Abstract
Virginiamycin M1 (VM1), produced by Streptomyces virginiae, is a polyunsaturated macrocyclic lactone antibiotic belonging to the virginiamycin A group. S. virginiae possesses an activity which stereospecifically reduces a 16-carbonyl group of VM1, resulting in antibiotically inactive 16R-dihydroVM1. The corresponding VM1 reductase was purified to homogeneity from crude extracts of S. virginiae in five steps, with 5,650-fold purification and 23% overall yield. The N-terminal amino acid sequence was determined to be MAIKLVIA. The purified enzyme showed an apparent Mr of 73,000 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and an Mr of 280,000 by native molecular sieve high-performance liquid chromatography, indicating the tetrameric nature of the native enzyme. NADPH served as a coenzyme for the reduction, with a Km value of 0.13 mM, but NADH did not support the reaction, even at a concentration of 5 mM, indicating the NADPH-specific nature of the enzyme. The Km for VM1 was determined to be 1.5 mM in the presence of 2 mM NADPH. In the reverse reaction, only 16R-dihydroVM1, not the 16S-epimer, served as a substrate, with a less than 0.1% overall reaction rate compared to that of the forward reaction, confirming that the VM1 reductase participates solely in VM1 inactivation in vivo.
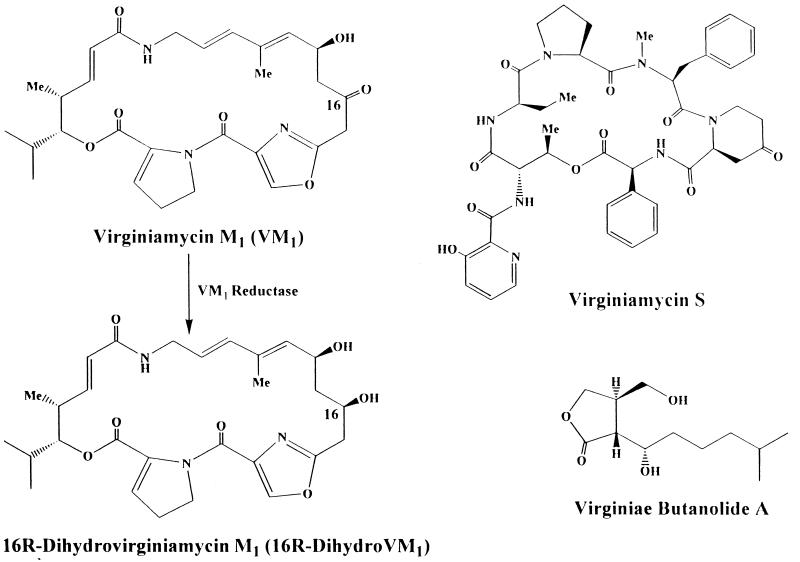
The antibiotic virginiamycin, produced by Streptomyces virginiae, is a member of the virginiamycin family (alternatively designated as the streptogramin family or the mikamycin family) (2). The characteristic feature of the family is that each member is produced as a mixture of two structurally different compounds exhibiting a synergistic antibacterial activity. They belong to one of the two following distinct groups: type A, exemplified by virginiamycin M1 (VM1), corresponding to polyunsaturated macrocyclic peptolides (Fig. 1); and type B, exemplified by virginiamycin S, corresponding to branched cyclic hexadepsipeptide. Each compound reversibly blocks protein synthesis and displays only bacteriostatic activity. However, in combination, they demonstrate a synergistic bactericidal activity at much lower concentrations. The virginiamycin family, which has been known for more than 30 years (3), has been used successfully as a performance promoter in animal husbandry (1).
FIG. 1.
Structures of VM1, virginiamycin S, and a VB and conversion of VM1 to 16R-dihydroVM1. Me, methyl.
Studies of antibiotic inactivation by producer microorganisms are important for understanding the producer’s defense mechanism against antibiotics and providing clues for locating the gene clusters necessary for antibiotic biosynthesis. However, despite the long history of virginiamycin production by S. virginiae, no enzymatic study of the inactivation of virginiamycin has been reported. Previously, we have confirmed that S. virginiae inactivates VM1 by reducing its 16-carbonyl group, stereospecifically forming 16R-dihydroVM1 (Fig. 1) (8), while leaving virginiamycin S intact. In this study, we have purified the corresponding enzyme, VM1 reductase, 5,650-fold from the crude cell extract of S. virginiae and characterized it.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
S. virginiae MAFF 10-06014 (National Food Research Institute, Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan) was used throughout this study. It was grown at 28°C for 40 h as previously described (4, 5).
Chemicals.
To obtain pure VM1, fodder additive STAFAC 500 (0.5 g; Smith Kline-RIT, Rixensart, Belgium) was suspended in 100 ml of acetonitrile, and the suspension was incubated for 16 h at 120 rpm and 30°C. After filtration with a glass filter (5-μm-pore diameter; Whatman), the filtrate was evaporated to dryness, and VM1 in the residue was purified by C18 reverse-phase high-performance liquid chromatography (HPLC) (Cosmosil 5C18-AR; 10 by 250 mm; Nacalai Tesque, Kyoto, Japan) with 50% CH3CN in water as the mobile phase at room temperature and at a flow rate of 3 ml/min with detection at 305 nm. VM1 and virginiamycin S were eluted at 11.0 and 18.0 min, respectively. Authentic 16R-dihydroVM1 and its 16S-epimer were prepared by chemical reduction with NaBH4 and purified by C18 reverse-phase HPLC as described previously (8). Their identity was confirmed by mass spectrometry and 600-MHz 1H-nuclear magnetic resonance spectrometry.
Assay of VM1 reductase activity.
The VM1 reductase activity during the purification was routinely monitored by the addition of 1 μl of VM1 (10 mg/ml in ethanol) to the reaction mixture containing an appropriate amount of enzyme and 2 mM NADPH in 0.05 M triethanolamine-HCl (TEA) (pH 7.5) for a total volume of 100 μl. For measuring total activity, pH optimum, and Km for NADPH, the concentration of VM1 was raised to 9.1 mM (5-μl addition of a 100-mg/ml VM1 solution) to ensure maximum activity. Due to the relatively nonpolar nature of VM1, 9.1 mM is nearly a saturating concentration in the buffer. The activities in 0.05 M potassium phosphate (pH 7.5), 0.05 M Tris-HCl (pH 7.5), and 0.05 M 3-(N-morpholine)propanesulfonic acid–KOH (pH 7.5) were 72, 123, and 91%, respectively, relative to that in 0.05M TEA (pH 7.5). Because Tris-HCl buffer gave high background A210 during HPLC detection, TEA was selected as the buffer. Incubation was carried out at 28°C and at a rate of 40 strokes per min in a reciprocating water bath. At appropriate times, the reaction was stopped by the addition of 400 μl of ethanol, and denatured proteins were removed by centrifugation (11,000 × g, 5 min). 16R-DihydroVM1 in the supernatant was measured by C18 reverse-phase HPLC (Cosmosil 5C18 column; 4.6 by 250 mm, detection at 210 nm, flow rate at 1.0 ml/min, isocratic elution with 40% CH3CN in water) with authentic 16R-dihydroVM1 as a standard. The elution times for VM1 and 16R-dihydroVM1 were 5.8 and 9.5 min, respectively. One unit of enzyme activity is defined as the amount of enzyme necessary to produce 1 μmol of 16R-dihydroVM1 per min at 28°C.
Protein content was determined with a Bio-Rad protein assay kit, unless otherwise stated, with bovine serum albumin as a standard.
Purification of VM1 reductase.
All of the purification procedures were carried out at 0 to 4°C unless otherwise indicated.
(i) Step 1.
Preparation of cell-free extract. In step 1, cells (100 g [wet weight]) from 8 liters of culture were suspended in 500 ml of 0.05 M TEA (pH 7.5) and disrupted by sonication. Cell debris was removed by centrifugation at 6,000 × g for 20 min.
(ii) Step 2. Ammonium sulfate fractionation.
To the supernatant from step 1, solid ammonium sulfate was added with gentle stirring to attain 60% saturation while keeping the pH at 7.0. After standing overnight, the precipitate was collected by centrifugation at 12,000 × g for 20 min and dissolved in 200 ml of 0.02 M TEA (pH 7.0). The solution was dialyzed against two changes of the same buffer (5 liters) to remove the ammonium sulfate.
(iii) Step 3. DEAE-Sephacel chromatography.
The dialyzed solution from step 2 was applied to a DEAE-Sephacel column (4.5 by 65 cm) previously equilibrated with 0.02 M TEA (pH 7.0). After being washed with 2 liters of the same buffer, the protein was successively eluted with 3 liters of 0.02 M TEA (pH 7.0) containing 0.1 M KCl, 5 liters of 0.02 M TEA (pH 7.0) containing 0.2 M KCl, and finally 3.3 liters of 0.02 M TEA (pH 7.0) containing 0.3 M KCl. The column was eluted at a flow rate of 340 ml/h, and 18-ml fractions were collected. VM1 reductase activity was eluted at 0.2 M KCl, and fractions 65 to 120 were pooled and concentrated by ultrafiltration (UK-20; Advantech Toyo).
(iv) Step 4. Gel filtration on Sepharose CL-6B column.
The concentrated solution from step 3 (30 ml) was applied to a Sepharose CL-6B column (5.5 by 65 cm), preequilibrated with 0.05 M TEA (pH 7.0) containing 0.3 M KCl, and eluted with the same buffer in fractions of 10 ml at a flow rate of 340 ml/h. Active fractions (fractions 78 to 105) were pooled and concentrated to 50 ml by ultrafiltration.
(v) Step 5. Affinity chromatography on a HiTrap Blue column.
The concentrated solution from step 4 was purified in aliquots of 2 ml (7.5 mg protein) per run on a HiTrap Blue column (1.0 by 3.0 cm; Pharmacia LKB) with a linear gradient of NADPH concentration from 0 to 10 mM (0.5 mM/min) in 0.05 M TEA (pH 7.0) containing 0.1 M KCl at a flow rate of 0.5 ml/min. Protein in the eluate was monitored with a fluorescence detector (excitation, 280 nm; emission, 340 nm; model FP-210, Japan Spectroscopic Co., Ltd.). VM1 reductase activity appeared at around 26 min.
The samples’ purity at each purification step was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a ready-made 4 to 20% gradient gel (Daiichi Pure Chemicals Co., Ltd.) with a mini-gel apparatus. Proteins in the gel were detected by a silver stain kit (Wako Pure Chemicals) according to the manufacturer’s protocol. Marker proteins were obtained from Pharmacia LKB.
Molecular mass determination of the VM1 reductase.
Purified sample was injected into a molecular sieve HPLC column (TSK G3000 SWXL; Mr, ≤500,000; Toso Manufacturing Co., Ltd.), and eluted with 0.1 M potassium phosphate buffer (pH 7.0) containing 0.2 M NaCl at a flow rate of 0.5 ml/min. Fractions were collected every 15 s and assayed for VM1 reductase. The calibration curve was made from the elution positions of HPLC marker proteins (rabbit muscle phosphorylase b, Mr, 94,000; bovine serum albumin, Mr, 67,000; egg ovalbumin, Mr, 43,000) purchased from Oriental Yeast Co., Ltd.
N-terminal amino acid sequencing.
The N-terminal amino acid sequence of purified enzyme was analyzed with an automated protein sequencer (Applied Biosystems 476A protein sequencer). The N-terminal amino acid sequence was compared to other protein sequences in the SwissProt, PIR, and PRF databases, but no significant homology was found.
RESULTS AND DISCUSSION
Purification of VM1 reductase.
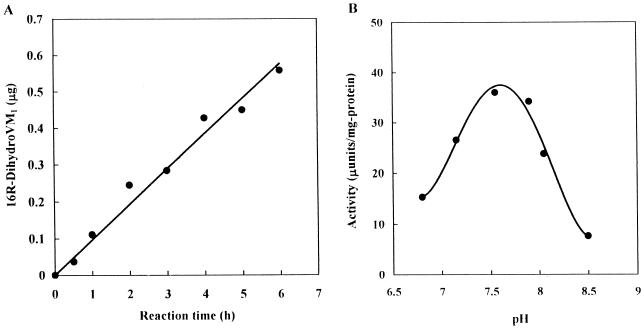
To characterize an enzyme that catalyzes the reduction of the 16-carbonyl group in VM1, we first had to establish a sensitive assay procedure, because preliminary experiments indicated that VM1 reductase activity in S. virginiae is very weak, with an activity of only several microunits per milligram of protein. This low level of activity prevented us from using conventional assays monitoring the VM1-dependent decay of NADPH or NADH. We have developed an easy but sensitive assay (detection limit of 1.6 μU or 0.05 μg of 16R-dihydroVM1 per h) based on the separation and detection of 16R-dihydroVM1 by C18 reverse-phase HPLC as described in Materials and Methods. Under the assay conditions, the enzyme reaction proceeded linearly for up to 6 h, as shown in Fig. 2A. Similarly, the practical pH optimum was determined to be at around 7.5 to 7.8 (Fig. 2B), although the instability and quick degradation of both VM1 and 16R-dihydroVM1 under acidic conditions could be the reason for the low activity at a pH lower than 7.
FIG. 2.
Dependence of VM1 reductase activity on reaction time (A) and pH (B). (A) The reaction was performed for the indicated time in the presence of 5 mM NADPH and 2 mM VM1 in 0.05 M TEA-HCl (pH 7.5). (B) The reaction was performed in 0.1 M TEA-HCl at the indicated pH in the presence of 2 mM NADPH. The pH after the reaction was confirmed to be the same as that before the reaction.
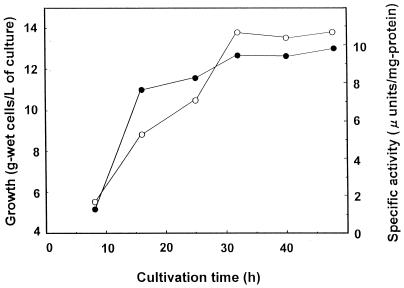
During cultivation of S. virginiae, the VM1 reductase activity was not detected in the culture broth (less than 0.2 μU/ml), while that in the mycelia increased with increasing cell mass and reached a plateau after 32 h of cultivation in parallel with the cessation of growth (Fig. 3). These characteristics suggest that the enzyme is intracellular and is produced constitutively, although we cannot exclude the possibility that some of the medium component might be the inducer of the enzyme. S. virginiae possesses a type of Streptomyces hormones called virginiae butanolides (VBs) which trigger virginiamycin production at nanomolar concentrations (7, 9, 10). Under our cultivation conditions, the production of VB starts at 11 h of cultivation, which in turn induced virginiamycin production with antibiotics first detected at 12.5 h of cultivation (6). Because the VM1 reductase activity was already present in the 9-h mycelia when neither VB nor virginiamycin was present (Fig. 3) and the external addition of VB did not increase the amount of the enzyme activity (our unpublished data), VM1 reductase seems to be produced constitutively, irrespective of VB or virginiamycin. To ensure the maximum amount of enzyme activity, mycelia harvested at 40 h were selected as the source of the enzyme.
FIG. 3.
Time course of VM1 reductase activity in the mycelia of S. virginiae. At the indicated time, a 500-ml culture was harvested. After sonication to disrupt cells, VM1 reductase activity in the crude cell extract was measured as described in Materials and Methods. Growth (•) is expressed as grams (wet weight) of cells per liter of culture. VM1 reductase activity (○) is expressed as microunits per milligram of protein.
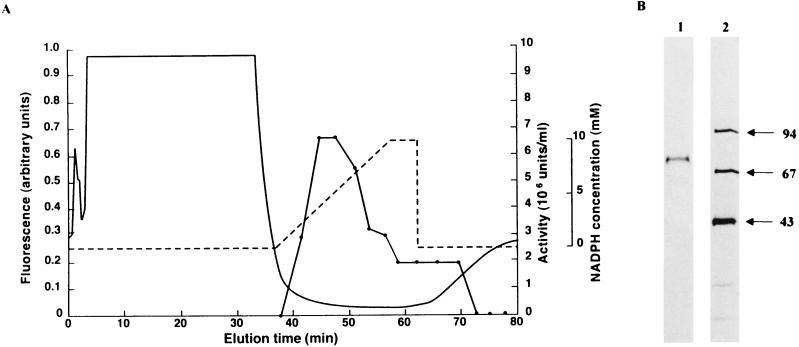
Starting from 100 g of 40-h mycelia, 0 to 60% ammonium-sulfate precipitation, DEAE-Sephacel ion-exchange chromatography, Sepharose CL-6B gel-filtration chromatography, and, finally, affinity chromatography on a HiTrap Blue column (Fig. 4A) gave a single band of protein (Fig. 4B). Affinity chromatography with the group-specific resin was very efficient, giving a more than 600-fold increase in enzyme specific activity (Table 1). It was so efficient that no protein peak was detected for the active fractions by means of the fluorescence detector. The protein content of the active fraction was later determined by reverse-phase HPLC on a C8 column with detection at 210 nm with bovine serum albumin as a standard.
FIG. 4.
Affinity chromatography HPLC of VM1 reductase on a HiTrap Blue column (A) and purity analysis of the sample by SDS-PAGE (B). (A) VM1 reductase activity (•). A typical elution profile with a 400-μl injection of the concentrate from step 3 was demonstrated. The gradient profile of NADPH concentration is indicated by a broken line. The protein concentration in the eluate was monitored by fluorescence (excitation, 280 nm; emission, 340 nm) (solid line). (B) Lane 1, purified enzyme (0.2 μg); lane 2, molecular mass standards. Numbers on the right indicate the size of standard proteins in kilodaltons.
TABLE 1.
Purification of VM1 reductase from S. virginiae starting from 100 g (wet weight) of mycelia
| Purification step | Total protein (mg) | Total activity (mU) | Sp act (mU/mg) | Purifica-tion factor (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude | 4,850 | 195 | 0.0402 | 1.00 | 100 |
| Ammonium sulfate | 3,010 | 115 | 0.0382 | 0.95 | 59.0 |
| DEAE-Sephacel | 353 | 68 | 0.198 | 4.93 | 35.9 |
| Sepharose 6B | 187 | 67 | 0.358 | 8.91 | 34.3 |
| HiTrap Blue | 0.19 | 45 | 227 | 5,650 | 23.1 |
Characterization of VM1 reductase.
The molecular mass of the enzyme under native conditions was determined to be 280,000 Da by gel filtration on a G3000SWXL HPLC column according to the elution position of the enzyme activity (data not shown). With the molecular mass of 73,000 Da by SDS-PAGE, VM1 reductase under native conditions is concluded to be a homotetramer.
The purified enzyme was analyzed for the amino acid sequence with an automated protein sequencer. Its N-terminal sequence was Met-Ala-Ile-Lys-Leu-Val-Ile-Ala. Only one amino acid was detected for each sequencing cycle, which further confirmed the purity of the enzyme.
To characterize its coenzyme specificity, the purified enzyme was assayed in the presence of 5 mM NADH or NADPH. No activity was detected with 5 mM NADH, while very clear activity was observed with NADPH, indicating that VM1 reductase was specific to NADPH. A conventional double reciprocal plot determined the Km for NADPH to be 0.13 mM in the presence of 9.1 mM VM1. Similarly, the Km values for VM1 and Vmax were determined to be 1.50 mM and 265 mU/mg of protein in the presence of 2 mM NADPH.
VM1 reductase catalyzes reduction of the 16-carbonyl group of VM1 in the forward reaction, but in the reverse reaction, it should catalyze the formation of VM1 from the inactive 16R-dihydroVM1. Therefore, it can be argued that major role of the enzyme may be in the biosynthesis of VM1, not in the self-defense against the antibiotics produced. To assess this possibility, the efficiency of the enzyme in the reverse reaction was measured with 16-dihydroVM1 and NADP+. Two epimers of 16-dihydroVM1 (16R-epimer and 16S-epimer) were prepared by the chemical reduction of VM1 with NaBH4, as described in Materials and Methods. With NADP+ concentrations ranging from 0.2 to 20 mM and a 16-dihydroVM1 concentration of 19 mM, only a trace amount of VM1 was produced from the 16R-epimer in the presence of 20 mM NADP+, but nothing was produced from the 16S-epimer (data not shown). Considering our detection limit for VM1, the forward reaction producing 16R-dihydroVM1 was favored more than 910-fold over the reverse reaction producing VM1, indicating that under physiological conditions, the enzyme should act in the self-defense pathway to inactivate VM1.
ACKNOWLEDGMENTS
This study was supported, in part, by the Proposal-Based Advanced Industrial Technology Development Organization (NEDO) of Japan, by the Research for the Future Program of the Japan Society for the Promotion of Science (JSPS), and by the Ministry of Agriculture, Forestry, and Fisheries of Japan (BMP-97-V-4-1-b).
REFERENCES
- 1.Biot A M. Virginiamycin: properties, biosynthesis, and fermentation. In: Vandamme E J, editor. Biotechnology of industrial antibiotics. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 695–720. [Google Scholar]
- 2.Cocito C. Antibiotics of the virginiamycin family, inhibitors which contain synergistic compounds. Microbiol Rev. 1979;43:145–198. doi: 10.1128/mr.43.2.145-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delpierre G R, Eastwood F W, Graeam G E, Kingston D G I, Sarin P S, Todd L, Williams D H. Antibiotics of the ostreogrycin complex. Part II. Structure of ostreogrycin A2. J Chem Soc. 1966;1966:1653–1669. doi: 10.1039/j39660001653. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto K, Nihira T, Yamada Y. Distribution of virginiae butanolides and IM-2 in the genus Streptomyces. J Ferment Bioeng. 1992;73:61–65. [Google Scholar]
- 5.Kim H S, Nihira T, Tada H, Yanagimoto M, Yamada Y. Identification of binding protein of virginiae butanolide C, an autoregulator in virginiamycin production, from Streptomyces virginiae. J Antibiot. 1989;42:769–778. doi: 10.7164/antibiotics.42.769. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J Bacteriol. 1997;179:6986–6993. doi: 10.1128/jb.179.22.6986-6993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo K, Higuchi Y, Sakuda S, Nihira T, Yamada Y. New virginiae butanolides from Streptomyces virginiae. J Antibiot. 1989;42:1873–1876. doi: 10.7164/antibiotics.42.1873. [DOI] [PubMed] [Google Scholar]
- 8.Lee C-K, Minami M, Sakuda S, Nihira T, Yamada Y. Stereospecific reduction of virginiamycin M1 as the virginiamycin resistance pathway in Streptomyces virginiae. Antimicrob Agents Chemother. 1996;40:595–601. doi: 10.1128/aac.40.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada Y, Sugamura K, Kondo K, Yanagaimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 10.Yamada Y, Nihira T, Sakuda S. Butyrolactone autoregulators, inducers of virginiamycin in Streptomyces virginiae: their structures, biosynthesis, receptor protein and induction of virginiamycin biosynthesis. In: Strohl W R, editor. Biotechnology of antibiotics. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 63–80. [Google Scholar]