Abstract
This study describes the first isolation and characterization of spontaneous mutants conferring natural resistance to an antibiotic for any Bartonella species. The Bartonella bacilliformis gyrB gene, which encodes the B subunit of DNA gyrase, was cloned and sequenced. The gyrB open reading frame (ORF) is 2,079 bp and encodes a deduced amino acid sequence of 692 residues, corresponding to a predicted protein of ∼77.5 kDa. Sequence alignment indicates that B. bacilliformis GyrB is most similar to the GyrB protein from Bacillus subtilis (40.1% amino acid sequence identity) and that it contains the longest N-terminal tail (52 residues) of any GyrB characterized to date. The cloned B. bacilliformis gyrB was expressed in an Escherichia coli S30 cell extract and was able to functionally complement a temperature-sensitive E. coli Cour gyrB mutant (strain N4177). We isolated and characterized spontaneous mutants of B. bacilliformis resistant to coumermycin A1, an antibiotic that targets GyrB. Sequence analysis of gyrB from 12 Cour mutants of B. bacilliformis identified single nucleotide transitions at three separate loci in the ORF. The predicted amino acid substitutions resulting from these transitions are Gly to Ser at position 124 (Gly124→Ser), Arg184→Gln, and Thr214→Ala or Thr214→Ile, which are analogous to mutated residues found in previously characterized resistant gyrB genes from Borrelia burgdorferi, E. coli, Staphylococcus aureus, and Haloferax sp. The Cour mutants are three to five times more resistant to coumermycin A1 than the wild-type parental strain.
Recent taxonomic reclassifications involving bacteria formerly constituting the Rochalimaea and Grahamella genera have rapidly expanded the number of species in the Bartonella genus (5, 8, 10, 23, 47). Of these 12 species, 5 are presently considered to be etiologic agents of emerging infectious disease in humans: Bartonella bacilliformis, B. clarridgeiae, B. elizabethae, B. henselae, and B. quintana (22, 23, 33). Hemotrophy and arthropod vector-mediated transmission are common parasitic strategies utilized by these small, gram-negative, facultatively intracellular pathogens.
Due to the lack of a system for site-specific genetic manipulation, few reports have been published concerning the molecular mechanisms involved in the pathogenesis, growth, and antibiotic resistance of Bartonella species (3, 15, 16, 24, 27, 29, 31, 34, 42, 46, 49). Therefore, we initially address this problem by molecularly characterizing the pathogens’ gyrB gene. DNA gyrase is the bacterial type II topoisomerase responsible for introducing negative supercoiling into DNA (reviewed in references 20 and 37), and it is the target of several types of antimicrobial agents. The holoenzyme is an A2B2 complex encoded by the gyrA and gyrB genes; the A subunit is responsible for DNA breakage and reunion, whereas the B subunit harbors the ATP binding site. The coumarin antibiotics coumermycin A1, novobiocin, and chlorobiocin impede DNA replication by inhibiting the ATP binding and hydrolysis catalyzed by GyrB (28). Several reports have demonstrated that single point mutations in the gyrB gene confer resistance to coumarin antibiotics (11, 13, 19, 36, 39, 44) providing a locus and selectable phenotype for allelic exchange experiments.
In this study, we describe the isolation and characterization of the first spontaneous mutants of any Bartonella species, as well as the first characterization of an antibiotic-resistant mutant. Analysis of coumermycin A1-resistant mutants revealed single nucleotide lesions corresponding to specific amino acid substitutions in the N-terminal domain of GyrB. These mutations confer an approximately three- to fivefold increase in the MIC of coumermycin A1 relative to the wild type. In addition, we show that the B. bacilliformis gyrB can functionally complement an E. coli gyrB mutant. Finally, we discuss the positions of the amino acid substitutions in B. bacilliformis GyrB as they relate to recently solved high-resolution crystal structures and enzyme function (26, 48).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strains were grown overnight at 37°C in Luria-Bertani (LB) medium with standard antibiotic supplements when required (12). B. bacilliformis was grown and harvested as previously described (34).
To isolate coumermycin A1-resistant mutants, suspensions of B. bacilliformis KC583 were plated on heart infusion agar supplemented with 5% erythrocytes and coumermycin A1 (0.1 μg/ml; Sigma Chemical Co., St. Louis, Mo.). Coumermycin A1-resistant mutants were usually observed after 5 days of growth and were harvested after 7 days. Resistant colonies were picked and resuspended in 150 μl of heart infusion broth. Resistant mutants were maintained in the presence of 0.04 μg of coumermycin A1 per ml. Strains of B. bacilliformis and Escherichia coli used or generated in this study are summarized in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| B. bacilliformis | ||
| KC583 | Wild-type strain | 7 |
| CR1, 2, 6, 8, 9 | KC583 GyrB(Gly124→Ser) Cour | This study |
| CR4, 7, 11, 12 | KC583 GyrB(Arg184→Gln) Cour | This study |
| CR3 | KC583 GyrB(Thr214→Ala) Cour | This study |
| CR5, 10 | KC583 GyrB(Thr214→Ile) Cour | This study |
| E. coli | ||
| HB101 | Host strain used for cloning | 6, Promega |
| TOP10F′ | TOPO TA Cloning Kit host strain | Invitrogen |
| N99 | Complementation analysis strA galK | 30 |
| N4177 | Isogenic to N99 except gyrB221 (Cour) and gyrB203 (TS) | 30 |
| Plasmids | ||
| pBK-CMV | Phagemid cloning vector | Stratagene |
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pGYRB1 | pBK-CMV recombinant containing 5′ portion of B. bacilliformis gyrB in an ∼2,000-bp Sau3AI fragment; derived from λ-ZAP library | This study |
| pGYRB2 | pBK-CMV recombinant with an ∼13-kb SacI fragment containing the B. bacilliformis gyrB; derived from λ-GEM 11 library | This study |
| pGYRB3 | pCR2.1-TOPO recombinant containing entire gyrB gene in a 2,410-bp BamHI fragment; derived from TA cloning strategy | This study |
Preparation and manipulation of DNA.
Chromosomal DNA from B. bacilliformis for use in DNA hybridization or PCR analyses was prepared with CTAB (hexadecyltrimethyl ammonium bromide) by the methods of Ausubel et al. (2). Plasmid DNA extraction and isolation from E. coli for cloning were performed by the alkaline lysis procedure of Birnboim and Doly (4), and plasmid preparations for sequencing were made with either a Midi-Prep kit (Qiagen, Chatsworth, Calif.) or a Perfect Prep kit (5 PRIME-3 PRIME, Boulder, Colo.) as per the manufacturer’s instructions. Cloning of individual DNA fragments was accomplished by two distinct methods. First, both λ-ZAP Express (Stratagene Cloning Systems, La Jolla, Calif.) and λ-GEM 11 (Promega, Madison, Wis.) genomic cloning systems were used as per the manufacturer’s recommendations to obtain phagemid clones containing the B. bacilliformis gyrB gene for sequence analysis. Second, the TOPO TA Cloning Kit (Invitrogen, Carlsbad, Calif.) was used as per the manufacturer’s instructions to obtain a plasmid clone containing the entire wild-type gyrB open reading frame (ORF) for gene expression and functional complementation analyses. When required, DNA was purified from ethidium bromide-stained agarose gels or PCRs with either a GeneClean kit (Bio 101, Inc., La Jolla, Calif.) or by a QIAquick kit (Qiagen). Plasmids and recombinants used or constructed in this study are summarized in Table 1.
PCR and oligonucleotides.
PCR amplifications were achieved by using a GeneAmp 2400 Thermocycler (Perkin-Elmer, Norwalk, Conn.) following procedures developed by Mullis et al. (35). Reaction mixtures contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, a 200 μM concentration of each deoxynucleotide triphosphate, 4 mM MgCl2, 2.5 U of AmpliTaq DNA polymerase (Roche Molecular Systems, Branchburg, N.J.), 1 to 100 ng of template DNA, and 0.1 μg of each primer. The reaction proceeded for 30 cycles of 1 min at 94°C, 1 min at 50 to 60°C (depending on calculated primer melting temperature), and 1 min at 72°C, with an initial 5-min denaturation at 94°C and a final 7-min extension at 72°C. Single-stranded degenerate oligonucleotide primers (based on regions of conserved homology [21]), GYRB5 (5′-AARMGNCCNGGNATGTAYATHGG-3′) and GYRB3 (5′-CCNACNCCRTGNARNCCNCC-3′), were synthesized by Gibco-BRL. Single-stranded oligonucleotide primers specific for the B. bacilliformis gyrB gene included the following: GYRB-F, nucleotide (nt) −219 to −187 (5′-CGCGGATCCCTGCGGAATAACAAATCATGGTG-3′); GYRB-R, nt 132 to 100 (5′-CGCGGATCCTATCGATAAAACGATCCATCTGGC-3′); LESION-F, nt 307 to 331 (5′-GCTGATTTGATTGATATAACATTGG-3′), and LESION-R, nt 711 to 688 (5′-TATAAATTTTTTCTGGGTCAAAAGC-3′).
DNA hybridization analysis.
Total DNAs from B. bacilliformis KC583 and KC584 and E. coli HB101 were isolated, digested to completion with BamHI, and then separated on an ethidium bromide-stained 1% (wt/vol) agarose gel. The gel was then blotted onto a nitrocellulose membrane (0.45-μm pore size; Schleicher & Schuell, Keene, N.H.) by the method of Southern (43) and baked for 1 h at 80°C. The 2,410-bp fragment used as the probe in this analysis was derived by PCR amplification using the amplimer set GYRB-F–GYRB-R and B. bacilliformis KC583 as template DNA. This 2,410-bp PCR fragment was subsequently labeled by random primer extension (14) with the Klenow fragment of E. coli polymerase I (Gibco-BRL) and [α-32P]dCTP (New England Nuclear, Boston, Mass.). The blot was probed overnight at 50°C with the 32P-labeled 2,410-bp PCR fragment and then was washed and visualized as previously described (32).
DNA hybridization was also used for probing two separate λ genomic libraries to clone and sequence the gyrB gene. In these experiments, either the 300-bp PCR product derived from the degenerate amplimer set GYRB5-GYRB3 or the internal 1,101-bp HindIII fragment was labeled by random primer extension and used to screen the libraries.
In vitro transcription-translation.
Expression of gyrB was done using an E. coli S30 cell in vitro transcription-translation system per the manufacturer’s instructions (Promega). 35S-labelled proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (25) and autoradiography as previously described (34).
In vivo complementation analysis.
A plasmid containing the cloned gyrB (pGYRB3) and the respective cloning vector (pCR2.1-TOPO) were separately introduced into strains N99 and N4177 by modifying the transformation procedure of Chung et al. (9) such that the culture temperature of N99 and N4177 was held below 30°C throughout the transformation procedure. Transformed clones of N99 or N4177 containing either pGYRB3 or pCR2.1-TOPO plasmids were selected by incubation at 30°C for 16 h in the presence of ampicillin (100 μg/ml). Immediately thereafter, clones of each of the four transformants (N99[pCR2.1-TOPO], N99[pGYRB3], N4177[pCR2.1-TOPO], and N4177[pGYRB3]) were simultaneously replica plated onto LB (supplemented with ampicillin [100 μg/ml]), and incubated at either 30°C (permissive temperature) or 42°C (restrictive temperature) for 20 h. Both E. coli host strains (N99 and N4177) were replica plated onto LB and LB-ampicillin (100 μg/ml) simultaneously for additional positive and negative controls, respectively. Replica-plated clones were scored after 20 h of growth by estimating relative colony size.
Antibiotic susceptibility testing.
MICs were determined by two methods. Initial determination of the MIC of coumermycin A1 for the wild type was achieved by plating 100 μl of Bartonella suspensions containing 105 CFU/ml on heart infusion agar supplemented with 5% erythrocytes and coumermycin A1 concentrations ranging from 0.01 to 1.0 μg/ml. Second, determination of the MICs for Cour mutants was accomplished by an agar dilution technique previously described for Bartonella (27). Briefly, resistant strains were harvested after 5 days of incubation, washed, and resuspended in phosphate-buffered saline (pH 7.5). The suspensions were then equilibrated to a McFarland 0.5 standard at an optical density at 600 nm. Aliquots (10 μl) were applied to heart infusion agar supplemented with 5% erythrocytes and coumermycin A1 concentrations of 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1.0 μg/ml. The MIC is defined as the concentration of coumermycin A1 at which no growth is detected following 7 days of routine incubation. MICs were obtained by three independent determinations.
Nucleotide sequencing and computer analysis.
The inserts of overlapping clones derived from λ-ZAP Express (pGYRB1) and λ-GEM 11 (pGYRB2) genomic libraries were sequenced separately to obtain the nucleotide sequence for the entire gyrB gene. Templates were primed with M13 universal primers or with synthetic oligonucleotides prepared with a DNA synthesizer (model 394; Applied Biosystems, Foster City, Calif.). The nucleotide sequences for both DNA strands of the gyrB gene were then determined by the dideoxy chain-termination method of Sanger et al. (41) using a Taq DyeDeoxy Terminator Cycle Sequencing Kit as per the manufacturer’s instructions (Applied Biosystems). Sequencing was done on an Applied Biosystems Automated DNA Sequencer (model 373A). Sequence data were compiled and analyzed by using PC/GENE 6.8 software (Intelligenetics, Mountain View, Calif.) for restriction site determination and ORF identification, BLAST (1) for database searches, CLUSTAL W 1.6 (45) for multiple sequence alignments, and BOXSHADE 3.21 (18) for sequence alignment formatting.
Nucleotide sequence accession number.
The GenBank accession number for the Bartonella bacilliformis gyrB nucleotide sequence is U82225.
RESULTS
Cloning the gyrB gene.
Two clones were required for sequence analysis of this gene. A positive plaque with the cloned B. bacilliformis gyrB gene was isolated from a λ-ZAP Express library (Stratagene) by probing with a [α-32P]dCTP-labeled 300-bp PCR product generated from B. bacilliformis KC583 template DNA by using the degenerate oligonucleotide primers GYRB5 and GYRB3. A pBK-CMV phagemid clone was excised from the λ-ZAP Express clone and termed pGYRB1. Nucleotide sequence analysis revealed that only the 5′ portion (1094 bp) of the gyrB gene was present in the ∼2,000-bp Sau3AI insert of pGYRB1.
To obtain the remainder of the sequence for gyrB, the 1,101-bp HindIII fragment of pGYRB1 containing the 5′ portion of the gyrB gene was labeled by random primer extension and used to probe a λ-GEM 11 genomic library (Promega) in hopes of obtaining a λ clone with a larger insert containing the entire gyrB gene. A second λ clone was identified and found to contain the entire gyrB gene in an ∼13-kbp SacI fragment by DNA hybridization. The SacI fragment was excised and cloned into pBK-CMV to generate pGYRB2. The insert in pGYRB2 was used to complete the nucleotide sequencing of the wild-type B. bacilliformis gyrB gene.
The complete gyrB gene (2,410 bp) was amplified from B. bacilliformis KC583 DNA by using the amplimer set GYRB-F–GYRB-R and cloned into pCR2.1-TOPO. This gyrB recombinant was designated pGYRB3.
Nucleotide sequence of the gyrB gene.
The nucleotide sequence of the wild-type (coumermycin A1-sensitive) B. bacilliformis gyrB gene was determined from both DNA strands and is presented in Fig. 1. Computer-assisted analysis of the gyrB gene showed a 2,079-bp ORF. This ORF is characterized by a common initiation codon, ATG, that is preceded by putative −35 (TTCAAA) and −10 (GATAAT) consensus regulatory elements and a potential ribosomal binding site (AGTA) (Fig. 1).
FIG. 1.
Nucleotide and predicted amino acid sequence of B. bacilliformis gyrB. The nucleotide sequence of a 2,250-bp fragment containing the wild-type coumermycin A1-sensitive B. bacilliformis gyrB is shown. Nucleotides within the 2,079-bp ORF are given in uppercase letters, and the deduced 692-residue amino acid sequence is shown below each corresponding codon. Putative consensus regulatory elements are indicated (−35, −10, ribosomal binding site [RBS]). The stop codon is marked with an asterisk. The three codons (and their corresponding amino acids) in which single nucleotide substitutions resulting in coumermycin A1 resistance were found are boxed. The unusually long 52-residue N terminus is shown in boldface type. The predicted molecular mass of the mature protein is 77.5 kDa. The GenBank accession number for the gyrB gene is U82225.
Further analysis of the ORF indicated that the encoded protein had a deduced length of 692 amino acid residues and a predicted molecular mass of approximately 77.5 kDa. BLAST (1) homology searches indicate that B. bacilliformis GyrB is most similar to Bacillus subtilis GyrB, with an amino acid sequence identity of 40.1%, whereas B. bacilliformis GyrB has only 18.4% identity with B. subtilis ParC, a GyrB homolog. The B. bacilliformis subunit has 34.1% identity with E. coli GyrB. Alignment of the deduced amino acid sequence from B. bacilliformis gyrB with the known amino acid sequences of GyrBs from E. coli, B. subtilis, and Mycobacterium tuberculosis (using CLUSTAL W 1.6) indicates multiple areas of strong homology (Fig. 2) and reveals that the B. bacilliformis GyrB has an unusually long N terminus. Sequence analysis of ∼600 bp of flanking sequence indicate a possible gene upstream of gyrB with homology to lipoate-protein ligase B, whereas 3′ flanking sequence produces no areas of strong homology to database sequences (data not shown).
FIG. 2.
Multiple alignment of B. bacilliformis GyrB with B. subtilis, E. coli, and M. tuberculosis GyrB. Multiple alignment of B. bacilliformis GyrB (Barba) with B. subtilis GyrB (Bacsu), E. coli GyrB (Ecoli), and M. tuberculosis (Myctu) generated with CLUSTAL W 1.6 (45) and formatted with BOXSHADE 3.21 (18). Identical amino acid residues are shown as white on black, conserved residues are shown as black on grey, and introduced gaps are shown as dots. Note the unusual 53-residue N-terminal extension that is similar in length to the N terminus of M. tuberculosis GyrB. The first universally conserved residue (E. coli of Tyr5) is indicated by the arrowhead. GenBank accession numbers for these GyrB sequences are U82225 (B. bacilliformis) D26185 (B. subtilis), AE000447 (E. coli), and X78888 (M. tuberculosis).
DNA hybridization analysis.
In order to verify that the gyrB-containing fragment was of Bartonella origin, DNA hybridization analysis was done with BamHI-digested DNA from B. bacilliformis strains KC583 and KC584 and from E. coli HB101. As shown in Fig. 3B, Southern blots probed at high stringency (7% mismatch) with a 32P-labeled 2,410-bp PCR fragment derived from B. bacilliformis KC583 template (by using amplimers GYRB-F and GYRB-R) clearly demonstrated single hybridization bands from both strains of B. bacilliformis (Fig. 3B, lanes 3 and 4). No signal was observed in BamHI-digested E. coli HB101 DNA (Fig. 3, lane 2). In addition, the G+C content of the ORF (38.4 mol%) is in good agreement with the overall G+C content (39 mol%) of B. bacilliformis (7).
FIG. 3.
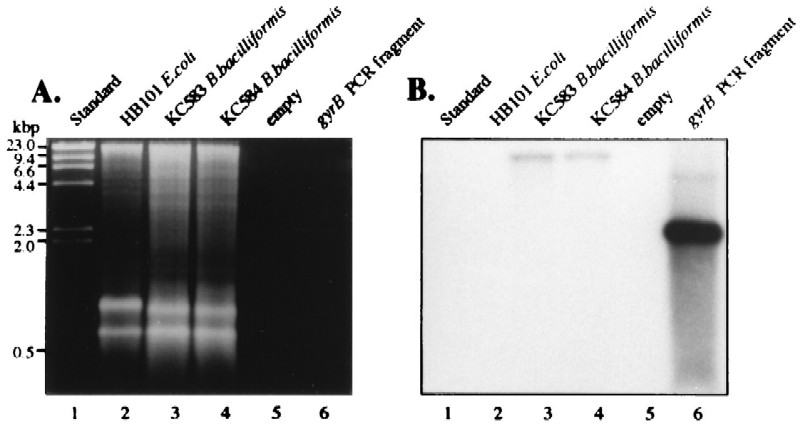
Detection of the gyrB gene in the B. bacilliformis chromosome by DNA hybridization. (A) Ethidium bromide-stained agarose gel (1%, wt/vol) containing λ HindIII size standards (lane 1), BamHI-digested chromosomal DNA of E. coli HB101 (lane 2), BamHI-digested chromosomal DNA of B. bacilliformis KC583 (lane 3), BamHI-digested chromosomal DNA of B. bacilliformis KC584 (lane 4), no DNA (lane 5), and 2,410-bp PCR fragment containing the entire B. bacilliformis gyrB ORF. (B) The corresponding autoradiograph following DNA hybridization with the described 2,410-bp PCR fragment labeled with [32P]dCTP. The lanes are the same as those panel A. Note the hybridization signal in both B. bacilliformis strains (lanes 3 and 4).
In vitro expression of gyrB.
To determine if E. coli transcription-translation machinery would express the cloned B. bacilliformis gyrB, an E. coli S30 cell DNA expression kit (Promega) was used to produce polypeptides in vitro. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of proteins expressed from pGYRB3 revealed a protein product consistent with the predicted molecular mass for GyrB of 77.5 kDa that was not expressed from the pCR2.1-TOPO control by this system (data not shown). The 77.5-kDa protein was the largest protein encoded, although additional insert-specific protein bands of approximately 68, 65, 52, and 38 kDa were observed and may have been produced by the E. coli S30 extract from anomalous ORFs on the noncoding strand of pGYRB3 or may be degradation products.
Functional complementation analysis.
Since the S30 extract expressed the cloned gyrB, an isogenic pair of E. coli strains first described by Menzel and Gellert (30) was used to evaluate the in vivo function of the cloned B. bacilliformis gyrB. E. coli N99 carries a wild-type gyrB, and strain N4177 has two gyrB mutations, which together confer a coumermycin A1-resistant (Cour) and temperature-sensitive (TS) phenotype. Growth of strain N4177 is permissive at 30°C but is restricted at 42°C unless a functional gyrB is supplied in trans to complement the Cour TS mutation. Therefore, we wanted to determine whether B. bacilliformis gyrB could functionally complement strain N4177. To address this question, the B. bacilliformis gyrB recombinant pGYRB3 was introduced into strains N99 and N4177, selected at 30°C, and subsequently replica plated and separately incubated at both permissive (30°C) and restrictive (42°C) temperatures. The B. bacilliformis gyrB recombinant, pGYRB3, was shown to increase the growth rate of strain N4177 at 42°C by approximately threefold relative to negative controls (Table 2). The presence of plasmids or varied incubation temperature did not affect the relative growth rates of host strain N99. The pattern of growth for this analysis was consistent and reproducible and shows that B. bacilliformis gyrB can functionally complement the Cour TS mutation of E. coli N4177.
TABLE 2.
Complementation with B. bacilliformis gyrB
| E. coli strain | Growth at 42°C with the following plasmida:
|
||
|---|---|---|---|
| None | pCR2.1-TOPO | pGYRB3 | |
| N99 | +++ | +++ | +++ |
| N4177 | +/− | +/− | +++ |
Growth is based upon colony size and logarithmic growth kinetics as measured by optical density at 600 nm. The temperature chosen (42°C) is the restrictive temperature for N4177 (30). Symbols: +++, robust growth; +/−, slight growth.
Isolation of coumermycin A1-resistant mutants.
Spontaneous coumermycin A1-resistant mutants were observed 7 days after inoculation and occurred at a frequency of ∼6 × 10−9 when selected in the presence of 0.1 μg of coumermycin A1 per ml. After initial selection, mutant strains were cultured on heart infusion agar supplemented with 0.04 μg of coumermycin A1 per ml. A total of 12 B. bacilliformis KC583 coumermycin-resistant mutants were selected in this manner and designated CR1 through CR12 (Table 1). In the absence of coumermycin A1, the growth rate and gross morphology of the Cour colonies were indistinguishable from those of wild-type strains.
Coumermycin A1 resistance is correlated with mutations in the gyrB gene.
Genomic DNA was isolated from wild-type B. bacilliformis KC583 and the 12 coumermycin A1-resistant mutants. The region of the gyrB gene encoding the N-terminal domain was amplified by PCR with LESION-F and LESION-R primers and subsequently sequenced with the LESION-F primer. Further analysis of these sequences revealed single nucleotide transitions at three separate loci that resulted in four distinct amino acid substitutions. First, in 5 of the 12 coumermycin A1-resistant strains (CR1, CR2, CR6, CR8, and CR9), identical G-to-A transitions at base 370 of the 2,079-bp ORF resulted in a deduced Gly124-to-Ser (Gly124→Ser) substitution. Second, 4 of the 12 resistant strains (CR4, CR7, CR11, and CR12) carried a G-to-A transition at base 550 that resulted in a deduced Arg184→Gln substitution. The third loci at which lesions were detected occurred in the Thr214 codon, in which two different transitions were observed with two distinct deduced substitutions; the ACA-to-GCA transition resulted in a Thr214→Ala substitution (CR3), whereas the ACA-to-ATA transition resulted in a Thr214→Ile substitution (CR5, CR10). These data demonstrate that spontaneous coumermycin A1-resistant mutants are correlated with specific and localized lesions in the gyrB gene. Table 3 summarizes several genotypic and phenotypic attributes of the coumermycin A1-resistant strains.
TABLE 3.
Genotypic and phenotypic analysis of B. bacilliformis gyrB mutants
| Residue | Substitution | Frequencya | MIC (μg/ml) of coumermycin A1b | Homologous GyrB lesions (reference[s]) |
|---|---|---|---|---|
| Gly124 | Ser | 41.6 | 0.2 | B. burgdorferi Gly74→Ser (40) |
| S. aureus Gly85→Ser (44) | ||||
| Arg184 | Glu | 33.3 | 0.2 | E. coli Arg136→Leu, Cys, His, Ser (11, 13) |
| B. burgdorferi Arg133→Gly, Ile (39) | ||||
| S. aureus Arg144→Ile (44) | ||||
| Haloferax sp. Arg137→His (19) | ||||
| Thr214 | Ala | 8.3 | 0.2 | B. burgdorferi Thr162→Ile (40) |
| S. aureus Thr173→Asn (44) | ||||
| Thr214 | Iso | 16.6 | 0.3 |
The frequency is expressed as a percentage of the 12 total isolates.
The MIC for wild-type KC 583 was 0.06 μg/ml. The MIC was determined for CR3, CR4, CR5, and CR9, which are representative of each of the mutant types.
In vitro coumermycin A1 susceptibilities.
We assessed the antibiotic susceptibility of wild-type B. bacilliformis KC583 to coumermycin A1 by using agar dilution techniques. At coumermycin A1 concentrations above 0.03 μg/ml, growth rates were noticeably decreased, and at those above 0.06 μg/ml, growth appeared to be completely inhibited. Thus, the MIC for KC583 was determined to be 0.06 μg/ml. One representative of each of the four different gyrB mutant types was assayed for coumermycin A1 susceptibility. MICs for mutant strains CR3, CR4, and CR9 were 0.2 μg/ml, whereas CR5 demonstrated a slightly higher level of resistance, with a MIC of 0.3 μg/ml (Table 3).
DISCUSSION
We have described the first isolation and molecular characterization of spontaneous mutant strains conferring natural resistance to an antibiotic for any Bartonella species. Generation of the mutant strains was accomplished by exposure to inhibitory (0.1-μg/ml) levels of the DNA gyrase inhibitor coumermycin A1 and occurred at a frequency of ∼6 × 10−9. Based upon amino acid sequence alignments, B. bacilliformis GyrB belongs to the shorter, 650-amino-acid size class represented by homologs of enzymes from B. subtilis, Mycoplasma pneumoniae, Staphylococcus aureus, Borrelia burgdorferi, and Haloferax sp. (20). In the larger, 800-amino-acid size class, represented by E. coli, an extra 150-amino-acid block is found in the C-terminal domain of the protein (20) (Fig. 2). The commonly recognized ATP binding motif GXXGXG is found at positions 162 to 167 of B. bacilliformis GyrB, corresponding to positions 114 to 119 of E. coli GyrB.
The structure of the B. bacilliformis GyrB is unusual in two ways. First, in GyrBs sequenced to date, the first N-terminal amino acid that demonstrates universal conservation throughout bacteria is a Tyr residue represented by E. coli Tyr5, corresponding to Tyr53 of B. bacilliformis (Fig. 2). The side chain of Tyr5 hydrogen bonds to the bound ATP analog (48). The number of amino acids preceding this conserved Tyr is less than 13 for nearly all bacteria examined to date. B. bacilliformis GyrB is unusual in this respect in that 52 amino acid residues precede the E. coli Tyr5 homolog, making it the longest N-terminal extension reported to date. Only M. tuberculosis has an N-terminal extension of this magnitude, with 50 amino acids (Fig. 2); however, the two extensions are not homologous. The crystal structure of the E. coli GyrB N-terminal domain complexed with a nonhydrolyzable ATP analog shows that the N-terminal 13 residues form a protrusion that interacts with the other GyrB protomer (48). This interaction stabilizes the dimer interface and forms part of the ATP binding site (48). However, the N terminus is apparently not ordered in the cocrystal structure with the coumarin inhibitor novobiocin (26). The function of the unusually long N-terminal extensions of M. tuberculosis and B. bacilliformis GyrBs is intriguing and remains to be determined. A second primary structural feature of B. bacilliformis GyrB that we have noted is Glu128 (E. coli equivalent, Gly81). In all wild-type GyrBs reported thus far, this residue is either glycine or aspartate, with the exception of those found in the Mycobacteria, which have alanine or glutamate at this position. In this respect, B. bacilliformis GyrB is also more similar to the mycobacterial GyrB. This position is one of three loci that is mutated in a novobiocin-resistant Haloferax (Asp82→Gly) (19), although it is distant from the coumarin binding site (26). Although both B. bacilliformis and M. tuberculosis are slow-growing bacteria and have several similar GyrB structural features, the effect of these properties on interactions with ATP or coumarins is unknown.
The mechanism of coumermycin A1 resistance in B. bacilliformis mutants was identified by sequencing PCR fragments generated with primers amplifying the portion of the gyrB gene that encodes the N-terminal domain. We have isolated 12 coumermycin A1-resistant mutants and have identified single nucleotide transitions at three separate loci resulting in single amino acid substitutions in the N-terminal domain of the GyrB protein. Lesions detected in the resistant B. bacilliformis gyrB genes are analogous in location and residue substitution to previously characterized resistant gyrB genes (11, 13, 19, 39, 40, 44). The crystal structure has revealed important interactions for each of the lesion sites. First, the side group of the E. coli Arg136 residue (B. bacilliformis Arg184) makes critical hydrogen bonds with the coumarins and with E. coli Tyr5 (B. bacilliformis Tyr53) on the other protomer (which is involved in ATP binding) (26). The second and third residues associated with coumarin resistance, E. coli Gly77 (B. bacilliformis Gly124) and E. coli Thr165 (B. bacilliformis Thr214), specifically interact with each other as well as stabilize interactions with ATP and coumarins (26).
These data demonstrate that the B. bacilliformis DNA gyrase B protein is a target for coumarin antibiotics. Wild-type B. bacilliformis (MIC, 0.06 μg/ml) was shown to be more susceptible to growth inhibition by coumermycin A1 than almost all other bacteria tested (50) and is 250 times more susceptible than E. coli (17). These data are consistent with the finding that Bartonella is extremely susceptible to a variety of antibacterial agents in vitro (27). The mutant strains demonstrated an approximately fivefold increase in resistance levels. The MICs for GyrB mutants represented by strains CR3, CR4, and CR9 were determined to be 0.2 μg/ml, whereas the MIC for CR5 was 0.3 μg/ml. This suggests that a Thr214→Ile substitution confers a higher level of resistance than Thr214→Ala, Gly124→Ser, or Arg184→Gln, consistent with findings in B. burgdorferi (40).
The transition between the diverse thermal environments of the arthropod vector and the human host, as well as the presentation of the verruga peruana on the extremities (<37°C), suggests that there is a close relationship between temperature and gene expression in B. bacilliformis. Yersinia enterocolitica DNA gyrase mutants simulate thermoinduced alterations of DNA supercoiling with coincident phenotypic changes (38). Likewise, DNA topology regulated by DNA gyrase may play an important role in the survival or virulence of B. bacilliformis in both the vector and host, and DNA gyrase mutants may provide a method for analysis of thermoregulation.
ACKNOWLEDGMENTS
We thank Joan Strange (The University of Montana Murdock Molecular Biology Facility) for her excellent technical assistance with nucleic acid sequencing and Marty Gellert and Mary O’Dea for their contribution of strains N99 and N4177. D.S.S. thanks Tony Maxwell for useful discussions.
This work was supported by Public Health Service grants AI34050 and RR10169 (to M.F.M.) and AI39695 (to D.S.S.) from the National Institutes of Health (NIAID) and National Science Foundation grant MCB-9722408 (to D.S.S.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Benson L A, Kar S, McLaughlin G, Ihler G M. Entry of Bartonella bacilliformis into erythrocytes. Infect Immun. 1986;54:347–353. doi: 10.1128/iai.54.2.347-353.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Brenner D J, O’Connor S P, Hollis D G, Weaver R E, Steigerwalt A G. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J Clin Microbiol. 1991;29:1299–1302. doi: 10.1128/jcm.29.7.1299-1302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner D J, O’Connor S P, Winkler H H, Steigerwalt A G. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb. nov., Bartonella henselae comb. nov., and Bartonella elizabethae comb. nov., and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol. 1993;43:777–786. doi: 10.1099/00207713-43-4-777. [DOI] [PubMed] [Google Scholar]
- 9.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarridge J E, Raich T J, Pirwani D, Simon B, Tsai L, Rodriguez-Barradas M C, Regnery R, Zollo A, Jones D C, Rambo C. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contreras A, Maxwell A. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol Microbiol. 1992;6:1617–1624. doi: 10.1111/j.1365-2958.1992.tb00886.x. [DOI] [PubMed] [Google Scholar]
- 12.Davis R W, Bostein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.del Castillo I, Vizan J L, Rodriguez-Sainz M C, Moreno F. An unusual mechanism for resistance to the antibiotic coumermycin A1. Proc Natl Acad Sci USA. 1991;88:8860–8864. doi: 10.1073/pnas.88.19.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Garcia F U, Wojta J, Broadly K N, Davidson J M, Hoover R L. Bartonella bacilliformis stimulates endothelial cells in vitro and is angiogenic in vivo. Am J Pathol. 1990;136:1125–1135. [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia F U, Wojta J, Hoover R L. Interactions between live Bartonella bacilliformis and endothelial cells. J Infect Dis. 1992;165:1138–1141. doi: 10.1093/infdis/165.6.1138. [DOI] [PubMed] [Google Scholar]
- 17.Gellert M, O’Dea M H, Itoh T, Tomizawa J-I. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann K, Baron M D. BOXSHADE 3.21. 1996. http://www.isrec.isb-sib.ch/software/BOX_form.html http://www.isrec.isb-sib.ch/software/BOX_form.html. . [Google Scholar]
- 19.Holmes M L, Dyall-Smith M L. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J Bacteriol. 1991;173:642–648. doi: 10.1128/jb.173.2.642-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W M. Type II DNA topoisomerase genes. In: Liu L F, editor. DNA topoisomerases, biochemistry and molecular biology. San Diego, Calif: Academic Press; 1994. pp. 201–222. [DOI] [PubMed] [Google Scholar]
- 21.Huang W M. Multiple DNA gyrase-like genes in eubacteria. In: Andoh T, Ideda H, Oguro M, editors. Molecular biology of DNA topoisomerases and its application to chemotherapy: proceedings of the International Symposium on DNA Topoisomerases in Chemotherapy, Nagoya, Japan. CRC Press, Boca Raton, Fla. 1992. pp. 39–48. [Google Scholar]
- 22.Koehler J E. Bartonella infections. Adv Pediatr Infect Dis. 1996;11:1–27. [PubMed] [Google Scholar]
- 23.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch fever) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreier J P, Ristic M. The biology of hemotrophic bacteria. Annu Rev Microbiol. 1981;35:325–338. doi: 10.1146/annurev.mi.35.100181.001545. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lewis R J, Singh O M P, Smith C V, Skarzynski T, Maxwell A, Wonacott A J, Wigley D B. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15:1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 27.Maurin M, Gasquet S, Ducco C, Raoult D. MICs of 28 antibiotic compounds for 14 Bartonella (formerly Rochalimea) isolates. Antimicrob Agents Chemother. 1995;39:2387–2391. doi: 10.1128/aac.39.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxwell A. The interaction between coumarin drugs and DNA gyrase. Mol Microbiol. 1993;9:681–686. doi: 10.1111/j.1365-2958.1993.tb01728.x. [DOI] [PubMed] [Google Scholar]
- 29.McGinnis Hill E, Raji A, Valenzuela M S, Garcia F, Hoover R. Adhesion to and invasion of cultured human cells by Bartonella bacilliformis. Infect Immun. 1992;60:4051–4058. doi: 10.1128/iai.60.10.4051-4058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 31.Mernaugh G, Ihler G M. Deformation factor: an extracellular protein synthesized by Bartonella bacilliformis that deforms erythrocyte membranes. Infect Immun. 1992;60:937–943. doi: 10.1128/iai.60.3.937-943.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minnick M F, Heinzen R A, Frazier M E, Mallavia L P. Characterization of the cbbE′ gene of Coxiella burnetii. J Gen Microbiol. 1990;136:1099–1107. doi: 10.1099/00221287-136-6-1099. [DOI] [PubMed] [Google Scholar]
- 33.Minnick, M. F. Bartonella species. In M. Sussman (ed.), Molecular medical microbiology, in press. Academic Press, London, United Kingdom.
- 34.Mitchell S J, Minnick M F. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect Immun. 1995;63:1552–1562. doi: 10.1128/iai.63.4.1552-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 36.Muñoz R, Bustamante M, de la Campa A G. Ser-127-to-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J Bacteriol. 1995;177:4166–4170. doi: 10.1128/jb.177.14.4166-4170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reece R J, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 38.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 39.Samuels D S, Marconi R T, Huang W M, Garon C F. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J Bacteriol. 1994;176:3072–3075. doi: 10.1128/jb.176.10.3072-3075.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuels, D. S., J. Alverson, S. W. Knight, C. H. Eggers, C. F. Garon, W. M. Huang, and B. J. Kimmel. Unpublished data.
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherer D C, DeBuron-Connors I, Minnick M F. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect Immun. 1993;61:4962–4971. doi: 10.1128/iai.61.12.4962-4971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 44.Stieger M, Angehrn P, Wohlgensinger B, Gmünder H. GyrB mutations in Staphylococcus aureus strains resistant to cyclothialidine, coumermycin, and novobiocin. Antimicrob Agents Chemother. 1996;40:1060–1062. doi: 10.1128/aac.40.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker T S, Winkler H H. Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect Immun. 1981;31:480–486. doi: 10.1128/iai.31.1.480-486.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss E, Dasch G A. Differential characteristics of strains of Rochalima: Rochalima vinsonii sp. nov., the Canadian vole agent. Int J Syst Bacteriol. 1982;32:305–314. [Google Scholar]
- 48.Wigley D B, Davies G J, Dodson E J, Maxwell A, Dodson G. Crystal structure of the N-terminal domain of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y-H, Lu Z-Y, Ihler G M. Purification of deformin, an extracellular protein synthesized by Bartonella bacilliformis which causes deformation of erythrocyte membranes. Biochim Biophys Acta. 1995;1234:173–183. doi: 10.1016/0005-2736(94)00271-p. [DOI] [PubMed] [Google Scholar]
- 50.Zimmer C, Storl K, Storl J. Microbial DNA topoisomerases and their inhibition by antibiotics. J Basic Microbiol. 1990;30:209–224. doi: 10.1002/jobm.3620300312. [DOI] [PubMed] [Google Scholar]




