Abstract
Resistance to fluconazole is becoming an increasing problem in the management of oropharyngeal candidiasis in human immunodeficiency virus-infected patients. Strains obtained from five patients developed decreased fluconazole susceptibility over time. DNA strain typing confirmed the high degree of relatedness among isolates from one patient and the variability among isolates from different patients. Expression of genes involved in development of fluconazole resistance was monitored in each isolate using probes specific for ERG11 (lanosterol 14α-demethylase), MDR1 (a major facilitator), and CDR (ATP-binding cassette or ABC transporter) genes. Increased expression of CDR genes was detected in the series of isolates from two patients. Isolates from one of the two patients also demonstrated increased ERG11 expression, whereas isolates from the other patient did not. Increased levels of MDR1 mRNA correlated with increased resistance in sequential isolates from another patient. Initial overexpression of MDR1 with subsequent overexpression of CDR genes and a final isolate again overexpressing MDR1 were detected in serial isolates from another patient. In another patient, overexpression of these genes was not detected despite an eightfold increase in fluconazole MIC. In this patient, sequence data of the ERG11 gene revealed no point mutations associated with decreased susceptibility. Five different patterns of gene expression were observed in isolates recovered from five patients who developed resistance. Therefore, these experiments demonstrate that a variety of mechanisms or combinations of mechanisms are associated with the development of fluconazole drug resistance. Additional studies are needed to estimate the frequency and clinical impact of these mechanisms of resistance.
Resistance to fluconazole and other azole antifungal drugs has become an important clinical problem in the management of human immunodeficiency virus (HIV)-infected patients with recurrent oropharyngeal candidiasis (OPC) (17, 20). Recent advances in our understanding of the molecular mechanisms leading to azole resistance in Candida albicans, the main etiologic agent of OPC, suggest the multifactorial nature of resistance. Alterations in the target enzyme (lanosterol 14α-demethylase), including point mutations (10, 11, 22, 28, 32) and overexpression (31), lead to decreased susceptibilities to azole drugs. Increased efflux of drug, mediated by multidrug pumps belonging to two different families, the major facilitators and the ATP-binding cassette (ABC) transporters, also confers resistance to azole antifungal agents (1, 23–25, 29, 31). The genes coding for several ABC transporters in C. albicans have been identified, including several CDR genes (2, 16, 24, 25, 30). These ABC transporters, which have been associated with drug resistance in a variety of eukaryotic cells, include a membrane pore composed of transmembrane segments and two ATP-binding cassettes on the cytosolic side of the membrane, which provide the energy source for the pump (3, 7). The MDR1 gene (also called BENr) is the only gene coding for a major facilitator that has been identified in C. albicans so far (5), and its overexpression leads to fluconazole resistance exclusively among azole drugs (23, 25, 31). The major facilitators contain a transmembrane pore but use proton motive force as their energy source (12).
In the present study, we have investigated the expression of C. albicans ERG11 (encoding lanosterol 14α-demethylase, formerly designated ERG16 [8, 9]), MDR1, and CDR genes in sequential clinical isolates obtained from HIV-infected patients with OPC in order to assess the distribution and frequency of various mechanisms responsible for the development of antifungal drug resistance and their clinical impact.
MATERIALS AND METHODS
Clinical samples and isolates.
C. albicans isolates were obtained by direct swab or by oral saline rinses from five HIV-infected patients with recurrent OPC enrolled in a longitudinal study to assess significance of fluconazole resistance (Table 1). Patients were treated initially with fluconazole at 100 mg/day. Doses were increased up to 800 mg/day in an effort to achieve therapeutic response after development of clinical resistance. In all five patients, sequential isolates showed decreased susceptibility to fluconazole, and in all five patients, therapeutic response was achieved by increasing the dose of fluconazole to a range of 200 to 800 mg/day (Table 1). In this study, clinical resistance was defined as the clinical requirement for increasing fluconazole doses for response. Resistance refers to mycological, in vitro resistance, which was the detection of increased MICs. The identity of these clinical isolates as C. albicans was confirmed by standard biochemical and microbiological procedures, including carbohydrate assimilation patterns (API 20C; Analytab Products, BioMerieux, France), germ tube formation in serum-containing medium, and color of colonies in chromogenic medium (CHROMagar Candida, CHROMagar, Paris, France). Isolates were stored at room temperature as suspensions in sterile water and subcultured onto plates containing Sabouraud dextrose agar 48 h prior to propagation in YEPD medium (2% yeast extract, 1% peptone, 2% glucose).
TABLE 1.
Serial isolates of C. albicans from patients with OPCa
| Patient | Isolate | OPC episode | Day | FLU dose (mg/day) | Cumulative FLU dose (g) | Previous azole use | MIC (μg/ml)
|
DNA type | ||
|---|---|---|---|---|---|---|---|---|---|---|
| FLU | ITRA | AMB | ||||||||
| 7 | 412 | 1 | 1 | 100 | 8.5 | ITRA, KETO | 0.5 | 0.03 | 0.5 | A |
| 1907 | 3 | 210 | 200 | 11.2 | 8 | 0.5 | 0.25 | A | ||
| 2307 | 6 | 280 | 400 | 18.1 | >128 | 0.5 | 0.25 | A | ||
| 9 | 437 | 1 | 1 | 100 | 3.0 | 1 | 0.015 | 0.25 | B | |
| 1002 | 3 | 61 | 100 | 5.7 | 0.25 | 0.015 | 0.25 | B | ||
| 1442 | 4 | 119 | 100 | 7.5 | 0.125 | 0.015 | 0.25 | B | ||
| 2271 | 5 | 273 | 100 | 8.4 | 16 | 0.03 | 0.25 | B | ||
| 2823 | 8 | 411 | 800 | 44.4 | 16 | 0.06 | 0.25 | B | ||
| 14 | 580 | 1 | 1 | 100 | 31.7 | CLO | 4 | 0.03 | 0.25 | C |
| 649 | 1 | 7 | 200 | 32.6 | 16 | 0.25 | 0.25 | C | ||
| 2438 | 3 | 296 | 200 | 90.1 | 32 | 0.25 | 0.25 | C | ||
| 40 | 1490 | 1 | 1 | 100 | 4.6 | 0.5 | 0.125 | 0.25 | D | |
| 1587 | 2 | 15 | 100 | 5.5 | 8 | 0.125 | 0.25 | D | ||
| 1622 | 2 | 15 | 100 | 5.5 | 0.5 | 0.015 | 0.25 | D | ||
| 1780 | 4 | 58 | 100 | 8.2 | 16 | 0.5 | 0.25 | D | ||
| 2225 | 6 | 149 | 200 | 11.8 | 4 | 0.125 | 0.25 | D | ||
| 2512 | 8 | 179 | 800 | 20.8 | 32 | 0.03 | 0.25 | D | ||
| 43 | 1649 | 1 | 1 | 100 | 0 | CLO | 0.25 | 0.06 | 0.25 | E |
| 1831 | 2 | 44 | 100 | 0.9 | 8 | 0.03 | 0.25 | E | ||
| 2183 | 3 | 100 | 100 | 1.8 | >128 | 0.5 | 0.25 | E | ||
Drug abbreviations: FLU, fluconazole; ITRA, itraconazole; KETO, ketoconazole; CLO, clotrimazole; AMB, amphotericin B.
Strain identification.
Strain identity was established by karyotyping, restriction fragment length polymorphism (RFLP), and DNA fingerprinting using the moderately repetitive probe Ca3 (a gift from D. Soll, University of Iowa) as previously described (18, 26). Briefly, chromosomes from the different isolates were prepared in agarose plugs and separated by pulsed-field gel electrophoresis (Bio-Rad, Hercules, Calif.). RFLP patterns were generated by digestion of genomic DNA with SfiI (Boehringer-Mannheim, Indianapolis, Ind.). After documentation, the materials present in the RFLP gels were transferred to nylon membranes (Nytran; Schleicher & Schuell, Keene, N.H.) and hybridized with a Ca3 probe radioactively labeled by random priming (Random Primers DNA Labeling System; Gibco-BRL, Gaithersburg, Md.). The membranes were then washed and exposed to autoradiography film (Du Pont, Wilmington, Del.).
Drug susceptibility testing and MIC determinations.
Initial fluconazole susceptibility at the time of primary isolation was determined by an agar dilution method as described previously by our group (14, 15). Antifungal susceptibilities to fluconazole, itraconazole, and amphotericin B were determined by the National Committee for Clinical Laboratory Standards standard procedures using macrobroth techniques and reading the endpoints at 48 h (13).
Northern (RNA) blot analysis.
Total RNA from the different isolates grown to mid-logarithmic phase in YEPD medium was obtained by using the RNAeasy mini kit (Qiagen Inc., Santa Clarita, Calif.) following the manufacturer’s instructions. Equal amounts (approximately 5 μg) of RNA as determined by A260 measurements were separated by electrophoresis (21) and subsequently transferred to nylon membranes (Nytran; Schleicher & Schuell) using the Turboblotter apparatus (Schleicher & Schuell). Probes for ERG11, MDR1, and CDR genes were purified from plasmids containing inserts of the respective genes as described before (31). The resulting CDR probe is based on the whole sequence of CDR1 and has been shown to cross-hybridize with other members of this gene family (24, 25, 31). Probes specific for CDR1 and CDR2 genes were prepared as described by Sanglard and coworkers (24) by PCR amplification from plasmids containing these sequences (pDS243 and pDS246 for CDR1 and CDR2, respectively), with the following primers: primers 5′-GAG ATC TAC CCT TTA AGA TA (forward) and 5′-TCT GAA TCG GGA TTC AAT TG (reverse) for CDR1 and primers 5′-GGTATATAAACTGGACAACA (forward) and 5′-CGGAATCTGGGTCTAATTGT (reverse) for CDR2. The identity between the two resulting probes at the level of nucleotide sequence was below 50%. All probes were labeled by random priming (Random Primers DNA Labeling System; Gibco-BRL), and hybridizations were performed with Rapid-hyb buffer (Amersham Life Science Inc., Arlington Heights, Ill.) following the manufacturer’s instructions. After hybridization, blots were washed and exposed to autoradiography film (Du Pont). Nylon membranes were probed sequentially with the different probes following stripping the previously bound probe with 95°C-heated 1× SSC buffer (1× SSC is 0.15 M sodium chloride plus 15 mM sodium citrate [pH 7.0]) with 0.5% sodium dodecyl sulfate twice for 15 min. For densitometric analysis, autoradiograms were scanned with the Adobe Photoshop program (Adobe Systems Inc., Mountain View, Calif.), and signals were quantitated with Dendron (Solltech Inc., Oakdale, Iowa). Relative values were adjusted for differences in sample loading based on quantification of 18S rRNA levels. For preparation of figures, digital images were processed by using the Adobe Photoshop program.
PCR amplification and sequencing.
The ERG11 genes encoding lanosterol 14α-demethylase from several isolates were amplified by PCR. Genomic DNA from the different isolates was extracted with YeaStar Genomic DNA (Zymo Research, Orange, Calif.) and used as a template for amplification of ERG11 genes. PCR was carried out with high-fidelity Pwo DNA polymerase (Boehringer Mannheim) using the following primers: 5′-GTT GAA ACT GTC ATT GAT GG (forward) and 5′-TCA GAA CAC TGA ATC GAA AG (reverse). Amplicons were purified with Wizard PCR Preps (Promega, Madison, Wis.), and their nucleotide sequences for both strands were determined by primer elongation with an ABI automated DNA sequencer (Applied Biosystems, Foster City, Calif.).
RESULTS
Clinical response.
Patients were treated initially with fluconazole at 100 mg/day and with increased doses of up to 800 mg/day if necessary for clinical resolution after development of refractory disease. In all five patients, clinical resistance to initial doses occurred, requiring increased doses of fluconazole (Table 1). However, therapeutic responses were seen in all patients with increased fluconazole doses. Table 1 shows the sequential isolates from each patient, the elapsed period between times of isolation, the dose of fluconazole required, the cumulative dose of fluconazole, prior regimens with other azoles, and the MICs for fluconazole, itraconazole, and amphotericin B as determined by a macrodilution method by National Committee for Clinical Laboratory Standards techniques. The initial isolate of the series for each patient was fluconazole susceptible (MIC of ≤8 μg/ml) (13, 19). Cross-resistance between itraconazole and fluconazole was detected in isolates from patients 7, 14, and 43 with an 8- to 16-fold increase in itraconazole resistance in the final isolates. Only one isolate from patient 40 showed decreased susceptibility to itraconazole, whereas all other isolates had the same or increased susceptibility to itraconazole relative to that of the first isolate obtained. On the other hand, all isolates from patient 9 remained susceptible to itraconazole (MIC of ≤0.125 μg/ml) despite increasing fluconazole MICs. All isolates remained susceptible to amphotericin B.
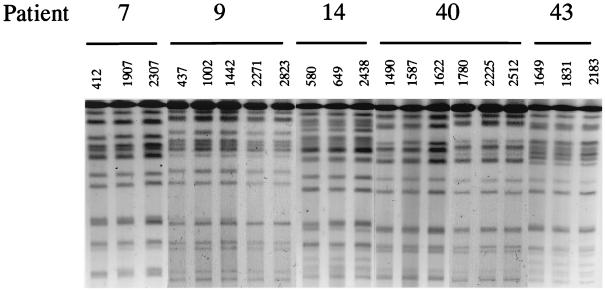
The identities of strains from a given patient were confirmed by karyotyping, RFLP, and fingerprinting with the C. albicans Ca3 probe. In each of these methods, sequential isolates from each patient were demonstrated to be highly related and distinct from isolates recovered from other patients by all typing methods employed (Table 1). RFLP patterns generated by digestion with SfiI of genomic DNA from the different isolates followed by separation by pulsed-field electrophoresis is shown in Fig. 1.
FIG. 1.
RFLP patterns generated by digestion with SfiI of genomic DNA from the different clinical C. albicans isolates recovered from successive episodes of OPC. The image is a composite of image scans from two different gels run under the same electrophoretic conditions.
Expression of ERG11, MDR1, and CDR genes in clinical isolates.
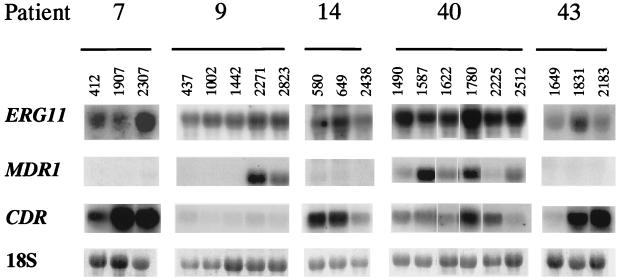
Total RNA extracted from the different isolates growing in YEPD medium in the absence of antifungal drug was analyzed by a Northern blot technique with probes specific for ERG11, MDR1, and CDR genes (Fig. 2).
FIG. 2.
Northern blots of total RNA from clinical C. albicans isolates probed with ERG11, MDR1, and CDR probes. Hybridizations were performed as described in Materials and Methods. The bottom series show the amounts of 18S rRNA used to standardize signal levels according to lane loading parameters.
In patient 7, with sequential isolates showing decreased susceptibility to fluconazole, mRNA levels for CDR genes were dramatically increased along the series of isolates. Densitometric analysis showed overexpression of CDR genes by a factor of 7 in the second isolate of the series (isolate 1907; fluconazole MIC = 8 μg/ml) and by a factor of 10 in the third highly resistant isolate (isolate 2307; fluconazole MIC > 128 μg/ml). The third isolate also demonstrated moderate overexpression of ERG11. MDR1 levels were negligible for all three isolates.
In contrast, study of five sequential isolates from patient 9, also requiring increasing fluconazole MICs, revealed the overexpression of the MDR1 gene and no increase in CDR gene expression. Levels of MDR1 mRNA were undetectable in the first three susceptible isolates of the series, but MDR1 was markedly overexpressed in the fourth and fifth less susceptible isolates. Levels of expression for ERG11 remained constant throughout the series. Expression of CDR genes was detected at very low levels for all isolates in this series, without significant changes as resistance developed.
In patient 14, although antifungal susceptibility testing revealed the decreasing susceptibility of isolates, the Northern blot technique failed to detect overexpression for any of the genes in the study. However, high constitutive levels of expression of CDR genes were noted for all isolates of the series, although decreased CDR message was detected in the third, most resistant isolate. Because the third isolate recovered from patient 14 showed decreasing susceptibility to fluconazole that did not correlate with overexpression of any of the genes studied (whereas the first and second isolates demonstrated elevated levels of CDR message), the ERG11 gene coding for lanosterol 14α-demethylase was sequenced to detect point mutations which may have resulted in decreased fluconazole susceptibility. ERG11 genes from these isolates were obtained by PCR. Single fragments of the expected length (1.6 kb) were obtained in each case. Compared to a published ERG11 sequence (9), a total of seven silent (not resulting in amino acid substitutions) nucleotide changes were found in isolate 580. ERG11 sequences from isolates 649 and 2438 were identical and showed a total of 10 base differences compared to the published sequence. For these two isolates, the only nucleotide change that had an effect in the amino acid sequence was the single amino acid substitution V437I. However, we have detected this same substitution in ERG11 genes from susceptible C. albicans isolates (data not shown). In addition, Sanglard and colleagues (22) have shown by functional expression of these genes in Saccharomyces cerevisiae that this mutation does not confer resistance to fluconazole. Therefore, the mechanism(s) of increased drug resistance for the third isolate (isolate 2438) is unknown.
Six sequential isolates from patient 40 were included in the study. Northern blot analysis revealed that levels of ERG11 mRNA remained constant for all isolates examined, including the first and third isolates that were sensitive to fluconazole (isolates 1490 and 1622; MIC = 0.5 μg/ml). Strong expression of MDR1 in the second (isolate 1587; fluconazole MIC = 8 μg/ml) and fourth (1780; MIC = 16 μg/ml) isolates with less expression in the first (1490), third (1622), and sixth (2512) isolates mirrored MIC results with the exception of the sixth isolate, which required the highest fluconazole MIC (32 μg/ml) but did not exhibit the strongest MDR1 expression. In isolate 1780 (fluconazole MIC = 16 μg/ml), increased message for MDR1 was accompanied by overexpression of CDR genes (by a factor of 2.5). A twofold increase in CDR message with no overexpression of MDR1 was detected in the fifth isolate (2225; fluconazole MIC = 4 μg/ml).
Isolates from patient 43 showed overexpression of CDR genes (by a factor of approximately 10 for both the second and third isolates in the series relative to the first isolate). In the case of the second isolate (isolate 1831; fluconazole MIC = 8 μg/ml), but not the third, more resistant isolate (2183; fluconazole MIC > 128 μg/ml) of the series, this change was accompanied by a moderate increase in ERG11 message. For all isolates from patient 43, MDR1 expression was below the detection limit.
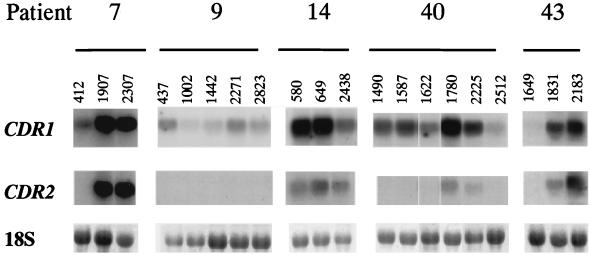
Differentiation between levels of expression of CDR1 and CDR2.
In order to discriminate between CDR1 and CDR2 gene expression, specific probes for CDR1 and CDR2 were used in Northern blots of total RNA extracted from sequential isolates from the different patients (Fig. 3). These experiments revealed that these two genes were simultaneously overexpressed in all cases in which CDR overexpression was observed. CDR1 was expressed constitutively but at low levels in azole-susceptible strains and overexpressed as azole resistance developed (isolates 1907 and 2307 from patient 7, isolates 1780 and 2225 from patient 40, and isolates 1831 and 2183 from patient 43). Levels of CDR2 mRNA were below detection limits in the same azole-susceptible isolates (with the exception of isolate 580, from patient 14, which required a fluconazole MIC of 4 μg/ml), and overexpressed in more-resistant isolates from patients 7 (isolates 1907 and 2307), 40 (isolates 1780 and 2225), and 43 (isolates 1831 and 2183). The relative CDR2 mRNA levels were equal to those of CDR1 in isolate 1907 (MIC = 8 μg/ml) and exceeded those of CDR1 in highly resistant isolates 2307 and 2183 (MICs >128 μg/ml) by factors of 1.5 and 1.2, respectively. Conversely, CDR1 mRNA levels exceeded those of CDR2 in all other, less resistant, isolates.
FIG. 3.
Northern blot analysis of total RNA from clinical C. albicans isolates using probes specific for CDR1 and CDR2 (see Materials and Methods). The bottom series show the amounts of 18S rRNA used as a control for lane loading in order to normalize signal levels.
DISCUSSION
Increasing reports of clinical and mycological fluconazole resistance in C. albicans have lead to the examination of the molecular mechanisms responsible for development of decreased fluconazole susceptibility. Under selective pressure from treatment with fluconazole, yeast cells can generate resistance through a variety of mechanisms (27, 33). Factors contributing to resistance include alterations in the target enzyme, including overexpression and point mutations, and increased efflux of drug mediated by ABC transporters and major facilitators (33). These studies have been performed in vitro, and their clinical significance is still unknown (1, 4, 10, 30). A limited number of studies support the role of these mechanisms in the development of C. albicans resistance in a small number of clinical isolates (11, 22, 23, 25, 31, 32). The aim of this study was to assess the frequency in which specific, known mechanisms of resistance occurred in series of isolates with decreasing fluconazole susceptibility recovered from OPC patients followed longitudinally.
In all five patients studied, isolation of strains with decreased fluconazole susceptibility was associated with clinical resistance requiring increased fluconazole doses for clinical response. However, it should be noted that clinical response to fluconazole was achieved in all five patients with increased doses of drug. Similar dose-dependent responses have been reported with OPC and decreased fluconazole susceptibility (17, 19).
Increased mRNA levels for ERG11, MDR1, and CDR genes are associated with resistance (1, 24, 25, 31). In the present study, we have evaluated expression of these genes in C. albicans isolates from HIV-infected patients with OPC. A total of 20 isolates from five different patients were included in this study. Successive isolates from one patient were determined to represent the same strain by three molecular typing methods. Antifungal susceptibility testing of isolates recovered from successive OPC episodes in each patient indicated that progressive decrease in fluconazole susceptibility occurred during the longitudinal study. No one mechanism was predominant for the development of drug resistance in all isolates. The mode of acquisition of drug resistance by a particular mechanism or mechanisms for isolates from the different patients was complex. In a series of isolates from two patients (patients 7 and 43), overexpression of CDR genes (both CDR1 and CDR2) was associated with the development of resistance. This increase was obvious even in the second isolate of each series, with fluconazole MICs still in the susceptible range (8 μg/ml), which could possibly indicate a tendency toward development of resistance. In one of these patients (patient 7), increased CDR expression was accompanied by increased ERG11 expression, which was not the case in patient 43. In the case of patient 43, we speculate that other undetermined mechanisms of resistance could have combined with CDR overexpression in the third, highly resistant isolate (as compared to the second, still susceptible isolate), since CDR mRNA levels were similar for both isolates and could not explain the dramatic increase in fluconazole MIC observed for the third isolate.
Increased mRNA levels for MDR1 were detected in two isolates with decreased fluconazole susceptibility (MIC = 16 μg/ml) from patient 9 compared to prior susceptible isolates. These isolates, however, remained itraconazole susceptible. An alternation in overexpression of MDR1 and CDR genes was detected in isolates from another patient (patient 40), suggesting the presence of unstable phenotypes in the same strain with progression of infection due to the selective pressure of the antifungal treatment.
No clear correlation between expression of these genes and development of fluconazole resistance was observed in isolates from patient 14, where expression of CDR genes (mainly CDR1) seemed to decrease, rather than increase, in the final, less susceptible isolate. Sequencing of the ERG11 genes from isolates from this patient revealed the absence of point mutations that could result in decreased susceptibility to azole drugs; and thus, another, yet uncharacterized, mechanism(s) may be responsible for their decreased susceptibility. Thus, for these five patients, five different patterns of resistance were detected, suggesting that C. albicans isolates develop resistance through a variety of molecular mechanisms and that the acquisition of fluconazole resistance is complex.
In at least one case (patient 7), cumulative changes in the expression of genes related to fluconazole resistance appeared to be responsible for decreased susceptibilities to the drug during progression of infection. However, in other cases, a single mechanism seemed to be clearly dominant and intimately associated with development of resistance (e.g., MDR overexpression in patient 9). Overall, these experiments confirm previous results suggesting a major role for efflux pumps in the development of resistance to azoles. In addition, others have demonstrated that ERG11 overexpression alone does not account for high-level azole resistance (1, 25, 31). Since the present study is limited to the study of genes known to be involved in fluconazole resistance, it should be noted that other yet unrecognized mechanisms may be operational in this series of isolates and may contribute to the overall decrease in fluconazole susceptibilities (22).
In C. albicans, the CDR genes constitute a large multigene family coding for highly related proteins (2, 24, 25, 30). Some, but not all, members of this family are associated with resistance to antifungal drugs. CDR1 and CDR2 were the first two members of this family associated with drug resistance in C. albicans, and both CDR1 and CDR2 have been reported to play a role in fluconazole resistance (24, 25, 31). The probe used in the initial experiments to monitor expression of CDR genes was based on the whole sequence of CDR1 and has been shown to cross-hybridize with other members of this gene family, including CDR2 (24, 25, 31). Preparation of probes specific for CDR1 and CDR2 allowed differentiation of the expression of these two highly related genes belonging to the same multigene family. Increased mRNA levels for both CDR1 and CDR2 were detected in those resistant isolates found to overexpress CDR genes (compare Fig. 1 and Fig. 2). CDR1 was constitutively expressed at low levels in susceptible isolates, but CDR2 mRNA was absent in the same susceptible isolates (i.e., isolate 412 from patient 7 and isolates 1490 and 1622 from patient 40). Moreover, as previously reported (24), levels of expression of CDR2 were greater than or equal to those of CDR1 in highly resistant isolates. These results confirmed those of Sanglard and coworkers on the participation of both CDR1 and CDR2 in the development of fluconazole resistance in clinical isolates, as well as the different patterns of expression observed for both genes (24, 25).
Consistent with previous observations (23, 25, 31), overexpression of CDR genes resulted in decreased susceptibility to other azole drugs (i.e., patients 7 and 43), whereas increased MDR1 levels conferred resistance to fluconazole only, as demonstrated by the susceptibility of isolates from patient 9. None of the changes observed in these isolates resulted in increased resistance to amphotericin B, which has been associated with changes in the ergosterol biosynthetic pathway (33).
Finally, fluconazole resistance may affect virulence, as has been demonstrated using some of the isolates described in the present study in a murine model of systemic candidiasis (6). However, the correlation between increased resistance and decreased virulence was not always observed and may be dependent on the underlying mechanism of resistance, thus indicating a more complex relationship between antifungal susceptibility and clinical outcome.
Overall, the present study indicates a high degree of complexity in the molecular mechanisms responsible for fluconazole resistance, with five different patterns of resistance observed in five different series of isolates with decreasing susceptibilities to this antifungal drug. Additional studies are needed to fully understand how specific mechanisms of antifungal drug resistance are induced and to determine the impact of clinical management on the development of resistance to antifungal agents.
ACKNOWLEDGMENTS
This work was supported in part by a grant from Pfizer Inc., and by Public Health Service grants 1 R01 DE11381 (to T.F.P.), 1 R29 AI42401 (to J.L.L.-R.), and M01-RR-01346 for the Frederic C. Bartter General Clinical Research Center.
Chromogenic media were provided by the CHROMagar Company. We thank the Fungus Testing Laboratory at UTHSCSA for performing antifungal susceptibility testing.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzi E, Goffeau A. Genetics and biochemistry of yeast multidrug resistance. Biochim Biophys Acta. 1994;1187:152–162. doi: 10.1016/0005-2728(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 4.Calvet H M, Yeaman M R, Filler S G. Reversible fluconazole resistance in Candida albicans: a potential in vitro model. Antimicrob Agents Chemother. 1997;41:535–539. doi: 10.1128/aac.41.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fling M E, Kopf J, Tamrkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 6.Graybill J R, Montalbo E, Kirkpatrick W R, Luther M F, Revankar S G, Patterson T F. Fluconazole versus Candida albicans: a complex relationship. Antimicrob Agents Chemother. 1998;42:682–697. doi: 10.1128/aac.42.11.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 8.Kirsch D R, Lai M H, O’Sullivan J. Isolation of the gene for cytochrome P450L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Gene. 1988;68:229–237. doi: 10.1016/0378-1119(88)90025-x. [DOI] [PubMed] [Google Scholar]
- 9.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb D C, Kelly D E, Schunck W-H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B D, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole-resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 11.Löffler J, Kelly S L, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 12.Marger M D, Sair M H. A major superfamily of transmembrane facilitators that catalyse uniport, symport, and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard. NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Patterson T F, Kirkpatrick W R, Revankar S G, McAtee R K, Fothergill A W, McCarthy D I, Rinaldi M G. Comparative evaluation of macrodilution and chromogenic agar screening for determining fluconazole susceptibility of Candida albicans. J Clin Microbiol. 1996;34:3237–3239. doi: 10.1128/jcm.34.12.3237-3239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson T F, Revankar S G, Kirkpatrick W R, Dib O, Fothergill A W, Redding S W, Sutton D A, Rinaldi M G. Simple method for detecting fluconazole-resistant yeasts with chromogenic agar. J Clin Microbiol. 1996;34:1794–1797. doi: 10.1128/jcm.34.7.1794-1797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 17.Revankar S G, Kirkpatrick W R, McAtee R K, Dib O P, Fothergill A W, Redding S W, McGough D A, Rinaldi M G, Patterson T F. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in HIV-infected patients. J Infect Dis. 1996;174:821–827. doi: 10.1093/infdis/174.4.821. [DOI] [PubMed] [Google Scholar]
- 18.Revankar S G, Kirkpatrick W R, McAtee R K, Fothergill A W, Redding S W, Rinaldi M G, Patterson T F. Interpretation of trailing endpoints in antifungal susceptibility testing by the National Committee for Clinical Laboratory Standards methods. J Clin Microbiol. 1998;36:153–156. doi: 10.1128/jcm.36.1.153-156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel-Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Berry A L. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 20.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J F, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 25.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 28.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Moereels H, Janssen P A J. Mutation in cytochrome P450-dependent 14α demethylase results in decreased affinity for azole antifungals. Biochem Soc Trans. 1990;18:56–59. doi: 10.1042/bst0180056. [DOI] [PubMed] [Google Scholar]
- 29.Venkateswarlu K D, Denning W, Manning N J, Kelly S L. Resistance to fluconazole in Candida albicans from AIDS patients correlated with reduced intracellular accumulation of drug. FEMS Microbiol Lett. 1995;131:337–341. doi: 10.1111/j.1574-6968.1995.tb07797.x. [DOI] [PubMed] [Google Scholar]
- 30.Walsh T J, Kasai M, Francesconi A, Landsman D, Chanock J. New evidence that Candida albicans possesses additional ATP-binding cassette MDR-like genes: implications for antifungal azole resistance. J Med Vet Mycol. 1997;35:133–137. [PubMed] [Google Scholar]
- 31.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White T C, Bowden R A, Marr K A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]