Abstract
Of 24 high-level gentamicin-resistant clinical isolates of Enterococcus faecalis, 20 carried gentamicin resistance (Gmr) plasmids. The plasmids ranged from 65.0 to 80.0 kb in size. Three of these plasmids were nonconjugative, and 17 transferred by conjugation to an E. faecalis recipient at low frequency (10−5 to 10−6 transconjugants per donor). The remaining four strains had a nonconjugative chromosomal Gmr determinant. On the basis of restriction enzyme and DNA-DNA hybridization profiles, Tn4001-like α elements were located on the chromosome and three types of Tn4001-truncated structures, I, II, and III, were found to be carried by the Gmr plasmids. Structure I lacked IS256 in the right-hand flanking extremity of Tn4001. Structure II was the same as structure I except that it also had a partial deletion of IS256 in the left-hand flanking extremity of Tn4001. Structure III lacked both the right- and left-hand flanking extremities of Tn4001. One of the wild-type strains carried the Gmr determinant both on the chromosome, as a Tn4001-like α element, and on a conjugative plasmid, as a Tn4001-truncated type I structure.
Enterococcus faecalis is increasingly implicated as an important cause of nosocomial infections, particularly in surgical intensive care units (17, 21). The natural tolerance of E. faecalis strains for antibiotics that interfere with cell wall synthesis has led to the current use of synergistic and bactericidal combinations of a beta-lactam or a glycopeptide with an aminoglycoside, usually gentamicin (16). The emergence and spread of E. faecalis strains with multiple antibiotic resistance, including resistance to penicillins, glycopeptides, and high levels of aminoglycosides, have greatly reduced the efficacy of such combinations. For this reason, nosocomial and severe E. faecalis infections continue to be a major medical problem (20).
The bifunctional enzyme, 6′-acetyltransferase-2"-phosphotransferase, which mediates high-level gentamicin resistance (Gmr) in enterococci and streptococci is encoded by the fused aac6-aph2 gene (6). This gene is plasmid borne in most gentamicin-resistant strains of E. faecalis (11, 18, 22) and other enterococcal strains studied so far (23, 25) and has been reported to be situated, on these plasmids, on elements similar to the α or β forms of Tn4001, originally identified in Staphylococcus aureus (15). The β element is a form of Tn4001 that contains a tandem duplication of IS256, whereas the α form does not. The Gmr determinant is carried on plasmids by Tn4001-like elements in strains of E. faecalis (7, 8, 22) and Enterococcus avium (23). There are other plasmid-borne structures, including the Tn4001-truncated elements in E. faecalis (22), Enterococcus hirae (23), and Enterococcus raffinosus (23), and, in E. faecalis (8), the hybrid element Tn4001-IS257, originally found in S. aureus (2). The Gmr determinant has been found to be carried on the chromosome in two E. faecalis (19, 24), one group B Streptococcus (strain B128) (1), and several Streptococcus mitis (13) clinical isolates. The chromosomal transposons so far reported that carry the Gmr determinant are Tn3706 (5.8 kb), the Tn4001-like β element in B128 (10), and, in E. faecalis, Tn5384 (26.0 kb), which consists in part of a Tn4001-like α structure (19), and Tn924 (27.0 kb), a novel type of transposable element (24).
The purpose of the present report was to provide information on the genetic and molecular basis of high-level gentamicin resistance in E. faecalis strains isolated in France. Twenty-four E. faecalis clinical isolates were examined with respect to the conjugative transfer, location, and structure of the genetic elements carrying the Gmr determinant.
MATERIALS AND METHODS
The 24 independent wild-type E. faecalis clinical strains used in this study (see Table 1) were isolated from the blood and urine of patients with septicemia, endocarditis, or genital and urinary tract infections. In addition to being resistant to high levels of gentamicin-kanamycin (MIC >2,000 μg/ml), most of them were also resistant to chloramphenicol, erythromycin, and tetracycline-minocycline. In mating experiments, carried out on membrane filters (9), the wild-type strains, used as donors, were crossed with E. faecalis recipient strain JH2-2 (12), which is resistant to rifampin and fusidic acid, and the resulted transconjugant clones were crossed with E. faecalis recipient strain BM133 (11), which is resistant to high levels of streptomycin. The antibiotics used were 1,000 μg of gentamicin per ml for the selection of transconjugants and 25 μg of fusidic acid per ml plus 100 μg of rifampin per ml or 2,000 μg of streptomycin per ml for the counterselection of the donors.
TABLE 1.
Relevant characteristics of the 24 wild-type strains and their transconjugantsa
| Strain no. (designation) | Hospital (city) (yr of isolation) | Hemolysis | Antibiotic resistance markers of wild-type strain | Frequency of conjugative transfer of Gmr marker into JH2-2 | Antibiotic resistance markers of transconjugantsb | Gmr plasmid in transconjugants or wild-type strains |
|---|---|---|---|---|---|---|
| 1 (D362)c | Hotel Dieu (Paris) (1978) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 7 × 10−2 | Cmr Gmr-Kmr | pIP655 |
| 2 (D367) | Hotel Dieu (Paris) (1978) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 2 × 10−1 | Cmr Gmr-Kmr | pIP687 |
| 3 (D366) | Boucicaut (Paris) (1978) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 3 × 10−2 | Cmr Gmr-Kmr | pIP683 |
| 4 (8942) | Broussais (Paris) (1981) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−1 | Cmr Gmr-Kmr | pIP1722 |
| 5 (8943) | Broussais (Paris) (1981) | NH | Cmr Emr Gmr-Kmr | <1 × 10−9 | NA | pIP1723d |
| 6 (D396) | Broussais (Paris) (1983) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 3 × 10−6 | Cmr Gmr-Kmr | pIP1724 |
| 7 (9636) | Broussais (Paris) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 2 × 10−5 | Gmr-Kmr | pIP1730 |
| 8 (10028) | Broussais (Paris) (1985) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−5 | Gmr-Kmr | pIP1731 |
| 9 (10029) | Broussais (Paris) (1985) | NH | Gmr-Kmr Tcr-Mnr | 3 × 10−6 | Gmr-Kmr | pIP1732 |
| 10 (10035) | Broussais (Paris) (1985) | NH | Emr Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | pIP1733d |
| 11 (10042) | Broussais (Paris) (1985) | NH | Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | NA |
| 12 (10628) | Broussais (Paris) (1988) | NH | Emr Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | NA |
| 13 (10586) | Broussais (Paris) (1988) | β | Emr Gmr-Kmr Tcr-Mnr | 1 × 10−1 | Gmr-Kmr | pIP1734 |
| 14 (10619) | Broussais (Paris) (1988) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−2 | Emr Gmr-Kmr | pIP1735 |
| 15 (10629) | Broussais (Paris) (1988) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 2 × 10−3 | Cmr Emr Gmr-Kmr | pIP1736 |
| 16 (10630) | Broussais (Paris) (1988) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 2 × 10−3 | Cmr Emr Gmr-Kmr | pIP1737 |
| 17 (10282) | Saint-Joseph (Paris) (1986) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | NA |
| 18 (9850) | Pontchaillou (Rennes) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | pIP1725d |
| 19 (9851) | Pontchaillou (Rennes) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−5 | Gmr-Kmr | pIP1726 |
| 20 (9852) | Pontchaillou (Rennes) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−5 | Gmr-Kmr | pIP1727 |
| 21 (9853) | Pontchaillou (Rennes) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−5 | Gmr-Kmr | pIP1728 |
| 22 (9854) | Pontchaillou (Rennes) (1984) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | 1 × 10−5 | Gmr-Kmr | pIP1729 |
| 23 (10703) | (Chateauroux) (1989) | β | Cmr Emr Gmr-Kmr Tcr-Mnr | 2 × 10−2 | Emr Gmr-Kmr | pIP1738 |
| 24 (10907) | (Longjumeaux) (1990) | NH | Cmr Emr Gmr-Kmr Tcr-Mnr | <1 × 10−9 | NA | NA |
Abbreviations: β, beta-hemolysis; Gmr-Kmr, high-level gentamicin-kanamycin resistance; NA, not applicable; NH, nonhemolytic.
All the transconjugants studied were nonhemolytic.
Reference 9 for strains 1 to 3; this study for strains 4 to 24.
Plasmid isolated from the wild-type strains.
Isolation of cellular and plasmid DNA from E. faecalis strains, digestion by restriction enzymes (EcoRI, HaeIII, HincII, HindIII, and ScaI), gel electrophoresis, DNA blotting, DNA-DNA hybridization (under stringent conditions), and labeling of the probes with [α-32P]dCTP were carried out as previously described (14). The probes used for hybridization were pSF815A, containing the aac6-aph2 gene (Gmr probe) (6); pIP1551, containing a 468-bp DNA fragment of IS256 which is a part of Tn4001 (IS256 probe) (see Fig. 1) (4); and pIP1644, containing a 629-bp DNA internal fragment of IS257 (IS257 probe) (5).
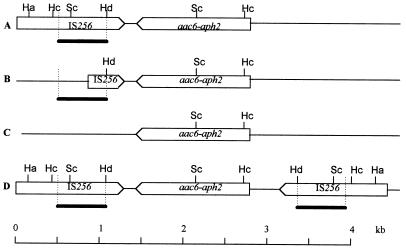
FIG. 1.
Restriction maps of the structures carrying the Gmr marker. (A) Tn4001-truncated type I element carried by the plasmids from strains 1 to 5, 7 to 10, and 13; lack of IS256 in the right-hand flanking region of Tn4001. (B) Tn4001-truncated type II element carried by the plasmids from strains 18 to 22; lack of IS256 in the right-hand flanking region of Tn4001 and a partial deletion of IS256 in its left-hand flanking region. (C) Tn4001-truncated type III element, carried by the plasmids from strains 6, 14 to 16, and 23; lack of IS256 in both right- and left-hand flanking regions of Tn4001. (D) Tn4001-like α element borne on the chromosome of strains 11 to 13, 17, and 24. The open horizontal arrows indicate the direction of transcription of the gene aac6-aph2 and of IS256. Restriction endonuclease abbreviations: Ha, HaeIII; Hc, HincII; Hd, HindIII; Sc, ScaI. The location of the probe IS256 is indicated by a thick black line.
Macrorestriction analysis of the SmaI-digested cellular DNA of the wild-type strains and pulsed-field gel electrophoresis were done as we described recently (22).
RESULTS
Conjugative transfer of the Gmr marker and plasmid isolation.
Each of the 24 wild-type E. faecalis strains was mated with JH2-2. The results are presented in Table 1. The Gmr determinant transferred by conjugation from the nine β-hemolytic wild-type strains (1 to 4, 13 to 16, and 23) at high frequency (1 × 10−1 to 2 × 10−3 transconjugants per donor cell), and in each cross, cell aggregates were observed on the mating filters (9). The frequency of transfer of the Gmr marker was low (1 × 10−5 to 3 × 10−6) for 8 (6 to 9 and 19 to 22) of the 15 nonhemolytic wild-type strains; no detectable transconjugants were obtained (transfer frequency <10−9) for the remaining seven strains (5, 10 to 12, 17, 18, and 24). Analysis of the transconjugants for the presence of unselected antibiotic resistance markers showed that the clones obtained by using as donors the wild-type strains 7 to 9, 13, and 19 to 22 were resistant to only gentamicin-kanamycin. Each of these clones carried a single plasmid, estimated to be 65.0 to 80.0 kb, after digestion with HindIII. The transconjugants that were resistant to gentamicin-kanamycin and also to chloramphenicol, erythromycin, or both carried at least two large plasmids. None of the transconjugant clones carried the tetracycline-minocycline marker. The Gmr plasmids retransferred into BM133 from the nonhemolytic transconjugants, whether obtained from β-hemolytic or from nonhemolytic wild-type strains, at low frequencies (10−5 to 10−6) and without the formation of cell aggregates on mating filters.
Location of the aac6-aph2 gene and of IS256 and mapping of the structures carrying the Gmr determinant.
Plasmid and cellular DNA isolated from all the wild-type strains was digested with the restriction enzymes used to characterize Tn4001-like structures: HaeIII, HincII, HindIII, ScaI, and ScaI plus HindIII (double digestion). The digested DNA was subjected to gel electrophoresis and to DNA-DNA hybridization experiments using the gene aac6-aph2 as well as IS256 and IS257 as probes.
The results obtained by DNA-DNA hybridization are presented in Table 2. On the Gmr plasmids in strains 1 to 5, 7 to 10, 13, and 18 to 22, the aac6-aph2 gene was located on HindIII fragments of 4.5 to 10.0 kb and sequences homologous to IS256 were found on HindIII fragments of 2.5 to 8.3 kb. On the Gmr plasmids in strains 6, 14 to 16, and 23, the aac6-aph2 gene was located on HindIII fragments of 11.0 kb. In these strains, no hybridization with the IS256 probe was detected. In strains 11, 12, 17, and 24, the aac6-aph2 gene was located on chromosomal HindIII fragments of 2.5 kb and the IS256 probe was located on chromosomal HindIII fragments of 4.0 to 11.0 kb; no homology with the two probes was detected on any of the plasmids carried by the wild-type strains 11, 12, 17, and 24. No sequences homologous to IS257 were found on any plasmid or cellular DNA.
TABLE 2.
Location of the Gmr marker and of IS256 on plasmid and cellular DNA
| Strain no. (designation) | Gmr plasmid designation | Conjugative transfer (Tra)a | Hybridization with Gmr and IS256 probe (HindIII digestion)
|
Structure of the element carrying the Gmr marker (Fig. 1) | |||
|---|---|---|---|---|---|---|---|
| Plasmid DNA
|
Cellular DNA
|
||||||
| Gmr | IS256 | Gmr | IS256 | ||||
| 1 (D362) | pIP655 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 2 (D367) | pIP687 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 3 (D366) | pIP683 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 4 (8942) | pIP1722 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 5 (8943) | pIP1723 | Tra− | + | + | + | + | Tn4001-truncated I |
| 7 (9636) | pIP1730 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 8 (10028) | pIP1731 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 9 (10029) | pIP1732 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 10 (10035) | pIP1733 | Tra− | + | + | + | + | Tn4001-truncated I |
| 13 (10586)b | pIP1734 | Tra+ | + | + | + | + | Tn4001-truncated I |
| 18 (9850) | pIP1725 | Tra− | + | + | + | + | Tn4001-truncated II |
| 19 (9851) | pIP1726 | Tra+ | + | + | + | + | Tn4001-truncated II |
| 20 (9852) | pIP1727 | Tra+ | + | + | + | + | Tn4001-truncated II |
| 21 (9853) | pIP1728 | Tra+ | + | + | + | + | Tn4001-truncated II |
| 22 (9854) | pIP1729 | Tra+ | + | + | + | + | Tn4001-truncated II |
| 6 (D396) | pIP1724 | Tra+ | + | − | + | − | Tn4001-truncated III |
| 14 (10619) | pIP1735 | Tra+ | + | − | + | − | Tn4001-truncated III |
| 15 (10629) | pIP1736 | Tra+ | + | − | + | − | Tn4001-truncated III |
| 16 (10630) | pIP1737 | Tra+ | + | − | + | − | Tn4001-truncated III |
| 23 (10703) | pIP1738 | Tra+ | + | − | + | − | Tn4001-truncated III |
| 11 (10642) | NAc | Tra− | − | − | + | + | Tn4001-like α |
| 12 (10628) | NA | Tra− | − | − | + | + | Tn4001-like α |
| 17 (10282) | NA | Tra− | − | − | + | + | Tn4001-like α |
| 24 (10907) | NA | Tra− | − | − | + | + | Tn4001-like α |
Tra+, conjugative transfer; Tra−, nonconjugative transfer.
Strain 13 carried the Gmr marker both on pIP1734 and on a nonconjugative chromosomal Tn4001-like α structure.
NA, not applicable.
Using the hybridization profiles obtained with HaeIII, HincII, HindIII, ScaI, and ScaI-HindIII digestions, we mapped the structures that carried the Gmr determinant in each wild-type strain. We detected (Table 2 and Fig. 1) three Tn4001-truncated structures, designated I, II, and III, as well as the previously described Tn4001 α element (15). Tn4001-truncated type I elements (Fig. 1A) were found on the plasmids from strains 1 to 5, 7 to 10, and 13; these were characterized by the absence of IS256 in the right-hand flanking region of the element. In the 10 wild-type strains carrying this type of structure, three macrorestriction patterns were detected by pulsed-field gel electrophoresis of SmaI-digested cellular DNA. Each pattern differed from the others by at least four bands (data not shown). Tn4001-truncated type II elements (Fig. 1B) were detected on the plasmids from strains 18 to 22; these elements also lacked IS256 in the right-hand flanking region but differed from the Tn4001-truncated type I structures in that there was a partial deletion of IS256 in the left-hand flanking region of the element. The macrorestriction patterns of the SmaI-digested genomic DNA of strains 18 to 22 were similar, if not identical (data not shown). In fact, these strains had been isolated in the same hospital in the same year (Table 1) and are probably epidemiologically related. The Tn4001-truncated type III elements (Fig. 1C) found on the plasmids from strains 6, 14 to 16, and 23 lacked IS256 in both the right- and the left-hand flanking regions of the element. This result suggests that the insertion sequences originally flanking the gene aac6-aph2 may have been lost, may differ from IS256 and IS257, or may never have been present. The SmaI profile of strain 6 differed from that of strains 14 to 16 and 23 by at least four bands, whereas the macrorestriction patterns of strains 14, 15, 16, and 23 differed from each other by only one or two bands (data not shown). Tn4001-like α elements (Fig. 1D), found in the cellular DNA of wild-type strains 11 to 13, 17, and 24, were identified by their hybridization with the gene aac6-aph2 and with IS256 on fragments that were equivalent in size to those of the corresponding fragments of Tn4001 (15). The macrorestriction patterns of the SmaI-digested cellular DNA of strains 12, 13, 17, and 24 differed from each other by one to three bands and from that of strain 11 by at least six bands (data not shown).
The use of EcoRI to cleave the cellular and plasmid DNA revealed that only one copy of the element was found in all wild-type strains, except strain 13, in which two copies were detected. In fact, the Gmr determinant of strain 13 is located on both the conjugative plasmid pIP1734, as a Tn4001-truncated type I element, and the chromosome of the wild-type host, as a Tn4001-like α element.
DISCUSSION
The results presented here suggest that the conjugative transfer frequency of the Gmr plasmids carried by the β-hemolytic wild-type or transconjugant E. faecalis strains may have been high because these plasmids cotransferred along with the β-hemolysin plasmids carried by these strains. In fact, the Gmr plasmids studied here, when they occur in nonhemolytic strains, either are nonconjugative or transfer at low frequency; apparently, the conjugative transfer of these plasmids is not mediated by bacterial sex pheromones (3).
Structures that carry the aac6-aph2 gene in E. faecalis and other enterococcal strains are highly diverse (8, 19, 22–24). The Gmr plasmids from E. faecalis strains isolated in France (this study) carry either Tn4001-truncated type I elements, as reported for several Gmr plasmids harbored by E. faecalis strains isolated in Romania (22) and for pIP1701 from an E. raffinosus strain isolated in Portugal (23), or Tn4001-truncated type II and III structures which, to our knowledge, have not yet been described. In addition to the truncated structures I, II, and III, pICC8, the Gmr plasmid from an E. hirae strain isolated in Romania (23), carries an element characterized by the lack of IS256 in the left-hand flanking region of the aac6-aph2 gene, designated now as a Tn4001-truncated type IV structure.
The Tn4001-truncated elements were detected in this study only on the Gmr plasmids (20 strains). By contrast, the Tn4001-like α element has been found to be carried only on the chromosome (four strains). The chromosomal location of this transposon may serve to stabilize it while the location on a plasmid may contribute to its instability, as a great variety of truncated forms of Tn4001 were found on the plasmids examined here. Certain regions of the Tn4001 transposon may have undergone natural molecular rearrangements, generating the truncated structures that we observed. As we examined the DNA of the wild-type strains, these rearrangements did not occur in the course of mating experiments, although they may have occurred during in vivo conjugative transfer prior to the isolation of the wild-type strains.
Except for the Tn4001-truncated type II structure, the other three types of elements, detected in the present study (Table 2 and Fig. 1), have been found in both epidemiologically related and unrelated strains. The spread of high-level gentamicin resistance in E. faecalis isolates in France could be due to the dissemination of such elements in unrelated strains as well as to the spread of endemic and epidemic gentamicin-resistant strains. However, for about 15 years the incidence of high-level gentamicin resistance has been stable (<10%) in France. It is possible that the genetic instability of the structures carrying the Gmr determinant on plasmids in E. faecalis, associated with a decreased selective pressure, may act as a natural regulation mechanism to maintain the incidence of gentamicin resistance in France at a low but stable level.
ACKNOWLEDGMENTS
We thank Karen Pepper and Névine El Solh for criticism of the manuscript. We are grateful to N. El Solh for giving us strains harboring pIP1551 and pIP1644 and to J. J. Ferretti for the strain bearing pSF815A.
REFERENCES
- 1.Buu-Hoï A, Le Bouguénec C, Horaud T. High-level chromosomal gentamicin resistance in Streptococcus agalactiae (group B) Antimicrob Agents Chemother. 1990;34:985–988. doi: 10.1128/aac.34.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne M E, Gillespie M T, Skurray R A. Molecular analysis of a gentamicin transposon-like element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother. 1990;34:2106–2113. doi: 10.1128/aac.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clewell D B. Sex pheromones, plasmids and conjugation in Streptococcus faecalis. In: Halvorson H O, Monroy A, editors. The origin and evolution of sex. New York, N.Y: Alan R. Liss, Inc.; 1985. pp. 13–18. [Google Scholar]
- 4.Dyke K G H, Aubert S, El Solh N. Multiple copies of IS256 in staphylococci. Plasmid. 1992;28:235–246. doi: 10.1016/0147-619x(92)90055-f. [DOI] [PubMed] [Google Scholar]
- 5.El Solh, N., and K. G. H. Dyke. 1991. Unpublished results.
- 6.Ferretti J J, Gilmore K S, Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986;167:631–638. doi: 10.1128/jb.167.2.631-638.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodel-Christian S L, Murray B E. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to the staphylococcal transposons, Tn4001 and Tn4031. Antimicrob Agents Chemother. 1991;35:1147–1152. doi: 10.1128/aac.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodel-Christian S L, Murray B E. Comparison of the gentamicin resistance transposon Tn5281 with regions encoding gentamicin resistance in Enterococcus faecalis isolates from diverse geographic locations. Antimicrob Agents Chemother. 1992;36:2259–2264. doi: 10.1128/aac.36.10.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horaud T, Delbos F, Pepper K. Does a tetracycline resistance determinant of class N exist? Antimicrob Agents Chemother. 1990;34:1447–1449. doi: 10.1128/aac.34.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horaud T, de Cespédès G, Trieu-Cuot P. Chromosomal gentamicin resistance transposon Tn3706 in Streptococcus agalactiae B128. Antimicrob Agents Chemother. 1996;40:1085–1090. doi: 10.1128/aac.40.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horodniceanu T, Bougueleret L, El Solh N, Bieth G, Delbos F. High-level plasmid-borne resistance to gentamicin in Streptococcus faecalis subsp. zymogenes. Antimicrob Agents Chemother. 1979;16:686–689. doi: 10.1128/aac.16.5.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufhold A, Potgieter E. Chromosomally mediated high-level gentamicin resistance in Streptococcus mitis. Antimicrob Agents Chemother. 1993;37:2740–2742. doi: 10.1128/aac.37.12.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bouguénec C, de Cespédès G, Horaud T. Molecular analysis of a composite chromosomal conjugative element (Tn3701) in Streptococcus pyogenes. J Bacteriol. 1988;170:3930–3936. doi: 10.1128/jb.170.9.3930-3936.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon B R, May J W, Skurray R A. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol Gen Genet. 1984;193:554–556. doi: 10.1007/BF00382099. [DOI] [PubMed] [Google Scholar]
- 16.Moellering, R. C., Jr., C. Wennersten, and A. N. Weinberg. 1971. Synergy of penicillin and gentamicin against enterococci. J. Infect. Dis. 124(Suppl.):S207–S209. [DOI] [PubMed]
- 17.Murray B E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson J E, Zervos M J. High level gentamicin resistance in Enterococcus: microbiology, genetic basis, and epidemiology. Rev Infect Dis. 1990;12:644–652. doi: 10.1093/clinids/12.4.644. [DOI] [PubMed] [Google Scholar]
- 19.Rice L B, Carrias L L, Marshall S H. Tn5384, a composite enterococcal element conferring resistance to erythromycin and gentamicin whose ends are directly repeated copies of IS256. Antimicrob Agents Chemother. 1995;39:1147–1153. doi: 10.1128/aac.39.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seetulsingh P S, Tomayko J F, Coudron P E, Markowitz S M, Skinner C, Singh K V, Murray B E. Chromosomal DNA restriction endonuclease digestion patterns of β-lactamase-producing Enterococcus faecalis isolates collected from a single hospital over a 7-year period. J Clin Microbiol. 1996;34:1892–1896. doi: 10.1128/jcm.34.8.1892-1896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91(Suppl. 3B):S72–S75. [DOI] [PubMed]
- 22.Straut M, de Cespédès G, Delbos F, Horaud T. Molecular typing of Enterococcus faecalis strains resistant to high levels of gentamicin isolated in Romania. J Antimicrob Chemother. 1997;39:483–491. doi: 10.1093/jac/39.4.483. [DOI] [PubMed] [Google Scholar]
- 23.Straut M, de Cespédès G, Horaud T. Plasmid-borne high-level resistance to gentamicin in Enterococcus hirae, Enterococcus avium, and Enterococcus raffinosus. Antimicrob Agents Chemother. 1996;40:1263–1265. doi: 10.1128/aac.40.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thal L A, Chaw J W, Clewell D B, Zervos M J. Tn924, a chromosome-borne transposon encoding high-level gentamicin resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:1152–1156. doi: 10.1128/aac.38.5.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodworf N, McNamara E, Smyth E, George R C. High-level resistance to gentamicin in Enterococcus faecium. J Antimicrob Chemother. 1992;29:395–403. doi: 10.1093/jac/29.4.395. [DOI] [PubMed] [Google Scholar]