Summary
Background
Inter-individual differences in response to immune checkpoint inhibitors (ICI) remain a major challenge in cancer treatment. The composition of the gut microbiome has been associated with differential ICI outcome, but the underlying molecular mechanisms remain unclear, and therapeutic modulation challenging.
Methods
We established an in vivo model to treat C57Bl/6j mice with the type-I interferon (IFN-I)-modulating, bacterial-derived metabolite desaminotyrosine (DAT) to improve ICI therapy. Broad spectrum antibiotics were used to mimic gut microbial dysbiosis and associated ICI resistance. We utilized genetic mouse models to address the role of host IFN-I in DAT-modulated antitumour immunity. Changes in gut microbiota were assessed using 16S-rRNA sequencing analyses.
Findings
We found that oral supplementation of mice with the microbial metabolite DAT delays tumour growth and promotes ICI immunotherapy with anti-CTLA-4 or anti-PD-1. DAT-enhanced antitumour immunity was associated with more activated T cells and natural killer cells in the tumour microenvironment and was dependent on host IFN-I signalling. Consistent with this, DAT potently enhanced expansion of antigen-specific T cells following vaccination with an IFN-I-inducing adjuvant. DAT supplementation in mice compensated for the negative effects of broad-spectrum antibiotic-induced dysbiosis on anti-CTLA-4-mediated antitumour immunity. Oral administration of DAT altered the gut microbial composition in mice with increased abundance of bacterial taxa that are associated with beneficial response to ICI immunotherapy.
Interpretation
We introduce the therapeutic use of an IFN-I-modulating bacterial-derived metabolite to overcome resistance to ICI. This approach is a promising strategy particularly for patients with a history of broad-spectrum antibiotic use and associated loss of gut microbial diversity.
Funding
Melanoma Research Alliance, Deutsche Forschungsgemeinschaft, German Cancer Aid, Wilhelm Sander Foundation, Novartis Foundation.
Keywords: Desaminotyrosine, Melanoma, Immune checkpoint inhibitors, Gut microbiome, Microbial metabolites, Antibiotics
Research in context.
Evidence before this study
Immunotherapy utilizing immune checkpoint inhibitors (ICI) has been a transformative breakthrough in cancer treatment. However, only a limited cohort of patients are responsive to this therapeutic modality. Multiple preclinical and clinical observation studies suggested that the composition of the microbial population inhabiting an individual's intestinal tract—the gut microbiota—may play a pivotal role in regulating the immune system, and thus the efficacy of cancer immunotherapy. Disruption of the gut microbiota by use of broad-spectrum antibiotics has been associated with poor outcome to ICI cancer immunotherapy. How intestinal bacteria are linked to systemic anti-cancer immune responses is still a matter of active research. Certain metabolites that are unique to bacterial metabolism but foreign to our eukaryotic cells have recently gained attention as potential modulators of the immune response during ICI therapy. However, while some bacterial metabolites such as inosine can promote tumour-reactive T cells, others such as short-chain fatty acids can actively suppress cancer immunotherapy.
Added value of this study
Our study explores the translational potential of the type I interferon-modulating bacterial metabolite desaminotyrosine (DAT) to enhance ICI-mediated tumour immunosurveillance and -therapy. We found that oral supplementation with DAT can improve the response to immunotherapy with anti-CTLA-4 or anti-PD-1 in murine models of malignant melanoma and pancreatic adenocarcinoma. DAT supplementation in mice compensated for the negative effects of broad-spectrum antibiotic-induced dysbiosis on anti-CTLA-4-mediated antitumour immunity. Oral administration of DAT altered the gut microbial composition in mice with increased abundance of bacterial taxa that are associated with beneficial response to ICI immunotherapy.
Implications of all the available evidence
The available evidence indicates that the gut microbiota exerts an influential role in shaping the response to cancer immunotherapy, and bacterial-derived metabolites may represent a mechanism how they impact on systemic immunity. The findings from this study on the metabolite DAT contribute to the growing body of evidence supporting the notion that bacterial metabolites can beneficially modulate the immune response to cancer and may even mitigate adverse effects of therapy. Despite these promising findings, further research is needed to determine the safety and efficacy of utilizing bacterial metabolites to enhance ICI immunotherapy in humans. In summary, this study underscores the potential of utilizing the gut microbiota and its metabolites as a means of improving the efficacy of cancer immunotherapy, particularly for patients in need of antibiotic treatment.
Introduction
Cancer immunotherapy has become an important tool in cancer therapy with immune checkpoint inhibitors (ICI) being commonly used in patients with a variety of advanced cancer entities. The most prevalent ICI are monoclonal antibodies (mAbs) targeting cytotoxic T lymphocyte antigen-4 (CTLA-4), programmed cell death protein-1 (PD-1), or programmed death-ligand 1 (PD-L1) as negative regulators of the immune system.1,2 ICI—first introduced for the treatment of malignant melanoma—have fundamentality changed the landscape of cancer treatment. However, while many patients experience long-term tumour control, large interindividual differences in immune-mediated tumour response remain a major clinical challenge.1,3, 4, 5
Cancer immunosurveillance and antitumour immunity are dependent on type I interferons (IFN-I).6, 7, 8 At the same time, chronic IFN-I signalling can promote cancer therapy resistance,6,8 suggesting a delicate balance between pro- and anti-tumourigenic effects of IFN-I. Apart from that, IFN-I are an immunologically broadly active defence mechanism against microbial and viral infections.9 Signs of tissue damage or the presence of pathogens—collectively coined danger-associated molecular patterns (DAMPs)—can be sensed by host cells via pattern recognition receptors (PRRs), which trigger release of IFN-I and other pro-inflammatory factors. Thus, IFN-I have crucial functions in inflammatory diseases and cancer, in which they can inhibit tumour progression and promote immunosurveillance.10 We and others have recently shown that ICI treatment efficacy against malignant melanoma is completely abrogated in mice that are genetically deficient for IFN-I signalling.11 In line with these findings, IFN-I gene signatures in bulk tumour samples have been associated with favourable clinical responses to ICI in melanoma patients.12,13 However, the exact stimuli that contribute to induce IFN-I signalling in this context remain to be determined.
Recent mouse studies suggested that specific components of the enteric commensal microbiota can have distal effects on systemic immune responses through modulation of IFN-I signalling.14,15 Hereby, microbial metabolites such as desaminotyrosine (DAT) have been shown to protect from influenza or Sars-CoV-2 infection through augmentation of IFN-I signalling.15,16 DAT is produced by human and mouse enteric bacteria like Flavonifractor plautii through the degradation of flavonoids.17,18 These findings are of particular interest to onco-immunology research, as several reports have proposed that the composition of gut and tumour microbiota modulates tumour response to ICI19,20 and that this partly relies on intact IFN-I signalling21,22 Germ-free mice or animals treated with antibiotics (i.e., less diverse gut microbiome) failed to respond to blockade of either CTLA-4 or PD-1. Faecal microbial transplants (FMT) from healthy individuals restored the positive anti-tumour effect of ICI.23 Later studies showed that resident gut bacteria can also affect ICI immunotherapy in human cancer patients.23, 24, 25 Metagenomic studies revealed functional differences in gut bacteria in responders, including enrichment of certain metabolic pathways.26 Generally, antibiotic consumption–and thus disruption of the gut microbiota with resulting dysbiosis–was associated with poor response to ICI in cancer patients.27
In addition, some bacteria of the gut microbiota might be associated with a better response to ICI than others.20,26, 27, 28 Although certain bacterial taxa such as Faecalibacterium or Bacillota were associated with a beneficial clinical response, they were more abundant in the context of Ipilimumab-induced colitis, whereas Bacteroidaceae was more prevalent in melanoma patients resistant to ICI-induced colitis.25,29 Currently, no obvious pattern among the phylogeny has emerged that would correlate with bacterial species and how they influence cancer therapeutic responses.
Mechanistically, some studies suggested that specific microbiota can augment dendritic cell (DC) function leading to enhanced cytotoxic T cell priming and their accumulation in the tumour microenvironment.19,20 However, the molecular factors that drive DC and subsequent T cell activation modulated by microbiota remain to be determined. Immunotherapy-induced decreased gut epithelial barrier function, translocation of microbes from the gut lumen to the intestinal lamina propria (where many immune cells reside) and associated inflammation have been implicated as a reason why ICI therapy can be affected by the gut microbiome.19,30 A current subject of debate is to which degree bacterial-derived metabolites are able to alter antitumour effects of ICI therapy. Systemic short-chain fatty acids (SCFAs) such as butyrate and propionate have been associated with resistance to anti-CTLA-4 therapy in a patient cohort.31 In mouse models, butyrate prevented anti-CTLA-4-induced maturation of DCs and accumulation of tumour-specific T cells and memory CD4+ T cells in the TME.31 In contrast, the microbial metabolite inosine was shown to enhance the anti-tumour capacity of TH1 cells dependent on expression of the adenosine A2A receptor.30
Based on the established role of IFN-I in initiating anti-tumour immune responses, we here explore the translational potential of the IFN-I-modulating bacterial metabolite DAT to enhance ICI-mediated tumour immunosurveillance and -therapy.
Methods
Animals and ethics
C57Bl/6j mice (RRID: IMSR_JAX:000664) were purchased from Janvier-Labs. Mice genetically deficient for the interferon-α receptor 1 (B6(Cg)-Ifnar1tm1.2Ees/J; RRID: IMSR_JAX:028,288; here referred to as Ifnar1−/−) have been described previously,32 and were bred in the animal facility of the University Hospital Regensburg. Mice were between 6 and 10 weeks old at the beginning of the experiments and were kept under specific pathogen-free (SPF) conditions in a controlled environment. Mice were maintained on a 12-h light–dark cycle, with 12 h of light followed by 12 h of darkness, a constant temperature of 24 °C and a humidity level of 55%–65%. Ad libitum access to autoclaved standard laboratory chow and (special treatment) water was guaranteed at any time. Environmental enrichment, including nesting material, houses, and chew toys, was provided to promote their welfare and well-being. The animals were housed in the facility where the experiments took place for at least one week prior to the start of an experiment for acclimatisation. Mice were not co-housed to avoid microbial transfer through coprophagy between groups. Animal studies were approved by the local regulatory agency (Regierung von Oberbayern, Munich, Germany, ROB-55.2-2532.Vet_02-19-159).
Media and reagents
DMEM (Invitrogen) and RPMI-1640 (Invitrogen) were supplemented with 10% (v/v) FCS (Gibco), 3 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma–Aldrich). Double-stranded in vitro-transcribed 3pRNA (sense, 5′- UCA AAC AGU CCU CGC AUG CCU AUA GUG AGU CG -3′) was generated as described.33 Complete RPMI contained 10% (v/v) FCS, 3 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM β-mercaptoethanol (Gibco), 1x non-essential amino acids (Gibco), 1 mM sodium pyruvate (Gibco). T cell media was prepared supplementing complete RPMI with 1 mM HEPES (Gibco). Gavage solutions were prepared with 17 mg/ml 3-(4-hydroxyphenyl)-propionic acid (DAT, 98%, Sigma–Aldrich) in 5% EtOH (Carl Roth) in water. Drinking water was prepared with 100 mM DAT, NaOH (Merck) for buffering and 2.5 mg/ml aspartame (Fragon) in tap water. The respective solvents served as controls. Gavage solutions and drinking water were filtered through 100 μm sterile filters and were prepared fresh every 3–5 days. Broad-spectrum antibiotics with 1 mg/ml ampicillin (Carl Roth), 1 mg/ml neomycin (Carl Roth), 0.5 mg/ml vancomycin (Thermo Fisher), 1 mg/ml metronidazole (Fisher Scientific), 0.01 mg/ml amphotericin B (Merck) were administered via the drinking water (ad libitum), which was additionally supplemented with 2.5 mg/ml aspartame. Drinking water supplemented with antibiotics was replaced every 2 days to maintain constant antibiotic activity. DAT (100 mM) and broad-spectrum antibiotics were provided in the same drinking water bottle.
Tumour cell lines
The full-length chicken ovalbumin expressing B16 murine melanoma cell line (here referred to as B16.OVA; RRID: CVCL_WM78) was cultured in complete DMEM medium (see media and reagents). Panc02 cells (RRID: CVCL_D627) were cultured in RPMI medium (see media and reagents). Cells were cultured at 37 °C and 5% CO2. Cells used for experiments were at early passages 5–10. Prior to in vivo experiments, the cells were thawed one week in advance and passaged three times to ensure optimal viability and consistency throughout experiments. All cells were routinely tested for mycoplasma. All cell lines have been validated using STR analysis with 18 STR markers.
Tumour challenge und treatment
Mice were inoculated subcutaneously (s.c.) with 1.5 × 106 Panc02 cells or 2.4 × 105 B16.OVA cells in the right flank on day 0. Supplementation with DAT was initiated on day −7, 0 or 5 (as indicated) and was continued until the end of the experiment. Drinking water ± DAT was provided ad libitum. In some experiments, oral gavage of 200 μl DAT solution was performed once daily. When tumours were palpable anti-CTLA-4 (clone 9H10), anti-PD-1 (clone RMP1-14) or an appropriate isotype control (all BioXCell) were injected intraperitoneally (i.p.) on days 7 (200 μg), 10 (100 μg) and 13 (100 μg) for the B16.OVA model. For the Panc02 model, injections were performed on days 4 (200 μg), 7 (100 μg), and 10 (100 μg). For some experiments a mixture of broad-spectrum antibiotics was administered (see media and reagents), starting 4 days prior to anti-CTLA-4 treatment until day 21. Drinking bottles and watery DAT solutions were refreshed every 2 days. Antibiotic activity was assessed by cultivating resuspended faecal pellets plated on blood agar plates (BD) for 48 h under aerobic or anaerobic conditions at 37 °C. In some experiments blood was collected in EDTA microvette tubes (Sarstedt), diluted in PBS, and transferred into 96-well round bottom plates (Hartenstein). After centrifugation (400×g, 5 min), cells were resuspended in red blood cell lysis buffer (Invitrogen), incubated for 5 min at RT, and PBS was added before centrifugation (400×g, 5 min). This step was repeated before cells were stained for flow cytometry. Mice were euthanized when the maximum tumour diameter exceeded 15 mm or other determination criteria were met according to standard legal procedure and were scored daily (responsible state office Regierung von Oberbayern).
Preparation of cell suspensions from tumours and tumour-draining lymph nodes
Mice were treated as described above and were sacrificed 2 days after the last ICI treatment. Tumours as well as inguinal tumour-draining lymph nodes were removed using surgical scissors and forceps. Tumour weight was measured directly after extraction. Tumours and lymph nodes were minced and homogenized by filtering through 100 μm and 70 μm nylon cell strainers (BD Bioscience) with PBS on ice. The obtained single cell suspensions were washed in PBS before staining for flow cytometry.
Vaccination and lung pseudo-metastases model
A vaccine was prepared by mixing 10 μg 3pRNA liposomes (pre-incubated for 15 min with in vivo-jetPEI (Polyplus)) and 50 μg ovalbumin protein (Sigma) in 5% glucose. Mice were injected s.c. with 20 μl vaccine mix or the vehicle (in vivo-jetPEI in water with 5% glucose) into the upper thigh. Blood collection was performed on day 7 after vaccination, and the expansion of OVA-specific T cells was analysed via flow cytometry. For some experiments mice were challenged with injection of 106 B16.OVA cells s.c. or 3 × 106 B16.OVA cells i.v. on day 7 after vaccination. S.c. tumours were measured daily. At day 14, mice were sacrificed, and their lungs were flushed with 20 ml PBS through intracardial injection, and then extracted. Superficial pulmonary pseudo-metastases of each lung were counted to estimate overall tumour load.
Neutrophil granulocyte influx
Neutrophil granulocyte influx as a surrogate marker for the degree of intestinal inflammation was performed as previously described.34 In brief, mice were sacrificed, their distal third of the small intestine was removed and flushed with cold PBS, opened, and cut into 1 cm pieces. The intestinal pieces were washed with cold PBS and then incubated in PBS supplemented with 10% FBS and 0.1 mM EDTA, and incubated for 20 min at 37 °C. Tissues were washed and filtered through a 100 μm strainer and transferred into complete RPMI. Next, intestines were incubated in complete RPMI supplemented with 200 U/ml collagenase II (Worthington) and 0.05 mg/ml DNase (Sigma) for 60 min on a shaker at 37 °C. Cells suspensions were filtered through a 100 μm strainer, washed with PBS and purified on a 20/40/80% Percoll (Sigma) gradient (30 min, 25 °C, 3000 rpm, Eppendorf 5810R with A-4-81 rotor). Immune cells were collected from the interphase and washed with PBS before subsequent flow cytometry analysis.
In vitro T cell and DC assays
T cells were isolated from spleens of donor C57Bl/6j mice. Spleens were minced through a 100 μm strainer using cold PBS and were then centrifuged (all centrifugation steps for 5 min, 4 °C, 400×g). All following steps were performed with cold reagents and on ice. After discarding the supernatant cells were incubated in 2 ml red blood cell lysis buffer (Invitrogen) per spleen for 2 min, and PBS was added before filtering through a 70 μm strainer and centrifugation. The cell pellet was resuspended in PBS, filtered through a 40 μm strainer and centrifuged. After discarding the supernatant, T cells were isolated with the Pan T Cell Isolation Kit II, mouse (130-095-130, Miltenyi Biotec) according to the manufacturers protocol. 5 × 105 T cells/ml were seeded in a 48-well cell culture treated flat bottom well in T cell media and supplemented with 30 IU/ml IL-2 (Clinigen) and 5 μl of Dynabeads™ Mouse T-Activator CD3/CD28 beads (Thermo Fisher). DAT (neutralized solution) was added at concentrations of 100 μM or 1 mM or cells were left untreated. After 24 h incubation at 37 °C and 5% CO2, cells were harvested and stained for subsequent flow cytometry analysis. DCs were generated from bone marrow of donor C57Bl6/J mice. Bone marrow was flushed out of bone shafts with complete RPMI, filtered through a 100 μm strainer and centrifuged. After centrifugation, the cell pellet was resuspended in 2 ml red blood cell lysis buffer (Invitrogen). Complete RPMI was added, and cells were centrifuged and washed once again. 5 × 106 cells were seeded in 10 ml complete RPMI supplemented with 20 ng/ml GM-CSF in 10 cm cell culture dishes, and were cultured for 7 days with addition of 10 ml media after 3 days and media change on day 6. 5 × 105 DCs/ml were then seeded in complete RPMI in 48-well flat bottom plates. DCs were either treated with DAT alone (100 μM or 1 mM) or DAT in the presence of 100 ng/ml LPS, or were left completely untreated. Both the vehicle controls were treated with the respective amount of sterile water. After 24 h incubation at 37 °C and 5% CO2, cells were harvested and stained for subsequent flow cytometry analysis.
Flow cytometry
Cell suspensions were stained in PBS. Fluorochrome-coupled antibodies were purchased from eBioscience or BioLegend (see Table S1). Data were acquired on a FACSCanto II (BD Biosciences) and analysed using FlowJo software (TreeStar). All staining protocols included 20 min staining with a live/dead marker and CD16/CD32 antibody (Biolegend) at 4 °C and surface marker staining for 50 min at RT. For intracellular cytokine staining the Foxp3 Transcription Factor Fixation/Permeabilization Kit (eBioscience) was used. Tumour model antigen OVA-specific T cells from the blood were defined as alive CD3+, CD4− and SIINFEKL-H-2Kb+ using iTAg MHC-I murine tetramers detecting SIINFEKL-specific CD8+ T cells (MBL). In this context, negative selection via CD4 was chosen, due to known interaction of anti-CD8 (particularly clone 53–6.7) with murine MHC-I tetramers. Neutrophils were defined as alive CD45+, Ly6G+ and CD11b+. For tumour and tumour-draining lymph node analysis, cells suspensions were incubated for 3 h in RPMI containing 1x cell stimulation cocktail plus protein transport inhibition (Thermo Fisher). DCs were defined as CD45+, CD11c+, and MHC-II+ cells. Activation in DCs was assessed by the MFI of CD86 expression. NK cells were defined as CD45+, CD3−, NK1.1+ cells. Cytotoxic T cells were defined as CD45+, CD3+, CD4−, and CD8+ cells, and conventional T helper cells were defined as CD45+, CD3+, CD8−, and CD4+ cells. Regulatory T cells were defined as CD45+, CD3+, CD8−, CD4+, and FoxP3+ cells. Naïve T cells were identified as CD4+ or CD8+ T cells as CD62L+ and CD44− cells, whereas effector memory T cells were defined as CD62L− and CD44+ T cells. For respective T cell subsets, frequencies of T-bet+, IFNγ+, or CD25+ cells were analysed.
16S-rRNA gene amplicon sequencing
Murine faecal samples were collected at different time points during the experiments and snap frozen in liquid nitrogen. Samples were processed as previously described with some changes.35 Briefly, after DNA isolation, targeted 16S rRNA gene amplicons were produced by fusing barcodes plus adapters using a 2-strep PCR. Primers used have been previously described: 341F 5′-CCT ACG GGN GGC WGC AG-3′ and 785R 5′-GAC TAC HVG GGT ATC TAA TCC-3’.36 For DNA isolation, the MaxWell RSC Faecal Microbiome DNA Kit (Promega) was used with bead beating. For the latter, the FastPrep-24 (MP Biomedicals Germany GmbH) was used, supplied with a CoolPrep adapter (MP Biomedicals, cooled with dry ice); thrice for 20 s of beating at a speed of 6 m/s, each followed by a 30 s break for cooling. Cycle times for the first and the second PCR had been adjusted for denaturation to 10 s, for annealing to 20 s and for extension to 45 s. After library preparation and equimolar pooling of the amplicons, the samples were sequenced using a MiSeq (Illumina) as described.32 For the DAT supplementation data, a total of 60 samples with a read length of 301 bp was analysed. For the responder/non-responder data a total of 17 samples with a read length of 301 bp was analysed. For the Abx dataset, a total of 80 samples with a read length of 301 bp was analysed. Raw sequencing data were analysed in a UPARSE-based pipeline clustering sequences down to zOTUs (denoised operational taxonomic units) with www.imngs2.org,37 which includes the TIC algorithm.38 Downstream analysis of zOTU tables was conducted using Rhea with default settings.39 Briefly, reads were normalized39 and spurious taxa were removed.40
Targeted metabolomic measurement
Approximately 20 mg of mouse faeces was weighed in a 2 ml bead beater tube (FastPrep-Tubes, Matrix D, MP Biomedicals Germany GmbH) and resuspended with 1 ml of methanol. The samples were extracted with a bead beater FastPep-24TM 5G (MP Biomedicals Germany GmbH) supplied with a CoolPrepTM (MP Biomedicals Germany; cooled with dry ice) for 3 times, each for 20 s of beating at a speed of 6 m/s and each followed by a 30 s break. The 3-NPH method was used for the quantitation of SCFAs, tryptophan derivates (ICA) or DAT.41 Briefly, 40 μl of the faecal extract and 15 μl of isotopically labelled standards (ca 50 μM) were mixed with 20 μl 120 mM EDC HCl-6% pyridine-solution and 20 μl of 200 mM 3-NPH HCL solution. After 30 min at 40 °C and shaking at 1000 rpm using an Eppendorf Thermomix (Eppendorf), 900 μl acetonitrile/water (50/50, v/v) was added. After centrifugation at 13,000 U/min for 2 min the clear supernatant was used for analysis. The measurement was performed using a QTRAP 5500 triple quadrupole mass spectrometer (Sciex) coupled to an ExionLC AD (Sciex) ultrahigh performance liquid chromatography system. The electrospray voltage was set to −4500 V, curtain gas to 35 psi, ion source gas 1 to 55 psi, ion source gas 2 to 65 psi and the temperature to 500 °C. The MRM-parameters were optimized using commercially available standards. The chromatographic separation was performed on a 100 × 2.1 mm, 10 nm, 1.7 μm, Kinetex C18 column (Phenomenex) column with 0.1% formic acid (eluent A) and 0.1% formic acid in acetonitrile (eluent B) as elution solvents. An injection volume of 1 μl and a flow rate of 0.4 ml/min was used. The gradient elution started at 23% B which was held for 3 min, afterward the concentration was increased to 30% B at 4 min, with another increase to 40% B at 6.5 min, at 7 min 100% B was used which was hold for 1 min and at 8.5 min, the column was equilibrated to starting conditions. The column oven was set to 40 °C and the autosampler to 15 °C. Data acquisition and instrumental control were performed with Analyst 1.7 software (Sciex) and data analysis with MultiQuant 3.0.3 (Sciex).
Immunohistochemistry
Mouse colon tissues were fixed in 10% neutral-buffered formalin solution (Sigma) for 48 h, dehydrated under standard conditions (Leica ASP300S) and embedded in paraffin. Serial 2 μm-thin sections prepared with a rotary microtome (HM355S, ThermoFisher Scientific) were collected and subjected to histological and immunohistochemical analysis. Immunohistochemistry was performed using a Bond RXm system (Leica, all reagents from Leica) with a primary antibody against cleaved caspase-3 (Clone 5A1E, dilution 1:150, Cell Signaling; RRID: AB_2070042). Briefly, slides were deparaffinized using deparaffinization solution, pretreated with Epitope retrieval solution 1 (corresponding to citrate buffer pH6) for 20 min. The primary antibody was incubated for 15 min at room temperature. Antibody binding was detected with a polymer refine detection kit without post primary reagent and visualized with DAB as a dark brown precipitate. Counterstaining was conducted with haematoxylin. Slides were cover slipped and digitalized with an AT2 scanner system (Leica Biosystems), and the staining reaction was evaluated with Imagescope (12.4.0.2018, Leica Biosystems).
Censoring
Follow-up of all tumour-bearing animals was generally stopped at the predetermined end of in vivo experiments (day 80 post tumour induction), as per local regulatory standards. Reasons for censoring include no engraftment of tumours in the control groups, tumours reaching end point criteria before the first ICI therapy time point (which could be attributed to the potential injection of an excessive number of tumour cells caused by cellular clustering), and tumours being too small for TME analysis. Respective censoring proportions per experiment are listed in Table S2. For survival analysis the censoring proportion until day 80 (maximum observation time) was calculated.
Confounding factors
Efforts were made to minimize potential confounding factors. Housing conditions were maintained consistent during all experiments. For experiments using mice bred in-house (Ifnar1−/−), equal distribution of male and female among treatment groups was guaranteed. Generally, all mice included in experiments were within the specific age range of six to ten weeks. Variations in immune response and tumour heterogeneity were mitigated through randomization (cages were randomly assigned to treatment groups before tumour sizes were known), an acclimation period, conducting experiments with replicates, varying cage and treatment order, and implementing proper control groups. Stress and pain were reduced by appropriate handling by well trained personnel, not disturbing the mice during the dark-cycle and daily monitoring by the experimenter, animal care takers, and veterinarians. The study was not blinded for practical reasons as application and treatment were more consistent with just one operator, which also reduced the stress for the animals.
Statistical analysis
The respective sample sizes were determined based on internal pilot studies, other projects, and previous research. For the B16 tumour model, a group size of at least 5 mice per group was demonstrated to be feasible for detecting differences in tumour growth after therapy.11 Effects of DAT were observed in another internal project with similar group sizes. Additionally, ethical guidelines were considered, and efforts were made to use the minimum number of animals to strike a balance between obtaining statistically valid results and reducing the overall number of animals used. The sample size for further downstream analyses was adjusted to accommodate resource constraints. All graphs for which parametric tests could be employed are presented as mean with 95% confidence interval (CI). Table S3 gives an overview of the differences of mean with 95% CI for all biologically relevant comparisons for all experimental groups in all graphs. All data for which non-parametric tests were employed are depicted as median with its corresponding CI at a requested confidence level of 95%. Confidence intervals of the median have been computed as previously described.42 Data was assessed for normality via visual inspection using a QQ plot and for homoscedasticity by applying the Brown–Forsythe test in addition to visual inspection of plotted values. Data for which only pairwise comparisons were performed always generally fulfilled normality criteria in our study, so an unpaired parametric t-test was applied in all cases. If data exhibited normal distribution but showed differences in the standard deviation (SD), an unpaired parametric t-test with Welch's correction was employed. Three or more different groups were analysed using ANOVA with Tukey's multiple comparison test. In cases where specific pre-selected pairs were analysed, Bonferroni's multiple comparison test was employed. When the data did not meet the ANOVA criteria, a Kruskal–Wallis test with Dunn's post-hoc test was utilized. Tumour growth curves were analysed with the two-way ANOVA with Tukey's post-hoc test, the alternative hypothesis being that the tumour volume in the DAT treated group is different (smaller) compared to the control groups over time. The reported p-values in the graphs refer to the latest possible time point (day) for conducting the test, depending on the groups included. Additionally, as a summary measure the area under curve (AUC) for tumour volume growth was calculated, standardised, and compared as previously described.43 Overall survival was analysed using the Log-rank (Mantel–Cox) test. The survival experiments were initiated at day 0 and the survival was monitored until day 80. The event of interest was monitored from day 0 on but was more likely to start at around day 10. All statistical calculations were performed using Prism (GraphPad Software).
Role of funders
The funders had no role in study design, data collection, data analysis, interpretation, or writing of the manuscript.
Results
Treatment with bacterial-derived metabolite DAT delays tumour growth and enhances anti-CTLA-4 immunotherapy
To assess a potential effect of the microbial metabolite DAT on tumour immunosurveillance and -therapy, C57Bl/6J mice were provided with DAT via the drinking water ad libitum as described previously.15 If not stated otherwise, supplementation with DAT was initiated on the day of subcutaneous tumour inoculation with the syngeneic melanoma cell line B16 expressing the model antigen ovalbumin (B16.OVA) (Fig. 1A). We observed a statistically not significant trend for delayed tumour growth in animals supplemented with DAT with reduced mean tumour volume on day 7 (Fig. S1A). Notably, 50% of DAT-treated mice had not developed a palpable tumour on day 7 (Fig. 1B). Visible tumours in these mice appeared 2–3 days later. Further on, tumours in mice receiving DAT supplementation but no additional treatment were growing almost as fast as tumours in the vehicle-treated control group (Fig. 1C). Accordingly, sole DAT supplementation did not prolong long-term survival in melanoma-bearing mice (Fig. 1D). However, a significant additive effect could be observed when combining DAT supplementation and anti-CTLA-4 immunotherapy, with markedly improved tumour control and improved survival compared to any single treatment group (Fig. 1C and D). Endogenous levels of DAT in murine stool samples collected from mice undergoing anti-CTLA-4 immunotherapy without additional supplementation were at the technical detection level of targeted metabolomics via mass spectrometry. Nonetheless, mice that later proved to be non-responders to anti-CTLA-4 retrospectively showed a statistically not significant trend for low endogenous DAT levels in stools samples obtained during immunotherapeutic treatment (Fig. S1B). In summary, our findings demonstrate that supplementation of the microbial metabolite DAT can enhance the efficacy of anti-CTLA-4 cancer immunotherapy.
Fig. 1.
Treatment with bacterial-derived metabolite DAT delays tumour growth and enhances anti-CTLA-4 immunotherapy via increased T cell activation. (A) Experimental setup: Mice were provided with DAT via the drinking water ad libitum beginning on the day of tumour cell inoculation (day 0) until the end of the experiment. Anti-CTLA-4 or isotype control antibodies were administered i.p. on days 7 (200 μg), 10 (100 μg), and 13 (100 μg). Tumours were extracted on day 15. (B) Percentage of mice without a palpable tumour on day 7 before treatment onset from n = 6 independent experiments [p-values correspond to an unpaired t-test with Welch's correction]. (C) Mean tumour growth from n = 3–6 independent experiments [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 14]. (D) Overall survival of n = 3 independent experiments [p-values correspond to a Log-rank (Mantel–Cox) test]. In n = 4 independent experiments, tumours were extracted for analysis of immune cells in the tumour microenvironment (TME) via flow cytometry (E) Representative dot plots and (F) scatter plots of the frequency of IFNγ+ CD4+ cells, (G) mean fluorescence intensity (MFI) of IFNγ expression in CD4+ cells normalized to ‘Ctrl + isotype’ group, (H) frequency of IFNγ+ CD8+ cells, (I) MFI of IFNγ expression in CD8+ cells normalized to ‘control + isotype’ group, and (J) frequency of IFNγ+ natural killer (NK) cells in the TME [p-values correspond to the respective one-way ANOVA with Tukey's multiple comparison test]. All graphs show mean with 95% confidence interval (CI). For an analysis of area under the curve (AUC) of tumour volume growth see Fig. S8.
DAT supplementation alters the tumour microenvironment and enhances anti-CTLA-4-mediated T cell activation
To investigate effects of DAT on the tumour microenvironment (TME), mice were treated as described above (Fig. 1A), and tumour tissue was analysed on day 15. Supplementation with DAT enhanced anti-CTLA-4-induced T cell activation, with increased frequency (mean difference 14.06%, 95% CI 0.60–27.53) and activity of IFNγ-producing CD4 T cells (Fig. 1E–G, Table S3). In contrast, DAT supplementation alone did not significantly impact on T cell activation in the TME. Similarly, in CD8+ cytotoxic T cells, the activation level was enhanced by DAT and anti-CTLA-4 combination treatment (mean difference to anti-CTLA-4 monotherapy of 16.29%, 95% CI 0.17–32.41) (Fig. 1H and I, Fig. S1C, Table S3). As expected, anti-CTLA-4 ICI immunotherapy strongly modulated the TME, but we did not observe any additional effects by DAT supplementation regarding expansion and activation of OVA antigen-specific T cells (Fig. S1C and D), frequency of Foxp3+ regulatory T cells (Tregs, Fig. S1E) and T cell memory phenotype (Fig. S1F–I). Also, the frequency and maturation of conventional CD11c+ MHC-II+ DCs in the TME as important antigen-presenting cells during T cell priming were not altered by oral DAT supplementation (Fig. S1J and K). Interestingly, the combinatorial treatment with anti-CTLA-4 and DAT increased the frequencies of activated, IFNγ-producing natural killer (NK) cells in the tumour (Fig. 1J). Taken together, these data show that DAT supplementation by itself does not significantly impact on the TME, but together with anti-CTLA-4 therapy DAT can potently enhance T and NK cell activation.
DAT directly enhances T cell and DC activation in vitro
To confirm whether these findings were mediated by direct effects of DAT on immune cells, we exposed T cells or DCs to varying concentrations of DAT in vitro. In the presence of a T-cell receptor (TCR) stimulus and costimulatory signals in the form of CD3/CD28-coated beads and recombinant IL-2, DAT effectively enhanced T cell activation (Fig. 2). DAT exposure increased the percentage of TCR-stimulated CD8+ T cells expressing the pro-inflammatory cytokine IFNγ (mean difference between 1 mM DAT and untreated T cell 5.46%, 95% CI 0.21–10.70) and the early activation marker CD25 (Fig. 2A and B, Table S3). In the absence of TCR stimulation, DAT did not impact on CD8+ T cell activation (Fig. S2A and B). In CD4+ T cells, exposure to DAT enhanced the frequency of cells expressing the Th1-favouring transcription factor T-bet (Fig. 2C), but not of IFNγ+ or CD25+ CD4 T cells (Fig. S2C and D). In DCs, engagement of DAT induced maturation with enhanced surface expression of the co-stimulatory molecules CD80 or CD86 only when DCs were simultaneously exposed to lipopolysaccharides (LPS), a component of the outer membrane of gram-negative bacteria and molecular pattern activating Toll-like receptor (TLR)4 (Fig. 2D and E). These data demonstrate that DAT can indeed directly promote activation of both T cells and DCs, but that this may require additional signals such as TCR engagement or microbial co-stimulatory signals in DCs.
Fig. 2.
DAT enhances T cell and DC activation in vitro. (A–C) Splenic T cells were stimulated with CD3/CD28-coated beads and IL-2 for 24 h, and were then exposed to different concentrations of DAT. T-cell activation and differentiation were analysed via flow cytometry from n = 4 independent experiments. Frequency of (A) IFNγ+ CD8+ cells, (B) CD25+ CD8+ cells, and (C) T-bet+ CD4+ cells. (D–E) Bone marrow-derived DCs were incubated with different concentrations of DAT, and in some cases additionally with LPS. After 24 h, surface expression of (D) CD80 or (E) CD86 on DCs from n = 3 independent experiments was analysed via flow cytometry. Data show CD80 or CD86 MFI. [p-values correspond to the respective one-way ANOVA with Tukey's multiple comparison test]. All graphs show mean with 95% CI.
The additive effect of DAT and ICI immunotherapy is time- and application-dependent
We next evaluated whether the timing of DAT administration could have impact on ICI immunotherapy in murine melanoma. We therefore initiated DAT administration via the drinking water on day 5 after tumour inoculation (referred to as “late” treatment; Fig. 3A). Compared to “early” DAT supplementation from day 0 on, late DAT administration from day 5 on resulted in a similar non-significant trend of delayed early tumour growth (Fig. 3B). Yet, combining the late DAT treatment with anti-CTLA-4 did not achieve the same beneficial effects on long-term tumour control and associated animal survival as the “early” DAT treatment (Fig. 3C). We then investigated if a lower dose of DAT (3.4 mg/mouse/day) but with controlled application via oral gavage starting on day 0 could also promote a therapeutic anti-tumour response (Fig. 3D). We found the oral gavage DAT regimen to be less therapeutically active in combination with anti-CTLA-4, with only non-significant mean tumour growth delay but some survival benefit for melanoma-bearing mice (Fig. 3E–G). To address whether the immunomodulatory effect of DAT was limited to the highly immunogenic B16.OVA malignant melanoma model, DAT supplementation via oral gavage was conducted in a subcutaneous model of murine pancreatic adenocarcinoma and was combined with anti-PD-1 treatment. Even administered at low doses (3.4 mg/mouse/day), we observed improved tumour control associated with significantly prolonged survival in mice treated in the DAT and anti-PD-1 combination group (Fig. S3A and B). Taken together, these data demonstrate that oral DAT supplementation increases the efficacy of both anti-CTLA-4 or anti-PD-1 ICI immunotherapy of different cancer entities in a time- and application-dependent manner.
Fig. 3.
The additive effect of DAT and ICI immunotherapy is time and dose dependent. (A) Scheme of experimental setup of B-C: Mice were inoculated s.c. with B16.OVA cells on day 0, and anti-CTLA-4 or isotype control antibodies were administered i.p. on days 7 (200 μg), 10 (100 μg), and 13 (100 μg) as described for Fig. 1, and DAT was continuously provided via the drinking water beginning on day 5. (B) Tumour volume on day 7 before onset of treatment with anti-CTLA-4 from n = 3 independent [the reported p-values correspond to a one-way ANOVA with Tukey's multiple comparison test]. (C) Continuous tumour growth from n = 2–3 independent experiments over time [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 14]. (D) Scheme of experimental setup of E–G: Mice were inoculated s.c. with B16.OVA cells on day 0, and anti-CTLA-4 or isotype control antibodies were administered i.p. on days 7, 10 and 13 as described for Fig. 1, and DAT was provided by daily oral gavage beginning on the day of tumour cell inoculation until the end of the experiment. (E) Tumour volume on day 7 before onset of treatment with anti-CTLA-4 [p-values correspond to an unpaired t-test]. (F) Continuous tumour growth over time from n = 3 independent experiments [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 12]. (G) Overall survival of tumour-bearing animals [p-values correspond to a Log-rank (Mantel–Cox) test]. All graphs show mean with 95% CI. For an integrated analysis of AUC of tumour volume growth see Fig. S8.
DAT enhances antigen-specific priming and expansion of cytotoxic T cells in vivo
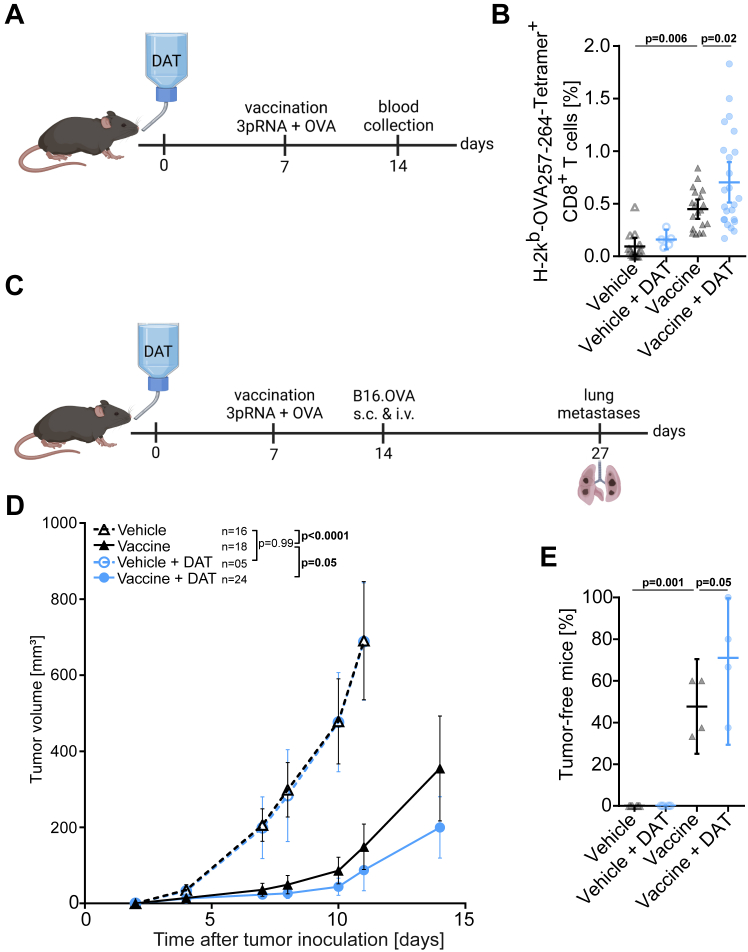
Even though our analysis of the TME demonstrated an increased abundance of activated IFNγ-producing T cells in response to DAT supplementation during anti-CTLA-4 immunotherapy, our data did not allow to differentiate whether DAT impacts on T-cell priming and/or effector function. Hence, we evaluated the potential of DAT to modulate T-cell priming in a controlled vaccination approach in the absence of tumour growth, using purified OVA protein and the RIG-I ligand 3pRNA44 as adjuvant in naïve mice. DAT application via the drinking water was initiated at day 0, and mice were vaccinated at day 7 with OVA and 3pRNA s.c (Fig. 4A). In line with our previous data, sole DAT supplementation was not sufficient to increase the OVA protein effect, but significantly enhanced the expansion of circulating OVA antigen-specific T cells in combination with the 3pRNA adjuvant (Fig. 4B). Vaccinated mice were then challenged with a bimodal (subcutaneous and intravenous) B16.OVA tumour cell inoculation (Fig. 4C). Mice immunized with the combinatorial vaccine incorporating 3pRNA adjuvant and oral DAT subsequently showed significantly better subcutaneous (Fig. 4D) and systemic tumour control with reduced numbers of pulmonary pseudo-metastases (Fig. 4E, Fig. S4). These data demonstrate that DAT treatment enhances antigen-specific priming and expansion of cytotoxic T cells with potent anti-tumour activity.
Fig. 4.
DAT enhances antigen-specific priming and expansion of cytotoxic T cells. Mice were provided with DAT via the drinking water from day 0 until the end of the experiment. On day 7 mice were injected s.c. with a vaccine/adjuvant mix of 50 μg OVA protein and 10 μg 3pRNA into the medial part of the upper tight. (A) Scheme for experimental setup I: For some mice, blood samples were taken on day 14, and (B) the frequency of OVA-specific CD8+ T cells in the peripheral blood was analysed from n = 5 independent experiments [p-values correspond to a one-way ANOVA with Bonferroni's multiple comparison test]. (C) Scheme for experimental setup II: Some mice were challenged with B16.OVA cells injected s.c. and i.v. on day 14. (D) Continuous growth of s.c. tumours from n = 4 independent experiments [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 11 and an additional unpaired t-test with Welch's correction for ‘vaccine’ and ‘vaccine + DAT’ group on day 14]. (E) The formation of lung pseudo-metastases on day 27 was analysed. Lungs were extracted on day 27, pseudo-metastases per lung were counted and the percentage of tumour-free mice per experiment (defined as animal with less than 5 metastases per lung) were determined [p-values correspond to a one-way ANOVA with Bonferroni's multiple comparison test]. All graphs show mean with 95% CI. For an integrated analysis of AUC of tumour volume growth see Fig. S8.
Host-IFN-I signalling is required for the additive effect of DAT and anti-CTLA-4 immunotherapy
Given its known function as an IFN-modulator,15 we next investigated the role of host cell-intrinsic IFN-I signalling for DAT-mediated enhanced anti-tumour immunity. As described previously,11 mice with genetic deficiency for the common IFN-α receptor 1 (Ifnar1−/−)15 showed a generally poor response to anti-CTLA-4 ICI treatment (Fig. 5). Oral supplementation of DAT in Ifnar1−/− mice did not impact on early tumour control as we did not observe any DAT-mediated tumour growth delay (Fig. 5A and B). Furthermore, the combinatorial effect of DAT and anti-CTLA-4 was completely abrogated in Ifnar1−/− mice with rapid tumour growth and poor survival (Fig. 5C and D). Oral supplementation with DAT did not rescue the deleterious effects of defective host IFN-I signalling on the therapeutic effect of anti-CTLA-4. Accordingly, when analysing the TME in Ifnar1−/− mice, we did not observe increased abundance of activated T and NK cells or other immune cells in response to anti-CTLA-4 neither as monotherapy nor in combination with oral DAT supplementation (Fig. 5E–H, Fig. S5). These data demonstrate that DAT-mediated enhancement of anti-CTLA-4 immunotherapy is critically dependent on functional host IFN-I signalling. However, these data do not distinguish whether DAT actively modulates IFN-I signalling or impacts on immunomodulatory functions downstream of IFNaR1 in host (immune) cells.
Fig. 5.
Host IFN-I signalling is required for the additive effect of DAT and ICI. Mice with genetic deficiency for the interferon-alpha receptor 1 were (Ifnar1−/−) were provided with DAT via the drinking water, inoculated with B16 melanomas and were treated with anti-CTLA-4 or isotype control antibodies as described for Fig. 1. (A) Mean tumour volume on day 7 before onset of ICI treatment from n = 6 independent experiments [the reported p-values correspond to an unpaired t-test.] (B) Percentage of mice without a palpable tumour on day 7 before treatment onset of ICI treatment from 6 independent experiments [the reported p-values correspond to an unpaired t-test]. (C) Mean tumour growth per experimental group from n = 6 independent experiments [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 13]. (D) Overall survival from n = 3 independents [p-values correspond to a Log-rank (Mantel–Cox) test]. In n = 3 independent experiments, tumours were extracted for analysis of immune cells in the TME via flow cytometry on day 15: (E) IFNγ+ CD4+ cells, (F) IFNγ+ CD8+ cells, (G) OVA antigen-specific T cells, and (H) IFNγ+ NK cells in the TME [p-values correspond to the respective one-way ANOVA with Tukey's multiple comparison test]. All graphs show mean with 95% CI. For an integrated analysis of AUC of tumour volume growth see Fig. S8.
DAT compensates for the adverse effects that result from antibiotic treatment during anti-CTLA-4 immunotherapy
A major clinical challenge in the therapy with ICI are previous treatments with broad-spectrum antibiotics, associated gut dysbiosis and often deleterious effects on immunotherapy.45, 46, 47, 48 To evaluate whether oral DAT supplementation may ameliorate the negative effects of gut dysbiosis, mice were treated with a mixture of broad-spectrum antibiotics (Abx) consisting of ampicillin, neomycin, vancomycin, metronidazole, and the anti-fungal compound amphotericin B prior to B16.OVA tumour cell inoculation and subsequent treatment. Indeed, Abx-treated mice showed severe changes in their intestinal microbiota with poor bacterial growth both under standard aerobic as well as anaerobic culture conditions of plated stool samples (Fig. S6A). Complementary 16S-rRNA analysis confirmed that the bacterial gut microbiome was largely depleted following broad-spectrum Abx treatment in mice. This bacterial depletion was not prevented by concurrent oral DAT supplementation (Fig. S6B). Consistent with previous reports,19 anti-CTLA-4 treatment failed to improve tumour growth control or host survival in Abx-treated mice when compared to isotype control mock treatment (Fig. 6A and B, Fig. S6C and D). Despite largely depleted intestinal bacteria, oral supplementation of DAT led to a partial recovery of anti-CTLA-4 efficacy with moderately improved mean tumour control and markedly prolonged survival of Abx-exposed mice undergoing combinatorial treatment with DAT and anti-CTLA-4 (Fig. 6A and B, Fig. S6C and D). Control groups without Abx treatment were run in parallel to confirm the effectiveness of the anti-CTLA-4 treatment and the additive DAT effect and are displayed in additional graphs for clarity (Fig. S6C and D). DAT supplementation allowed for the anti-CTLA-4-induced expansion of circulating OVA tumour antigen-specific T cells, even in Abx-treated mice (Fig. 6C). Taken together, these data demonstrate that oral supplementation with DAT cannot prevent intestinal dysbiosis after broad-spectrum antibiotic treatment, but can ameliorate its deleterious effects on the anti-tumour efficacy of anti-CTLA-4 ICI immunotherapy.
Fig. 6.
DAT compensates for the adverse effects that result from antibiotic treatment during anti-CTLA-4 immunotherapy. Mice were treated with a mixture of broad-spectrum antibiotics (ampicillin, neomycin, vancomycin, metronidazole) and amphotericin B via the drinking water starting on day −4 before s.c. B16.OVA cell inoculation on day 0. The Abx-containing drinking water was provided until day 21. Some mice were additionally supplemented with DAT via the drinking water starting on day 0. Anti-CTLA-4 or isotype control antibodies were administered i.p. on days 7 (200 μg), 10 (100 μg), and 13 (100 μg). (A) Mean tumour growth per experimental group from at least n = 3 independent experiments [p-values correspond to a two-way ANOVA with Tukey's multiple comparison test on day 12 for comparisons of ‘Ctrl + isotype’ and other groups and a two-way ANOVA with Tukey's multiple comparison test on day 14 excluding ‘Ctrl + isotype’ for all other comparisons; the statistical analysis was performed on the whole data set from Fig. S6C]. (B) Overall survival in Abx-treated tumour-bearing mice from at least n = 3 independent experiments [p-values correspond to a Log-rank (Mantel–Cox) test; the statistical analysis was performed on the whole data set from Fig. S6D]. (C) Expansion of OVA antigen-specific T cells in the peripheral blood on day 15 with Ctrl + Abx + isotype n = 25, Ctrl + Abx + anti-CTLA-4 n = 15, DAT + Abx + isotype n = 12, DAT + Abx + anti-CTLA-4 n = 17, Ctrl + isotype n = 15, Ctrl + anti-CTLA-4 n = 17, DAT + isotype n = 16, and DAT + anti-CTLA-4 n = 17 [p-values correspond to a Kruskal–Wallis test with Dunn's multiple comparison test]. Graph (A) shows mean with 95% CI and graph (C) shows median with its corresponding CI at a requested confidence level of 95%. For an integrated analysis of AUC of tumour volume growth see Fig. S8.
Oral DAT supplementation alters the microbial composition of the gut in mice
In multiple studies, certain bacterial taxa were found to be enriched in responder versus non-responder patients.20,23,24,29 Bacterial-derived metabolites were suggested to act as potential mediators of varying anti-tumour immune responses.30,31 We thus addressed whether the beneficial immunomodulatory effects of oral DAT application were solely mediated by compensating insufficient endogenous DAT levels and/or actively modulated the composition of the gut microbiota. Therefore, mice were supplemented with DAT via the drinking water for 7 days, and 16S-rRNA from faecal samples was analysed using next generation sequencing (NGS). We found that the effective richness and the Shannon effective diversity were significantly decreased in DAT treated mice, meaning that the number of species and their relative abundances were lower in stool samples of DAT-treated mice (Fig. 7A and B). For beta-diversity of stool microbiota, we found a significant difference in similarity between DAT-treated and naive mice (Fig. 7C). Next, we analysed potential changes in taxonomic composition. Application of DAT in mice altered the composition of their intestinal microbiota at the class level. The relative abundance of Bacteroidia was significantly higher following DAT application, and the relative abundance of Clostridia—the taxonomic class of the known DAT producer Flavonifractor plautii (NCBI:txid292800)49—decreased (Fig. 7D). On order level, we found that the relative abundances of Bacteroidales and Burkholderiales were increased and the relative abundance of Lachnospirales was decreased following DAT supplementation (Fig. 7E). Burkholderiales were the only bacterial taxon retrospectively found to be enriched before treatment onset in anti-CTLA-4 responder versus non-responder melanoma-bearing mice in the absence of DAT supplementation (Fig. 7F). For this purpose, we analysed 16S-rRNA in stool samples of mice before tumour inoculation and onset of treatment with anti-CTLA-4, and retrospectively categorized these mice into responder versus non-responder to ICI therapy. The results were also narrowed down to family level. The relative abundance of Tannerellaceae was increased whilst the abundances of Lachnospiraceae and Oscillospiraceae were decreased following DAT supplementation (Fig. 7G). Nonetheless, these differences in class, order or family level are too broad to imply contribution by specific bacterial genera or species to ICB-related outcomes. When looking at genus level, we were able to identify only few defined genera, none of which were altered by oral DAT supplementation (data not shown). In summary, oral DAT supplementation may change the gut microbial composition in mice towards a beneficial response to ICI immunotherapy.
Fig. 7.
Oral DAT supplementation alters the microbial composition of the gut in mice. Mice were provided with DAT via the drinking water ad libitum for 7 days. Stool samples of DAT treated (n = 10) or untreated (n = 10) mice were collected on day 7 and were analysed via NGS for 16S-rRNA. (A) Effective richness [p-values correspond to an unpaired t-test] and (B) Shannon effective representing alpha-diversity [p-values correspond to an unpaired t-test]. (C) Non-metric multidimensional scaling (NMDS) biplot showing distance between DAT and Ctrl groups. Taxonomic binning on (D) class and (E) order level [p-values correspond to an unpaired t-test]. (F) Taxonomic binning on order level of mice on day 0 which were retrospectively categorized into responder and non-responder to anti-CTLA-4 therapy [p-values correspond to an unpaired t-test with Welch's correction]. (G) Taxonomic binning on family level on day 7 after DAT treatment [p-values correspond to an unpaired t-test with Welch's correction]. All graphs show mean with 95% CI.
DAT supplementation does not aggravate immune-related adverse events during anti-CTLA-4 treatment
A major clinical limitation of ICI are immune-related adverse events (irAEs). With any combinatorial approach, concern for such over-stimulated and errant immune responses are apparent. A common irAE with significant morbidity in cancer patients undergoing ICI is colitis.50 Generally, murine models are poorly sensitive to irAEs. Accordingly, in our experimental mice, we did not observe any obvious clinical signs of ICI-induced colitis, such as (bloody) diarrhoea or weight loss. Previous reports described a subclinical form of colitis in mice undergoing anti-CTLA-4 immunotherapy.19 We were able to reproduce these observations with increased cleavage of caspase-3 in large intestinal tissue sections from mice undergoing anti-CTLA-4 treatment as a sign of therapy-induced apoptosis of intestinal cells (Fig. S7A and B). As these findings were not statistically significant, we additionally used our established assay of neutrophil influx into the gut lamina propria as a very sensitive surrogate marker for epithelial damage, loss of barrier function and intestinal inflammation.34 We could indeed recapitulate the previously published finding19 that anti-CTLA-4 treatment results in gut epithelial damage and subclinical colonic inflammation, here associated with massive neutrophil influx (mean difference of 1.11%, 95% CI 0.22–2.00); Fig. S7C, Table S3). Interestingly, oral supplementation with DAT ameliorated anti-CTLA-4-mediated gut damage and subsequent subclinical tissue inflammation (Fig. S7A–C). Taken together, these data suggest that oral supplementation of DAT in combination with ICI does not aggravate irAEs such as colitis, but in contrast may reduce colonic inflammation.
Discussion
The gut microbiome can modulate the response to ICI cancer immunotherapy, and bacterial-derived metabolites have emerged as one missing link between systemic host immunity and microbes at the epithelial interface. We here show that the bacterial metabolite DAT can enhance checkpoint inhibitor-mediated anti-tumour T-cell immunity. Oral supplementation of DAT not only resulted in more activated cytotoxic T and NK cells in the TME during immunotherapy, but also promoted priming and expansion of tumour antigen-specific CD4 and CD8 T cells. However, the exact cellular subsets that respond to DAT and mediate its immunomodulatory function remain to be determined. At this time, it is also unclear whether supplemented DAT primarily acts in the intestinal tract on the interaction of luminal gut microbiota, the epithelial barrier and local immune cells to fine-tune systemic immunity, or directly on cells in the TME. As shown previously, both endogenous (produced from gut microbiota) or administered DAT can enter the blood circulation and reach distant organ sites.15 In a murine model of viral infection, the same study suggested that tissue-resident phagocytes mediate the immunomodulatory DAT effect. Even though, we did not observe DAT-mediated maturation of DCs as an important type of phagocytes in the TME, we demonstrated that DAT in combination with concurrent (microbial) co-stimulatory signals was principally capable of stimulating DC maturation in vitro. Generally, the transient effects of DAT monotherapy on early tumour growth and its additive effect with anti-CTLA-4 or anti-PD-1 immunotherapy on anti-tumour T cell immunity implicate multilayered immunomodulatory DAT effects.
Several studies have suggested direct impact of bacterial metabolites on the function of T cells. The metabolite inosine has been described to augment immunotherapy with checkpoint inhibition (particularly anti-CTLA-4) via activation of the adenosine receptor A2A on T cells.30 Very recently, the microbial tryptophan catabolite indole-3-aldehyde (I3A) has been shown to promote immune checkpoint inhibitor treatment in murine melanoma via engagement of the aryl hydrocarbon receptor (AhR) on T cells.51 Interestingly, in this model, I3A derived from tumour-resident bacteria. However, the immunomodulatory effects of bacterial metabolites seem to be highly context-dependent and may thus appear contradictory. In contrast to the above-mentioned study, in a murine model of pancreatic ductal adenocarcinoma, engagement of AhR in tumour-associated macrophages by microbiota-derived indoles suppressed T-cell anti-tumour immunity.52 While direct stimulatory effects on T cells via epigenetic reprogramming have been described for the SCFA butyrate,53 its systemic administration was found to limit the antitumour effect of CTLA-4 blockade in mice via impaired DC maturation and subsequent T-cell priming.31 We found that DAT was capable of directly enhancing activation and differentiation of T cells undergoing TCR stimulation. Along these lines, the immune-promoting effects of oral DAT supplementation were robustly observed in two different tumour models, with different checkpoint inhibitors, and also following Abx treatment. Generally, the response to bacterial metabolites can be influenced by various host factors, including genetics, immune status, and the existing microbial community in the gut and also in the tumour tissue.21,22 These factors can impact the expression of receptors or signalling pathways involved in metabolite recognition and response, leading to differences in the immune response.54,55 Furthermore, different (tumour) tissues and organs have distinct immune microenvironments with different cellular composition and local cytokine milieu and thus the response to bacterial metabolites may vary.56,57
Our data suggest that DAT can directly act on immune cells including T cells and DCs, but the exact molecular mechanisms how DAT is detected in mammalian hosts remain unclear. DAT has been suggested to modulate and enhance IFN-I signalling, primarily by augmenting the positive feedback loop of IFNAR1 via STAT1.15 Accordingly, we found that DAT potently enhanced T-cell priming in response to a known IFN-I-inducer but failed to do so as monotherapy. In line with our previous findings,11 IFNAR1-deficient mice showed a severe defect of anti-CTLA-4-mediated anti-tumour immunity, which could not be circumvented by oral DAT supplementation. This again suggests that DAT enhances anti-tumour immunity via IFN-I signalling, and that DAT could be one of the microbial mediators that is required to help sustain basal IFN-I production in this context. Along these lines, previous reports identified intestinal microbiota to maintain constitutive host IFN-I expression via nucleic-acid sensing PRRs such as cGAS/STING or RIG-I, thereby allowing for anti-viral and anti-tumour immunity.21,58 The fact that DAT augmented anti-tumour immunity also in Abx-treated mice with severely reduced gut microbiota, suggests that microbial-derived metabolites may at least in part compensate the missing microbiota signals required to trigger basal IFN-I signalling. However, the immunostimulatory effects of DAT seem to require some level of co-stimulation of an intact (beneficial) microbiome, as i) in vitro maturation of DCs by DAT required co-exposure to LPS, and ii) DAT supplementation in Abx-treated mice did not reach the same additive efficacy with anti-CTLA-4 as in non-Abx groups. This is different to previous studies, in which the immunotherapy-promoting effects of inosine were totally dependent on inflammatory stimuli of the gut microbiota or exogenously added DAMPs to activate TLR9, and in their absence could even revert to immuno-suppressive function.30 At this stage, it is unclear which molecular co-stimulatory components (of the gut microbiome) contribute to antitumor efficacy of DAT. We and others have previously shown that tumour cell-derived nucleic acids can trigger cGAS/STING or RIG-I signalling for potent IFN-I production.11,59,60 In our current study, in vitro transcribed RNA as an artificial RIG-I ligand and “microbial mimicry” did indeed exhibit a strong additive effect with oral DAT supplementation to induced expansion of tumour antigen-specific T cells.
The interaction between immune-modulating cancer treatments and the host microbiota is dynamic and often bidirectional. Checkpoint inhibition with anti-CTLA-4 antibodies relies on the gut microbiota, but vice versa also modulates intestinal microbial composition in both mice and men.19 We here found that oral supplementation of mice with DAT also altered their intestinal microbiota, associated with a reduced bacterial richness and diversity. While high α-diversity is often considered as an indicator for a healthy gut microbiota, several meta-analyses could not find an association between gut microbial diversity and patient response to immunotherapy with checkpoint inhibitors.28,61,62 DAT supplementation resulted in reduced abundance of Clostridia, a class of obligate anaerobes in which several member species such as Flavonifractor plautii (Clostridium orbiscindens) have been shown to be producers of DAT.15 However, the role of endogenously produced DAT derived from the prevalent gut microbiota (in comparison to therapeutically supplemented DAT) remains unclear. Even though our data suggested a possible association between endogenous DAT levels in the stool and subsequent response to anti-CTLA-4 immunotherapy, this has to be interpreted with caution as the DAT levels in murine stools samples were at the detection limit of our mass spectrometry approach. Furthermore, due to ambiguous results, the predictive value for the abundance of certain bacterial species and their metabolites regarding clinical responses still requires further experimental validation. For example, SCFA-producing bacteria have been linked to favourable responses to checkpoint inhibitor therapy,63 while high blood butyrate and propionate levels were associated with resistance to CTLA-4 blockade in mice and patients.31
In our study, oral DAT supplementation resulted in increased abundance of Bacteroidales and Burkholderiales, the latter being the only bacterial class found to be enriched in anti-CTLA-4 responders. This is in line with previous studies, that linked bacterial species belonging to these classes to anti-CTLA-4-induced anti-tumour immunity.19 However, DAT-mediated changes of the gut microbiota in our study were only obvious at the order level. Meta-analysis from patients under ICI treatment have revealed commonalities among the bacteria identified not only at the level of individual species but also in larger taxonomical branches (on the level of family or higher).28,63 Generally, at this point, it remains unclear whether DAT-induced changes in host gut microbial composition impact on DAT-modulated anti-tumour immunity (in addition to its direct effects on CD8 T-cell and DC function), or are just a coincidental effect. Furthermore, modulation of microbial communities by exogenous supplementation seems to be dynamic and context-dependent. The intestinal microbial composition of mice with gut dysbiosis due to a constant high-fat diet was not significantly altered by oral DAT supplementation.64 However, in these mice, DAT treatment could attenuate chemically-induced colonic inflammation in an IFN-I-dependent manner, by replenishing endogenous DAT levels. Our data, that oral DAT supplementation could augment anti-CTLA-4-induced anti-tumour immunity during broad-spectrum antibiotic treatment without preventing Abx-induced intestinal bacterial depletion, hint to a possible similar mechanism.
Although oral DAT supplementation potently augmented anti-CTLA-4-induced anti-tumour T-cell immunity, we did not observe aggravated irAEs but rather reduced subclinical colitis. The microbial tryptophan catabolite indole-3-carboxaldehyde (3-IAld) has previously shown to ameliorate chemically-induced colitis in mice.65 Both engagement of the AhR and modulation of the gut microbiota contributed to improved epithelial barrier function and immune homeostasis in the gut. In a model of chemically-induced colitis in mice with metabolic syndrome, oral supplementation with DAT similarly protected epithelia barrier integrity and attenuated mucosal inflammation.64 Others have shown that certain microbiota-targeting interventions can uncouple anti-tumour efficacy from toxicity effects.19 Here, administration of Bacteroides and Burkolderia species in mice resulted in improved anti-CTLA-4 efficacy but reduced histopathological signs of colitis. The exact mechanisms how DAT supplementation seems to dissociate therapy efficacy from irAEs remains to be determined. At this stage, it is unclear as to what extent sex, gender, or diversity account for this study. Nevertheless, where possible, we have included equal ratios of female and male mice. We have monitored our data regularly to consider sex as a biological variable.
In summary, our results identify the IFN-I-modulating bacterial metabolite DAT to efficiently promote anti-tumour T-cell immunity. Given its low cytotoxicity, high tolerability, and oral bioavailability, DAT is a promising candidate as a new microbial-based therapeutic to improve the efficacy of cancer immunotherapy with checkpoint inhibitors, particularly in patients with prior antibiotic treatment. Nonetheless, a comprehensive and functional assessment of the beneficial or adversary effects of specific microbial consortia and their associated metabolites on the immune and anti-tumour responses elicited by immune checkpoint inhibitors will be necessary before defined metabolite-producing consortia or metabolite mixtures can be envisioned as new personalized microbial-based anticancer therapeutics.
Contributors
L.J., S.H., and H.P. designed the research, analysed, and interpreted the results. L.J., S.G. and A.S. did cell culture and animal experiments. K.N., L.J. and E.T.O. analysed 16S RNA-sequencing data. K.S. performed immunohistochemistry analyses. K.K. performed targeted metabolomic analyses. L.J., S.H. and H.P. prepared the manuscript. S.G., P.H., E.T-O., K.F., J.R., F.B, W.H. and C.P. gave methodological support and conceptual advice. S.H., and H.P. guided the study. L.J., S.H. and H.P. have accessed and verified the underlaying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Primary experimental data will be shared upon personal request by the corresponding authors. 16S RNA sequencing data of stool samples from mice undergoing DAT supplementation, anti-CTLA-4 immunotherapy, or Abx treatment were deposited in the European Nucleotide archive (ENA, reference #PRJEB61041, #PREJB61405, #PRJEB65308), and are publicly available.
Declaration of interests
S.H. is a consultant for Bristol Myers-Squibb, Novartis, Merck, Abbvie, and Roche. S.H. has received research funding from Bristol Myers-Squibb and Novartis. H.P. is a consultant for Gilead, Abbvie, Pfizer, Novartis, Servier, and Bristol Myers-Squibb. K.S. is consultant for TRIMT GmbH and has filed a patent for radiopharmaceutical target. H.P. and S.H. have received research funding from Bristol Myers-Squibb. E.T.O. has received honoraria from BeiGene. C.P. received research funding from Almirall and honoraria from MSD, BMS, Pierre-Fabre, Sanofi, Novartis, Almirall, Pelpharma, Pritzer, Merck, Leo Pharma, Sun Pharma, Janssen, Abbvie, Amazentis, and Scarletred. S.H. is an employee of and holds equity interest in Roche/Genentech. The remaining authors declare no financial conflict of interest.
Acknowledgements
This study was supported by a Young Investigator Award (Melanoma Research Alliance to S.H.), Deutsche Forschungsgemeinschaft—Projektnummer 360372040—SFB 1335 (to F.B., H.P. and K.S.), Projektnummer 395357507—SFB 1371 (to H.P), Projektnummer 324392634—TRR 221 (to H.P and W.H.), Bavarian Cancer Research Centre (BZFK; to H.P., W.H., and F.B.), and Projektnummer BA 2851/6–1 (to F.B.), the German Cancer Aid (70114547 to H.P.), the Wilhelm Sander Foundation (2021.041.1 to S.H., 2021.040.1 to H.P.), the Else-Kröner-Fresenius-Stiftung (funding line: Else-Kröner Forschungskolleg), the Bavarian State Ministry for Science and Art (to H.P.), the German José Carreras Foundation (DJCLS 07 R/2020 to S.H.), the TUM Innovation Network Fund (NextGenDrugs to F.B.) and the InCa research prize 2020 by the Novartis Foundation (to S.H). H. P. is supported by the EMBO Young Investigator Program. This work is part of the doctoral thesis of L.J. at the Technical University of Munich. We thank Tatiana Nedelko, Lena Klostermeier, and Maria Krieger (AG Poeck and AG Heidegger) as well as Olga Seelbach and Marion Mielke (Comparative Experimental Pathology, School of Medicine, TUM) for excellent technical support.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104834.
Contributor Information
Simon Heidegger, Email: simon.heidegger@tum.de.
Hendrik Poeck, Email: hendrik.poeck@ukr.de.
Appendix A. Supplementary data
References
- 1.Callahan M.K., Wolchok J.D. At the Bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. J Leukoc Biol. 2013;94:41–53. doi: 10.1189/jlb.1212631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodi F.S., O'Day S.J., McDermott D.F., et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkins M.B., Larkin J. Immunotherapy combined or sequenced with targeted therapy in the treatment of solid tumors: current perspectives. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv414. [DOI] [PubMed] [Google Scholar]
- 4.Pires da Silva I., Ahmed T., Reijers I.L.M., et al. Ipilimumab alone or ipilimumab plus anti-PD-1 therapy in patients with metastatic melanoma resistant to anti-PD-(L)1 monotherapy: a multicentre, retrospective, cohort study. Lancet Oncol. 2021;22:836–847. doi: 10.1016/S1470-2045(21)00097-8. [DOI] [PubMed] [Google Scholar]
- 5.Jessurun C.A.C., Vos J.A.M., Limpens J., Luiten R.M. Biomarkers for response of melanoma patients to immune checkpoint inhibitors: a systematic review. Front Oncol. 2017;7:233. doi: 10.3389/fonc.2017.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budhwani M., Mazzieri R., Dolcetti R. Plasticity of type I interferon-mediated responses in cancer therapy: from anti-tumor immunity to resistance. Front Oncol. 2018;8:322. doi: 10.3389/fonc.2018.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y., Hannan R., Fu Y.X. Type I IFN activating type I dendritic cells for antitumor immunity. Clin Cancer Res. 2021;27:3818–3824. doi: 10.1158/1078-0432.CCR-20-2564. [DOI] [PubMed] [Google Scholar]
- 8.Boukhaled G.M., Harding S., Brooks D.G. Opposing roles of type I interferons in cancer immunity. Annu Rev Pathol Mech Dis. 2021 2020;16:167–198. doi: 10.1146/annurev-pathol-031920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker B.S., Rautela J., Hertzog P.J. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 11.Heidegger S., Wintges A., Stritzke F., et al. RIG-I activation is critical for responsiveness to checkpoint blockade. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aau8943. [DOI] [PubMed] [Google Scholar]
- 12.Duarte C.W., Willey C.D., Zhi D., et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weichselbaum R.R., Ishwaran H., Yoon T., et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abt M.C., Osborne L.C., Monticelli L.A., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steed A.L., Christophi G.P., Kaiko G.E., et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stutz M.R., Dylla N.P., Pearson S.D., et al. Immunomodulatory fecal metabolites are associated with mortality in COVID-19 patients with respiratory failure. Nat Commun. 2022;13:6615. doi: 10.1038/s41467-022-34260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saura-Calixto F., Pérez-Jiménez J., Touriño S., et al. Proanthocyanidin metabolites associated with dietary fibre from in vitro colonic fermentation and proanthocyanidin metabolites in human plasma. Mol Nutr Food Res. 2010;54:939–946. doi: 10.1002/mnfr.200900276. [DOI] [PubMed] [Google Scholar]
- 18.Schoefer L., Mohan R., Schwiertz A., Braune A., Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol. 2003;69:5849–5854. doi: 10.1128/AEM.69.10.5849-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vétizou M., Pitt J.M., Daillère R., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivan A., Corrales L., Hubert N., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam K.C., Araya R.E., Huang A., et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–5356.e21. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nejman D., Livyatan I., Fuks G., et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan V., Spencer C.N., Nezi L., et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matson V., Fessler J., Bao R., et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubin K., Callahan M.K., Ren B., et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7 doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Routy B., Le Chatelier E., Derosa L., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 27.Derosa L., Routy B., Desilets A., et al. Microbiota-centered interventions: the next breakthrough in immuno-oncology? Cancer Discov. 2021;11:2396–2412. doi: 10.1158/2159-8290.CD-21-0236. [DOI] [PubMed] [Google Scholar]
- 28.Fernandes M.R., Aggarwal P., Costa R.G.F., Cole A.M., Trinchieri G. Targeting the gut microbiota for cancer therapy. Nat Rev Cancer. 2022;22:703–722. doi: 10.1038/s41568-022-00513-x. [DOI] [PubMed] [Google Scholar]
- 29.Chaput N., Lepage P., Coutzac C., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 30.Mager L.F., Burkhard R., Pett N., et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 31.Coutzac C., Jouniaux J.-M., Paci A., et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. 2020;11:2168. doi: 10.1038/s41467-020-16079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller U., Steinhoff U., Reis L.F.L., et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 33.Poeck H., Besch R., Maihoefer C., et al. 5′-triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 34.Fischer J.C., Wintges A., Haas T., Poeck H. Assessment of mucosal integrity by quantifying neutrophil granulocyte influx in murine models of acute intestinal injury. Cell Immunol. 2017;316:70–76. doi: 10.1016/j.cellimm.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Reitmeier S., Kiessling S., Clavel T., et al. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe. 2020;28:258–272.e6. doi: 10.1016/j.chom.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Klindworth A., Pruesse E., Schweer T., et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagkouvardos I., Joseph D., Kapfhammer M., et al. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6 doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kioukis A., Pourjam M., Neuhaus K., Lagkouvardos I. Taxonomy informed clustering, an optimized method for purer and more informative clusters in diversity analysis and microbiome profiling. Front Bioinform. 2022;2 doi: 10.3389/fbinf.2022.864597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagkouvardos I., Fischer S., Kumar N., Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ. 2017;5 doi: 10.7717/peerj.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reitmeier S., Hitch T.C.A., Treichel N., et al. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. ISME Commun. 2021;1 doi: 10.1038/s43705-021-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han J., Lin K., Sequeira C., Borchers C.H. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography–tandem mass spectrometry. Anal Chim Acta. 2015;854:86–94. doi: 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Zar J.H. 5th ed. Pearson; 2010. Biostatistical analysis. [Google Scholar]
- 43.Matthews J.N., Altman D.G., Campbell M.J., Royston P. Analysis of serial measurements in medical research. BMJ. 1990;300:230–235. doi: 10.1136/bmj.300.6719.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornung V., Ellegast J., Kim S., et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 45.Derosa L., Hellmann M.D., Spaziano M., et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29:1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tinsley N., Zhou C., Tan G., et al. Cumulative antibiotic use significantly decreases efficacy of checkpoint inhibitors in patients with advanced cancer. Oncologist. 2020;25:55–63. doi: 10.1634/theoncologist.2019-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elkrief A., El Raichani L., Richard C., et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinato D.J., Howlett S., Ottaviani D., et al. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol. 2019;5:1774–1778. doi: 10.1001/jamaoncol.2019.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoch C.L., Ciufo S., Domrachev M., et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020;2020 doi: 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martins F., Sofiya L., Sykiotis G.P., et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol. 2019;16:563–580. doi: 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
- 51.Bender M.J., McPherson A.C., Phelps C.M., et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. 2023;186:1846–1862.e26. doi: 10.1016/j.cell.2023.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hezaveh K., Shinde R.S., Klötgen A., et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. 2022;55:324–340.e8. doi: 10.1016/j.immuni.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luu M., Riester Z., Baldrich A., et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat Commun. 2021;12:4077. doi: 10.1038/s41467-021-24331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanco-García R.M., López-Álvarez M.R., Garrido I.P., et al. CD28 and KIR2D receptors as sensors of the immune status in heart and liver transplantation. Hum Immunol. 2011;72:841–848. doi: 10.1016/j.humimm.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Dil N., Qureshi M.A. Differential expression of inducible nitric oxide synthase is associated with differential Toll-like receptor-4 expression in chicken macrophages from different genetic backgrounds. Vet Immunol Immunopathol. 2002;84:191–207. doi: 10.1016/s0165-2427(01)00402-0. [DOI] [PubMed] [Google Scholar]
- 56.Conway J.W., Rawson R.V., Lo S., et al. Unveiling the tumor immune microenvironment of organ-specific melanoma metastatic sites. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2022-004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fridman W.H., Remark R., Goc J., et al. The immune microenvironment: a major player in human cancers. Int Arch Allergy Immunol. 2014;164:13–26. doi: 10.1159/000362332. [DOI] [PubMed] [Google Scholar]
- 58.Schaupp L., Muth S., Rogell L., et al. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell. 2020;181:1080–1096.e19. doi: 10.1016/j.cell.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 59.Woo S.R., Fuertes M.B., Corrales L., et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rose D., Ranoa E., Parekh A.D., et al. Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs. Oncotarget. 2016;7:26496–264515. doi: 10.18632/oncotarget.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gharaibeh R.Z., Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2019;68:385–388. doi: 10.1136/gutjnl-2018-317220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Limeta A., Ji B., Levin M., Gatto F., Nielsen J. Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCulloch J.A., Davar D., Rodrigues R.R., et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat Med. 2022;28:545–556. doi: 10.1038/s41591-022-01698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei Y., Gao J., Kou Y., et al. The intestinal microbial metabolite desaminotyrosine is an anti-inflammatory molecule that modulates local and systemic immune homeostasis. Faseb J. 2020;34:16117–16128. doi: 10.1096/fj.201902900RR. [DOI] [PubMed] [Google Scholar]
- 65.Renga G., Nunzi E., Pariano M., et al. Optimizing therapeutic outcomes of immune checkpoint blockade by a microbial tryptophan metabolite. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.