Abstract
Gene therapy using recombinant adeno-associated virus (rAAV) relies on safe, efficient, and precise in vivo gene delivery that is largely dependent on the AAV capsid. The proteinaceous capsid is highly amenable to engineering using a variety of approaches, and most resulting capsids carry substitutions or insertions comprised of natural amino acids. Here, we incorporated a non-canonical amino acid (ncAA), Nε-2-azideoethyloxycarbonyl-L-lysine (also known as NAEK), into the AAV5 capsid using genetic code expansion, and serendipitously found that several NAEK-AAV5 vectors transduced various cell lines more efficiently than the parental rAAV5. Furthermore, one NAEK-AAV5 vector showed lung-specific transduction enhancement following systemic or intranasal delivery in mice. Structural modeling suggests that the long side chain of NAEK may impact on the 3-fold protrusion on the capsid surface that plays a key role in tropism, thereby modulating vector transduction. Recent advances in genetic code expansion have generated synthetic proteins carrying an increasing number of ncAAs that possess diverse biological properties. Our study suggests that ncAA incorporation into the AAV capsid may confer novel vector properties, opening a new and complementary avenue to gene therapy vector discovery.
Keywords: rAAV5, capsid structures, non-canonical amino acids, NAEK, lung, alveolar cells, adeno-associated virus
Graphical abstract

Wang and colleagues report that non-canonical amino acid incorporation into the AAV capsid may confer novel vector properties, opening a new and complementary avenue to gene therapy vector discovery.
Introduction
Recombinant adeno-associated virus (rAAV) is currently the leading in vivo gene therapy delivery platform for preclinical and clinical applications.1,2 Attributing to its high transduction efficiency, broad tissue tropism, durable transgene expression, and clinically proven safety profile, rAAV gene therapy has shown encouraging outcomes in clinical trials targeting monogenic disorders,3 leading to regulatory approval of several AAV-based gene therapy drugs.4,5,6
Notwithstanding the encouraging safety and efficacy outcomes for many genetic indications, rAAV-based gene therapy is facing several hurdles toward broader utilization, such as immunogenicity,7,8 durability of transgene expression,9 and difficulties in achieving tissue-specific delivery and requirement of high doses for certain applications. To overcome these limitations, AAV capsid development has been a major focus in gene therapy research. A variety of strategies are employed to generate novel AAV capsids with desired features,10 including isolation of natural serotypes from human and other species,11 rational design enabled by understanding of structure-function relationships,12,13 directed evolution by DNA shuffling and random peptide insertion,14,15,16,17 in silico design,18,19 and their combinatorial approaches.20,21 These efforts continue to produce AAV vectors with increasing specificity and potency.
Most engineered AAV capsids comprise structural viral proteins (VPs) of canonical amino acids encoded by a modified AAV capsid gene (Cap). Others carry additional chemical decorations to redirect tropism or shield from neutralizing antibodies, among which genetic code expansion provides a versatile platform to engineer AAV capsid.13,22,23,24,25 In this approach, a non-canonical amino acid (ncAA) is incorporated into VPs via recoding a nonsense mutation in the Cap gene; the ncAA (e.g., Nε-2-azideoethyloxycarbonyl-L-lysine, also known as NAEK) harbors a chemical moiety that enables bio-orthogonal click reaction with a ligand, thereby allowing site-specific ligand conjugation on capsid surface.13,22,23,25 As a result, an additional purification step following click reaction is necessary to remove unconjugated ligand, increasing the complexity and cost associated with vector manufacturing. Furthermore, the stoichiometry of conjugation needs careful optimization and characterization because it may impact on AAV capsid stability, integrity, and function.
In this study, we used genetic code expansion to generate a series of AAV5 vectors bearing NAEK. We chose to engineer the AAV5 capsid because recent studies showed that it has a favorable immunological profile in clinical applications.26 Surprisingly, we found that some NAEK-modified AAV5 (NAEK-AAV5) vectors exhibited enhanced cell transduction efficiency compared with the parental AAV5 vector without further conjugation to a ligand. In particular, NAEK substitution of VP residue 374D (i.e., 374NAEK) confers lung-specific transduction enhancement in mice. Structural modeling studies indicate that the long side chain of NAEK may alter a critical receptor binding region of the AAV5 capsid, thereby modulating tropism and transduction.
Recent advances in genetic code expansion technology allow a variety of ncAAs to be incorporated during protein synthesis.27 These ncAAs possess diverse physical and chemical properties that are not present in natural amino acids, and can functionalize the synthetic protein products for a range of research and therapeutic applications.28 Our study suggests that non-ncAA incorporation is a promising approach to AAV capsid engineering.
Results
Generation of NAEK-AAV5 vectors and screen for packaging yield
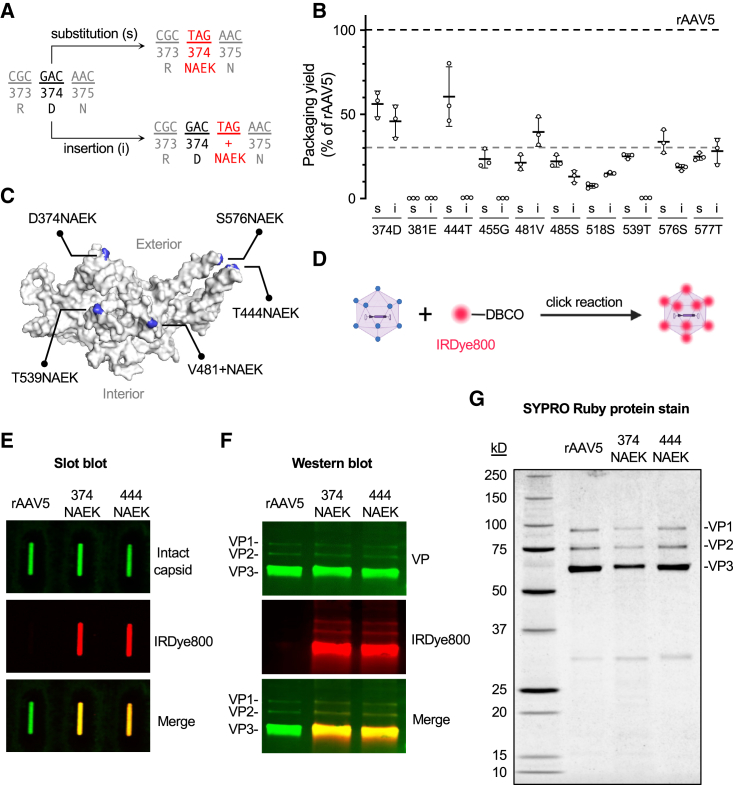
We used genetic code expansion to incorporate NAEK into specific sites of AAV5 capsid during vector production (Figure 1A). Specifically, a nonsense mutation was introduced into the AAV capsid gene (Cap∗), resulting in Cap∗ mRNA bearing a UAG premature termination codon. The UAG was decoded by an orthogonal suppressor tRNA system that comprises the pyrrolysyl-tRNA synthetase/pyrrolysyl-tRNA pair and NAEK, thereby incorporating NAEK into VPs during translation. In the presence of adenoviral helper genes, the NAEK-AAV5 capsid encapsidates a recombinant AAV vector genome that expresses enhanced green fluorescent protein (EGFP) as a reporter. NAEK-AAV5 vectors were purified in the same way as for standard rAAV5 (Figure 1B).
Figure 1.
Schematic diagram showing NAEK incorporation into the AAV capsid during vector production
(A) NAEK (blue dot) is incorporated into the capsid via suppression of a premature termination codon in the Cap mRNA (Cap∗), also known as genetic code expansion. With adenoviral gene products from pHelper, the transgene cassette flanked by inverted terminal repeats in pCis is packaged into the modified rAAV capsid harboring NAEK at a specific site of the capsid surface. (B) Workflow of the NAEK-AAV vector packaging process. Four plasmids are co-transfected into HEK293 cells, and NAEK is added to the culture medium. NAEK-AAV vectors are then purified using the same procedure for regular rAAV.
Ten VP amino acid residues across variable regions (VRs) III to VIII were selected for NAEK engineering (Figure S1) because VRs were shown to impact on rAAV tropism.29 NAEK was incorporated as either a substitution of a single residue or an insertion downstream (Figure 2A), generating a total of 20 NAEK-AAV5 vector designs. Next, we performed a small-scale vector production assay,30 and found that NAEK modification generally compromised or abolished packaging yield, consistent with a previous report.25 However, a few NAEK-AAV5 vectors exhibited packaging yield close to or more than 30% that of rAAV5 (Figure 2B); we therefore selected the top 5 candidates for further studies.
Figure 2.
Packaging yield screen and validation of NAEK incorporation
(A) NAEK was incorporated into the AAV5 capsid via suppression of a TAG nonsense mutation in the Cap gene as either a substitution (s) or insertion (i). Modification at residue 374D is shown as an example. (B) Packaging yield of 20 NAEK-AAV5 vectors in a small-scale packaging assay. Data are normalized to the yield of unmodified rAAV5. Gray dashed line indicates 30% of rAAV5 yield. (C) Space-filling rendering of AAV5 VP monomer (PDB: 7kp3). Five selected residues for NAEK modification are highlighted in blue and labeled. (D) Schematic diagram showing the conjugation between NAEK-AAV5 and DBCO-IRDye800. Blue dots indicate NAEK. (E) Slot-blotting image of purified rAAV5, or rAAV5 carrying D374NAEK substitution (374NAEK) or T444NAEK substitution (444NAEK). The green signal is generated from antibody that detects intact AAV5 capsid, and the red signal shows the fluorescence of IRDye800; a merged image is shown at the bottom. (F) Western blotting image of the purified rAAV shown in (E). The green signal is generated from B1 antibody that detects all three AAV VP isoforms (VP1, VP2, and VP3), and the red signal shows the fluorescence of IRDye800; a merged image is shown at the bottom. (G) SYPRO Ruby protein stain image of SDS-PAGE of the purified rAAV shown in (E).
These NAEK incorporation sites were mapped onto the cryogenic electron microscopy (cryo-EM) structure of the AAV5 capsid (PDB: 7KP3), which suggested that NAEK was exposed to the outside of assembled capsids (Figure 2C). To confirm the solvent-facing incorporation of NAEK, we took advantage of the azido moiety in NAEK that enables click reaction with a dibenzocyclooctyne (DBCO) group (Figure 2D). As expected, purified NAEK-AAV5 vectors were successfully conjugated with an infrared dye (IRDye800) via click reaction under non-denaturing condition as demonstrated in a slot-blotting assay (Figure 2E). Furthermore, western blotting assay revealed that all three VPs contained NAEK (Figure 2F). To examine whether truncated VPs generated by a Cap∗ gene were incorporated in purified rAAV (predicted molecular weight of truncated VPs: 20.5–41.3 kDa for D374∗; 28.6–49.5 kDa for T444∗, where asterisks indicate the premature stop codon), we performed SDS-PAGE followed by SYPRO Ruby protein staining. Compared with the full-length VPs, we did not observe appreciable truncated VP species of predicted molecular weights (Figure 2G). Together, these results showed that NAEK was incorporated into the AAV5 capsid, and that capsid integrity was not impaired with NAEK exposed to the outside of capsid.
In vitro transduction of cell culture by NAEK-AAV5 vectors
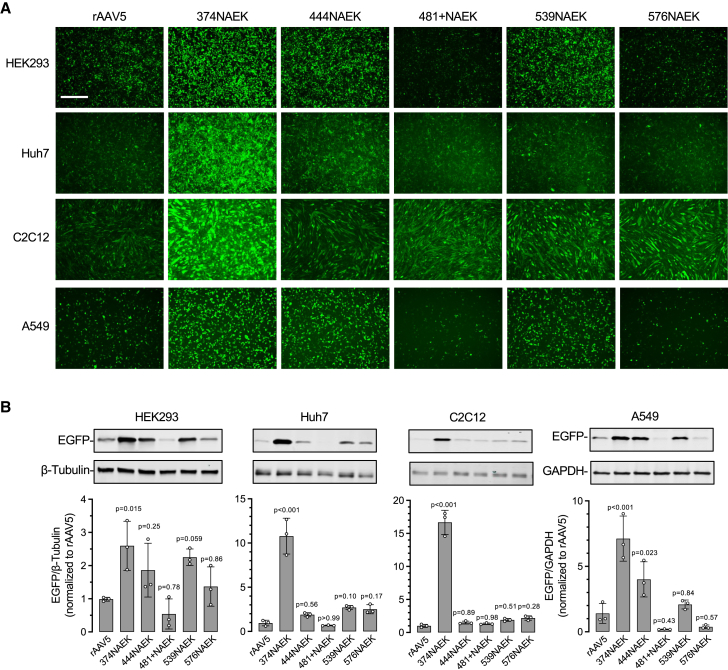
Next, we investigated the transduction efficiency of selected NAEK-AAV5 vectors in commonly used cell lines, including a human embryonic kidney cell line (HEK293), a human hepatocyte-derived carcinoma cell line (Huh7), a mouse myoblast cell line (C2C12), and a human lung epithelial cell line (A549), and compared them against parental rAAV5. Fluorescence imaging (Figure 3A) and western blotting (Figure 3B) revealed that 374NAEK-AAV5 exhibited the highest transduction efficiency among all cell lines tested, and was significantly more potent than rAAV5 by 2.5- to 10.5-fold. In addition, 444NAEK-AAV5 and 539NAEK-AAV5 also showed a trend of higher transduction than rAAV5 in HEK293 and A549 cells (Figure 3). Therefore, incorporating the ncAA NAEK into rAAV5 could modulate cellular transduction.
Figure 3.
Transduction of various cell lines by selected NAEK-AAV5 vectors
(A) Fluorescence microscopy images of various cell lines transduced by unmodified rAAV5 or rAAV5 carrying the indicated NAEK modifications. All rAAVs package the same transgene cassette that expresses enhanced green fluorescent protein (EGFP). Cells were infected with each rAAV at a multiplicity of infection (MOI) of 50,000 with co-infection of adenovirus type 5 (AdV5) at a MOI of 100. Scale bar, 500 μm. (B) Representative western blotting images of protein expression from cells as described in (A), and quantification of EGFP protein expression normalized to either β-tubulin or GAPDH. Data are normalized to the rAAV5 group. Data are mean ± SD.
Evaluation of NAEK-AAV5 transduction in vivo
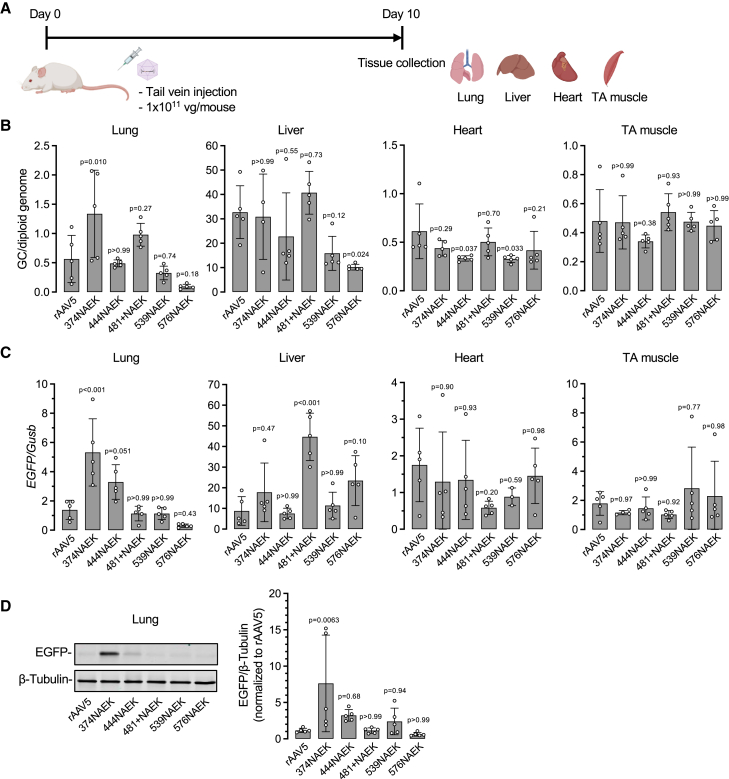
The in vitro transduction results prompted us to further study NAEK-AAV5 vectors for their tissue tropism in vivo. To this end, we injected adult mice with individual NAEK-AAV5 vectors at a dose of 1 × 1011 vector genomes (vg)/mouse via tail vein. Ten days later, mice were euthanized and lung, liver, heart, and tibialis anterior (TA) muscle were collected for molecular analysis (Figure 4A). Consistent with the in vitro results, 374NAEK-AAV5 emerged as the most potent vector targeting the lung, resulting in 2.5- and 4.5-fold higher vector DNA abundance and EGFP mRNA expression than rAAV5, respectively (Figures 4B and 4C). Interestingly, 374NAEK-AAV5 was comparable with rAAV5 for targeting the liver, heart, and skeletal muscle, indicating tissue-specific transduction enhancement upon in vivo delivery. To further confirm the enhanced lung-targeting capacity of 374NAEK-AAV5, total lung protein was extracted and subjected to western blotting. Indeed, 374NAEK-AAV5-treated mice showed nearly 8-fold higher EGFP protein expression in the lung than those receiving rAAV5 (Figure 4D).
Figure 4.
In vivo transduction by selected NAEK-AAV5 vectors in mice via systemic administration
(A) Schematic diagram of workflow. Six-week-old female mice were injected with rAAV at a dose of 1 × 1011 vector genomes (vg) per mouse via tail vein. Lung, liver, heart, and tibialis anterior (TA) muscle were collected 10 days post injection. (B) ddPCR quantification of rAAV transgene (i.e., EGFP) genome copy (GC) number in lung, liver, heart, and TA muscle of the mice described in (A). (C) ddPCR quantification of EGFP cDNA reverse-transcribed from its mRNA in lung, liver, heart, and TA muscle of the mice described in (A). (D) Representative western blotting images and quantification of transgene EGFP protein expression in lungs of the mice described in (A). Data are shown as mean ± SD; each white dot represents an individual mouse. n = 3–5 mice per group.
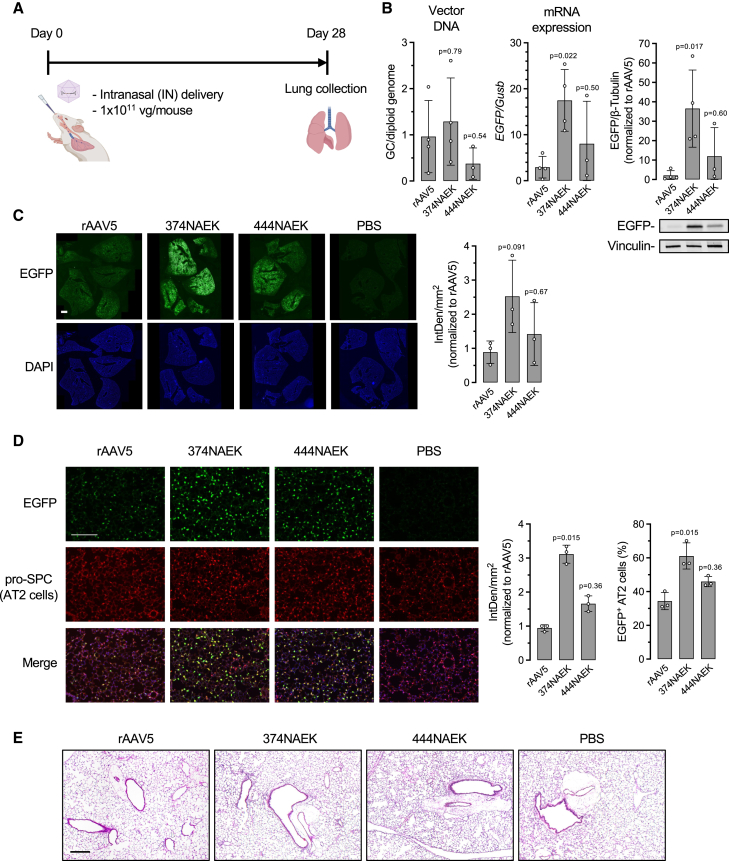
Encouraged by 374NAEK-AAV5’s superb lung transduction following systemic delivery, we next performed intranasal instillation to study its lung tropism in mice (Figure 5A) because intranasal delivery of rAAV was shown to target the lung more efficiently than intravenous delivery.31 Following this localized treatment regimen (Figure 5A), 374NAEK-AAV5 also outperformed the other vectors to target the lung, reaching 3- and 6-fold higher transgene EGFP expression than rAAV5 at mRNA and protein levels, respectively, although difference in transgene DNA abundance among all vectors did not reach statistical significance (Figure 5B). Taken together, these results demonstrate that NAEK incorporation at a specific site of the AAV5 capsid (i.e., residue 374) can enhance in vivo transduction in a tissue-specific manner.
Figure 5.
In vivo transduction by selected NAEK-AAV5 vectors in mice via intranasal administration
(A) Schematic diagram of workflow. Six-week-old female mice were injected with rAAV at a dose of 1 × 1011 vg per mouse via intranasal instillation. Lungs were collected 10 days post injection. (B) Quantification of rAAV vector DNA, transgene expression of mRNA, and EGFP protein expression in lung of the mice described in (A). Representative western blotting images of transgene EGFP protein expression are shown. Data are shown as mean ± SD; each white dot represents an individual mouse. n = 4–5 mice per group.
Characterization of lung tropism of leading NAEK-AAV5 vectors
To further characterize the lung tropism, another cohort of mice receiving intranasal instillation of rAAV5 or 374NAEK-AAV5 were euthanized at a longer time point of 4 weeks post treatment (Figure 6A), when rAAV transgene expression plateaued and stabilized.32 In addition, we also included 444NAEK-AAV5 in this experiment, because it repeatedly showed a trend of higher transgene expression than rAAV5 in prior experiments (Figures 3B, 4C, 4D, and 5B). Molecular analysis (Figure 6B) and immunostaining of lung sections (Figure 6C) corroborated the previous finding that 374NAEK-AAV5 led to the highest EGFP transgene expression in the lung.
Figure 6.
Characterization of lung transduction by two NAEK-AAV5 vectors via intranasal administration
(A) Schematic diagram of workflow. Six-week-old female mice were injected with rAAV at a dose of 1 × 1011 vg per mouse via intranasal instillation. Lungs were collected 28 days post injection. (B) Quantification of rAAV vector DNA, transgene expression of mRNA, and EGFP protein expression in lungs of mice described in (A). Representative western blotting images of transgene EGFP protein expression are shown. (C) Representative fluorescence microscopy images of lung sections and quantification of EGFP immunofluorescence intensity. Five lobes of one mouse are shown in each image. IntDen, integrated density of EGFP signal. Scale bar, 1 mm. (D) Representative fluorescence immunostaining images of lung sections and quantification. Alveolar type 2 (AT2) cells are labeled with antibody against pro-SPC (red). The fraction of EGFP+ AT2 cells is calculated as the percentage of EGFP+ pro-SPC+ double-positive cells among pro-SPC+ cells. Scale bar, 200 μm. (E) Representative images of hematoxylin and eosin (H&E) staining of lung sections from mice administered with the indicated rAAV. Data are shown as mean ± SD; each white dot represents an individual mouse. n = 3–4 mice per group. In (D) the Kruskal-Wallis test was used followed by Dunn’s test against the rAAV5 group, because the data did not pass normal distribution test.
Furthermore, we investigated the major lung cell types targeted by rAAV5 or the two NAEK-AAV5 vectors. Immunostaining results showed that most EGFP fluorescence was co-localized with pro-SPC, a specific marker of alveolar type 2 cells33 (Figure 6D). 374NAEK-AAV5 not only led to 3-fold higher transgene expression than rAAV5, but also transduced more alveolar type 2 cells than rAAV5 (60% vs. 35%) (Figure 6D). In contrast, EGFP expression from all vectors was rarely co-localized with the alveolar type 1 cell marker podoplanin (Figure S2). Importantly, neither 374NAEK-AAV5 nor 444NAEK-AAV5 triggered discernible inflammatory responses or histological changes to the lung (Figure 6E), suggesting that NAEK-AAV vectors were well tolerated despite the unnatural side chain of NAEK.
NAEK is a lysine (K) mimic (Figure 7A). To test whether the D374K substitution would confer the same lung tropism as D374NAEK, we performed intranasal instillation to another cohort of mice with various vectors (Figure 7B). However, 374K-AAV5 showed comparable lung transduction with rAAV5, both of which were inferior to 374NAEK-AAV5 (Figure 7C). Previous studies showed that rAAV6 and its two derivatives, rAAV6.2 and rAAV6.2FF, could transduce alveolar and airway epithelium in mice following intranasal or intratracheal delivery.34,35,36 Therefore, we also included these vectors for comparison and found that they were not as efficient as 374NAEK-AAV5 for lung transduction (Figure 7C). As expected, liver and heart contained much less vector DNA than the lung, presumably caused by vector leakage following intranasal delivery, and the transgene mRNA expression in the liver and heart was largely comparable among all tested vectors and much lower than that in the lung (Figures 7D and 7E).
Figure 7.
Comparison between NAEK-AAV5 vectors and other AAV vectors for in vivo transduction
(A) Chemical structures of canonical amino acids aspartic acid (D) and lysine (K) and the non-canonical amino acid NAEK. (D) The wild-type amino acid at residue 374 of AAV5 VP1. (B) Schematic diagram of workflow. Six-week-old female mice were injected with rAAV at a dose of 1 × 1011 vg per mouse via intranasal instillation. Lung, liver, and heart were collected 10 days post injection. (C) Quantification of rAAV vector DNA, transgene expression of mRNA, and EGFP protein expression in lungs of mice described in (B). Representative western blotting images of transgene EGFP protein expression are shown. (D) Quantification of rAAV vector DNA and transgene expression of mRNA in livers of mice described in (B). (E) Quantification of rAAV vector DNA and transgene expression of mRNA in hearts of mice described in (B). Data are shown as mean ± SD; each white dot represents an individual mouse. n = 3–5 mice per group.
Mechanistic studies on NAEK incorporation and vector properties
To explore the mechanisms underlying the enhanced lung transduction by 374NAEK-AAV5, we characterized its cellular trafficking in the human lung epithelial cell line A549 and compared it against rAAV5 and other NAEK-AAV5 vectors. In this set of in vitro experiments, 374NAEK-AAV5 exhibited the highest cell surface binding capacity and the most efficient internalization among all tested vectors (Figures 8A and 8B). After cell entry, 374NAEK-AAV5 delivered the greatest total number of vector DNA copies to nuclei, where transgene expression took place (Figure 8C). Notably, this cellular trafficking pattern was unique to 374NAEK-AAV5 but not the other NAEK-modified vectors, indicating that the structural context of NAEK incorporation played a key role in determining vector property.
Figure 8.
Mechanistic studies of the impact of NAEK incorporation on vector properties
(A) Comparison between rAAV5 and five NAEK-AAV5 vectors for their binding capacity to A549 cells (a human lung epithelial cell line). (B) Comparison between rAAV5 and five NAEK-AAV5 vectors for their internalization efficiency in A549 cells. (C) Comparison between rAAV5 and five NAEK-AAV5 vectors for their cytoplasmic and nuclear distribution in A549 cells. (D) Ribbon diagram of AAV5 VP1 monomer (PDB: 7KP3). The position of the 3-fold axis is indicated by a black triangle. Enlarged views show the VR-III-containing residue 374D and the neighboring β turn. (E) Ribbon diagrams (top panels) showing three amino acids at residue 374D (purple), 374K (green), and 374NAEK (cyan). In the bottom panels, electron density maps of 374D (purple), 374K (green), 374NAEK (cyan), and their neighboring amino acid 498E (yellow) are shown as a colored mesh. (F) Interatomic distances (in angstroms and indicated by dashed lines) between 374NAEK (cyan) and 498E (yellow) are labeled.
Residue 374 resides in VR-III, which is remote from the 3-fold protrusion, a capsid structural feature determining receptor binding and tropism.29 However, we noticed that VR-III is spatially close to a β turn near the 3-fold axis (Figure 8D). When various amino acids (D, K, and NAEK) were fitted at residue 374 by structural modeling, only 374NAEK was in proximity with 498E in the neighboring β turn owing to its long side chain (Figure 8E), potentially forming hydrogen bonds between the two residues (Figure 8F). Thus, we postulate that the potential interaction between 374NAEK and 498E induces conformational changes near the 3-fold axis, thereby modulating the tropism of the 374NAEK-AAV5 vector.
Discussion
In this study, we show that incorporation of a ncAA (i.e., NAEK) into the AAV5 capsid can modulate vector transduction. In particular, the 374NAEK-AAV5 vector exhibits lung-specific transduction enhancement in mice by either systemic injection or intranasal instillation, and significantly outperforms leading lung-tropic AAV capsids including AAV5, AAV6, and certain AAV6 variants. Our study suggests that ncAA incorporation is a viable approach to improving AAV vector tropism, expanding the arsenal of novel capsid discovery strategies.
A variety of ncAAs have been used to modify recombinant proteins as research tools. For example, Seidel et al. incorporated the photo-crosslinking amino acid p-azido-Phe (Azi) into various sites of class B G-protein-coupled receptors (GPCRs). When a peptide antagonist binds to the Azi-GPCR and locates in proximity to the Azi residue, crosslinking occurs upon UV light activation, which reveals the detailed interaction between GPCR and peptide antagonists including the structural footprints and contact sites.37 To our knowledge, ncAA incorporation into the proteinaceous AAV capsid for gene therapy vector development has been largely relying on NAEK as a chemical handle, followed by ligand conjugation via click reaction.13,22,24,25 Alternatively, the capsid surface-exposed amino acid side chains can be directly modified by chemical approaches.38,39,40 For example, Horowitz et al. modified AAV2 arginine residues to the neutral adduct hydroimidazolone by glycation, which redirected the vector tropism from liver to skeletal and cardiac muscle in mice by systemic administration.38
The simplicity of one-step ncAA incorporation without a downstream chemical reaction sets our study apart from the aforementioned methods for capsid engineering. Therefore, manufacturing ncAA-AAV vectors is very similar to standard rAAV production, whereas a two-step capsid modification method requires an additional purification procedure, which increases the complexity and cost of vector manufacture and may decrease yield. Importantly, ncAA incorporation occurs via genetic code expansion at a pre-defined premature termination codon, and therefore is precise and consistently generates a homogeneous batch of vectors. In contrast, decorating purified rAAV via either click reaction or other chemical approaches usually results in incomplete modification and/or unspecific modification at multiple residues, consequently producing inconsistent vector batches.
We identified 374NAEK-AAV5 as a potent vector to target lung alveolar type 2 cells in mice. AAV-based gene therapy is regarded as a promising therapeutic modality to treat a range of lung diseases, such as cystic fibrosis, and several lung-targeting AAV capsids have been reported.35,41 For example, Limberis et al. compared AAV serotypes 1–9 for their lung transduction efficiency in mice, and found that AAV5 and AAV6 were among the most efficient. Furthermore, they demonstrated that AAV6.2, an AAV6 F129L mutant, outperformed the other serotypes tested to transduce mouse lung epithelium.35 Transduction efficiency of rAAV6.2 was approximately 2-fold higher than the parental rAAV6 in both airways and alveoli.34 Introducing Y455F and Y731F substitutions to AAV6.2 yielded AAV6.2FF, which was shown to further increase transgene expression in muscle and lung, presumably due to reduced proteasomal degradation of the capsid.36 Notably, 374NAEK-AAV5 vector showed higher lung transduction efficiency than AAV5, AAV6, AAV6.2, and AAV6.2FF vectors 10 days after intranasal delivery, suggesting that 374NAEK-AAV5 is a promising gene therapy vector targeting the lung, especially for gene delivery to alveolar type 2 cells that are involved in diseases such as surfactant B deficiency.42 Future translational studies may evaluate cell-type-specific transgene expression over a longer time course and tropism in non-human primates. It is also important to assess potential immune responses against ncAA and possible new epitopes harboring ncAA. We note that other AAV capsids, such as AAV443 and an engineered AAV9-derived capsid,44 exhibit promising lung transduction in mice following intravenous delivery and hold promise in clinical translation.
Our comparative studies on several NAEK-AAV5 vectors indicate that both NAEK incorporation and its insertion site (and hence structural context) play a role in modulating transduction efficiency and intracellular trafficking. Consistent with this notion, structural modeling reveals the potential interaction between 374NAEK and 498E that resides near the 3-fold protrusion, a critical structural determinant of tropism. This interaction is enabled by the long side chain of NAEK that does not exist in canonical amino acids, including the wild-type aspartic acid (D) or lysine (K). Detailed interatomic interactions and conformational changes caused by 374NAEK substitution will require structural analysis such as cryo-EM and three-dimensional reconstruction. Moreover, how these interaction changes impart enhanced cellular transduction also warrants further investigation. Our in vitro study using A549 cells indicates that 374NAEK substitution confers stronger vector binding to the cell surface, eventually leading to higher nuclear vector DNA. However, we note that 374NAEK-AAV5 may exhibit enhanced lung transduction via a different mechanism once delivered in vivo. Nevertheless, these results support the conclusion that incorporation of ncAA with a synthetic side chain may confer novel AAV capsid properties that are not readily achievable by the limited canonical amino acids.
To identify ncAA-AAV vectors of desired properties more efficiently and rapidly, it is plausible to synergize with a library screening approach.45,46 Alternatively, rational design to incorporate specific ncAAs with desired biological characteristics may also functionalize the resulting vectors. As advances in genetic code expansion make an increasing number of ncAAs amenable to synthetic protein production, the diverse ncAA-AAVs may provide an untapped yet rich repertoire for gene therapy vector discovery.
Materials and methods
Cell culture
HEK293 cells (cat. no. CRL-1573), C2C12 cells (cat. no. CRL-1772), and A549 cells (cat. no. CRM-CCL-185) were purchased from ATCC. Huh7 cells were obtained from Wen Xue as a gift. All cells were maintained in Gibco Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, cat. no. 11965-084) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (culture medium hereafter) under 37°C and 5% CO2.
Small-scale AAV vector packaging assay
To generate pRep2/Cap5∗ plasmids, TAG nonsense mutations were introduced in the Cap gene by Gibson Assembly (New England BioLabs, cat. no. E2611) or site-directed mutagenesis (TaKaRa, cat. no. R045A) according to the manufacturer’s instructions. Four plasmids (pAAVsc.PI-CB6-EGFP, pRep2/Cap5∗, pHelper, and ptRNA/aaRS [Addgene, cat. no. 50832]) were co-transfected into HEK293 cells at a mass ratio of 1:1:1:2 using the calcium precipitation method (Promega, cat. no. E1200). After 12 h, culture medium was replaced with fresh medium containing 1 mM NAEK (Aquila Pharmatech, cat. no. SCI0043). At 72 h post transfection, cells were detached, suspended in the culture medium, and subjected to three successive freeze-thaw cycles. The crude lysates were centrifuged at 15,000 g/min for 15 min at 4°C to remove cell debris; supernatant was stored at −20°C. DNase-resistant rAAV titer in cleared crude lysate was determined by real-time quantitative PCR (qPCR) as described previously.30
Large-scale AAV vector production
Similar to the small-scale AAV vector packaging process, the aforementioned four plasmids were co-transfected into HEK293 cells. After 12 h, culture medium was replaced with fresh DMEM containing 1 mM NAEK (Aquila Pharmatech, cat. no. SCI0043). At 72 h post transfection, AAV vectors were purified from crude lysates by CsCl gradient centrifugation followed by dialysis.47 All AAV vectors packaged the same self-complementary vector genome expressing EGFP. Purified AAV vectors were quantified by droplet digital PCR (ddPCR) for vector genome titer and silver-stained SDS-PAGE analysis for purity assessment.
rAAV conjugation with DBCO-IRDye800 via click reaction
rAAV (2.5 × 1011 vg) and DBCO-IRDye800 (LI-COR Biosciences, cat. no. 929–50000) was mixed at a molar ratio of 1:1,000 in 200 μL of Dulbecco’s phosphate-buffered saline (DPBS) (pH 7.0) (Corning, cat. no. 21-031-CV) in a 1.5 mL microcentrifuge tube, and incubated at room temperature in darkness for 2 h. After click reaction, the mixture was purified using Amicon Ultra 100K filter (MilliporeSigma, cat. no. UFC510096) and reconstituted in 50 μL of DPBS.
rAAV transduction assay in vitro
Cells were seeded at a density of 2 × 105 to 4 × 105 cells/well in a 12-well plate. Next day, cells were co-infected with rAAV at a multiplicity of infection (MOI) of 50,000 and human adenovirus 5 (AdV5) at a MOI of 100. At 48 h (HEK293, Huh7, A549 cells) or 168 h (C2C12 cells) post infection, cells were subjected to fluorescence imaging to qualitatively detect EGFP expression. Another batch of cells were infected in the same way, and cell lysates were collected at 48 h post infection and subjected to western blotting to quantitatively measure transduction.
Animal use and treatment
Six-week-old female C57BL/6J mice were purchased from The Jackson Laboratory (Stock no. 000664). Mice were administered with AAV vectors at a dose of 1 × 1011 vg/mouse by either tail vein injection or modified intranasal instillation.31 After 10 or 28 days post treatment, mice were sacrificed for tissue collection. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of University of Massachusetts Chan Medical School.
rAAV vector DNA and transcript quantification
Flash-frozen tissues, including lung, liver, heart, and TA muscle, were subjected to total DNA and RNA extraction using AllPrep DNA/RNA Mini Kit (QIAGEN, cat. no. 80204). RNA was reverse-transcribed into cDNA using a cDNA Reverse Transcription Kit (Thermo Fisher Scientific, cat. no. 4374966). Vector DNA and cDNA were quantified using duplexing TaqMan ddPCR assays targeting EGFP (Thermo Fisher Scientific, Assay ID Mr00660654_cn) and Tfrc genomic sequences (Thermo Fisher Scientific, cat. no. 4458367), or EGFP and Gusb cDNA (Thermo Fisher Scientific, cat. no. 4448490), respectively. ddPCR was carried out using QX200 (Bio-Rad), and results were analyzed using QuantaSoft (Bio-Rad).
Western blotting
For rAAV5 conjugated with IRDye800, 1 × 1010 vg of rAAV purified using an Amicon filter was loaded in each lane for SDS-polyacrylamide gel electrophoresis (SDS-PAGE), followed by standard membrane transfer procedure. Membrane was then incubated with mouse anti-VP1/2/3 (B1) primary antibody (PROGEN, cat. no. 61058) at 4°C overnight and subsequently incubated with goat anti-mouse IgG polyclonal antibody-IRD-680RD secondary antibody (LI-COR, cat. no. 926–68070) at room temperature for 1 h. Membrane was imaged using a LI-COR Odyssey Imaging System and images were analyzed with Image Studio Lite Quantification Software (LI-COR).
To detect proteins in cells and tissues, total protein was extracted using Novex NP40 Cell Lysis Buffer (Thermo Fisher Scientific, cat. no. FNN0021) and T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific, cat. no. 78510), respectively. Primary antibodies include rabbit anti-EGFP antibody (1:3,000, Thermo Fisher Scientific, cat. no. A-11122), rabbit anti-GAPDH antibody (1:3,000, Abcam, cat. no. ab9485), anti-vinculin antibody (1:5,000, Abcam, cat. no. ab219649), and mouse anti-β-tubulin antibody (1:5,000, Abcam, cat. no. ab7291). Secondary antibodies include goat anti-mouse IgG polyclonal antibody-IRD-680RD and donkey anti-rabbit IgG polyclonal antibody-IRD-800RD (1:5,000, LI-COR, cat. no. 926–68070 and 926–32213, respectively).
Slot blotting
rAAV (1 × 1010 vg) was diluted in 150 μL of DPBS and then loaded on a nitrocellulose membrane in a Blotting Manifold (Hoefer, PR648). After the rAAV suspension filtered through by gravity (∼30 min), blocking buffer (LI-COR, cat. no. 927–70003) was added onto the membrane and allowed to filter through by gravity (∼1 h). After blocking, the membrane was washed with DPBS three times using a vacuum. Washed membrane was incubated with mouse anti-AAV5 intact particle antibody (PROGEN, cat. no. 610148) at 4°C overnight. Subsequently, the membrane was incubated with goat anti-mouse IgG IRD-680RD secondary antibody at room temperature for 1 h, and imaged using a LI-COR Odyssey Imaging System.
SYPRO Ruby protein stain
Purified rAAVs and molecular weight markers (Bio-Rad, cat. no. 1610363) were subjected to SDS-PAGE as mentioned above. The gel was fixed in 50% methanol and 7% acetic acid for 60 min (two 30-min rounds), and incubated in SYPRO Ruby gel stain (Thermo Fisher Scientific, cat. no. S12000) overnight. Next day, gel was washed in 10% methanol and 7% acetic acid, rinsed in MilliQ water, and imaged using a GelDoc Go Gel Imaging System (Bio-Rad).
rAAV binding, internalization, and subcellular distribution analysis
rAAV binding and internalization assay was performed as previously reported48 with minor modifications. In brief, A549 cells were seeded 18 h prior to assay. Cells were pre-chilled at 4°C for 30 min to stop endocytosis, infected with rAAV at a MOI of 67,000, and incubated at 4°C for 1 h. After incubation, cells were washed gently with cold DPBS three times to remove unbound rAAV. For binding assay, washed cells were harvested and subjected to total DNA isolation using QIAGEN AllPrep DNA/RNA Mini Kit. For internalization assay, after unbound vectors were removed, cells were incubated in warm culture medium at 37°C and 5% CO2 for 1 h to allow rAAV endocytosis to occur. After incubation, cells were detached in 0.05% trypsin (Cytiva, cat. no. SV3003101) for 5 min followed by quenching with culture medium. Cell suspension was centrifuged and cell pellets were washed with DPBS three times followed by total DNA isolation using the QIAGEN AllPrep DNA/RNA Mini Kit.
Subcellular distribution of rAAV was analyzed as reported previously49 with minor modifications. A549 cells were infected with rAAV at a MOI of 50,000 and harvested after 24 h. The cytoplasmic and nuclear fractions were isolated using an NE-PER Kit (Thermo Fisher Scientific, cat. no. 78833). Both fractions were subjected to duplexing TaqMan qPCR to quantify rAAV vector DNA distribution. Vector DNA was detected using a TaqMan reagent targeting the EGFP gene (Thermo Fisher Scientific, Assay ID Mr00660654_cn). For nuclear fraction, a TaqMan reagent targeting the human RNase P gene (Thermo Fisher Scientific, TaqMan copy number assay ID 4331182) was chosen as a normalization reference of A549 genomic DNA. For cytoplasmic fraction, a TaqMan reagent targeting mitochondrial DNA (forward primer: CACCCAAGAACAGGGTTTGT; reverse primer: TGGCCATGGGTATGTTGTTA; probe: /5HEX/TTACCGGGC/ZEN/TCTGCCATCT/3IABkFQ/; synthesized by Integrated DNA Technologies) was used as normalization reference. The qPCR assays were performed using TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, cat. no. 4369016) on a StepOne Real-Time PCR System (Thermo Fisher Scientific).
Immunofluorescence staining of lung sections
Mice were intratracheally perfused with fresh 4% paraformaldehyde (PFA). Lungs were dissected, fixed in 4% PFA for 24 h, and dehydrated in 15% sucrose in 1× PBS for 24 h. Dehydrated lung was embedded in a disposable mold containing OCT and flash frozen at −80°C for at least 24 h. Embedded lung was cut into 15 μm sections using a cryostat microtome and stored at −80°C. Sections on glass slides were permeabilized with 0.1% Triton X-100 in 1× PBS for 10 min and blocked with 5% donkey serum for 30 min, followed by primary antibody incubation at 4°C overnight and secondary antibody incubation at room temperature for 1 h. Primary antibodies used were anti-EGFP antibody (1:1,000, Aves Labs, cat. no. GFP1010), anti-podoplanin (1:500, Abcam, cat. no. ab11936), and rabbit anti-pro-SPC (1:1,000, MilliporeSigma, cat. no. AB3786). Secondary antibodies include goat anti-chicken IgY H&L Alexa Fluor 488 (1:1,000, Abcam, cat. no. ab150169) and goat anti-rabbit IgG (H + L) Alexa Fluor Plus 594 (1:1,000, Thermo Fisher Scientific, cat. no. 32740). Immunostained sections were mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, cat. no. P36970) and subjected to imaging using a THUNDER Imaging System (Leica Microsystems, Germany). Fluorescence intensity was quantified and analyzed using ImageJ (National Institutes of Health).
Structural modeling of AAV capsid
The structure of intact 60-mer AAV5 capsid (PDB: 7KP3) was retrieved from RCSB PDB and VP monomer was displayed using PyMOL 2.4.1. The amino acid residues selected for NAEK substitution or insertion were pseudo-colored and mapped onto the VP monomer structure. The NAEK chemical structure was created using the Builder module of PyMOL. The Mutagenesis module of PyMOL was used to replace the wild-type 374 aspartic acid (D) to either lysine (K) or NAEK. Interatomic distance was measured using the measurement module of PyMOL.
Statistical analysis
Data are presented as mean ± standard deviation (SD). Comparison among multiple groups was analyzed by one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test against rAAV5 group, corrected for multiple comparison. GraphPad Prism 9 was used for statistical analysis and data plotting.
Acknowledgments
We wish to thank Qin Su for providing training on intratracheal perfusion, Mengtian Cui for generously sharing the TaqMan reagents detecting mitochondrial DNA, the Morphology Core Facility at UMass Chan for tissue sectioning and immunostaining, and the Viral Vector Core at UMass Chan for producing the AAV vectors used in this study. The Wang Lab is supported by a grant from the National Institutes of Health (NIH) (P01HL158506). The Gao Lab is supported by grants from the NIH (R01NS076991-01, P01AI100263-01, P01HL131471-02, 35 R01AI121135, UG3HL147367–01, R01HL097088, and U19AI149646-01) and the Cystic Fibrosis Foundation. Some illustrations were created with biorender.com.
Author contributions
H.C. and D.W. designed the research. H.C. carried out the majority of the experiments. H.C., A.D., X.Z., and J.L. performed mouse injection and other mouse work. J.J. provided NAEK and advice on its use. L.R., H.C., and N.L. did molecular cloning. L.R. and N.L. produced rAAV. H.C., D.W., and G.G. analyzed data and wrote the manuscript.
Declaration of interests
This work was funded in part through a sponsored research agreement with Luye Pharma. J.J. was a paid employee of Luye Pharma. None of the other authors received personal compensation from Luye Pharma or hold financial interest in the company. H.C., A.D., G.G., and D.W. are inventors of a patent application filed by the University of Massachusetts Chan Medical School concerning the work described in this study. G.G. is a scientific co-founder of Voyager Therapeutics, Adrenas Therapeutics, and Aspa Therapeutics, and holds equity in the companies.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.101129.
Contributor Information
Guangping Gao, Email: guangping.gao@umassmed.edu.
Dan Wang, Email: dan.wang@umassmed.edu.
Supplemental information
Data and code availability
All study data are included in the article and/or supplemental information.
References
- 1.Hastie E., Samulski R.J. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success--a personal perspective. Hum. Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendell J.R., Al-Zaidy S.A., Rodino-Klapac L.R., Goodspeed K., Gray S.J., Kay C.N., Boye S.L., Boye S.E., George L.A., Salabarria S., et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021;29:464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Büning H. Gene therapy enters the pharma market: the short story of a long journey. EMBO Mol. Med. 2013;5:1–3. doi: 10.1002/emmm.201202291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeler A.M., Flotte T.R. Recombinant Adeno-Associated Virus Gene Therapy in Light of Luxturna (and Zolgensma and Glybera): Where Are We, and How Did We Get Here? Annu. Rev. Virol. 2019;6:601–621. doi: 10.1146/annurev-virology-092818-015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heo Y.A. Etranacogene Dezaparvovec: First Approval. Drugs. 2023;83:347–352. doi: 10.1007/s40265-023-01845-0. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton B.A., Wright J.F. Challenges Posed by Immune Responses to AAV Vectors: Addressing Root Causes. Front. Immunol. 2021;12:675897. doi: 10.3389/fimmu.2021.675897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ertl H.C.J. Immunogenicity and toxicity of AAV gene therapy. Front. Immunol. 2022;13:975803. doi: 10.3389/fimmu.2022.975803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muhuri M., Levy D.I., Schulz M., McCarty D., Gao G. Durability of transgene expression after rAAV gene therapy. Mol. Ther. 2022;30:1364–1380. doi: 10.1016/j.ymthe.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 11.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y., Wang J., Liu Y., Qu Y., Wang K., Zhang Y., Chang Y., Yang Z., Wan J., Liu J., et al. Variants of the adeno-associated virus serotype 9 with enhanced penetration of the blood-brain barrier in rodents and primates. Nat. Biomed. Eng. 2022;6:1257–1271. doi: 10.1038/s41551-022-00938-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., Yao T., Zheng Y., Li Z., Zhang Q., Zhang L., Zhou D. Development of next generation adeno-associated viral vectors capable of selective tropism and efficient gene delivery. Biomaterials. 2016;80:134–145. doi: 10.1016/j.biomaterials.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Jiang J., Drouin L.M., Agbandje-McKenna M., Chen C., Qiao C., Pu D., Hu X., Wang D.Z., Li J., Xiao X. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. USA. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonnenmacher M., Wang W., Child M.A., Ren X.Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabebordbar M., Lagerborg K.A., Stanton A., King E.M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J.J., Messemer K.A., et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinn E., Pacouret S., Khaychuk V., Turunen H.T., Carvalho L.S., Andres-Mateos E., Shah S., Shelke R., Maurer A.C., Plovie E., et al. In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector. Cell Rep. 2015;12:1056–1068. doi: 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez T.J., Simon K.E., Blondel L.O., Fanous M.M., Roger A.L., Maysonet M.S., Devlin G.W., Smith T.J., Oh D.K., Havlik L.P., et al. Cross-species evolution of a highly potent AAV variant for therapeutic gene transfer and genome editing. Nat. Commun. 2022;13:5947. doi: 10.1038/s41467-022-33745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shay T.F., Sullivan E.E., Ding X., Chen X., Ravindra Kumar S., Goertsen D., Brown D., Crosby A., Vielmetter J., Borsos M., et al. Primate-conserved carbonic anhydrase IV and murine-restricted LY6C1 enable blood-brain barrier crossing by engineered viral vectors. Sci. Adv. 2023;9:eadg6618. doi: 10.1126/sciadv.adg6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelemen R.E., Mukherjee R., Cao X., Erickson S.B., Zheng Y., Chatterjee A. A Precise Chemical Strategy To Alter the Receptor Specificity of the Adeno-Associated Virus. Angew. Chem., Int. Ed. Engl. 2016;55:10645–10649. doi: 10.1002/anie.201604067. [DOI] [PubMed] [Google Scholar]
- 23.Yao T., Zhou X., Zhang C., Yu X., Tian Z., Zhang L., Zhou D. Site-Specific PEGylated Adeno-Associated Viruses with Increased Serum Stability and Reduced Immunogenicity. Molecules. 2017;22:1155. doi: 10.3390/molecules22071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katrekar D., Moreno A.M., Chen G., Worlikar A., Mali P. Oligonucleotide conjugated multi-functional adeno-associated viruses. Sci. Rep. 2018;8:3589. doi: 10.1038/s41598-018-21742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puzzo F., Zhang C., Powell Gray B., Zhang F., Sullenger B.A., Kay M.A. Aptamer-programmable adeno-associated viral vectors as a novel platform for cell-specific gene transfer. Mol. Ther. Nucleic Acids. 2023;31:383–397. doi: 10.1016/j.omtn.2023.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pipe S.W., Leebeek F.W.G., Recht M., Key N.S., Castaman G., Miesbach W., Lattimore S., Peerlinck K., Van der Valk P., Coppens M., et al. Gene Therapy with Etranacogene Dezaparvovec for Hemophilia B. N. Engl. J. Med. 2023;388:706–718. doi: 10.1056/NEJMoa2211644. [DOI] [PubMed] [Google Scholar]
- 27.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 28.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 29.Agbandje-McKenna M., Kleinschmidt J. AAV capsid structure and cell interactions. Methods Mol. Biol. 2011;807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 30.Ai J., Ibraheim R., Tai P.W.L., Gao G. A Scalable and Accurate Method for Quantifying Vector Genomes of Recombinant Adeno-Associated Viruses in Crude Lysate. Hum. Gene Ther. Methods. 2017;28:139–147. doi: 10.1089/hgtb.2016.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santry L.A., Ingrao J.C., Yu D.L., de Jong J.G., van Lieshout L.P., Wood G.A., Wootton S.K. AAV vector distribution in the mouse respiratory tract following four different methods of administration. BMC Biotechnol. 2017;17:43. doi: 10.1186/s12896-017-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm D., Zhou S., Nakai H., Thomas C.E., Storm T.A., Fuess S., Matsushita T., Allen J., Surosky R., Lochrie M., et al. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood. 2003;102:2412–2419. doi: 10.1182/blood-2003-02-0495. [DOI] [PubMed] [Google Scholar]
- 33.Liebler J.M., Marconett C.N., Juul N., Wang H., Liu Y., Flodby P., Laird-Offringa I.A., Minoo P., Zhou B. Combinations of differentiation markers distinguish subpopulations of alveolar epithelial cells in adult lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;310:L114–L120. doi: 10.1152/ajplung.00337.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halbert C.L., Allen J.M., Miller A.D. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Limberis M.P., Vandenberghe L.H., Zhang L., Pickles R.J., Wilson J.M. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 2009;17:294–301. doi: 10.1038/mt.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Lieshout L.P., Domm J.M., Rindler T.N., Frost K.L., Sorensen D.L., Medina S.J., Booth S.A., Bridges J.P., Wootton S.K. A Novel Triple-Mutant AAV6 Capsid Induces Rapid and Potent Transgene Expression in the Muscle and Respiratory Tract of Mice. Mol. Ther. Methods Clin. Dev. 2018;9:323–329. doi: 10.1016/j.omtm.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidel L., Zarzycka B., Zaidi S.A., Katritch V., Coin I. Structural insight into the activation of a class B G-protein-coupled receptor by peptide hormones in live human cells. Elife. 2017;6:e27711. doi: 10.7554/eLife.27711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horowitz E.D., Weinberg M.S., Asokan A. Glycated AAV vectors: chemical redirection of viral tissue tropism. Bioconjugate Chem. 2011;22:529–532. doi: 10.1021/bc100477g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mével M., Bouzelha M., Leray A., Pacouret S., Guilbaud M., Penaud-Budloo M., Alvarez-Dorta D., Dubreil L., Gouin S.G., Combal J.P., et al. Chemical modification of the adeno-associated virus capsid to improve gene delivery. Chem. Sci. 2019;11:1122–1131. doi: 10.1039/c9sc04189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Fang Y., Zhou Y., Zandi E., Lee C.L., Joo K.I., Wang P. Site-specific modification of adeno-associated viruses via a genetically engineered aldehyde tag. Small. 2013;9:421–429. doi: 10.1002/smll.201201661. [DOI] [PubMed] [Google Scholar]
- 41.Meyer-Berg H., Zhou Yang L., Pilar de Lucas M., Zambrano A., Hyde S.C., Gill D.R. Identification of AAV serotypes for lung gene therapy in human embryonic stem cell-derived lung organoids. Stem Cell Res. Ther. 2020;11:448. doi: 10.1186/s13287-020-01950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang M.H., van Lieshout L.P., Xu L., Domm J.M., Vadivel A., Renesme L., Mühlfeld C., Hurskainen M., Mižíková I., Pei Y., et al. A lung tropic AAV vector improves survival in a mouse model of surfactant B deficiency. Nat. Commun. 2020;11:3929. doi: 10.1038/s41467-020-17577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westhaus A., Cabanes-Creus M., Rybicki A., Baltazar G., Navarro R.G., Zhu E., Drouyer M., Knight M., Albu R.F., Ng B.H., et al. High-Throughput In Vitro, Ex Vivo, and In Vivo Screen of Adeno-Associated Virus Vectors Based on Physical and Functional Transduction. Hum. Gene Ther. 2020;31:575–589. doi: 10.1089/hum.2019.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goertsen D., Goeden N., Flytzanis N.C., Gradinaru V. Targeting the lung epithelium after intravenous delivery by directed evolution of underexplored sites on the AAV capsid. Mol. Ther. Methods Clin. Dev. 2022;26:331–342. doi: 10.1016/j.omtm.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., Herrmann A.K., Pham Q.H., Borowski C., Hille S., et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 2020;11:5432. doi: 10.1038/s41467-020-19230-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu H.L., Brown A., Loveland A.B., Lotun A., Xu M., Luo L., Xu G., Li J., Ren L., Su Q., et al. Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties. Nat. Commun. 2020;11:3279. doi: 10.1038/s41467-020-17047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su Q., Sena-Esteves M., Gao G. Purification of Recombinant Adeno-Associated Viruses (rAAVs) by Cesium Chloride Gradient Sedimentation. Cold Spring Harb. Protoc. 2020;2020:095604. doi: 10.1101/pdb.prot095604. [DOI] [PubMed] [Google Scholar]
- 48.Berry G.E., Tse L.V. Virus Binding and Internalization Assay for Adeno-associated Virus. Bio. Protoc. 2017;7:e2110. doi: 10.21769/BioProtoc.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madigan V.J., Yuziuk J.A., Chiarella A.M., Tyson T.O., Meganck R.M., Elmore Z.C., Tse L.V., Hathaway N.A., Asokan A. Ring finger protein 121 is a potent regulator of adeno-associated viral genome transcription. PLoS Pathog. 2019;15:e1007988. doi: 10.1371/journal.ppat.1007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supplemental information.