Abstract
The availability of an ever-increasing diversity of prokaryotic genomes and metagenomes represents a major opportunity to understand and decipher the mechanisms behind the functional diversification of microbial biosynthetic pathways. However, it remains unclear to what extent a pathway producing a specific molecule from a specific precursor can diversify. In this study, we focus on the biosynthesis of ubiquinone (UQ), a crucial coenzyme that is central to the bioenergetics and to the functioning of a wide variety of enzymes in Eukarya and Pseudomonadota (a subgroup of the formerly named Proteobacteria). UQ biosynthesis involves three hydroxylation reactions on contiguous carbon atoms. We and others have previously shown that these reactions are catalyzed by different sets of UQ-hydroxylases that belong either to the iron-dependent Coq7 family or to the more widespread flavin monooxygenase (FMO) family. Here, we combine an experimental approach with comparative genomics and phylogenetics to reveal how UQ-hydroxylases evolved different selectivities within the constrained framework of the UQ pathway. It is shown that the UQ-FMOs diversified via at least three duplication events associated with two cases of neofunctionalization and one case of subfunctionalization, leading to six subfamilies with distinct hydroxylation selectivity. We also demonstrate multiple transfers of the UbiM enzyme and the convergent evolution of UQ-FMOs toward the same function, which resulted in two independent losses of the Coq7 ancestral enzyme. Diversification of this crucial biosynthetic pathway has therefore occurred via a combination of parallel evolution, gene duplications, transfers, and losses.
Keywords: biosynthetic pathway, enzyme diversification, neofunctionalization, subfunctionalization, flavin monooxygenase
Introduction
Over billions of years, bacteria have evolved a tremendous diversity in their metabolic capacities. The enormous diversity of the natural products they synthesize is the result of a multitude of enzymes which have diversified over time (Crits-Christoph et al. 2018; Paoli et al. 2022). The evolution of individual enzymes and functional innovation among homologous protein families have been extensively studied, sometimes providing an atomic level of details of a family functional diversification, along with the documentation of the evolutionary events leading to such diversification (Zou et al. 2015; Kalluraya et al. 2023). The role of several phenomena, including enzymatic promiscuity, gene transfers, and gene duplications, has been extensively discussed and reviewed (Baier et al. 2016; Glasner et al. 2020; Jayaraman et al. 2022).
Deciphering the evolution of individual biochemical pathways is of high interest for evolutionary biology and for understanding the rules that might enable their efficient engineering (Raman et al. 2014; Bachmann 2016). Of particular interest are the biosynthetic pathways of coenzymes, which produce small organic molecules that are essential to the function of a wide variety of enzymes (Kirschning 2022). The ever-increasing availability of genomic sequences provides valuable material for in-depth investigations into the origins and evolution of these biosynthetic pathways. However, such studies are often complicated by the fact that the pathways have only been characterized experimentally in a few model organisms and that homologs with distinct functions can be difficult to disambiguate (Zallot et al. 2016). Moreover, despite significant progress, the accuracy of automated annotation pipelines is still limited (Zallot et al. 2016; Salzberg 2019). Therefore, studying the evolution of biosynthetic pathways requires combining expert biochemical knowledge, detailed genomic and phylogenetic analyses, and experimental validation. For instance, biosynthetic pathways of coenzymes like cobalamins, tetrahydrofolate, or pyridoxal 5′-phosphate were shown to be diverse, with nonhomologous iso-functional enzymes acting at several steps and with the presence of alternate branches in specific clades (de Crécy-Lagard 2014; Balabanova et al. 2021; Denise et al. 2023). Overall, the contribution of nonorthologous displacements and convergent evolution to shaping extant biochemical pathways is well appreciated (Michael 2017). The role of the diversification of homologous protein families is also well recognized, notably to increase the diversity of biochemical pathways allowing to expand inputs (substrates) and outputs (products) (Forsberg et al. 2018; Vu et al. 2019; Hansen et al. 2021). However, to what extent a protein family can diversify within the frame of an evolutionary constrained biosynthetic pathway (i.e. a pathway that does not change its input and output) is currently an open question. A few pathways could serve as case studies, as they involve enzymes from the same family modifying various positions of a molecule in a sequential manner. For example, three O-methyltransferases sequentially add methyl groups on contiguous oxygen atoms on sugar moieties leading to the synthesis of specialized metabolites (Patallo et al. 2001; Kim et al. 2010; Simeone et al. 2013). However, the evolution of these enzymes could not be studied, owing to the narrow phylogenetic distribution of the pathways. Here, we show that the biosynthetic pathway of ubiquinone (UQ) represents an ideal case to address this question of the diversification of a protein family in the framework of an evolutionary constrained pathway.
UQ is a central component of bioenergetic electron transfer chains that sustains ATP production in the eukaryotic mitochondria and in the large bacterial group of Pseudomonadota (a portion of the formerly named Proteobacteria) (Nowicka and Kruk 2010; Pelosi et al. 2019). UQ also acts as a coenzyme (coenzyme Q) for several biological processes (Aussel et al. 2014; Franza and Gaudu 2022). The pathway to produce UQ in bacteria is ancient as UQ innovation is thought to have coincided with the Great Oxidation Event that irreversibly changed Earth's atmosphere 2.4 billion years ago (Schoepp-Cothenet et al. 2009). Moreover, it has evolutionary ties with the plastoquinone pathway found in Cyanobacteria and is very likely to have given rise to the UQ pathway in eukaryotes that slightly differs from its bacterial counterpart (Degli Esposti 2017; Kawamukai 2018; Abby et al. 2020).
The main pathway to produce UQ is O2-dependent and involves, among other reactions, three hydroxylation steps on carbon atoms 5 (C5), 1 (C1), and 6 (C6) (Fig. 1). This pathway has been extensively studied in Escherichia coli, a gammaproteobacterium that employs three flavin-dependent monooxygenases (FMOs), UbiI, UbiH, and UbiF, to catalyze hydroxylation on C5, C1, and C6, respectively (Hajj Chehade et al. 2013) (Fig. 1). In a previous bioinformatic investigation of 67 Pseudomonadota genomes, we discovered UbiL and UbiM, 2 new UQ-FMOs related to UbiI, UbiH, and UbiF (Pelosi et al. 2016). In addition, an unrelated iron-dependent hydroxylase called Coq7 is known to hydroxylate C6 instead of UbiF in some bacterial species and in eukaryotes (Stenmark et al. 2001). Thus, in total, six enzymes—hereafter referred to as UQ-hydroxylases—are involved in the three hydroxylation steps of the O2-dependent UQ biosynthetic pathway. Various combinations of these enzymes were observed in different species of Pseudomonadota, but very few have been experimentally confirmed to date (Fig. 1). We previously showed that the Rhodospirillum rubrum genome encodes a UbiL enzyme that modifies C1 and C5 and a Coq7 enzyme that hydroxylates C6 (Pelosi et al. 2016). We also found that UbiM, the only UQ-hydroxylase encoded in the genome of Neisseria meningitidis, was able to hydroxylate all three positions C1, C5, and C6 (Fig. 1) (Pelosi et al. 2016). Taken together, these data raise interesting evolutionary questions as they suggest that the related UQ-FMOs (UbiI, UbiH, UbiF, UbiL, and UbiM) have different capacities to modify a particular position on a same molecule, i.e. different regioselectivities. However, due to a lack of experimental data, it is currently unclear whether Coq7 and each of the five UQ-FMOs have a conserved regioselectivity. Moreover, it is not known how and why Pseudomonadota evolved such a large repertoire of UQ-hydroxylases. Thus, UQ-FMOs represent a case of pathway diversification within a constrained framework, i.e. with the imperative to keep UQ as the pathway's end product and 4-hydroxybenzoic acid (4-HB) as the pathway's precursor. In the present study, we have examined the evolutionary history of the hydroxylases of the UQ biosynthetic pathway across the diversity of Pseudomonadota by combining phylogenetic and large-scale biochemical analyses. We propose a scenario for the diversification of the UQ-FMO protein family, which involves several gene duplications followed by neo- and subfunctionalizations, two parallel losses of the unrelated Coq7 enzyme, and the horizontal transfer of the generalist enzyme UbiM. We also propose the existence of UbiN, a new member of the UQ-FMO protein family with a specific hydroxylation selectivity.
Fig. 1.
Variety of O2-dependent hydroxylases mapped on the UQ biosynthetic pathway of E. coli. The three O2-dependent hydroxylation reactions of the UQ biosynthetic pathway employ different combinations of hydroxylases in E. coli (UbiI, UbiH, and UbiF, in pink), N. meningitidis (UbiM, in violet), or R. rubrum (UbiL and Coq7, in blue). The oxygen atoms added by the hydroxylation steps originate from dioxygen and are shown in red. The numbering of the carbon atoms of the phenyl ring used throughout this study is shown for the precursor 4-HB. The octaprenyl tail of UQ (in green) is represented by R on carbon 3 of the different biosynthetic intermediates. OPP, 3-octaprenylphenol; DMQ8, C6-demethoxyubiquinone.
Results
Overall Distribution of UQ-Hydroxylases in Pseudomonadata
All complete genomes of Pseudomonadata were downloaded from the National Center for Biotechnology Information (NCBI), and one genome per species was selected. In total, 2,373 genomes were selected, in which the genes coding for UQ-hydroxylases were annotated using previously designed hidden Markov model (HMM) protein profiles (Pelosi et al. 2019), except for that of Coq7, which was refined for the needs of this study (Table S1; Materials and Methods). The presence of the ubiA, ubiE, and ubiG genes was used as a proxy for the presence of the UQ biosynthetic pathway (Pelosi et al. 2019). From the 2,351 genomes harboring the three genes and thus considered as UQ-producers (99% of the data set), we extracted the sequences of the UQ-hydroxylases: the five UQ-FMOs UbiF, UbiH, UbiI, UbiL, and UbiM and the iron-dependent hydroxylase Coq7, a member of the ferritin-like superfamily. The relevance of these annotations was confirmed by phylogenetic analyses, from which we could observe that the enzymes were gathered in well-supported monophyletic groups and regrouped sequences from consistent taxonomic clades, except for UbiM (Fig. 2). These results are also in agreement with our previous analysis based on a smaller data set of 67 Pseudomonadota genomes (Pelosi et al. 2016). Overall, we could annotate 1,153 Coq7, 638 UbiF, 1,470 UbiH, 1,590 UbiI, 1,103 UbiL, and 243 UbiM sequences, amounting to a total of 5,044 UQ-FMOs.
Fig. 2.
Global UQ-FMO phylogeny based on a data set of representative sequences (CD-Hit, 50% sequence identity clustering). This maximum likelihood phylogenetic tree shows that the annotated sequences gather by protein families (inner colored ring) in well-supported groups. These families are, at the exception of UbiM, taxonomically consistent at the class level (outer colored ring). Red dots represent branches that harbor a high support (UFBoot ≥ 95%). The tree was rooted between UbiM and other UQ-FMO according to UbiM deep position in phylogenies rooted with members of the FMO superfamily (Text S1, Fig. S1). IQ-Tree v2.2 tree with LG + F + R9 selected as best model of protein sequence evolution. Tree scale is expressed in substitutions per site.
We then analyzed the distribution of the UQ-hydroxylases across Pseudomonadota. Among the taxonomic orders presumably containing a complete UQ biosynthetic pathway, only one did not harbor any of the UQ-hydroxylases: the Magnetococcales. This was consistent with previous results, as we showed that the only complete genome available for the order, Magnetococcus marinus MC-1, had instead the proteins of the O2-independent pathway: UbiT, UbiU, and UbiV (Pelosi et al. 2019). The possible combinations of UQ-hydroxylases found in genomes at the “order” taxonomic level were extracted, and their respective occurrences were calculated (Table S2). We mapped the fraction of the majority combinations of UQ-hydroxylases in each order, provided that the combination was found in more than five genomes (Fig. 3). To this end, a simplified representation of the species tree based on a survey of the literature was used, as reconstructing a species tree for Pseudomonadata is notoriously challenging (see Discussion). It was observed that the distribution of UQ-hydroxylases followed the species tree. In particular, the enzymes UbiL and UbiF are restricted to Alphaproteobacteria and a subset of monophyletic Gammaproteobacteria orders, respectively. The enzymes UbiH and UbiI are found exclusively outside of the Alphaproteobacteria, across the monophyletic group formed by the Acidithiobacillia and Beta-, Gamma-, and Zetaproteobacteria. Coq7 is more widespread, with occurrences found across Acidithiobacillia and Alpha-, Beta-, Gamma-, and Zetaproteobacteria. The UbiM enzyme, which was proposed to have endured lateral gene transfer (LGT) (Pelosi et al. 2016), is restricted to a few scattered lineages from Alpha-, Beta- and Gammaproteobacteria (mostly Rhodospirillales, Neisseriales, and Xanthomonadales) and was found in only 240 out of 2,373 genomes.
Fig. 3.
The majority repertoire of UQ-hydroxylases along a tentative species tree of Pseudomonadota. The most represented combinations of UQ-hydroxylases are displayed for all the orders represented by more than five genomes, except for the only order of Zetaproteobacteria (Mariprofundales, only two genomes). Pie charts indicate the proportion of the genomes within the order displaying the majority combination, and the total number of genomes analyzed for each order is indicated in the last column “#genomes”. The species tree is drawn as a consensus from previous studies (Williams et al. 2010; Williams and Kelly 2013; Roger et al. 2017; Munoz-Gomez et al. 2019). See Discussion for more details and Fig. S2 for a representation including minor combinations of UQ-hydroxylases.
Besides individual enzyme distribution, we observed that the repertoire of UQ-hydroxylases combinations is itself taxonomically well-conserved. Alphaproteobacteria harbor two main UQ-hydroxylase combinations: the UbiL/Coq7 pair for most orders or UbiL in two copies for the Hyphomicrobiales and the Rhodobacterales (Fig. 3). The Betaproteobacteria mainly display three hydroxylases: UbiH, UbiI, and Coq7. Several lineages of Gammaproteobacteria also harbor the UbiH, UbiI, and Coq7 combination, while others possess the UbiH, UbiI, and UbiF combination.
Based on the few previously reported activities (Table S3), these majority sets of UQ-hydroxylases seemed to correspond to combinations of enzymes with complementary hydroxylation activities on C1, C5, and C6 (Fig. 1). One exception was the combinations involving UbiM, which were neither consistent nor conserved across Pseudomonadota (Fig. S2). Indeed, UbiM was found as the only UQ-hydroxylase in some genomes but coexisted with diverse sets of UQ-hydroxylases in other genomes (Fig. 3, Fig. S2, Table S1). These elements could not provide a clear indication of a conserved hydroxylation activity for UbiM.
Despite indications for complementary hydroxylation activities within the major UQ-hydroxylase combinations, the scarcity of experimental data prevents any conclusion regarding the possible conservation of specific activities in the various UQ-hydroxylases. Moreover, the combination with two UbiL proteins, exclusively found in Rhodobacterales and Hyphomicrobiales (Alphaproteobacteria), has never been tested before. Thus, we decided to jointly explore the evolutionary events leading to the diversification of the UQ-hydroxylase repertoire and the functional diversification of the UQ-FMO family in terms of hydroxylation activities.
The Iron-Dependent Hydroxylase Coq7 Shows Conserved C6-Hydroxylation Activity and Endured Repeated Losses across Pseudomonadota
We first investigated the evolution of the Coq7 family. Coq7 is the only UQ-hydroxylase which spreads across Pseudomonadota. Moreover, when present in genomes, it is found in a single copy (Fig. 3, Text S2, Fig. S3A). Such a distribution could be the result of different events, including vertical gene transmission, but also LGTs and/or duplications mixed with losses. We built a phylogenetic tree of Coq7 sequences (Fig. S3B). Even though not well-supported (see Text S2), the tree showed consistent grouping of sequences by taxa at several taxonomic levels: classes, orders, and families. The overall tree topology is in agreement with the species tree of Pseudomonadata; we thus advocate for a vertical transmission of the corresponding gene. Yet, Coq7 is absent from several lineages: the Rhodobacterales and Hyphomicrobiales within Alphaproteobacteria, and the Aeromonadales, Alteromonadales, Enterobacterales, Methylococcales, Pasteurellales, and Vibrionales within Gammaproteobacteria (Fig. 3). Interestingly, it was previously proposed that five of these six lineages of Gammaproteobacteria could be monophyletic, Methylococcales excluded (Williams et al. 2010). Thus one loss event might explain Coq7 missing from these five lineages of Gammaproteobacteria. Similarly, a single loss of Coq7 in Alphaproteobacteria might suffice to explain its absence from Rhodobacterales and Hyphomicrobiales, which are closely related lineages (see also Discussion).
Given its broad conservation across Pseudomonadota and its likely vertical transmission, we hypothesized that Coq7 might harbor a conserved C6-hydroxylation activity. In accord with this hypothesis, six bacterial Coq7 sequences were previously confirmed to possess a C6-hydroxylation activity (Kwon et al. 2000; Stenmark et al. 2001; Pelosi et al. 2016; Jiang et al. 2019; Zhou et al. 2019; Kazemzadeh et al. 2021) (Table S3). Their capacity to hydroxylate C1 or C5 in addition to C6 was nevertheless not systematically tested. Here, we chose to evaluate the activity and specificity of Coq7 orthologs across the whole diversity of Pseudomonadota by selecting 20 representative sequences and by testing them experimentally according to a previously developed heterologous complementation assay (Pelosi et al. 2016). In this assay, the UQ-hydroxylases of interest are expressed in E. coli strains lacking either ubiI, ubiH, ubiF, or combinations of those genes. The production of DMQ8 and UQ8 provides a readout of the C1-, C5-, or C6-hydroxylation activities, depending on the strains tested (Fig. 1, Table S4). We quantified DMQ8 and UQ8 in lipid extracts of the strains of interest by high-performance liquid chromatography (HPLC) coupled to electrochemical detection (ECD) and mass spectrometry (MS). As expected and as illustrated by ECD chromatograms (Fig. 4a), the ΔubiF strain containing an empty plasmid accumulated DMQ8 and was unable to produce UQ8 (Fig. 4b). In contrast, plasmids containing Coq7 orthologs from Alpha-, Beta-, Gamma-, and Zetaproteobacteria allowed for robust production of UQ8 (Fig. 4a and b), demonstrating that these proteins possess C6-hydroxylase activity. Simultaneous MS detection confirmed the identity of UQ8 (Fig. S3B). Among the 20 sequences tested, 15 showed significant C6-hydroxylase activity (Fig. 4b). Overall, the Coq7 protein sequences from Alphaproteobacteria showed lower activity when compared with those originating from Beta-, Gamma-, or Zetaproteobacteria, some of them allowing UQ8 synthesis at levels close to the wild-type E. coli strain. We assessed C5- and C1-hydroxylase activities in ΔubiIF and ΔubiH strains, respectively. We did not detect any C1-hydroxylase activity; however, a weak C5-hydroxylase activity was apparent for several Coq7 proteins (Table S5). Interestingly, C5 activity was detected only in Coq7 proteins displaying C6-hydroxylase activity and in all cases the C6 activity was much higher (Fig. 4c). Several reasons may explain the lack of activity of five sequences from Alphaproteobacteria (see Discussion). In any case, when considering the phylogenetic tree of experimentally tested Coq7 sequences, it was evident that Coq7 proteins sampled from all major bacterial classes possess C6 activity. We therefore conclude that Coq7 proteins display a selective C6-hydroxylase activity across the diversity of Pseudomonadota. Thus, our biochemical results fully support the usual annotation of Coq7 as a demethoxyubiquinone hydroxylase.
Fig. 4.
Experimental characterization of the hydroxylase activities of Coq7. a) HPLC ECD analysis of lipid extracts from 1 mg of E. coli MG1655 WT cells and ΔubiF cells containing an empty plasmid (vec) or a plasmid carrying the indicated coq7 genes (from Alpha-, Beta, Gamma-, or Zetaproteobacteria). The chromatograms are representative of at least three independent samples. The peaks corresponding to UQ8 (highlighted in gray), DMQ8, DMK8, MK8, and the UQ10 standard are indicated. Corresponding HPLC-MS profiles for UQ8 are presented in Fig. S3C. b) UQ8 content of ΔubiF cells containing pBAD24i or pBAD33i with the indicated Alpha-, Beta, Gamma-, or Zeta-proteobacterial coq7 genes or empty plasmids (vec). ****P < 0.0001 by unpaired Student's t-test comparing with vec. c) Experimental results reported along a phylogenetic tree of the tested Coq7 sequences (see Fig. S3B for broader Coq7 tree). The hydroxylase activity for each position (C1, C5, and C6) is displayed with a color gradient as a percentage of the maximal hydroxylation activity reported in the E. coli MG1655 WT strain (see Materials and Methods; NS, not significant). The taxonomy of the selected sequences (class and order) is shown with a color code along the tree. Branches displaying a UFBoot support above 95% are indicated with a red dot. The tree was obtained using IQ-tree with LG + I + G4 as best selected model of protein sequence evolution. Tree scale is expressed in substitutions per site.
Duplication of a UQ-FMO Explains the Distribution and Activity of UQ-Hydroxylases in Alphaproteobacteria
We next investigated the reasons for the presence of the two major combinations of hydroxylases within the Alphaproteobacteria: UbiL and Coq7 for most orders and two UbiL proteins for Hyphomicrobiales and Rhodobacterales (Fig. 3). In these two orders, Coq7 loss(es) were invoked above. We then built a phylogenetic tree for UbiL to understand the origins of the two copies. We observed before that the proteins annotated as UbiL were monophyletic (Fig. 2), in agreement with a previous phylogenetic analysis (Pelosi et al. 2016). In addition, the tree shows that UbiL sequences generally cluster according to taxonomic groups, even though the phylogenetic tree is not fully resolved (Fig. 5a, Fig. S4, Text S1). Interestingly, the UbiL tree harbors two subtrees of sequences for each of the two orders Rhodobacterales and Hyphomicrobiales (Fig. 5a), consistent with genomes having two copies of the gene. One pair of subtrees grouped together, while the two others were not grouped but close in the tree, as forming a group together with Caulobacterales and Hyphomonadales sequences. The former subtree grouping Rhodobacterales and Hyphomicrobiales was not highly supported in this version of the tree but was highly supported on the trees displayed on Fig. 5b and Fig. S4. We decided to call the underlying protein sequences “UbiN.” Based on the tree topology for UbiL, we firstly propose that UbiL was vertically transmitted in all Alphaproteobacteria lineages, resulting in one UbiL in all lineages. Secondly, we propose that the Rhodobacterales and Hyphomicrobiales acquired a second copy from the duplication of the ubiL gene, giving rise to ubiN (Fig. 5a).
Fig. 5.
Phylogenies of UbiL and UbiN proteins and experimental testing of their hydroxylase activities across the diversity of Alphaproteobacteria. a) An overview of the phylogeny obtained for the UQ-FMO specific to Alphaproteobacteria is provided, with UbiFHI and UbiM as outgroups (collapsed). This tree is based on a subset of representative protein sequences of UbiL and UbiN obtained from the CD-HIT clustering at the 50% identity level of all annotated UQ-FMO proteins. Sequences selected for experimental testing were picked across the diversity of UbiLN and are highlighted in yellow. b) Experimental results obtained for tested UbiLN sequences are mapped on the corresponding phylogenetic tree, as in Fig. 4 (NT, not tested; NS, not significant). For both panels, the taxonomy of the selected sequences (order level) is shown with a color code along the trees. Branches displaying a UFBoot support above 95% are indicated with a red dot. The trees were obtained using IQ-tree with LG + F + R10 selected as the best model of sequence evolution for tree on panel a and Q.pfam + R6 for the tree on panel b. The subtrees gathering the outgroup sequences were collapsed (UbiM and UbiFHI in panel a and UbiFHI in panel b). A tree of UbiLN without outgroup sequences is displayed in Fig. S4. The tree scale is expressed in substitutions per site.
Based on previous findings that UbiL from R. rubrum harbors C1- and C5-hydroxylation activity (Pelosi et al. 2016) and on the observation that Coq7 hydroxylates C6 in Alphaproteobacteria lacking UbiN (Fig. 4c), we formulated two hypotheses: (i) UbiN evolved a C6-hydroxylation activity, thereby replacing the missing Coq7 protein in Hyphomicrobiales and Rhodobacterales, and (ii) the ancestral UbiL possessed both C1- and C5-hydroxylation activities, which should be conserved in extant UbiL sequences across orders where UbiL did not undergo duplication.
In order to verify these hypotheses, we tested experimentally 38 UbiL and 12 UbiN paralogs. These proteins showed conserved sequence motifs characteristic of UQ-FMOs (Fig. S5). All UbiN sequences showed moderate to strong C6-hydroxylation activity in our heterologous complementation assay (Fig. 5b and Table S5). Interestingly, each UbiN also displayed a C5-hydroxylation activity, albeit much lower than for C6. A low C1 activity was detected for only 3 UbiN sequences (<2% of WT activity; Fig. 5b). Out of the 38 UbiL sequences tested, 17 did not show any hydroxylase activity, the majority belonging to Caulobacterales and Sphingomonadales (Fig. 5b and Table S5). Nevertheless, one sequence of each clade (WP_066774902.1 from “Caulobacteraceae bacterium OTSz_A_272” and WP_013934691.1 from Zymomonas mobilis subsp. pomaceae ATCC 29192) showed C1 and C5 activities. Most UbiL sequences from Hyphomicrobiales and Rhodobacterales displayed a major C5 activity associated with a lower C1 activity, and a minor C6 activity was detected in a few cases (Fig. 5b). In the unsupported group formed by sequences from Rhodospirillales, Rickettsiales, Pelagibacterales, and Holosporales, the UbiL proteins displayed mostly comparable levels of C1 and C5 activities, with minor C6 activity detected in a few cases. Overall, these data strongly support a scenario in which an ancestral UbiL protein with C1 and C5 activities gave rise by duplication and neofunctionalization to UbiN proteins possessing a rather unselective C6 activity. The emergence of UbiN would have allowed the functional replacement of Coq7 in Hyphomicrobiales and Rhodobacterales.
From One to Three UQ-FMOs: Two Duplications Occurred within the Clade Grouping Acidithiobacillia and Beta-, Gamma-, and Zeta-proteobacteria
We next focused on the dynamics of the UQ-hydroxylase repertoire within the rest of the Pseudomonadota, i.e. the clade consisting of the Acidithiobacillia and Beta-, Gamma-, and Zetaproteobacteria. These four bacterial classes possess UbiH, UbiI, and Coq7 as the major combination of hydroxylases, with the exception of (i) the order of Neisseriales (mostly UbiM alone, followed by the combination UbiH, UbiI, and Coq7); (ii) a subclade of Gammaproteobacteria—comprising the Aeromonadales, Alteromonadales, Enterobacterales, Pasteurellales, and Vibrionales—that showed the UbiH, UbiI, and UbiF combination (Fig. 3 and Fig. S2); and (iii) the order of Methylococcales, presenting UbiH and UbiF as the most frequent combination (see later and Discussion).
The evolutionary history of the UQ-FMO enzymes UbiH, UbiI, and UbiF was then considered. Interestingly, the two enzymes UbiH and UbiI formed two highly supported sister subtrees in the tree of UQ-FMO (Fig. 6a). Moreover, the grouping of sequences within the subtrees agreed with the species tree of Pseudomonadota, showing a split between Acidithiobacillia and Beta-, Gamma-, and Zetaproteobacteria, and well-delineated taxonomic orders arranging in similar fashions between the two subtrees (Fig. S6). We therefore propose that UbiH and UbiI are two paralogs stemming from an ancestral UQ-FMO, which likely duplicated before the divergence of Acidithiobacillia and Beta-, Gamma-, and Zetaproteobacteria. On the other hand, a highly supported UbiF subtree branched from within the UbiI subtree and showed a branching pattern overall supportive of another duplication event, that of a subclade of UbiI. More precisely, given the gene tree and species tree topologies and their consistency with UbiF distribution, we propose that a duplication of UbiI gave rise to UbiF. This duplication likely occurred in an ancestor of the five monophyletic lineages having lost Coq7 and acquired UbiF, namely the ancestor of the Aeromonadales, Alteromonadales, Enterobacterales, Pasteurellales, and Vibrionales (Fig. 3, Fig. 6a, and S5).
Fig. 6.
Phylogenies of UbiFHI proteins and experimental testing of their hydroxylase activities across their diversity. a) An overview of the phylogeny obtained for the UQ-FMO specific to the Acidithiobacillia and Beta-, Gamma-, and Zetaproteobacteria is provided, with UbiLN and UbiM as outgroups (collapsed). This tree is based on a subset of representative protein sequences of UbiFHI obtained from the CD-HIT clustering at the 50% identity level of all annotated UQ-FMO proteins. Sequences selected for experimental testing were picked across the diversity of UbiFHI and are highlighted in yellow. b) Experimental results obtained for tested UbiFHI sequences are mapped on the corresponding phylogenetic tree, as in Fig. 4. For both panels, the taxonomy of the selected sequences (class and order) is shown with a color code along the trees. Branches displaying a UFBoot support above 95% are indicated with a red dot. The trees were obtained using IQ-tree, with LG + F + R10 selected as the best model for protein sequence evolution in panel a and Q.pfam + R6 in panel b. The subtrees gathering the outgroup sequences were collapsed (UbiM and UbiLN in panel a and UbiLN in panel b). Tree scale is expressed in substitutions per site.
This scenario implies that changes in activities may have occurred following the duplications, as evidenced by the UbiI, UbiH, and UbiF proteins in E. coli that are known to hydroxylate C5, C1, and C6, respectively (Hajj Chehade et al. 2013) (Fig. 1). However, our understanding of the activity of these protein families is currently limited, as only a few members outside of E. coli have been characterized to date (Table S3). To address this problem, we sampled several sequences for each protein across the diversity of Pseudomonadota (Fig. 6a) and assayed their hydroxylase activity. All three selected UbiF sequences had a robust C6 activity and a minor C5 activity, with no detectable C1 activity (Fig. 6b). These results fit perfectly with the minor C5 activity of E. coli UbiF that is found in addition to its main C6 activity (Hajj Chehade et al. 2013). Among the 13 UbiI sequences, all but 1 displayed a strong C5 activity, and a minor C1 activity was detected for 5 of them. The UbiI protein of Mariprofundus ferrinatatus CP-8 (WP_100265575.1) stood out as it showed equally robust activities at C1, C5, and C6 (Fig. 6b) and was able to restore high levels of UQ8 in the E. coli ΔubiIHF strain (Table S5). C1-hydroxylation was the main activity detected in UbiH proteins, and it was associated with minor C5 or C6 activities in a few cases (Fig. 6b). Overall, our data demonstrate that the hydroxylation activities are globally homogeneous within each subfamily, with UbiF displaying a strong unselective C6 activity, whereas UbiI and UbiH are characterized by strong selective C5 and C1 activities, respectively.
The Case of UbiM
The case of the UbiM enzyme is more challenging to approach than the ones investigated above. UbiM is sometimes the only UQ-hydroxylase found in genomes, as previously observed in Neisseriales (Fig. 3). Accordingly, it was shown to hydroxylate three C positions in N. meningitidis (Pelosi et al. 2016). But UbiM is also found in genomes with either a fully fledged UQ-hydroxylase repertoire (e.g. UbiM in combination with UbiH, UbiI, and Coq7 in Xanthomonadales) or an “incomplete” set of UQ-hydroxylases (e.g. in Acetobacter ascendens with UbiL alone—Coq7 being missing; see Table S1). In addition, UbiM is found in scattered lineages across Pseudomonadota (Figs. 3 and 7a). The UbiM phylogenetic tree presents mixed groupings of UbiM sequences from Alpha-, Beta-, and Gammaproteobacteria, evidencing multiple cases of LGTs (Fig. 7a). While the lack of resolution of the UbiM tree in its deep nodes prevents the inference of a precise number of LGT events, some appear clearly. This is the case of a LGT likely to have occurred from Neisseriales to Moraxellales (Fig. 7a). Clades of Burkholderiales sequences appear six times in the tree, evidencing a complex history of LGT between Pseudomonadota. Interestingly, despite its scattered distribution and propensity to LGT, UbiM places as a sister group to other UQ-FMO (Fig. 2, Fig. S1). Overall, this makes it difficult to predict whether UbiM has a conserved regioselectivity or even to predict a candidate regioselectivity at all. We thus tested two UbiM sequences from major combinations of UQ-hydroxylases found in Rhodospirillales and Xanthomonadales, in which UbiM is respectively associated either to UbiL or to UbiI, UbiH, and Coq7 (Fig. 3). The UbiM sequences from Xanthomonas campestris and Roseomonas gilardii allowed robust production of UQ8 in the E. coli ΔubiIHF strain (Fig. 7b and Table S5), demonstrating their capacity to hydroxylate C1, C5, and C6. To determine whether the UbiM protein from X. campestris pv. campestris ATCC 33913 is associated with a functional complete set of UQ-hydroxylases, the regioselectivity of the UbiI, UbiH, and Coq7 proteins from this species was tested. UbiI and Coq7 hydroxylated C5 and C6, respectively, but UbiH did not show a significant C1 activity (Table S5). However, the ubiH gene from the closely related X. campestris pv. campestris strain 8004 (see Table S6 for sequence homology between the two strains) was shown to be essential for UQ biosynthesis (Zhou et al. 2019). Overall, the data are coherent with UbiM harboring a consistent C1, C5, and C6 regioselectivity (Fig. 7) (Pelosi et al. 2016) and sometimes being associated with a complete set of UQ-hydroxylases, as in the case of X. campestris.
Fig. 7.
Phylogeny of UbiM and experimental testing of its activities. a) The phylogenetic tree of all UbiM sequences annotated in the genome data set is presented on the left along with the corresponding taxonomy (class and order levels). The tree was rooted using members of the FMO superfamily as an outgroup (class A flavin monooxygenases [FMOs]) (Mascotti et al. 2016). Well-supported groups of sequences from a same genus were collapsed, and the number of corresponding UbiM sequences was indicated in parenthesis. Names of clades containing experimentally validated UbiM sequences are written in blue (this study and (Pelosi et al. 2016)). A species tree of orders is presented on the right, while the connections of the UbiM sequences to the corresponding orders in the species tree are indicated by colored lines. Orders presenting no UbiM sequence are displayed in gray; otherwise, the number of UbiM sequences and the number of genomes within the order are presented in front of each UbiM-containing order. Branches displaying a UFBoot support above 95% are indicated with a red dot. The tree was obtained using IQ-tree, with LG + F + I + R7 selected as the best model for protein sequence evolution. b) UQ8 content of WT MG1655 cells or ΔubiIHF cells containing an empty plasmid (vec) or plasmids encoding the indicated UbiM sequences. WP_075797773.1 is from R. gilardii U14-5 and WP_011037777.1 from X. campestris pv. campestris str. ATCC 33913. ***P < 0.001; ****P < 0.0001 by unpaired Student's t-test comparing with vec.
Evolutionary Scenario of the UQ-Hydroxylase Repertoire
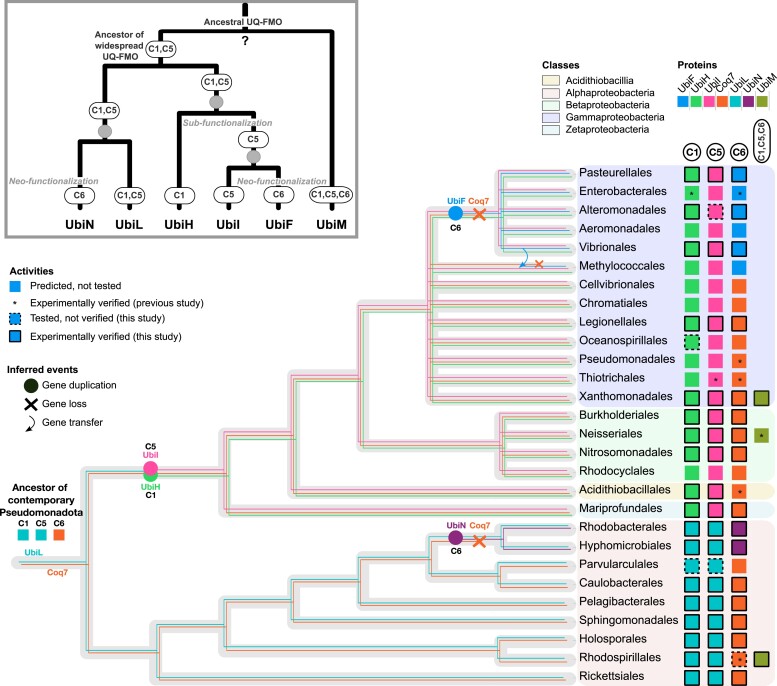
Combining the results of the evolutionary and experimental analyses described above, we propose the following global scenario (Fig. 8): (i) the last common ancestor of contemporary Pseudomonadota had one UbiL-like enzyme and one Coq7 as UQ-hydroxylases, which were transmitted to the ancestor of Alphaproteobacteria and to the ancestor of all the other Pseudomonadota (Aciditiobacillia and Beta-, Gamma-, and Zetaproteobacteria). (ii) Within Alphaproteobacteria, UbiL was duplicated in a common ancestor of Hyphomicrobiales and Rhodobacterales and gave rise to UbiN that evolved toward the functionality of Coq7, which was lost in the process. (iii) In the common ancestor of the remaining Pseudomonadota (Aciditiobacillia and Beta-, Gamma-, and Zetaproteobacteria), the inherited UbiL-like protein was duplicated and subfunctionalized in UbiH and UbiI. (iv) UbiI was then duplicated, giving rise to UbiF in a sublineage of Gammaproteobacteria that evolved toward the function of Coq7, which once again was lost.
Fig. 8.
Overall proposed scenario for the evolution of UQ-hydroxylases. The predicted and experimentally validated repertoire of UQ-hydroxylases is summarized alongside the phylogenetic tree of the Pseudomonadota, with the depiction in each taxonomic order of the proteins involved in each hydroxylation step (colored boxes). The most parsimonious scenario is represented on the form of the gene trees (thin, colored branches) embedded in a candidate species tree (thick, light gray branches). The inferred events of UQ-FMO duplications are represented along the corresponding branches with bubbles and Coq7 losses with crosses. The evolutionary history of UQ-FMO is also summarized in the inset panel, with duplications symbolized by gray dots. The dispersal of UbiM suggests multiple LGTs, while its phylogenetic positioning points at an ancient origin. Based on our combined experimental and phylogenetic analyses, we propose that the last common ancestor of contemporary Pseudomonadota was able to produce UQ via the use of two hydroxylases: one ancestral UbiL-like protein performing the C1- and C5-hydroxylation steps and Coq7 performing the C6-hydroxylation step. This configuration was kept in Alphaproteobacteria, until the loss of Coq7 and a duplication of UbiL that resulted in a neofunctionalization in one of the paralogs, UbiN, that performs mainly C6-hydroxylation and is specific to Hyphomicrobiales and Rhodobacterales. In the ancestor of other Pseudomonadota, there was a duplication of an ancestral UbiL-like protein which resulted in the subfunctionalization of two paralogs: UbiH (C1) and UbiI (C5). This repertoire of hydroxylases was well-conserved, until a duplication of UbiI occurred in the ancestor of a sublineage of Gammaproteobacteria, with the UbiF paralog evolving toward C6 activity and the other conserving the ancestral C5 activity of UbiI. This event was accompanied by the loss of Coq7. A LGT could explain the presence of UbiF in Methylococcales, but the donor lineage is unknown and the arrow is purely illustrative. Finally, whether the first original UQ-FMO was able to perform two (UbiL-like) or three (UbiM-like) hydroxylations remains unresolved. See also the variants of this proposed scenario in Discussion.
The reasoning for the proposed scenario is as follows: given that most UbiL proteins display a C1/C5-hydroxylation activity and that the sister lineage groups the two paralogs UbiH and UbiI with, respectively, C1- and C5-hydroxylation activity, we hypothesize that the enzyme ancestral to UbiH and UbiI had a dual C1/C5 activity similar to that found in UbiL. Upon duplication and subfunctionalization of the ancestor of UbiH and UbiI, this dual activity was split between the two paralogs. This implies that the ancestral UQ-FMO enzyme also had a C1/C5 activity. As for Coq7, it is shown to harbor a conserved C6 activity, has an evolutionary history compatible with its vertical transmission, and is found in all classes of Pseudomonadota, suggesting that a Coq7-type enzyme was also present in the ancestor of Pseudomonadota. Altogether, this leads us to the proposal of an ancestral repertoire of UQ-hydroxylases consisting of a UbiL-like enzyme with C1/C5-hydroxylation activity and of a Coq7 enzyme with C6-hydroxylation activity. In the Alphaproteobacteria, the ancestral repertoire consisted of UbiL and Coq7 with, respectively, C1/C5 and C6 as main activities. A secondarily repertoire evolved in Rhodobacterales and Hyphomicrobiales involving UbiL and the newly characterized UbiN enzyme that acquired C6 activity after duplication of UbiL. This duplication could also explain the fact that UbiN harbors a secondary C5 activity (Fig. 5b), which may derive from its UbiL ancestor. This means that upon UbiL duplication, there was a neofunctionalization toward C6 activity giving rise to UbiN, while Coq7 was lost in an ancestor of Rhodobacterales and Hyphomicrobiales (Fig. 8; the precise order of the events cannot be inferred, as discussed below). Interestingly, this duplication/neofunctionalization of a UQ-FMO and concomitant loss of Coq7 also occurred in the Gammaproteobacteria. Indeed, we propose that UbiF emerged in the ancestor of the Aeromonadales, Alteromonadales, Enterobacterales, Pasteurellales, and Vibrionales from a UbiI ancestor possessing a C5 activity and evolved a C6-hydroxylation activity. As for UbiN, the observed C5 secondary activity in UbiF (Fig. 6b) can be interpreted as residuals of an ancestral activity. Again, while UbiF evolved a C6 activity, Coq7 was lost. The presence of UbiF in the unrelated Methylococcales might be explained by a LGT (see Discussion). The case of UbiM, its origins, and role in ancestral Pseudomonadota remain a puzzle. Given its deep phylogenetic positioning, it is possible that a more ancient ancestor of Pseudomonadota harbored a single three-C-hydroxylating hydroxylase. Another possibility is that UbiM evolved the capacity to hydroxylate the C6 position in an ancient Pseudomonadota lineage from a UbiL-like protein able to hydroxylate C1 and C5 and then extended to several lineages of Pseudomonadota via LGT. Independently of this, the sparse distribution of UbiM (around 10% of the analyzed genomes) points toward multiple losses early in the diversification of Pseudomonadota, making it uncertain whether UbiM was present in the last common ancestor of contemporary Pseudomonadota.
Discussion
In this article, an original experimental approach coupled with comparative genomics and phylogenetics was used to study the overall underlying evolutionary history of UQ-hydroxylases. We showed that UQ-FMOs diversified into six subfamilies (UbiF, H, I, L, M, and N) with distinct hydroxylation selectivity via at least three duplication events associated with two cases of neofunctionalization (UbiF and UbiN) and one case of subfunctionalization (UbiI and UbiH).
UQ-FMO Subfamilies Are Characterized by Distinct Regioselectivities
On the experimental side, in vitro assays to test the activity of purified UQ-FMOs are currently unfeasible, mainly because of the extreme hydrophobicity of the UQ biosynthetic intermediates that are substrates for the C1-, C5-, and C6-hydroxylation reactions. Therefore, to gain insights in the hydroxylation activities of UbiF, UbiH, UbiI, UbiL, UbiM, and UbiN proteins, we resorted to an in vivo heterologous complementation assay, which is based on the expression of UQ-FMO sequences in E. coli strains deleted for one or several UQ-FMO genes (Pelosi et al. 2016). It should be noted that in cases where a particular activity was monitored in several mutants, for example, C1-hydroxylation in ΔubiH and ΔubiHF cells, the results were mostly consistent across strains, reinforcing the validity of our assay (Tables S4 and S5). Several sequences of UbiL and Coq7 from Alphaproteobacteria did not show any activity. A poor expression is unlikely as the codons of all sequences were optimized for E. coli expression. A more likely explanation could be a poor interaction of the tested hydroxylases with E. coli Ubi proteins, several of which form a multiprotein “Ubi complex” that is essential for UQ biosynthesis (Hajj Chehade et al. 2019). Disruption of the Ubi complex or inability to interact with it would translate into a lack of activity in our assay. Even though we had to sample UbiL sequences repeatedly in Caulobacterales and Sphingomonadales, we eventually obtained one active protein in each group (out of a total of nine and seven sequences tested, respectively). These two proteins showed an activity qualitatively similar to that of the other sequences in the UbiL clade, suggesting that they do not possess functional specificities. Taken together, the results of our experiments increase greatly our understanding of the regioselectivity of UQ-FMOs and demonstrate that distinct activities and selectivities are associated with each subfamily of UQ-FMOs: UbiM possesses a broad C1-, C5-, and C6-hydroxylation activity; UbiH and UbiI possess quite selective C1 and C5 activities, respectively; UbiF and UbiN share a major C6 activity and a minor C5 activity; and UbiL displays C1 and C5 activities, which can be of comparable efficiency in some lineages or with C5 surpassing C1 in others.
Possible Reasons for the Observation of Numerous “Minor Combinations” of UQ-Hydroxylases in Pseudomonadota
We believe that the complex pattern of “minor combinations” presented in Fig. S2 is a mixture of methodological and biological phenomena. Firstly, no annotation pipeline is perfect, and while our HMM protein profiles enabled to capture most of the specificities of each UQ-hydroxylase subfamily (Fig. 2), for some lineages, we could propose a revision of the automatic annotations using phylogenetics. This was the case, for example, for the Neisseriales, in which a UbiF was sometimes annotated in addition to the UbiH, UbiI, and Coq7 expected for Betaproteobacteria. Inspection of the phylogenetic trees of UQ-FMO showed that the proposed UbiF indeed robustly grouped within the UbiH subtree, leading us to reevaluate the set of UQ-hydroxylases as UbiH, UbiI, and Coq7 (Fig. S6). Another example is that of the Thiotrichales (Gammaproteobacteria), where UbiH was misannotated as UbiL in two genomes. More intriguing was the case of the Oceanospirillales, which displayed a variety of different sets of UQ-hydroxylases (Fig. S2, Table S2) and several of which harbored a set of hydroxylases typical of other orders (e.g. UbiFHI). Similarly to the difficulties with the phylogeny (and taxonomy) of Pseudomonadata discussed below, this may be partly explained by the fact that the Oceanospirillales are polyphyletic, with some representatives being more closely related to other lineages, e.g. the Gammaproteobacteria that have acquired UbiF and lost Coq7 (Liao et al. 2020). However, such a pattern could also result from the lateral transfer of the ubiF gene to some Oceanospirillales, as appears to be the case for UbiF in the Methylococcales (Fig. 8). Finally, it is interesting to note that some Hyphomicrobiales possess only UbiL (Fig. S2), which could therefore theoretically perform all three hydroxylation reactions. Further studies, both at the evolutionary and experimental levels, will be needed to investigate the full breadth and complexity of the diversification of UQ-hydroxylases.
Gene Duplications and Neofunctionalization with Respect to C6-Hydroxylation Led to Repeated Losses of Coq7
A striking feature of the evolutionary scenario proposed herein is the parallel evolution of UQ-FMO paralogs toward C6-hydroxylation activity in the Alphaproteobacteria and Gammaproteobacteria, accompanied by the loss of Coq7. In addition to phylogenetic analyses and primary hydroxylation activities, we used the observed secondary activities as further support for our evolutionary scenario, especially the secondary C5 activity displayed by the UbiF and UbiN paralogs. This could also be indicative of a certain promiscuity of the C5- and C6-hydroxylation activities across the UQ-FMO family that could favor the appearance of new homologs with C6 as main activity. Interestingly, the co-option of an unrelated family of FMO, with members involved in the hydroxylation of 2,4-dichlorophenol, was recently shown to have provided a new FMO for UQ biosynthesis in Viridiplantae (Latimer et al. 2021; Xu et al. 2021). Incidentally, this proved to be yet another example of the adaptive evolution of C6-hydroxylation activity. How many times this kind of co-option occurred throughout the evolution of the UQ biosynthetic pathway, both in its bacterial and eukaryotic versions, remains to be investigated. Finally, the fact that Coq7 was lost repeatedly in Alphaproteobacteria and Gammaproteobacteria could be a sign that replacing the iron protein with a FMO provides a selective advantage. A possible explanation is that the lineages having lost Coq7 may be highly dependent on iron for their growth and/or for thriving in iron-limited environments. Indeed, iron is a limiting resource for which bacteria compete in several environments, for instance, marine ones (Morel and Price 2003; Moore et al. 2013; Kramer et al. 2020), and various strategies to cope with iron limitation have been documented, ranging from active iron foraging (e.g. siderophores) to limiting iron dependability. In that case, the loss of Coq7 would fall into the latter category. The relative timing of the Coq7 losses with respect to the cases of UbiL and UbiI duplications could not be inferred from our analyses. However, given the fact that UQ is often the only respiratory quinone of Pseudomonadota, it is most likely that the duplications and neofunctionalizations toward C6 activity preceded the loss of the C6-hydroxylating Coq7 to avoid disruption of UQ biosynthesis.
UbiM Is a Highly Transferable Generalist UQ-Hydroxylase
Contrary to the other UQ-hydroxylases, which are widely distributed and are part of evolutionarily conserved combinations, UbiM has a scattered distribution and was found in association with various sets of UQ-hydroxylases. Biochemical characterization of three UbiM proteins from Alpha-, Beta- and Gammaproteobacteria revealed the capacity of these proteins to hydroxylate the C1, C5, and C6 positions (Fig. 7b, (Pelosi et al. 2016)). The genome of X. campestris pv. campestris harbors two complete sets of UQ-hydroxylases, as it encodes a UbiM protein together with UbiI, UbiH, and Coq7. Surprisingly, deletion of ubiI or ubiH was sufficient to completely abolish UQ biosynthesis in X. campestris pv. campestris strain 8004 (Zhou et al. 2019). This suggested that the endogenous UbiM protein was unable to compensate the C1- or C5-hydroxylation defects caused by deletions of ubiI or ubiH, despite having demonstrated robust C1, C5, and C6 activities in our heterologous complementation assay (Fig. 7b). An obvious explanation is that transcriptional regulation of the ubiM gene might prevent the expression of the UbiM protein under the conditions used to culture the X. campestris ubiI or ubiH mutants. In this case, UbiM may represent an alternative hydroxylation system that might be functional only under specific environmental conditions. Our study of a large and representative genome data set reports UbiM in ∼10% of the genomes, whereas it was earlier observed in nearly half of a selected set of 67 Pseudomonadota genomes (Pelosi et al. 2016). While our evolutionary scenario cannot exclude UbiM from being ancestrally found in Pseudomonadota, it is possible that it is a nonessential UQ-hydroxylase in many cases. This would explain why the distribution of UbiM remains sparse while the other UQ-hydroxylases have been widely vertically distributed and conserved. Nevertheless, lateral acquisition of UbiM has been selected for in some cases, indicating that it could be advantageous under some circumstances. Now that we have demonstrated the respective and conserved role of the UQ-hydroxylases, future analyses of the role of UbiM in the context of various UQ-hydroxylase repertoire will be facilitated.
Robustness of Proposed Evolutionary Scenario in the Context of a Hard-to-Solve Species Tree
We present our global scenario in the context of a candidate species tree of Pseudomonadota, which is the subject of intense debate. Slightly alternative scenarios might be preferred in the future if alternative species trees are proposed. The phylogeny of Alphaproteobacteria has been thoroughly investigated in several recent papers, often with a view to identifying the origins of the mitochondrion (Martijn et al. 2018; Munoz-Gomez et al. 2019; Fan et al. 2020). It has been shown that Alphaproteobacterial phylogeny is difficult to reconstruct due to a combination of rapidly evolving lineages, sequence compositional biases, and sites with heterogeneous evolutionary rates (Munoz-Gomez et al. 2019; Muñoz-Gómez et al. 2022). The question of the monophyly of Hyphomicrobiales and Rhodobacterales is of special interest to our global scenario, as the most parsimonious scenario of one UbiL duplication and one Coq7 loss in Alphaproteobacteria presented in Fig. 8 relies on their shared common ancestry. However, the two lineages were recently proposed to be paraphyletic, with Caulobacterales branching as a sister lineage to Rhodobacterales to the exclusion of Hyphomicrobiales, even if this branching did not get the maximal support (Munoz-Gomez et al. 2019). Genomes of the closely related candidate order Parvibaculales (Genome Taxonomy Database [GTDB], release 214) harbor the Coq7 and UbiL combination of UQ-hydroxylases (Table S1). Their phylogenetic positioning remains to be clarified, as they were observed either as a sister lineage to Hyphomicrobiales or as a more deeply branching lineage (Munoz-Gomez et al. 2019; Hördt et al. 2020; Cevallos and Degli Esposti 2022; Muñoz-Gómez et al. 2022). In the case where Caulobacterales were indeed a sister group of the Rhodobacterales and Parvibaculales a sister group of the Hyphomicrobiales, the scenario for UQ-hydroxylase evolution within Alphaproteobacteria would be more complex than proposed above. It would involve one duplication of UbiL in the common ancestor of the four lineages giving rise to UbiN, two independent losses of Coq7 in the respective ancestors of Rhodobacterales and Hyphomicrobiales, and two independent losses of UbiN in the respective ancestors of Caulobacterales and Parvibaculales.
Several studies have investigated the phylogeny of Gammaproteobacteria using different sets of markers but have tended to focus on the relationships of specific lineages within the class (Spring et al. 2015; Liao et al. 2020). In the end, we chose to include in the candidate species tree (Fig. 8), a split proposed in 2010 in one of the few articles covering the entire breadth of Gammaproteobacteria (Williams et al. 2010). It corresponded to the monophyly of the Aeromonadales, Alteromonadales, Enterobacterales, Pasteurellales, and Vibrionales. Interestingly, this split was consistent with the distribution of UQ-hydroxylases and the existence of a shared ancestor having endured a Ubi-FMO gene duplication and the loss of coq7. In addition, the GTDB now considers the Aeromonadales, Alteromonadales, Pasteurellales, and Vibrionales as part of a same order provisionally named “Enterobacterales_A” (Parks et al. 2022).
We argue that species trees obtained using purely phylogenetic approaches could get support from the study of the evolutionary history of traits, including the dynamics of the genome contents, and the fact that genes being shared specifically between lineages at the exception of others can constitute reliable synapomorphies. In particular, if the gene sets are shared because of a same evolutionary event, such as ancient LGTs or duplications, this can be considered as further pieces of evidence for past common history and descent from a common ancestor (Huang and Gogarten 2006; Abby et al. 2012). Accordingly, recent phylogenetic approaches aiming at reconstructing jointly species trees and gene trees while inferring events of duplications, transfers, and losses have shown promising results to tackle complex evolutionary questions (Szollosi et al. 2013; Williams et al. 2017). We suggest that the shared repertoire in hydroxylases for the crucial UQ biosynthetic pathway could represent relevant patterns to consider when assessing new candidate species trees of Pseudomonadata.
Diversification of Biochemical Pathways via Homologous Enzyme Specialization
To our knowledge, our study of UQ-hydroxylases provides a unique understanding of the evolution of enzymes catalyzing sequential regioselective reactions within a given biosynthetic pathway. The wide conservation of the UQ pathway in Pseudomonadata allowed us to collect a large number of sequences, which was instrumental in establishing the evolutionary scenario. Several pathways leading to the synthesis of specialized molecules are known to involve three regioselective O-methyltransferases to methylate contiguous O atoms on sugar moieties (Patallo et al. 2001; Kim et al. 2010; Simeone et al. 2013). Within each pathway, the methyltransferases are likely to be paralogs, but the narrow distribution of the pathways precludes a phylogenetic reconstruction of the evolutionary history of the enzymes. Similarly, the synthesis of xantholipin was recently shown to rely on XanM1-3, three distinct O-methyltransferases with substrate-dependent regiospecificity that evolved from a single ancestor via an innovation–amplification–divergence model (Kong et al. 2020). Together, these findings suggest that pathways implicating sequential biochemical reactions to modify specific positions of a metabolite evolved specialized regioselective enzymes rather than a single multifunctional enzyme. In this sense, most extant Pseudomonadota species indeed possess three distinct UQ-hydroxylases (Fig. 3) with relatively high regioselectivity, as demonstrated by our experimental results. However, most Alphaproteobacteria possess only two UQ-hydroxylases (Fig. 3) and we detected several genomes with a single UQ-hydroxylase gene (Fig. S2 and Table S2), suggesting that the corresponding proteins are generalist and hydroxylate C1, C5, and C6, as now demonstrated for three UbiM proteins.
Prospects
The objectives of future studies will be to understand why specialized UQ-hydroxylases evolved in some species whereas generalist UQ-hydroxylases evolved in others. They could also aim at revealing the molecular mechanisms and adaptations that control the regioselectivity of UQ-hydroxylases. This could be done in conjunction with the experimental testing of resurrected ancestral proteins, as a means to test and refine the proposed scenario for the evolution of UQ-hydroxylases with various regioselectivities. On a broader scale, it will be of interest to decipher the evolutionary relationships between the biosynthesis pathways of isoprenoid quinones. Indeed, homologous enzymes are found in the pathways of UQ, menaquinone, and plastoquinone, but a unifying evolutionary scenario remains elusive (Zhi et al. 2014; Degli Esposti 2017; Abby et al. 2020).
Materials and Methods
Genome Data Set
A total of 10,470 complete genomes from the clade of UQ-producing Pseudomonadata (i.e. the former Proteobacteria, excluding Epsilon- and Delta-proteobacteria) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/), last accessed in October 2020. Priority was given to the “RefSeq” over the “GenBank” assemblies when both were available. The taxonomy of the data set was downloaded from the NCBI (Schoch et al. 2020) in August 2023 and added to the genome annotation Table S1 together with the GTDB classification (release 214) (Parks et al. 2018; Parks et al. 2020). When taxonomy was conflicting between the two databases, priority was given to that of the GTDB that relies on homogeneous genome-based criteria. For instance, Moraxellaceae were kept as part of the Pseudomonadales and Parvibaculales in an order separate from Hyphomicrobiales (see also Discussion). In the end, 2,373 complete genomes were kept for analysis, including 726 Alphaproteobacteria, 414 Betaproteobacteria, 1,224 Gammaproteobacteria, 2 Zetaproteobacteria, 6 Acidithiobacillia, and 1 Hydrogenophilalia (see Table S1).
Annotation of UQ Proteins in Genomes
The genes involved in the UQ biosynthetic pathways were annotated as described previously (Pelosi et al. 2019): HMM protein profiles corresponding to three essential genes of the UQ biosynthesis pathway were used (UbiA, UbiE, and UbiG) as well as profiles for the hydroxylases involved in the O2-dependent pathway for UQ production: UbiF, UbiH, UbiI, UbiL, and UbiM, but also four specifically designed “decoy” profiles for the diverse set of homologs from the large FMO superfamily (Pelosi et al. 2016; Pelosi et al. 2019) (Text S3). For Coq7, we used the previously reported profile (Pelosi et al. 2019) and designed two new “decoy” profiles for homologs of the “ferritin-like” protein family, in order to improve annotation specificity (see Text S2 and Text S3).
Selected complete genomes were annotated with the abovementioned 17 HMM protein profiles using the hmmsearch program from the HMMER suite version 3.2.1 (Eddy 2009, 2011). To this end, a Snakemake workflow and Python scripts were designed, and the pandas and Biopython libraries were used (Cock et al. 2009; Köster and Rahmann 2012). Best matching profiles (best score) were selected when multiple profiles matched, and the hits were then extracted when having an i-evalue below 0.001 and a profile coverage (portion of the profile aligning with the hit) above 50%. The results of the annotation are displayed for all selected genomes in Table S1, and the HMM protein profiles are available in Data Set S1.
Selection of Representative Sequences
In most of the cases, sequence dereplication was applied to the set of annotated UQ-hydroxylases before running phylogenetic analyses. The CD-Hit program was used with varying parameters: 60% sequence identity clustering was applied for the Coq7 protein family and 50% sequence identity for the UQ-FMO protein family (Li and Godzik 2006). One sequence per cluster was then selected as representative sequence in include in phylogenies, with the priority given to experimentally validated sequences.
Phylogenetic Analyses
Annotated sequences were aligned by protein families using MAFFT (linsi), version 7.511 (Katoh et al. 2002; Katoh and Standley 2013). Sites were filtered from the obtained multiple sequence alignment using BMGE version 2 using the default parameters and the BLOSUM30 matrix (Criscuolo and Gribaldo 2010). Maximum likelihood phylogenetic trees were computed from the filtered multiple sequence alignments using the IQ-Tree software, version 2.2.0 (Hoang et al. 2018; Minh et al. 2020). Both aLRT and UFBoot supports were computed (1,000 replicates for each), and the best model was assessed for each alignment using the default parameters. Upon first phylogenetic tree reconstruction, sets of sequences provoking long branches (typically >1 substitutions per site) were removed and alignments and trees were reconstructed. For each final alignment analyzed, 11 iterations of IQ-Tree were ran, the first run being the one used for the selection of the best model of sequence evolution that was subsequently reused for the 10 additional runs. All obtained phylogenetic trees were manually inspected, and a representative one displaying high supports in its deep nodes was selected for illustration and annotated with iTOL (Letunic and Bork 2021) and Inkscape version 1.3.1 (https://inkscape.org/). Species trees on Fig. 3 and Fig. S2 were decorated with UQ-hydroxylase combinations and proportions using the ETE 3 Python library (Huerta-Cepas et al. 2016). All trees and iTOL annotation files are available in Data Set S2. All trees presented in main text figures are available annotated with the corresponding species names in Data Set S3.
FMO Outgroup
Sequences from the class A of the FMO superfamily were selected as the most closely related FMO according to a previous evolutionary study (Mascotti et al. 2016). All sequences from Table S2 of that article were included as outgroup in rooted UQ-FMO phylogenies, except for that of Mus musculus (Uniprot sequence ID “Q8VDP3,” [F-actin]-monooxygenase MICAL1) that presented a much longer sequence than the other FMOs. The list of the sequence identifiers of the included FMO sequences is the following: P00438, P20586, Q93NG3, A6T923, Q0SFK6, Q9HWG9, Q93L51, P38169, Q54530, Q93LY7, Q8KI25, P15245, and Q6SSJ6. Their full denomination (enzyme description and organism) is presented in Data Set S3.
Strain Construction
All bacterial strains used in this study are listed in Table S7. The ΔubiH ΔubiF::kan double mutant was constructed by P1 transduction using MG1655 ΔubiHc as the recipient strain and the JW0659 from the Keio collection (Baba et al. 2006) as the donor strain (https://barricklab.org/twiki/bin/view/Lab/ProtocolsP1Transduction). The presence of the ubiF::kan mutation was confirmed by colony polymerase chain reaction (PCR) with primers flanking the ubiF locus.
Cloning and Plasmid Construction
The plasmids and the primers used in this study are listed in Tables S8 and S9 (Supplementary material), respectively. pBAD24i was kindly constructed by Dr Laurent Loiseau (IMM, Marseille) by modifying the multiple cloning site (MCS) of pBAD24 (Guzman et al. 1995) from GGAATTCACCATGGTACCCG into GGAAATCACCATGGAATTCCCG in order to move the EcoRI site from 5′ to 3′ of the NcoI site. pBAD33i was constructed from pBAD33 (Guzman et al. 1995) by replacing its MCS with that of pBAD24i using EcoRV and HindIII restriction enzymes. In order to subclone the genes of interest in the MCS of pBAD33i by NcoI and HindIII restriction enzymes, a NcoI site was eliminated from chloramphenicol resistant (cat) gene of pBAD33i. To do so, a point mutation was introduced in the cat gene on pBAD33i by overlap PCR using two pairs of primer, CAT ss NCO 5/CAT ss NCO 3 and CAT BeaG 5/CAT Bpu10I (Table S9).
The hydroxylase genes experimentally tested in this study are listed in Table S5. The genes were synthesized by the “Proteogenix” and “Genecust” companies and cloned into pBAD24i or pBAD33i vector downstream of arabinose-inducible promotor using NcoI (5′ end) and HindIII (3′ end) restriction enzymes. The nucleotide sequences were optimized for expression in E. coli and are available in Data Set S4. The ATG start codon of the hydroxylase genes is comprised in the NcoI site sequence (CCATGG), which imposes a G as the first nucleotide of the second codon. In case the second amino acid did not correspond to a codon starting with a G, an alanine codon (GCG) was added before the second codon of the nucleotide sequence. Alanine was chosen as it shows minimal adverse effects (Bivona et al. 2010), and the sequences with an additional alanine in position 2 are listed in Table S8. The absence or presence of an extra alanine in position 2 of tested sequences is also reported in Table S2.
Growth Conditions for In Vivo Complementation Assays
The E. coli mutant strains inactivated for one, two, or three UQ-hydroxylase(s) (Table S7) were transformed by pBAD24i or pBAD33i empty vectors or pBAD carrying the genes of interest and selected on lysogeny broth (LB) plates supplemented with either Ampicillin (100 µg/mL) or chloramphenicol (30 µg/mL). Individual clones were inoculated in 1.5 mL LB medium with the appropriate antibiotic and grown overnight at 37°C in closed Eppendorf tubes. These precultures were used to inoculate, at OD600 ∼ 0.02, cultures of 5 mL LB medium containing 0.05% (wt/vol) arabinose and the appropriate antibiotic. The aerobic cultures were grown overnight in 30-mL glass tubes at 37°C with 180 rpm shaking. The 5 mL cultures were cooled on ice for at least 30 min before centrifugation at 3,200 × g at 4°C for 10 min. Cell pellets were washed in 1 mL ice-cold phosphate-buffer saline (PBS) and transferred to preweighted 1.5-mL Eppendorf tubes. After centrifugation at 12,000 × g at 4°C for 1 min, the supernatant was discarded, the cell wet weight was determined, and pellets were stored at −20°C until lipid extraction, if necessary.
Lipid Extraction and Quinone Analysis
Quinone extraction from cell pellets was performed as previously described (Hajj Chehade et al. 2013). The dried lipid extracts were resuspended in 100 µL ethanol, and a volume corresponding to 1 mg of cell wet weight was analyzed by HPLC ECD-MS with a BetaBasic-18 column at a flow rate of 1 mL/min with a mobile phase composed of 50% methanol, 40% ethanol, and 10% of a mix (90% isopropanol, 10% ammonium acetate [1 M], and 0.1% formic acid). When necessary, MS detection was performed with a MSQ spectrometer (Thermo Scientific) with electrospray ionization in positive mode (probe temperature, 400°C; cone voltage, 80 V). Single-ion monitoring detected the following compounds: UQ8 (M + NH4+), m/z 744 to 745, 6 to 10 min, scan time of 0.2 s; UQ10 (M + NH4+), m/z 880 to 881, 10 to 17 min, scan time of 0.2 s; DMQ8 (M + NH4+), m/z 714 to 715 to 10 min, scan time of 0.4 s; OPP (M + NH4+), 656 to 657, 5 to 9 min, scan time of 0.4 s; and 4-HP8 (M + NH4+), 670 to 671, 6 to 10 min, scan time of 0.4 s. MS spectra were recorded between m/z 600 and 900 with a scan time of 0.3 s.
ECD and MS peak areas were corrected for sample loss during extraction on the basis of the recovery of the UQ10 internal standard and were then normalized to cell wet weight. The peaks of UQ8 and the biosynthetic intermediates obtained with ECD were quantified with a standard curve of UQ10 as previously described (Hajj Chehade et al. 2013). The absolute quantification of UQ8 based on the m/z 744 to 745 signal at 7.8 min was performed with a standard curve of UQ8 ranging from 6.25 to 50 pmol UQ8.
Statistical Analyses of Experimental Results
We normalized the experimental data using a log transformation and performed quality checks. Then, for each gene and each genetic background, we assessed the significance of the encoded activity (DMQ8 or Q8) with a t-test (Table S5). We corrected for multiple tests using Bonferroni (Bonferroni 1936) and considered significance at a threshold of 0.05. We thus controlled the overall family-wise error rate. We then computed the fold change for each gene/genetic background and categorized the fold change in four categories: between 0% and 2%, between 2% and 25%, between 25% and 75%, and above 75%. We define 100% as (i) the level of Q8 in the wild-type strain and (ii) the level of DMQ8 in the ΔubiF strain containing an empty vector. Due to the different genetic backgrounds, a protein can be tested several times for hydroxylation at a specific position (C1, C5, or C6). We aggregated the results to obtain one corrected P-value/fold change per protein and position by considering the highest activity.
Supplementary material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Alexandre de Brevern and members of Isabelle André's group for discussion. We appreciate the constructive comments provided by Eduardo Rocha, Jérémy Esque, and Joël Gaffé on an earlier version of this manuscript. We are very grateful to John Willison for improving the language of the manuscript. This project was supported by the Université Grenoble Alpes and the French National Research Agency (ANR, Agence Nationale de la Recherche) in the framework of the “Investissements d’avenir” program (ANR-15-IDEX-02, “Grenoble Alpes Data Institute,” and IDEX-IRS 2020 call) and the CNRS (Centre National de la Recherche Scientifique) and by two grants from the ANR: QUINEVOL (grant agreement ANR-21-CE02-0018) and DEEPen (grant agreement ANR-19-CE45-0013).
Conflict of interest. None declared.
Contributor Information
Katayoun Kazemzadeh, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Ludovic Pelosi, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Clothilde Chenal, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Sophie-Carole Chobert, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Mahmoud Hajj Chehade, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Margaux Jullien, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Laura Flandrin, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
William Schmitt, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Qiqi He, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Emma Bouvet, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Manon Jarzynka, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Nelle Varoquaux, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Ivan Junier, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Fabien Pierrel, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Sophie S Abby, Univ. Grenoble Alpes, CNRS, UMR 5525, VetAgro Sup, Grenoble INP, TIMC, 38000 Grenoble, France.
Data Availability
The data underlying this article are available in the article and in its online Supplementary material. The HMM protein profiles are available as Data Set S1 on Figshare (DOI: 10.6084/m9.figshare.23230421 available at https://figshare.com/s/cada5807e393cc6888c8). The multiple sequence alignments and corresponding phylogenetic trees are available as Data Set S2 on Figshare (DOI: 10.6084/m9.figshare.23230562 available at https://figshare.com/s/fbe913faabb2ba08253b).
References
- Abby SS, Kazemzadeh K, Vragniau C, Pelosi L, Pierrel F. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim Biophys Acta Bioenerg. 2020:1861(11):148259. 10.1016/j.bbabio.2020.148259. [DOI] [PubMed] [Google Scholar]
- Abby SS, Tannier E, Gouy M, Daubin V. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A. 2012:109(13):4962–4967. 10.1073/pnas.1116871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta Bioenerg. 2014:1837(7):1004–1011. 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006:2(1):2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann BO. Applied evolutionary theories for engineering of secondary metabolic pathways. Curr Opin Chem Biol. 2016:35:133–141. 10.1016/j.cbpa.2016.09.021. [DOI] [PubMed] [Google Scholar]
- Baier F, Copp JN, Tokuriki N. Evolution of enzyme superfamilies: comprehensive exploration of sequence–function relationships. Biochemistry 2016:55(46):6375–6388. 10.1021/acs.biochem.6b00723. [DOI] [PubMed] [Google Scholar]
- Balabanova L, Averianova L, Marchenok M, Son O, Tekutyeva L. Microbial and genetic resources for cobalamin (vitamin B12) biosynthesis: from ecosystems to industrial biotechnology. Int J Mol Sci. 2021:22(9):4522. 10.3390/ijms22094522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona L, Zou Z, Stutzman N, Sun PD. Influence of the second amino acid on recombinant protein expression. Protein Expr Purif. 2010:74(2):248–256. 10.1016/j.pep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE. 1936. Teoria statistica delle classi e calcolo delle probabilità. Seeber Available from: https://books.google.fr/books?id=3CY-HQAACAAJ
- Cevallos MA, Degli Esposti M. New alphaproteobacteria thrive in the depths of the ocean with oxygen gradient. Microorganisms 2022:10(2):455. 10.3390/microorganisms10020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009:25(11):1422–1423. 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010:10(1):210. 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 2018:558(7710):440–444. 10.1038/s41586-018-0207-y. [DOI] [PubMed] [Google Scholar]
- de Crécy-Lagard V. Variations in metabolic pathways create challenges for automated metabolic reconstructions: examples from the tetrahydrofolate synthesis pathway. Comput Struct Biotechnol J. 2014:10(16):41–50. 10.1016/j.csbj.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli Esposti M. A journey across genomes uncovers the origin of ubiquinone in Cyanobacteria. Genome Biol Evol. 2017:9(11):3039–3053. 10.1093/gbe/evx225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denise R, Babor J, Gerlt JA, de Crécy-Lagard V. Pyridoxal 5'-phosphate synthesis and salvage in Bacteria and Archaea: predicting pathway variant distributions and holes. Microbial Genomics 2023:9(2):mgen000926. 10.1099/mgen.0.000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009:23:205–211. 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011:7(10):e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Wu D, Goremykin V, Xiao J, Xu Y, Garg S, Zhang C, Martin WF, Zhu R. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria. Nat Ecol Evol. 2020:4(9):1213–1219. 10.1038/s41559-020-1239-x. [DOI] [PubMed] [Google Scholar]
- Forsberg Z, Bissaro B, Gullesen J, Dalhus B, Vaaje-Kolstad G, Eijsink VGH. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J Biol Chem. 2018:293(4):1397–1412. 10.1074/jbc.M117.817130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franza T, Gaudu P. Quinones: more than electron shuttles. Res Microbiol. 2022:173(6-7):103953. 10.1016/j.resmic.2022.103953. [DOI] [PubMed] [Google Scholar]
- Glasner ME, Truong DP, Morse BC. How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation. FEBS J. 2020:287(7):1323–1342. 10.1111/febs.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995:177(14):4121–4130. 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj Chehade M, Loiseau L, Lombard M, Pecqueur L, Ismail A, Smadja M, Golinelli-Pimpaneau B, Mellot-Draznieks C, Hamelin O, Aussel L, et al. Ubii, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J Biol Chem. 2013:288(27):20085–20092. 10.1074/jbc.M113.480368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj Chehade M, Pelosi L, Fyfe CD, Loiseau L, Rascalou B, Brugière S, Kazemzadeh K, Vo CD, Ciccone L, Aussel L, et al. A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem Biol. 2019:26(4):482–492.e7. 10.1016/j.chembiol.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Hansen CC, Nelson DR, Møller BL, Werck-Reichhart D. Plant cytochrome P450 plasticity and evolution. Mol Plant. 2021:14(8):1244–1265. 10.1016/j.molp.2021.06.028. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018:35(2):518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hördt A, López MG, Meier-Kolthoff JP, Schleuning M, Weinhold L-M, Tindall BJ, Gronow S, Kyrpides NC, Woyke T, Göker M. Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of alphaproteobacteria. Front Microbiol. 2020:11:468. 10.3389/fmicb.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gogarten JP. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends Genet. 2006:22(7):361–366. 10.1016/j.tig.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016:33(6):1635–1638. 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman V, Toledo-Patiño S, Noda-García L, Laurino P. Mechanisms of protein evolution. Protein Sci. 2022:31(7):e4362. 10.1002/pro.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H-X, Wang J, Zhou L, Jin Z-J, Cao X-Q, Liu H, Chen H-F, He Y-W. Coenzyme Q biosynthesis in the biopesticide shenqinmycin-producing Pseudomonas aeruginosa strain M18. J Ind Microbiol Biotechnol. 2019:46(7):1025–1038. 10.1007/s10295-019-02179-1. [DOI] [PubMed] [Google Scholar]
- Kalluraya CA, Weitzel AJ, Tsu BV, Daugherty MD. Bacterial origin of a key innovation in the evolution of the vertebrate eye. Proc Natl Acad Sci U S A. 2023:120(16):e2214815120. 10.1073/pnas.2214815120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002:30(14):3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai M. Biosynthesis and applications of prenylquinones. Biosci Biotechnol Biochem. 2018:82(6):963–977. 10.1080/09168451.2018.1433020. [DOI] [PubMed] [Google Scholar]
- Kazemzadeh K, Hajj Chehade M, Hourdoir G, Brunet CD, Caspar Y, Loiseau L, Barras F, Pierrel F, Pelosi L. The biosynthetic pathway of ubiquinone contributes to pathogenicity of Francisella novicida. J Bacteriol. 2021:203(23):e00400-21. 10.1128/JB.00400-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, White-Phillip JA, Ogasawara Y, Shin N, Isiorho EA, Liu H. Biosynthesis of spinosyn in Saccharopolyspora spinosa : synthesis of permethylated rhamnose and characterization of the functions of SpnH, SpnI, and SpnK. J Am Chem Soc. 2010:132(9):2901–2903. 10.1021/ja910223x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschning A. On the evolution of coenzyme biosynthesis. Nat Prod Rep. 2022:39(11):2175–2199. 10.1039/D2NP00037G. [DOI] [PubMed] [Google Scholar]
- Kong L, Wang Q, Yang W, Shen J, Li Y, Zheng X, Wang L, Chu Y, Deng Z, Chooi Y-H, et al. Three recently diverging duplicated methyltransferases exhibit substrate-dependent regioselectivity essential for xantholipin biosynthesis. ACS Chem. Biol. 2020:15(8):2107–2115. 10.1021/acschembio.0c00296. [DOI] [PubMed] [Google Scholar]
- Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012:28(19):2520–2522. 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- Kramer J, Özkaya Ö, Kümmerli R. Bacterial siderophores in community and host interactions. Nat Rev Microbiol. 2020:18(3):152–163. 10.1038/s41579-019-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O, Kotsakis A, Meganathan R. Ubiquinone (coenzyme Q) biosynthesis in Escherichia coli: identification of the ubiF gene. FEMS Microbiol Lett. 2000:186(2):157–161. 10.1111/j.1574-6968.2000.tb09097.x. [DOI] [PubMed] [Google Scholar]
- Latimer S, Keene SA, Stutts LR, Berger A, Bernert AC, Soubeyrand E, Wright J, Clarke CF, Block AK, Colquhoun TA, et al. A dedicated flavin-dependent monooxygenase catalyzes the hydroxylation of demethoxyubiquinone into ubiquinone (coenzyme Q) in Arabidopsis. J Biol Chem. 2021:297(5):101283. 10.1016/j.jbc.2021.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021:49(W1):W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006:22(13):1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liao H, Lin X, Li Y, Qu M, Tian Y. Reclassification of the taxonomic framework of orders Cellvibrionales, Oceanospirillales, Pseudomonadales, and Alteromonadales in class Gammaproteobacteria through phylogenomic tree analysis. mSystems 2020:5(5):e00543-20. 10.1128/mSystems.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 2018:557(7703):101–105. 10.1038/s41586-018-0059-5. [DOI] [PubMed] [Google Scholar]
- Mascotti ML, Juri Ayub M, Furnham N, Thornton JM, Laskowski RA. Chopping and changing: the evolution of the flavin-dependent monooxygenases. J Mol Biol. 2016:428(15):3131–3146. 10.1016/j.jmb.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AJ. Evolution of biosynthetic diversity. Biochem J. 2017:474(14):2277–2299. 10.1042/BCJ20160823. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020:37(5):1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, Galbraith ED, Geider RJ, Guieu C, Jaccard SL, et al. Processes and patterns of oceanic nutrient limitation. Nature Geosci. 2013:6(9):701–710. 10.1038/ngeo1765. [DOI] [Google Scholar]
- Morel FMM, Price NM. The biogeochemical cycles of trace metals in the oceans. Science 2003:300(5621):944–947. 10.1126/science.1083545. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, Hess S, Burger G, Lang BF, Susko E, Slamovits CH, Roger AJ. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. eLife 2019:8:e42535. 10.7554/eLife.42535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, Susko E, Williamson K, Eme L, Slamovits CH, Moreira D, López-García P, Roger AJ. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria. Nat Ecol Evol. 2022:6(3):253–262. 10.1038/s41559-021-01638-2. [DOI] [PubMed] [Google Scholar]
- Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010:1797(9):1587–1605. 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Paoli L, Ruscheweyh H-J, Forneris CC, Hubrich F, Kautsar S, Bhushan A, Lotti A, Clayssen Q, Salazar G, Milanese A, et al. Biosynthetic potential of the global ocean microbiome. Nature 2022:607(7917):111–118. 10.1038/s41586-022-04862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Chaumeil P, Rinke C, Mussig AJ, Hugenholtz P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020:38(9):1079–1086. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022:50(D1):D785–D794. 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018:36(10):996–1004. [DOI] [PubMed] [Google Scholar]
- Patallo EP, Blanco G, Fischer C, Braña AF, Rohr J, Méndez C, Salas JA. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. J Biol Chem. 2001:276(22):18765–18774. 10.1074/jbc.M101225200. [DOI] [PubMed] [Google Scholar]
- Pelosi L, Ducluzeau AL, Loiseau L, Barras F, Schneider D, Junier I, Pierrel F. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 2016:1(4):e00091-16. 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi L, Vo CD, Abby SS, Loiseau L, Rascalou B, Hajj Chehade M, Faivre B, Goussé M, Chenal C, Touati N, et al. Ubiquinone biosynthesis over the entire O2 range: characterization of a conserved O2-independent pathway. MBio 2019:10(4):e01319-19. 10.1128/mBio.01319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA. 2014:111(50):17803–17808. 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Muñoz-Gómez SA, Kamikawa R. The origin and diversification of mitochondria. Curr Biol. 2017:27(21):R1177–R1192. 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Salzberg SL. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019:20(1):92. 10.1186/s13059-019-1715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020:2020:baaa062. 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, Lieutaud C, Baymann F, Vermeglio A, Friedrich T, Kramer DM, Nitschke W. Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci U S A. 2009:106(21):8549–8554. 10.1073/pnas.0813173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, Huet G, Constant P, Malaga W, Lemassu A, Laval F, Daffé M, Guilhot C, Chalut C. Functional characterisation of three O-methyltransferases involved in the biosynthesis of phenolglycolipids in Mycobacterium tuberculosis. PLoS One 2013:8(3):e58954. 10.1371/journal.pone.0058954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Scheuner C, Göker M, Klenk H-P. A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front Microbiol. 2015:6:281. 10.3389/fmicb.2015.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Grünler J, Mattsson J, Sindelar PJ, Nordlund P, Berthold DA. A new member of the family of Di-iron carboxylate proteins. J Biol Chem. 2001:276(36):33297–33300. 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- Szöllősi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. Efficient exploration of the space of reconciled gene trees. Syst Biol. 2013:62(6):901–912. 10.1093/sysbio/syt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu VV, Hangasky JA, Detomasi TC, Henry SJW, Ngo ST, Span EA, Marletta MA. Substrate selectivity in starch polysaccharide monooxygenases. J Biol Chem. 2019:294(32):12157–12166. 10.1074/jbc.RA119.009509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. Phylogeny of Gammaproteobacteria. J Bacteriol. 2010:192(9):2305–2314. 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol. 2013:63(Pt_8):2901–2906. 10.1099/ijs.0.049270-0. [DOI] [PubMed] [Google Scholar]
- Williams TA, Szöllősi GJ, Spang A, Foster PG, Heaps SE, Boussau B, Ettema TJG, Embley TM. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci U S A. 2017:114(23):E4602–E4611. 10.1073/pnas.1618463114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-J, Zhang X-F, Jiang Y, Fan H, Li J-X, Li C-Y, Zhao Q, Yang L, Hu Y-H, Martin C, et al. A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes. Sci Adv. 2021:7(50):eabl3594. 10.1126/sciadv.abl3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R, Harrison K, Kolaczkowski B, De Crécy-Lagard V. Functional annotations of paralogs: a blessing and a curse. Life 2016:6(3):39. 10.3390/life6030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi XY, Yao JC, Tang SK, Huang Y, Li HW, Li WJ. The futalosine pathway played an important role in menaquinone biosynthesis during early prokaryote evolution. Genome Biol Evol. 2014:6(1):149–160. 10.1093/gbe/evu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Li M, Wang X-Y, Liu H, Sun S, Chen H, Poplawsky A, He Y-W. Biosynthesis of coenzyme Q in the phytopathogen Xanthomonas campestris via a yeast-like pathway. Mol Plant Microbe Interact. 2019:32(2):217–226. 10.1094/MPMI-07-18-0183-R. [DOI] [PubMed] [Google Scholar]
- Zou T, Risso VA, Gavira JA, Sanchez-Ruiz JM, Ozkan SB. Evolution of conformational dynamics determines the conversion of a promiscuous generalist into a specialist enzyme. Mol Biol Evol. 2015:32(1):132–143. 10.1093/molbev/msu281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online Supplementary material. The HMM protein profiles are available as Data Set S1 on Figshare (DOI: 10.6084/m9.figshare.23230421 available at https://figshare.com/s/cada5807e393cc6888c8). The multiple sequence alignments and corresponding phylogenetic trees are available as Data Set S2 on Figshare (DOI: 10.6084/m9.figshare.23230562 available at https://figshare.com/s/fbe913faabb2ba08253b).