two-SNP model (primary, rs483769; secondary, rs1509661) provided informative results (Figure E5). Namely, the P values for SNPs in the primary LD “block” after conditioning on the secondary SNP (rs1509661) became approximately 10,000-fold smaller (P ≈ 7 × 10−14) than in the original single-variant analysis (P ≈ 2.6 × 10−9) (Table 2). Further investigation of the chr11p13 locus revealed that the minor alleles of the primary and secondary SNPs are positively associated (r2 = 0.28) but have associations in opposite directions with KNoRMA. In this scenario, most subjects in this study have at least one risk allele at the primary locus and at least one protective allele at the secondary locus, with combinations of risk alleles from either locus contributing to overall phenotype consequences (Figure E6). For the chr5p15 and chr11p13 regions, haplotype analyses that account for the linkage phase (see online supplement) were not more significant than the primary genotype-based analyses.
Causality at each significant locus has not been established because of LD structure, and analysis by Causal Variants Identification in Associated Regions (13) and annotation of the top SNPs in the six regions with the Ensembl Variant Effect Predictor (14) did not point to any obvious causal links (Table E2). The gene-level rare variant analyses did not identify any significant gene at q < 0.1, perhaps reflecting reduced power for rare variant detection compared with common variant analyses.
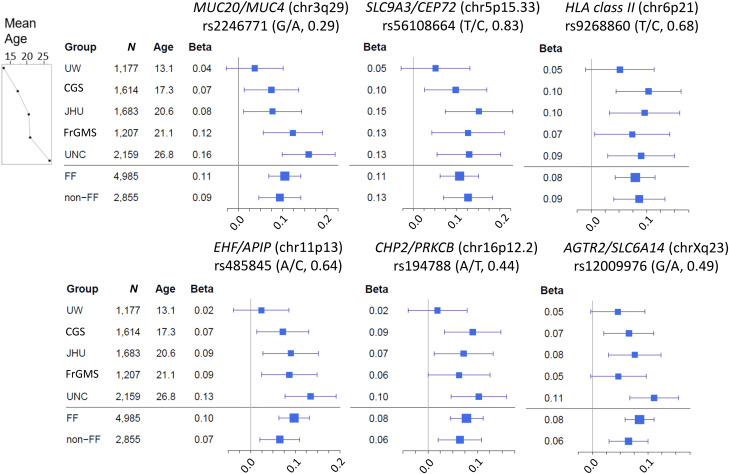
Our reverse regression model included terms for age at phenotyping and age × KNoRMA interaction and largely recapitulated our main findings, with five of the six reported loci achieving significance (rs194788 near CHP2 achieving only P = 3.23 × 10−7), and no new significant findings. The age and age × KNoRMA interaction terms were not significant for these regions. Nonetheless, substantial cohort variation was apparent. The effect sizes (magnitude of β-coefficients) for the peak SNP at the six significant loci were evaluated using forest plots for each cohort, ordered by mean age (Figure 3). There is a similar distribution of the effect (size) across cohorts, with the UNC cohort (the oldest) showing the largest effect size and the UW cohort (the youngest) showing the smallest. This concordance of effect sizes manifests as positive correlation in all pairwise comparisons (mean correlation, 0.70), as depicted in plots of effect sizes (Figure E7). A test of concordance of effect sizes demonstrated concordance among cohorts (P = 3.4 × 10−4), with the UW cohort consistently exhibiting the smallest effect size.
Figure 3.
Forest plots for SNP association effect size by cohort at significant loci. β (coefficient) refers to the average change in Kulich normal residual mortality-adjusted (KNoRMA) phenotype for each copy of the protective allele. Square sizes are proportional to the sample size (n) of each cohort, and the line segments are 95% confidence intervals of each β. The most significant SNP from each locus was chosen. For each SNP, the protective allele is listed on the left, and frequencies of the protective alleles are shown in parentheses. Cohorts are arranged by increasing mean age (at KNoRMA). CFGP = Cystic Fibrosis Genome Project; CGS = Canadian CF Gene Modifier Study (population-based); FF = patients with CF who are F508del homozygous in the CFTR gene; FrGMS = French CF Gene Modifier Consortium (population-based); JHU = Johns Hopkins University (twin-siblings design); non-FF = patients with CF who are not homozygous for F508del in the CFTR gene; UNC = University of North Carolina (extremes of phenotype); UW = University of Washington (longitudinal study for effect of Pseudomonas aeruginosa acquisition on lung disease).
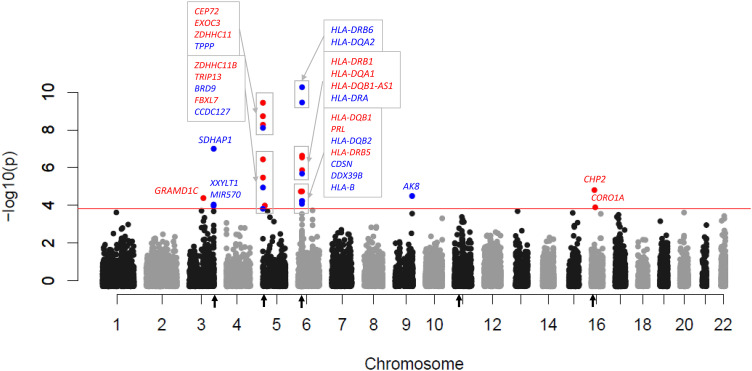
To further investigate genotype associations for all pwCF (N = 7,840), we imputed expression values to investigate TWAS association with lung phenotype using a modified approach (15) compared with a previous study of TWAS in CF (7). Twenty-nine annotated genes displayed a false-discovery q value lower than 0.1 (Figure 4 and Tables E3 and E4). Most genes with significant TWAS signals occurred in the six significant GWAS loci (Figure 2), congruent with a previous report (7). In this analysis, MUC4 was suggestive (q = 0.14).
Figure 4.
Significant genes based on transcriptome-wide association (TWAS) evidence for expression versus lung function (Kulich normal residual mortality-adjusted phenotype). Genome-wide Manhattan plot of TWAS associations. The red line corresponds to transcriptome-wide false-discovery q lower than 0.10, with significant genes labeled. Red-colored text corresponds to increased expression associated with improved lung function, and blue-colored text corresponds to increased expression associated with decreased lung function. Regions of significant genome-wide phenotype–genotype association are marked with black arrows on the x-axis.
Previous studies suggested that there may be GWAS loci associated more strongly with pwCF homozygous for the CFTR variant F508del (6). Analyses in this study identified three new suggestive loci (Figures E8–E10).
Significant results (q < 0.10) from the VEGAS2 gene-level association analyses are provided in Table E5. Because gene-level analyses can capture effects of long-range LD, we grouped significant regions into those separated by more than 5 Mb. Five of the six regions with individually significant SNPs (see above) were also significant in gene-level analysis (excepting the chr11p13 region). Among the remaining significant genes identified by VEGAS2, several achieved Bonferroni significance at a more stringent α = 0.05 (P < 2.5 × 10−6): ADAMTS8, LINC01844, and PTTG1IP.
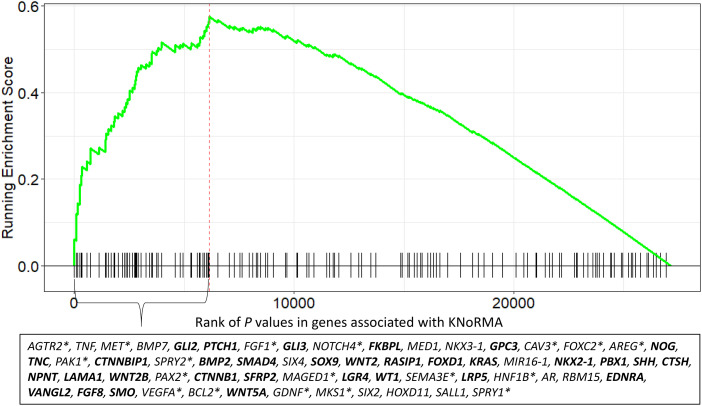
We performed GSEA on genes ranked using VEGAS2 P values (Table E5) (19) to explore pathways linked to lung disease severity. Pathways identified were largely related to pathogenic mechanisms linked to pulmonary host defense and genes at GWAS-significant loci (Table E6) (6, 7, 23, 24) involving inflammation, viral and bacterial infection and host responses, immunity and HLA-II pathways, endomembrane function, and microtubular/cytoskeletal function. In addition, multiple pathways related to organ development and morphogenesis were identified (Table E6). The most significant development/morphogenesis pathway (Gene Ontology [GO] BP0048754, branching morphogenesis of an epithelial tube; GSEA plot shown in Figure 5) includes 32 genes in the leading edge (in boldface in Figure 5) that relate to three signaling pathways (Shh [Sonic Hedgehog], TGFb [transforming growth factor β], and Wnt [wingless related-integration site]/β-catenin) that are necessary for lung development and branching morphogenesis (25–27). Thus, genetic variation that affects lung development in utero and early childhood has implications for the severity of CF lung disease.
Figure 5.
Genes that drive core enrichment-significant results for this branching morphogenesis pathway (Gene Ontology 0048754). This VEGAS2 analysis Gene Set Enrichment Analysis plot includes 32 genes that are in three key signaling pathways (Shh [25]; TGFb [26]; and Wnt [27]) for lung development (including branching morphogenesis) and/or interact with genes in those three signaling pathways and/or have other roles in lung development (shown in bold). The 18 genes that are associated with lung repair and/or play a role in molecular pathogenic aspects of lung disorders (e.g., chronic obstructive pulmonary disease, asthma, lung fibrosis, cellular morphogenesis) are shown with asterisks. The remaining 11 genes are reported to have a role in development and/or morphogenesis in other tissues.
Discussion
Variability of lung disease severity in CF reflects substantial non-CFTR genetic variation (3). Identifying the molecular basis of CF lung disease severity will provide pathobiological insights and identify new therapeutic targets. Studies using genotype array–based platforms and a standardized lung disease phenotype (KNoRMA) in different cohorts, study designs, and ages/birth cohorts have identified non-CFTR genetic variation of high mechanistic interest (6). By combining WGS with imputation from array-based genotypes across multiple cohorts, we provide the largest analyses associating genetic variation with CF lung disease severity in the premodulator era to date (7,840 pwCF; an estimated 19% of pwCF currently in North America and France) (2, 4).
One novel insight emerged from pathway analyses (i.e., GSEA) of genes ranked by VEGAS2, in which multiple significant pathways related to organ development were identified. Although not annotated specifically to the lung, the genes within these pathways, especially those related to three key signaling pathways (Shh, TGFb, and Wnt), are known to be critical for lung development and branching morphogenesis (25–27). There are at least 40 genes that relate to these three key signaling pathways in the top annotated pathway (Figure 5). The next challenge will be to decipher the mechanism by which these genes could influence CF lung disease severity. The issue is complex because not only do these genes play a role in lung development/morphogenesis, but it is also now appreciated that reactivation of developmental genes/pathways is a necessary component of lung repair after injury/inflammation (28). Several potential complementary mechanisms could be operative. First, variable early-life growth of the bronchial tree airway diameter relative to lung volume (i.e., dysanapsis) was proposed nearly 50 years ago (29). There is now anatomical evidence from computed tomography to confirm dysanapsis in conducting airways, and the presence of smaller-diameter bronchi is known to associate with chronic obstructive pulmonary disease and childhood asthma (30–33). Dysanapsis has not been previously recognized as a potential pathogenic driver of CF airway disease, but, given the periods of bronchial injury common in CF, dysanapsis could have profound effects on long-term outcomes. Second, CFTR itself is known to interact with lung development in several ways: tracheal and proximal bronchial diameter is altered in CF compared with normal pigs during embryonic development (34), CFTR plays a key role in fluid-mediated distension of airways during development (35), and lack of CFTR with consequent infections and inflammation are associated with tracheomalacia, which is linked to poorer outcomes (36, 37). Finally, because reactivation of developmental pathways is important for repair after airway injury (28), genetic variation in these pathways is expected to alter outcomes after CF-related inflammatory damage. Other significant pathways involve microtubular/cytoskeletal function (Gene Ontology BP0051494, “negative regulation of cytoskeleton organization”; Figure E11), which is of particular interest because of a recent potential therapeutic advance by restoration of microtubular dysfunction in CF cells (38).
The five genome-wide significant loci previously reported (6) are highly significant in the present analyses and contain genes of relevance (2, 4). In addition, another locus (chr16p12.2) is genome-wide significant, and four new loci are suggestive (P < 5 × 10−7; all with low minor allele frequency; range, 0.005–0.009). The newly significant locus on chr16p12.2 is intergenic between CHP2 and PRKCB (39, 40). CHP2 regulates airway pH through the apical membrane Na+/H+ exchanger (40). A SNP at this locus (rs11646605) is associated with Mycobacterium avium complex lung disease in non-CF patients and is an expression quantitative trait locus (eQTL) for CHP2 in the lung (41). TWAS analyses point to CHP2 expression as a key candidate in this region (Figure 4). PRKCB is a protein kinase that plays a role in multiple cellular functions, including apoptosis and autophagy (39). Finally, a nearby gene (ERN2) regulates airway mucin genes (MUC5B and MUC5AC) (42).
At the chr3q29 locus, MUC4 and MUC20 are highly relevant candidate genes because they play important roles in lung host defense and mucociliary clearance, which are abnormal in CF (2, 4). These WGS data now support MUC4 as the mechanistic link, with all significant SNPs intragenic to MUC4; plus, MUC4 is supported by a separate study integrating eQTLs and CF GWAS summary statistics using colocalization analysis (43).
Significant SNPs at the chr5p15.3 locus span approximately 300 kb and cover four pertinent genes expressed in respiratory epithelia. Airway surface liquid pH is abnormal in CF and regulated in part by SLC9A3, which codes for an Na+/H+ exchanger (44). Moreover, variable numbers of tandem repeats in this region are associated with expression of SLC9A3 in CF respiratory epithelia (45). The other three genes at this locus (EXOC3, CEP72, and TPPP) are involved in cellular microtubular function, which is abnormal in CF (46, 47). These three microtubule-related genes are consistently seen in TWAS-type studies (Figure 4) (7). Resveratrol is an antiinflammatory polyphenol that is known to activate several pathways relevant to microtubule stability, and it has been recently shown to restore microtubule function and intracellular transport in CF cells (38). Finally, an intergenic SNP (rs11738281) in CEP72 in our study is associated with airflow obstruction (i.e., reduced FEV1/FVC ratio) in the UK Biobank GWAS (48) and is in LD (r2 = 0.61) with the most significant regional SNP at this locus (chr5p13).
We observed strong gene-expression signatures at the chr6p21.3 (HLA class II) locus, which is associated with many inflammatory and respiratory conditions (49). In addition to TWAS (Figure 4), differential gene expression and biological pathway studies have identified several HLA-II genes associated with CF lung disease (23, 24), as did our pathway analyses (see online supplement). Functional interpretation of these data is confounded by many polymorphisms and allotypes of genes in this region (50).
Since the chr11p13 locus was first associated with CF lung disease, it has been extensively studied (51, 52). The most significant SNPs are intergenic between EHF, an epithelial transcription factor, and APIP, an enzyme involved in inflammation through roles in apoptosis and the methionine salvage pathway (4, 24). Conceptually, either of these genes could impact CF lung disease severity (2, 4). Regulatory regions are in the significant LD block that interacts with EHF and nearby ELF5 (52, 53), but extensive studies have not identified any eQTLs that might drive the phenotype (7, 51, 52). Further, our TWAS analysis (Figure 4) produced no signatures that suggest a mechanism. Interpretation of this region is further complicated by finding a second group of significantly associated SNPs over APIP after conditioning on the top-ranked SNP. The presence of two significant groups of SNPs at this locus implies that the risk for each pwCF can be viewed in terms of four (rather than two) alleles, minor alleles of the primary and secondary SNPs have opposite associations with KNoRMA, and the effect sizes (β-coefficients) for the primary and secondary SNPs are different (0.9 and 0.2, respectively). Taken together, these features create a potential complex molecular interplay among four alleles, whereby genotype associations of the primary SNP with KNoRMA are affected by the genotypes of the secondary SNP (Figure E6).
The chrXq22-q23 locus contains two genes (AGTR2, SLC6A14) that are expressed in respiratory epithelia, with functions relevant to pathophysiology of CF lung disease. AGTR2 functions in the RAS2 pathway (renin-angiotensin signaling), which is involved in several aspects of lung biology, including inflammation (4). The renin-angiotensin signaling pathway is altered in CF, and studies in genetically modified mice have therapeutic implications, as deletion and pharmacologic inhibition of AGTR2 improves several features of lung function in CF mice (54). AGTR2 is also prominent in pathway analyses (Figure 5 and online supplement). SLC6A14 encodes an amino acid transporter with pleiotropic effects in CF, as it has been linked to lung disease and neonatal intestinal obstruction, but the pathophysiologic mechanisms have not been defined (55, 56).
We noted significant concordance of effect sizes across significant loci among cohorts, with the youngest cohort (UW) showing the smallest effect size, despite medians and distribution of KNoRMA being similar across cohorts. This may reflect smaller effects of variants on lung function (i.e., FEV1) over a shorter time period in younger pwCF. In addition, age at phenotyping is confounded by year of birth cohort, as improvements in treatment (before modulators) may have blunted the decline in lung function in these younger pwCF. Therefore, it is challenging to define the specific mechanism(s) for smaller effect size in the youngest cohort.
There are several limitations of this study. First, although this is a large sample size for a study of a rare Mendelian disorder, it is likely underpowered to detect rare lung disease–associated variants. Second, a replication study was not performed because there is no adequate CF population readily available. Third, we were unable to establish causality at any locus, and identification of causal SNPs is complicated by multiple potential modifiers at each locus. Fourth, some potential variants were not fully queried, such as variable numbers of tandem repeats and structural variants. Finally, the population studied largely reflects European ancestry, and important modifier loci present in other populations therefore may have been missed.
In summary, WGS of pwCF enabled accurate genome-wide imputation, which allowed a premodulator association study of genetic variants with lung disease severity in 7,840 pwCF. This approach validated previously identified loci, provided better molecular understanding of significant loci, and enabled discovery of new biologically relevant candidate genes and biological pathways, particularly related to lung development. Taken together, these genomic, transcriptional, and pathway data will inform future mechanistic and postmodulator genetic studies and enable development of novel therapeutics for CF lung disease.
Acknowledgments
Acknowledgment
The authors thank the CF Foundation and CF Canada for the use of CF Foundation Patient Registry data and Canadian CF Registry data to conduct this study and the patients, care providers, and clinic coordinators at CF Centers throughout the United States, Canada, and France for their contributions to the CF Foundation Patient Registry, the Canadian CF Registry, and the French CF Registry.
Footnotes
Supported by the Cystic Fibrosis Foundation (CUTTIN18XX1, BAMSHA18XX0, KNOWLE18XX0, and BOUCHE19R0); partially supported by Dr. Zhou’s Start-up funding at NCSU; Canadian Institutes of Health Research (FRN 167282); Cystic Fibrosis Canada (2626); National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disorders P30 DK065988; National Heart, Lung, and Blood Institute (NHLBI) through the BioData Catalyst program (awards 1OT3HL142479-01, 1OT3HL142478-01, 1OT3HL142481-01, 1OT3HL142480-01, and 1OT3HL147154); government of Canada through Genome Canada (OGI-148); and a grant from the Government of Ontario. Any opinions are those of the authors and do not reflect the views of NHLBI, individual BioData Catalyst members, or affiliated organizations.
Author Contributions: R.G.P, E.E.B., J.M.C., and M.R.K: data acquisition/data analysis/interpretation/final manuscript review; M.A.A., A.V.F., F.M.O., and K.S.R.: data acquisition/data analysis/final manuscript review; G.R.C. and R.L.G.: data acquisition/interpretation/final manuscript review; K.J.B., M.R., H.C., L.J.S., M.J.B., and S.M.B: data acquisition/final manuscript review; W.W.G., K.N.H., H.L., W.L., K.P., Q.S., J.W., and Y.L.: data analysis/final manuscript review; Y.-H.Z., P.J.G., F.A.W., H.D., and E.W.P.: data analysis/interpretation/final manuscript review; Y.-H.Z., P.J.G., H.D., Y.L., and F.A.W.: statistical analysis; Y.-H.Z., P.J.G., R.G.P., H.D., F.A.W., W.K.O’N., and M.R.K.: manuscript drafting; Y.-H.Z., F.A.W., W.K.O’N., and M.R.K: study design; Y.-H.Z., H.C., L.J.S., M.J.B., S.M.B., G.R.C., R.L.G, W.K.O’N., and M.R.K.: obtained funding.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202209-1653OC on March 15, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet . 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paranjapye A, Ruffin M, Harris A, Corvol H. Genetic variation in CFTR and modifier loci may modulate cystic fibrosis disease severity. J Cyst Fibros . 2020;19:S10–S14. doi: 10.1016/j.jcf.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med . 2007;175:1036–1043. doi: 10.1164/rccm.200608-1164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Neal WK, Knowles MR. Cystic fibrosis disease modifiers: complex genetics defines the phenotypic diversity in a monogenic disease. Annu Rev Genomics Hum Genet . 2018;19:201–222. doi: 10.1146/annurev-genom-083117-021329. [DOI] [PubMed] [Google Scholar]
- 5. Egan ME. Cystic fibrosis transmembrane conductance receptor modulator therapy in cystic fibrosis, an update. Curr Opin Pediatr . 2020;32:384–388. doi: 10.1097/MOP.0000000000000892. [DOI] [PubMed] [Google Scholar]
- 6. Corvol H, Blackman SM, Boëlle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun . 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang H, Polineni D, Pace RG, Stonebraker JR, Corvol H, Cutting GR, et al. Mining GWAS and eQTL data for CF lung disease modifiers by gene expression imputation. PLoS One . 2020;15:e0239189. doi: 10.1371/journal.pone.0239189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raraigh KS, Aksit MA, Hetrick K, Pace RG, Ling H, O’Neal W, et al. Complete CFTR gene sequencing in 5,058 individuals with cystic fibrosis informs variant-specific treatment. J Cyst Fibros . 2022;21:463–470. doi: 10.1016/j.jcf.2021.10.011. [DOI] [PubMed] [Google Scholar]
- 9. Zhou Y, Gallins P, Pace R, Dang H, O’Neal W, Li Y, et al. Genetic variants that modify severity of CF lung disease: update from the CF genome project. J Cyst Fibros . 2021;20:S306. [Google Scholar]
- 10. Sun Q, Liu W, Rosen JD, Huang L, Pace RG, Dang H, et al. Cystic Fibrosis Genome Project Leveraging TOPMed imputation server and constructing a cohort-specific imputation reference panel to enhance genotype imputation among cystic fibrosis patients. HGG Adv . 2022;3:100090. doi: 10.1016/j.xhgg.2022.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knapp EA, Fink AK, Goss CH, Sewall A, Ostrenga J, Dowd C, et al. The Cystic Fibrosis Foundation Patient Registry. Design and methods of a national observational disease registry. Ann Am Thorac Soc . 2016;13:1173–1179. doi: 10.1513/AnnalsATS.201511-781OC. [DOI] [PubMed] [Google Scholar]
- 12. Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol . 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics . 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol . 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet . 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science . 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ligthart L, van Beijsterveldt CEM, Kevenaar ST, de Zeeuw E, van Bergen E, Bruins S, et al. The Netherlands Twin Register: longitudinal research based on twin and twin-family designs. Twin Res Hum Genet . 2019;22:623–636. doi: 10.1017/thg.2019.93. [DOI] [PubMed] [Google Scholar]
- 18. Mishra BH, Mishra PP, Raitoharju E, Marttila S, Mononen N, Sievänen H, et al. Modular genome-wide gene expression architecture shared by early traits of osteoporosis and atherosclerosis in the Young Finns Study. Sci Rep . 2021;11:7111. doi: 10.1038/s41598-021-86536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet . 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA . 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) . 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kingston H, Stilp AM, Gordon W, Broome J, Gogarten SM, Ling H, et al. Cystic Fibrosis Genome Project; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Accounting for population structure in genetic studies of cystic fibrosis. HGG Adv . 2022;3:100117. doi: 10.1016/j.xhgg.2022.100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O’Neal WK, Gallins P, Pace RG, Dang H, Wolf WE, Jones LC, et al. Gene expression in transformed lymphocytes reveals variation in endomembrane and HLA pathways modifying cystic fibrosis pulmonary phenotypes. Am J Hum Genet . 2015;96:318–328. doi: 10.1016/j.ajhg.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polineni D, Dang H, Gallins PJ, Jones LC, Pace RG, Stonebraker JR, et al. Airway mucosal host defense is key to genomic regulation of cystic fibrosis lung disease severity. Am J Respir Crit Care Med . 2018;197:79–93. doi: 10.1164/rccm.201701-0134OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Belgacemi R, Danopoulos S, Deutsch G, Glass I, Dormoy V, Bellusci S, et al. Hedgehog signaling pathway orchestrates human lung branching morphogenesis. Int J Mol Sci . 2022;23:5265. doi: 10.3390/ijms23095265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito A, Horie M, Nagase T. TGF-β signaling in lung health and disease. Int J Mol Sci . 2018;19:2460. doi: 10.3390/ijms19082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aros CJ, Pantoja CJ, Gomperts BN. Wnt signaling in lung development, regeneration, and disease progression. Commun Biol . 2021;4:601. doi: 10.1038/s42003-021-02118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev . 2019;99:513–554. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Green M, Mead J, Turner JM. Variability of maximum expiratory flow-volume curves. J Appl Physiol . 1974;37:67–74. doi: 10.1152/jappl.1974.37.1.67. [DOI] [PubMed] [Google Scholar]
- 30. Smith BM, Kirby M, Hoffman EA, Kronmal RA, Aaron SD, Allen NB, et al. MESA Lung, CanCOLD, and SPIROMICS Investigators Association of dysanapsis with chronic obstructive pulmonary disease among older adults. JAMA . 2020;323:2268–2280. doi: 10.1001/jama.2020.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vameghestahbanati M, Kirby M, Tanabe N, Vasilescu DM, Janssens W, Everaerts S, et al. Central airway tree dysanapsis extends to the peripheral airways. Am J Respir Crit Care Med . 2021;203:378–381. doi: 10.1164/rccm.202007-3025LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith BM, Traboulsi H, Austin JHM, Manichaikul A, Hoffman EA, Bleecker ER, et al. MESA Lung and SPIROMICS investigators Human airway branch variation and chronic obstructive pulmonary disease. Proc Natl Acad Sci USA . 2018;115:E974–E981. doi: 10.1073/pnas.1715564115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Casas M, den Dekker HT, Kruithof CJ, Reiss IK, Vrijheid M, Sunyer J, et al. The effect of early growth patterns and lung function on the development of childhood asthma: a population based study. Thorax . 2018;73:1137–1145. doi: 10.1136/thoraxjnl-2017-211216. [DOI] [PubMed] [Google Scholar]
- 34. Adam RJ, Abou Alaiwa MH, Bouzek DC, Cook DP, Gansemer ND, Taft PJ, et al. Postnatal airway growth in cystic fibrosis piglets. J Appl Physiol (1985) . 2017;123:526–533. doi: 10.1152/japplphysiol.00263.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brennan SC, Wilkinson WJ, Tseng HE, Finney B, Monk B, Dibble H, et al. The extracellular calcium-sensing receptor regulates human fetal lung development via CFTR. Sci Rep . 2016;6:21975. doi: 10.1038/srep21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fischer AJ, Singh SB, Adam RJ, Stoltz DA, Baranano CF, Kao S, et al. Tracheomalacia is associated with lower FEV1 and Pseudomonas acquisition in children with CF. Pediatr Pulmonol . 2014;49:960–970. doi: 10.1002/ppul.22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wallis C, Alexopoulou E, Antón-Pacheco JL, Bhatt JM, Bush A, Chang AB, et al. ERS statement on tracheomalacia and bronchomalacia in children. Eur Respir J . 2019;54:1900382. doi: 10.1183/13993003.00382-2019. [DOI] [PubMed] [Google Scholar]
- 38. Lu B, Corey DA, Kelley TJ. Resveratrol restores intracellular transport in cystic fibrosis epithelial cells. Am J Physiol Lung Cell Mol Physiol . 2020;318:L1145–L1157. doi: 10.1152/ajplung.00006.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newton AC. Protein kinase C: perfectly balanced. Crit Rev Biochem Mol Biol . 2018;53:208–230. doi: 10.1080/10409238.2018.1442408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Di Sole F, Vadnagara K, Moe OW, Babich V. Calcineurin homologous protein: a multifunctional Ca2+-binding protein family. Am J Physiol Renal Physiol . 2012;303:F165–F179. doi: 10.1152/ajprenal.00628.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Namkoong H, Omae Y, Asakura T, Ishii M, Suzuki S, Morimoto K, et al. Nontuberculous Mycobacteriosis and Bronchiectasis – Japan Research Consortium (NTM-JRC) Genome-wide association study in patients with pulmonary Mycobacterium avium complex disease. Eur Respir J . 2021;58:1902269. doi: 10.1183/13993003.02269-2019. [DOI] [PubMed] [Google Scholar]
- 42. Chen G, Ribeiro CMP, Sun L, Okuda K, Kato T, Gilmore RC, et al. XBP1S regulates MUC5B in a promoter variant-dependent pathway in IPF airway epithelia. Am J Respir Crit Care Med . 2019;200:220–234. doi: 10.1164/rccm.201810-1972OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang F, Panjwani N, Wang C, Sun L, Strug LJ. A flexible summary statistics-based colocalization method with application to the mucin cystic fibrosis lung disease modifier locus. Am J Hum Genet . 2022;109:253–269. doi: 10.1016/j.ajhg.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pereira SV, Ribeiro JD, Bertuzzo CS, Marson FAL. Association of clinical severity of cystic fibrosis with variants in the SLC gene family (SLC6A14, SLC26A9, SLC11A1 and SLC9A3) Gene . 2017;629:117–126. doi: 10.1016/j.gene.2017.07.068. [DOI] [PubMed] [Google Scholar]
- 45.Roshandel D, Mastromatteo S, Wang C, Gong J, Thiruvahindrapuram B, Sung WWL, et al. A cystic fibrosis lung disease modifier locus harbors tandem repeats associated with gene expression. 2022. https://www.medrxiv.org/content/10.1101/2022.03.28.22272580v1
- 46. Rymut SM, Kampman CM, Corey DA, Endres T, Cotton CU, Kelley TJ. Ibuprofen regulation of microtubule dynamics in cystic fibrosis epithelial cells. Am J Physiol Lung Cell Mol Physiol . 2016;311:L317–L327. doi: 10.1152/ajplung.00126.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rymut SM, Lu B, Perez A, Corey DA, Lamb K, Cotton CU, et al. Acetyl-CoA carboxylase inhibition regulates microtubule dynamics and intracellular transport in cystic fibrosis epithelial cells. Am J Physiol Lung Cell Mol Physiol . 2019;316:L1081–L1093. doi: 10.1152/ajplung.00369.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet . 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D’Antonio M, Reyna J, Jakubosky D, Donovan MK, Bonder MJ, Matsui H, et al. Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife . 2019;8:e48476. doi: 10.7554/eLife.48476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Osoegawa K, Mallempati KC, Gangavarapu S, Oki A, Gendzekhadze K, Marino SR, et al. HLA alleles and haplotypes observed in 263 US families. Hum Immunol . 2019;80:644–660. doi: 10.1016/j.humimm.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dang H, Gallins PJ, Pace RG, Guo XL, Stonebraker JR, Corvol H, et al. Novel variation at chr11p13 associated with cystic fibrosis lung disease severity. Hum Genome Var . 2016;3:16020. doi: 10.1038/hgv.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swahn H, Sabith Ebron J, Lamar KM, Yin S, Kerschner JL, NandyMazumdar M, et al. Coordinate regulation of ELF5 and EHF at the chr11p13 CF modifier region. J Cell Mol Med . 2019;23:7726–7740. doi: 10.1111/jcmm.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stolzenburg LR, Yang R, Kerschner JL, Fossum S, Xu M, Hoffmann A, et al. Regulatory dynamics of 11p13 suggest a role for EHF in modifying CF lung disease severity. Nucleic Acids Res . 2017;45:8773–8784. doi: 10.1093/nar/gkx482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Darrah RJ, Jacono FJ, Joshi N, Mitchell AL, Sattar A, Campanaro CK, et al. AGTR2 absence or antagonism prevents cystic fibrosis pulmonary manifestations. J Cyst Fibros . 2019;18:127–134. doi: 10.1016/j.jcf.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gong J, Wang F, Xiao B, Panjwani N, Lin F, Keenan K, et al. Genetic association and transcriptome integration identify contributing genes and tissues at cystic fibrosis modifier loci. PLoS Genet . 2019;15:e1008007. doi: 10.1371/journal.pgen.1008007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruffin M, Mercier J, Calmel C, Mésinèle J, Bigot J, Sutanto EN, et al. Update on SLC6A14 in lung and gastrointestinal physiology and physiopathology: focus on cystic fibrosis. Cell Mol Life Sci . 2020;77:3311–3323. doi: 10.1007/s00018-020-03487-x. [DOI] [PMC free article] [PubMed] [Google Scholar]