Summary
The germline TP53 p.R337H mutation is reported as the most common germline TP53 variant. It exists at a remarkably high frequency in the population of southeast Brazil as founder mutation in two distinct haplotypes with the most frequent co-segregating with the p.E134∗ variant of the XAF1 tumor suppressor and an increased cancer risk. Founder mutations demonstrate linkage disequilibrium with neighboring genetic polymorphic markers that can be used to identify the founder variant in different geographic regions and diverse populations. We report here a shared haplotype among Brazilian, Portuguese, and Spanish families and the existence of three additional distinct TP53 p.R337H alleles. Mitochondrial DNA sequencing and Y-STR profiling of Brazilian carriers of the founder TP53 p.R337H allele reveal an excess of Native American haplogroups in maternal lineages and exclusively European haplogroups in paternal lineages, consistent with communities established through male European settlers with extensive intermarriage with Indigenous women. The identification of founder and independent TP53 p.R337H alleles underlines the importance for considering the haplotype as a functional unit and the additive effects of constitutive polymorphisms and associated variants in modifier genes that can influence the cancer phenotype.
Keywords: R337H, founder mutation, haplotype, ancestry, Iberian Peninsula, Brazil, Sephardic Jewish, mitochondrial DNA, Y-STR, cancer predisposition
p.R337H is the most common germline TP53 variant that exists as a founder and as independent alleles. In this context, the haplotype as a functional unit needs to be considered due to the additive effects of constitutive polymorphisms and associated variants in modifier genes that can influence cancer susceptibility and phenotype.
Introduction
TP53 is a tumor suppressor that regulates the cell cycle, senescence, DNA repair, metabolism, and apoptosis in response to DNA damage and other cellular stresses. Given this central role in maintaining cellular homeostasis, it is not surprising that p53 is frequently selected against in human cancer by mutation, thus rendering this tumor suppressor inactive. Although most TP53 mutations are somatically acquired, individuals who inherit an inactivating TP53 variant are highly susceptible to developing cancer at an early age with diverse tumor types and multiple malignancies.
The TP53 p.R337H variant, the most common germline variant in The TP53 Database (formerly the International Agency for Research on Cancer; IARC), was first reported in a pediatric patient of Portuguese ancestry with adrenocortical cancer treated in France.1 Previous studies have established p.R337H as a hypomorphic, attenuated mutant allele that confers a highly variable cancer risk ranging from individuals who remain unaffected over their lifetime to those who meet the clinical criteria for classic Li-Fraumeni Syndrome (LFS).2,3,4 It is now recognized that the TP53 p.R337H variant is a founder mutation5 frequently found throughout south and southeastern Brazil, including the major states of São Paulo and Paraná (total population of 60 million) being detected in one in every 300 individuals.6,7 Carriers of the founder TP53 p.R337H variant share an identical DNA sequence for the TP53 locus,8 with the majority retaining homology that extends to the telomeric region of chromosome 17 encompassing a nonsense mutation (p.E134∗) in XAF1 (X-linked inhibitor of apoptosis-associated factor 1).8 Previous studies have established XAF1 as a proapoptotic tumor suppressor that functions within a positive autoregulatory loop with TP53.9 Consistent with these findings, individuals who inherit the compound mutant haplotype (TP53 p.R337H; XAF1 p.E134∗) are at an increased risk for cancer in general, sarcomas, and multiple tumors compared with those carriers with only the p.R337H mutation.8
Haplotype analysis makes it possible to discriminate between a founder mutation10 or a variant resulting from an independent mutational event. This is particularly important in the context of hypomorphic TP53 variants, as specific polymorphisms can impact p53 expression and function, and thus influence cancer risk.11,12 By integrating genotype, haplotype, and ancestry analyses we have assessed 38 TP53 p.R337H carriers from South America, North America, Europe, South Africa, and Asia to determine the diversity of the TP53 p.R337H alleles.
Materials and methods
TP53 p.R337H carriers
Genomic DNA samples from 38 unrelated individuals (33 patients with cancer, including 25 females) harboring the TP53 p.R337H variant from 11 different countries (Brazil, Portugal, Spain, France, Germany, England, United States, Japan, American Samoa, South Africa, and Argentina) were included in this study (Table 1). Personal and family histories of cancer were collected by the genetic counselor or physician at each participating institution. Genetic testing was offered as a TP53 single gene analysis or as part of a multi-gene hereditary cancer panel (Ambry Genetics, OvaNext; Myriad, Myrisk Hereditary Cancer Test; Illumina, TruSight Hereditary Cancer Panel) with additional genetic findings included in Table 1. The validation cohort included 86 unrelated (documented for at least three generations) and unaffected Brazilian males who carry the TP53 p.R337H allele. Appropriate institutional informed consent guidelines were followed for all participants or caregivers. This study was approved by local ethics committees and the institutional review board at St. Jude Children’s Research Hospital.
Table 1.
Geographic origin and demographics of 38 TP53 p.R337H carriers
| Country | Primary tumor (age/diagnosis)/Additional tumors | Status | Gendera | R337H | E134∗ | Haplotype |
|---|---|---|---|---|---|---|
| (1) Brazil | ACT (9) | Pediatric/Deceased | M | Pos (H) | Pos (H) | Hap1 |
| (2) Brazil | CPC (6) | Pediatric | M | Pos (H) | Pos (H) | Hap1 |
| (3) USA/Brazil | ACT (3) | Pediatric | M | Pos | Neg | Hap2 |
| (4) USA/Brazil | ACT (1) | Pediatric | F | Pos | Pos | Hap1 |
| (5) Brazil | ACT (3) | Pediatric | M | Pos | Pos | Hap1 |
| (6) Brazil | ACT (2) | Pediatric/Deceased | F | Pos | Pos | Hap1 |
| (7) Argentina | ACT (4) | Pediatric/Deceased | F | Pos | Pos | Hap1 |
| (8) Brazil | ACT (11) | Pediatric | F | Pos | Pos | Hap1 |
| (9) Argentina | ACT (9) | Pediatric | F | Pos | Neg | Hap3 |
| (10) Spain | ACT (3) | Pediatric | M | Pos | Pos | Hap1 |
| (11) Spain | ACT (2) | Pediatric | M | Pos | Pos | Hap1 |
| (12) Spain | Unaffected A&W (19) | Adult | M | Pos | Pos | Hap1 |
| (13) Portugal | ACT (4) | Pediatric | F | Pos | Neg | Hap2 |
| (14) Portugal/Brazil | ACT (4) | Pediatric | F | Pos | Pos | Hap1 |
| (15) Portugal | Liposarcoma (40) | Adult | F | Pos | Pos | Hap1 |
| (16) Portugal | Gastric (62) | Adult | M | Pos | Pos | Hap1 |
| (17) Portugal | Breast Her2+ (40) | Adult | F | Pos | Pos | Hap1 |
| (18) USA/Brazil | ACT (34) | Adult | M | Pos | Neg | Hap2 |
| (19) USA | Breast (41) | Adult | F | Pos | Neg | Hap4 |
| (20) USA | Breast (39), Thyroid (57), Leiomyosarcoma (62) | Adult | F | Pos | Neg | Hap4 |
| (21) USA | Thyroid (27), Breast (36), Lung (59), Melanoma (60) | Adult/Deceased | F | Pos | Neg | Hap4 |
| (22) USA | Unclassified spindle cell sarcoma (27), Breast (29), Sarcoma (33) | Adult | F | Pos | Neg | Hap4 |
| (23) USA | Basal cell carcinoma (53) | Adult | F | Pos | Neg | Hap3 |
| (24) USA | Unaffected A&W (39) | Adult | M | Pos | Neg | Hap3 |
| (25) USA | Breast (38) | Adult | F | Pos | Pos | Hap1 |
| (26) USA | Breast (45), Thyroid (55) | Adult | F | Pos | Neg | Hap4 |
| (27) USA | Breast (46), Breast (49), Head and neck (59), STS (61) | Adult | F | Pos | Neg | Hap4 |
| (28) USA/Brazil | Breast (45), Low grade mucinous appendiceal carcinoma (47) | Adult | F | Pos | Neg | Hap2 |
| (29) France | Breast (64), Endometrial (77) | Adult | F | Pos | Neg | Hap3 |
| (30) Germany | Colon (65), ACT (71), Prostate (74) | Adult/Deceased | M | Pos | Neg | Hap3 |
| (31) England | Unaffected A&W (45) | Adult | F | Pos | Neg | Hap3 |
| (32) Japan/Brazil | ACT (2) | Pediatric | M | Pos | Pos | Hap1 |
| (33) Samoa | ACT (9) | Pediatric | F | Pos (de novo) | Neg | Hap5 |
| (34) Germany | Tubal carcinoma (54), Peritoneal carcinomatosis (58) | Adult | F | Pos | Neg | Hap3 |
| (35) Germany | Unaffected A&W (29) | Adult | F | Pos | Neg | Hap3 |
| (36) Germany | Unaffected A&W (30) | Adult | M | Pos | Neg | Hap3 |
| (37) Germany | Gastric (62) | Adult | F | Pos | Neg | Hap3 |
| (38) South Africa | ACT (5) | Pediatric | F | Pos | Neg | ND |
#15, 16, 17, 19, 20, 21, 25, 26, 28 and 29 are BRCA1/2 negative by genetic panel testing.
#22 is positive for BRCA1 c.4065_4068del(TCAA) inherited from father.
A&W, alive and well (no current cancer); ACT, adrenocortical tumor; CPC, choroid plexus carcinoma; F, female; H, homozygous; M, male; ND, not determined; STS, soft tissue sarcoma.
Gender defined by biological attributes.
Haplotype determination of recurrent TP53 p.R337H alleles
Genomic DNA was isolated and used to determine TP53 sequence and the XAF1 p.E134∗ variant status as previously reported.8 Additionally, fluorescent-labelled PCR products for 10 polymorphic microsatellite markers located within (VNTRp53) or telomeric to the TP53 locus (Table S1) were genotyped by using the ABI 3500 sequencer (ThermoFisher Scientific) and sizes assigned to the different fragments using GeneMapper v 6.0 (ThermoFisher Scientific).8,13 Haplotypes were determined by segregation analysis of genomic DNA from parents and descendants, loss of heterozygosity in tumor samples, or inferred.
Databases
Information for the TP53 p.R337H variants was retrieved from The TP53 Database (https://tp53.isb-cgc.org/; resource for more than 30,000 somatic and germline TP53 variants in human cancer); ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/; public archive for human variants and associated clinical phenotypes); gnomAD (https://gnomad.broadinstitute.org/; repository for more than 140,000 exome and whole genome sequences selected from human disease-specific and population genetic studies); Global Biobank Engine (https://biobankengine.stanford.edu/; summary of genotype-phenotype correlations in large biobank cohorts representing 750,000 individuals); ABraOM (abraom.ib.usp.br; archive of exome and whole genome sequences from >1,000 unrelated, elderly (>70 years old) individuals from São Paulo, Brazil); the Precision Oncology Knowledge Base (OncoKB; https://www.oncokb.org/; housed by Memorial Sloan Kettering Cancer Center representing more than 7,000 genetic alterations in 134 cancer types); and the Catalog of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic).
Mitochondrial DNA control region
The nucleotide sequence of mitochondrial DNA (mtDNA) hypervariable segment 1 (HVI; between nucleotide positions 16024 and 16365), segment 2 (HV2; between nucleotide positions 73 and 340), and segment 3 (HV3, covering the position 438 to 574) of mtDNA control region was determined for 37 of 38 probands, and available family members (n = 26). We have also determined the mtDNA haplotype for all Brazilian male individuals harboring the p.R337H allele (validation cohort) inherited from their mother (n = 45).
Amplification of the mtDNA control region was performed by using L15781 5′-CCCTTTTACCATCATTGGACA-3′ and H727 5′-AGGGTGAACTCACTGGAACG-3′ primers. Amplified segments were sequenced by using L15781, H16478, L109, H408, and H727 primers as previously described.14 Resulting sequences were compared with the Homo sapiens mitochondrion, complete genome (NCBI reference sequence: NC_012920.1), known as revised Cambridge Reference Sequence (rCRS). Haplogrep software (https://haplogrep.i-med.ac.at/), EMPOP (https://empop.online/), and visual inspection analyses were used to define the mtDNA haplogroups.
Y chromosome markers
A set of 23 Y-STRs (short tandem repeats) was analyzed for 13 unrelated male p.R337H carriers and four first-degree male relatives in the main cohort of 38 individuals using the PowerPlex Y23 System (Promega) as described by the manufacturer. Y-STR haplotypes were also determined for Brazilian male individuals harboring the p.R337H allele (validation cohort) that was paternally inherited (n = 41). Alleles were separated and detected on an ABI 3500 sequencer (ThermoFisher Scientific) and sizes were assigned to the different fragments using GeneMapper v 6.0 (ThermoFisher Scientific). The alleles were named according to the number of repeated units, based on the sequenced allelic ladder following International Society for Forensic Genetics recommendations.15,16 The classification of Y chromosome haplogroup was done using the Haplogroup Predictor program FTDNA 2.0 (http://www.hprg.com/hapest5/index.html) and Y chromosome Haplotype Reference Database (https://yhrd.org/search).
Results
Haplotypes of TP53 p.R337H carriers
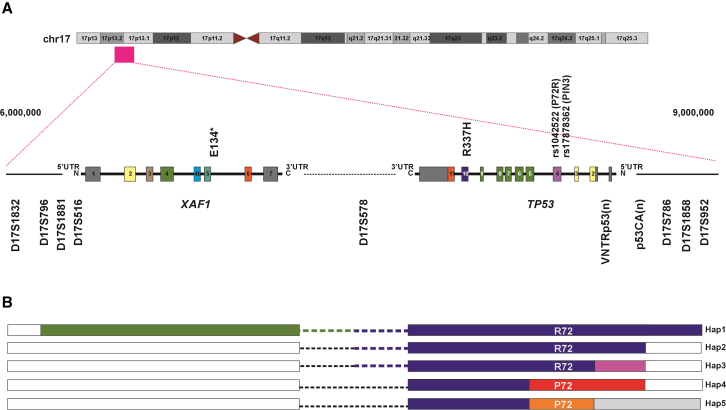
TP53 sequence and chromosome 17 polymorphic marker analyses of genomic DNA from 38 probands (Table 1) who carry the TP53 p.R337H variant revealed multiple distinct haplotypes (Figure 1; Table S2). TP53 p.R337H alleles were distinguished by the common polymorphisms at codon 72 (rs1042522) encoding proline (P72) or arginine (R72), and PIN3 (rs17878362) containing one (A1) or two copies (A2) of a 16-base pair sequence in intron 3. In this cohort of TP53 p.R337H carriers, R72 co-segregates with A1 (R72/A1) and P72 co-segregates with A2 (P72/A2). Excluding one undetermined case (#38), carriers with R72/A1 were more frequently observed (n = 30) compared with P72/A2 genotype (n = 7), establishing two distinct TP53 p.R337H alleles. Extending the analysis to include the polymorphic marker VNTRp53, a pentanucleotide (AAAAT)n repeat within the 6.1-kb-long TP53 intron 1,17,18 the p53CA(n) polymorphic marker,19 and the XAF1 p.E134∗ variant,8 we delineated five TP53 p.R337H haplotypes (Figure 1; Table S2).
Figure 1.
Diagram of five distinct TP53 p.R337H alleles
(A) Five distinct haplotypes were identified based on TP53 internal polymorphisms and microsatellite markers on chromosome 17p13.
(B) Identical DNA sequences shared among the five haplotypes are identified by solid blue bars and blue dashed lines. Polymorphic markers distinguishing the five haplotypes are highlighted in purple [Hap3, VNTRp53(n) and p53CA(n)], red [Hap4 P72R, PIN3, VNTRp53(n) and p53CA(n)], orange (Hap5, de novo, P72/A2), and green (Hap1, DNA sequence spanning D17S796 to D17S952 containing XAF1 p.E134∗ variant) solid bars.
The most common TP53 p.R337H haplotype (hereafter Hap1) (n = 16 carriers; two homozygous) spanned from D17S796 (chr17:6,251,522–6,251,775; GRCh37/hg19) to D17S952 (chr17:9,113,650–9,113,834; GRCh37/hg19) and contains the XAF1 p.E134∗ variant. An identical, but shorter, p.R337H haplotype (Hap2), that lacks XAF1 p.E134∗, extended from D17S578 (chr17:6,823,880; GRCh37/hg19) to the marker p53(CA)n (chr17:7,617,440 -7,617,490; GRCh37/hg19) (n = 4 individuals) (Figure 1; Table S2). These two TP53 p.R337H haplotypes (Hap1 and Hap2), which comprise the R72/A1 polymorphisms and consistent alleles for VNTRp53(n) and p53(CA)n have been characterized as a founder p.R337H variant, widespread in Brazil.5,8 Identical Hap1 (n = 7) and Hap2 haplotypes (n = 1) were also identified in the eight unrelated families from Spain and Portugal (Table 1; Table S2).
A third TP53 p.R337H haplotype containing the R72/A1 polymorphisms but distinct from Hap1 and Hap2 based on allelic differences in the VNTRp53(n) and p53CA(n) polymorphic markers (Hap3) was observed in 10 probands from Europe and the United States (Figure 1; Table S2). A TP53 p.R337H allele with P72/A2 (Hap4) was observed in six individuals from the United States. A documented de novo TP53 p.R337H variant with P72/A2 (Hap5) was identified in a pediatric ACT patient from an American Samoa family (Table 1).20 In addition, a heterozygous TP53 p.R337H proband with ACT from South Africa (#38)21 was identified, but the haplotype, and whether it was inherited or de novo, was not determined due to unavailability of parental DNA or tumor samples. Nonetheless, microsatellite analysis excluded this germline variant as being Hap 1 or Hap2.
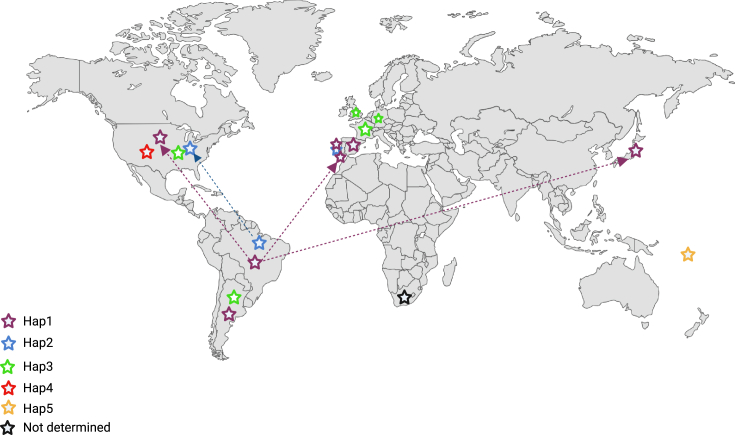
The country of birth and migration status when known for each proband was annotated (Figure 2; Table 1). Carriers of Hap1 (n = 3) and Hap2 (n = 2) identified in the United States and Japan are known to have immigrated from Brazil (Table 1). However, seven of the eight families from the Iberian Peninsula (Spain and Portugal) report no connection to Brazil.
Figure 2.
World map showing the distribution of TP53 p.R337H alleles and their constitutive haplotypes
Known migration routes of carriers from Brazil to the United States, Portugal, and Japan are highlighted by purple (Hap1) and blue (Hap2) arrows with dotted lines.
Frequency of reported TP53 p.R377H alleles in public databases
Of the 4,453 reported germline mutations (n = 1,655 families) in The TP53 Database (R20, July 2019 version; https://tp53.isb-cgc.org/), p.R337H was observed in 305 individuals (n = 117 families) representing 240 patients with cancer and 65 carriers unaffected by cancer. This database includes cases from Brazil (n = 282; 101 families), France (n = 7; 6 families), Germany (n = 4; 2 families), Japan (n = 5; 1 family), China (n = 1), Portugal (n = 1), and South Africa (n = 1) and an additional four cases whose country of origin was not identified. ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) also lists 21 entries for the TP53 p.R337H allele (variation ID 12379), with 18 being classified as pathogenic and three as likely pathogenic. Of note, p.R337H cases are often presumed to be the founder variant without recognizing differences in TP53 sequences (e.g., constitutive internal polymorphisms) or associated haplotypes.22,23
TP53 p.R337H was also reported as a somatic mutation in (1) The TP53 Database (n = 4), (2) The OncoKB database (n = 3), and (3) The COSMIC database (COSM43882, COSM4173039; n = 22). However, the germline status of these patients with p.R337H-positive tumors was not established. The TP53 p.R337H variant was not reported at Global Biobank Engine and ABraOM databases. However, it was observed in the Genome Aggregation Database (gnomAD) in three female individuals (two cancer patients, one unaffected by cancer), two identified as Latino/Admixed American individuals.
Interestingly, the TP53 p.R337H variant has also been observed in two ancient genomes (European Nucleotide Archive; ENA Browser [www.ebi.ac.uk]), one carrier from Mongolia and the other from Italy. Whether these ancient alleles are fixed in the human lineage or independent events was not determined.24
Family histories of cancer
The cancer history for the 38 p.R337H carriers in this study was collected (Table 1). All six Hap4 probands developed breast cancer at an early age (Table 1; range from 29 to 46 years old) with five developing multiple LFS core tumor types. Except for one proband (#22), all cases tested negative for BRCA1/2 mutations.
The cancer phenotype for Hap3 probands (n = 10) appears to be milder than Hap4, with four adults currently unaffected by cancer and the remaining six carriers having developed non-LFS core tumors with late onset (Table 1). Hap3 proband #24 (39 years old), presently unaffected by cancer, was self-reported to be of Ashkenazi Jewish descendant with no remarkable family history of cancer, except for a nephew with a diagnosis of choroid plexus carcinoma at age 2. Excluding two carriers who developed childhood ACT (proband #37, de novo p.R337H variant; proband #38, de novo or inherited not determined), nearly all pediatric tumors in the current cohort (n = 14) were associated with the Hap1 and Hap2 founder p.R337H mutation (n = 13; 93%).
Ancestry of the TP53 p.R337H variants
The maternal genetic composition of carriers of the TP53 p.R337H variant was investigated by characterizing the mitochondrial control region sequences (HV1-3) from the 37 probands (Table S3; proband #30 DNA unavailable). Altogether, TP53 p.R337H carriers fall into four major mtDNA haplogroups with the predominance being of European descent (H, J, K, R, U, V, X; n = 23, 61%) followed by Native American (A–D; n = 6, 16%), African (L; n = 6, 16%), and Asian descent (M7c; n = 2, 5%). The paternal composition was examined by genotyping 23 Y-STRs from 13 male probands in this cohort. These results indicate that the paternal lineage of carriers is associated with the following European haplogroups: R1b (n = 6), J1 (n = 3), I2a1 (n = 2), G2a (n = 1), and E1b1b (n = 1) (Table S4).
Ancestry of Hap 1 (n = 65) and Hap2 (n = 21) was analyzed in a validation cohort of 86 independent Brazilian male TP53 p.R337H carriers who were identified through a general population neonatal screening program (Tables S3 and S4).6 This cohort provided the opportunity to determine Y-STR profiling and mitochondrial DNA sequence analysis of carriers with paternal and maternal inheritance of TP53 p.R337H, respectively. Of those male Brazilian carriers who inherited the p.R337H variant from their fathers (n = 41; 33 Hap1 and 8 Hap2), all Y-STR haplotypes were found to be associated with European/Eurasian descent, primarily the major haplogroup R (n = 23, 56%; in particular R1b, n = 22) followed by E (n = 7; 17%), I (n = 5; 12%), J (n = 3; 7%), L (n = 2; 5%), and Q (n = 1; 2%) (Table S4). For those male Brazilian carriers who inherited the p.R337H variant from their mothers (n = 45; 32 Hap1 and 13 Hap2), mitochondrial haplotypes were predominantly associated with Native American ancestry (haplogroups A–D; n = 33, 73%), followed by European/Eurasia (n = 9, 20%) and African (n = 3, 7%) haplogroups (Table S3). Although the 86 male individuals selected for this study were recruited from different families and documented for at least three generations, we have identified relatedness in 15 cases (17%) based on shared mtDNA sequences (n = 11; A–D haplogroups) or Y-STR (n = 4; I2b1 and E1b1a haplogroups), consistent with a founder effect5,8 (Tables S3–S5).
Discussion
In the present study we have identified five distinct TP53 p.R337H haplotypes, with two (Hap1 and Hap2) sharing an identical Caucasian founder allele widespread in the Brazilian population.5,8 The TP53 p.R337H founder variant co-segregates with XAF1 p.E134∗ in the majority of Brazilian cases (Hap1) and as a single mutation (Hap2).8 Hap1 was also observed in the majority of cases from Spain and Portugal. The remaining three TP53 p.R337H haplotypes (Hap3-5) that have been identified outside of Brazil are distinct from the founder allele based on internal polymorphisms and polymorphic markers along chromosome 17p.
The p.R337H variant has been functionally characterized by studies in vitro and in vivo as hypomorphic,2,25,26,27 with individuals exibiting a wide range of cancer phenotypes. In many instances, however, the p.R337H variant has been classified as pathogenic and carriers, independent of haplotype and other possible pathogenic variants, are considered to be associated with classic LFS.28
The TP53 p.R337H haplotypes identified here diverge in internal TP53 polymorphisms, such as codon 72 (Pro72Arg) and PIN3 (intron 3), reported to affect p53 expression and function.11,12 Notably, codon 72 is part of the p53 proline-rich motif that is critical for interactions with MDM-2,29 ASPPs,30 XAF1,9 NF-κB,31 and proteins containing the SH3 domain.32 Specifically, the R72 variant is more active at stimulating cellular apoptosis and suppressing cellular transformation.33 In addition, the PIN3 variant (A2) has been associated with decreased p53 expression34 and several case-control studies have reported an increased risk of various cancer types associated with the A2 allele in Caucasians, with the most consistent association reported for breast and colorectal cancers.35 It is well established that the combination of specific variants in cis can potentially modify the penetrance of coding variants,36 with these variants exhibiting much smaller effects individually but with additive haplotypic effects. Phenotypic variability associated with another variant in cis is exemplified by the presence of XAF1 p.E134∗ in a subset of TP53 p.R337H carriers (Hap1) that are at higher risk of cancer in general, sarcomas, and multiple tumors compared with those with TP53 p.R337H alone (Hap2).8
Although the number of carriers of Hap3 and Hap4 were not sufficient to establish a clear haplotype-cancer phenotype association, we did observe a pattern whereby Hap4 carriers more closely resembled classical LFS, including breast cancers, sarcomas, and multiple primary tumors. In contrast, Hap3 carriers present generally with tumors not seen typically in LFS and at late onset with incomplete penetrance, suggestive of a lower risk of cancer.
To determine the ancestry and occurrence of TP53 p.R337H variant within the framework of the history of Brazil, our analysis of mitochondrial haplogroups of carriers defined an excess of Native American haplogroups (A–D; 73%) in maternal lineages. Although many native groups in Brazil have vanished, particularly in southeast regions,37 our observed 73% of Brazilian p.R337H carriers with the Native American matrilineal lineages is in clear contrast with present day Brazilians in the same region that still carry the genetic imprinting of early colonization phase but with much more uniform distribution: 39% European, 33% Amerindian, and 28% African lineages.37 Y-STR profiling of carriers from the paternal lineage revealed exclusively European haplogroups, particularly R1b. This composition indicates a remarkable signature among communities established by European male settlers with extensive intermarriage with Indigenous women.38 We postulate that Iberian Peninsula Europeans introduced the TP53 p.R337H variant in the Brazilian population in early colonial times. Our hypothesis is based on the knowledge that the European-Amerindian admixture started soon after the arrival of the first colonizers.38 In addition, we observed a high frequency of Y-STR haplogroup R1b in Brazilian carriers as well as in TP53 p.R337H carriers from Portugal and Spain. It is well established that R1b haplogroup is the most common Y chromosome branch of Atlantic Europe, the most frequent haplogroup in the Iberian Peninsula and with great representation among Sephardic Jews.39 Of note, our Spanish p.R337H carriers are from Extremadura, a Spanish border region adjacent to Portugal, with large Sephardic communities and a prominent source for migrants to the Americas during the 16th century.40
Although we cannot determine whether the p.R337H founder allele first arose within the context of Hap1 or Hap2, we have observed the uncoupling of XAF1 p.E134∗ and TP53 p.R337H in several Hap1 Brazilian families.8 These findings suggest that the region separating XAF1 and TP53 on chromosome 17 is susceptible to recombination and that gene conversion could promote haplotype diversity. These findings also highlight the importance of determining the haplotype of this hypomorphic mutation in each carrier even when occurring within the same family.8
In conclusion, we have identified five distinct TP53 p.R337H haplotypes that differ in specific intragenic and extragenic polymorphisms, as well as associated modifiers. Given the hypomorphic nature of p.R337H, in contrast to pathogenic DNA binding domain TP53 mutants, its tumor suppressor activity can be significantly influenced by associated polymorphisms and variants. One such example is the p.R337H founder mutation that is common throughout southern Brazil, but in the context of two haplotypes that differ in the status of XAF1 and tumor susceptibility (Hap1 and Hap2).8,41,42 As shown here, additional non-founder independent p.R337H alleles (Hap3-5) have been identified that also likely influence cancer risk. Therefore, genetic counselors and health care providers must be aware of these distinct p.R337H haplotypes and the potential implications of their associated differences on p53 expression, structure, and function, and consequently cancer risk.
Acknowledgments
The authors would like to thank the patients and their family members who have contributed to this study. We also thank Vani Shanker, Senior Scientific Editor St. Jude Children’s Research Hospital, for editing the article and Jessica Marquez, Senior Researcher St Jude Children’s Research Hospital for Y-STR profiling. Our gratitude is expressed to Nair Modolo Pinto and Gail Sacks Kirschner for their significant input and expert discussion.
This work was supported by NCI 5R01CA260175, Cancer Center Support Grant CA21765, Speer Charitable Trust, and ALSAC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. C.P.K. has been supported by the BMBF ADDRess (01GM1909A) and by the Deutsche Kinderkrebsstiftung (DKS2021.25).
Author contributions
Data conceptualization: E.M.P., G.Z. Designed and conducted experiments: E.M.P. Formal analysis of data: E.M.P., C.F. Provided resources: B.C.F., H.S., M.R.T., C.P., M.P., C.P.K., C.L., E.A.M.F.L., A.L., G.K., E.K., E.M.S., K.B., S.H., B.H., P.T., A.K., T.O., Y.F., M.F.W., H.Q.R., K.N.M., J.E.G., C.R.-G. Writing original draft: E.M.P., G.Z. Writing, review, and editing: all authors.
Declaration of interests
K.B. reports an immediate family member on the scientific advisory board of Emendo Biotherapeutics, Karyopharm Therapeutics, Imago BioSciences, and DarwinHealth; is co-founder of Isabl Technologies; and has equity interest in Imago BioSciences, Emendo Biotherapeutics, and Isabl Technologies.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100244.
Contributor Information
Emilia M. Pinto, Email: emilia.pinto@stjude.org.
Gerard P. Zambetti, Email: Gerard.zambetti@stjude.org.
Supplemental information
References
- 1.Chompret A., Brugières L., Ronsin M., Gardes M., Dessarps-Freichey F., Abel A., Hua D., Ligot L., Dondon M.-G., Bressac-de Paillerets B., et al. P53 germline mutations in childhood cancers and cancer risk for carrier individuals. Br. J. Cancer. 2000;82:1932–1937. doi: 10.1054/bjoc.2000.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro R.C., Sandrini F., Figueiredo B., Zambetti G.P., Michalkiewicz E., Lafferty A.R., DeLacerda L., Rabin M., Cadwell C., Sampaio G., et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc. Natl. Acad. Sci. USA. 2001;98:9330–9335. doi: 10.1073/pnas.161479898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latronico A.C., Pinto E.M., Domenice S., Fragoso M.C., Martin R.M., Zerbini M.C., Lucon A.M., Mendonca B.B. An inherited mutation outside the highly conserved DNA-binding domain of the p53 tumor suppressor protein in children and adults with sporadic adrenocortical tumors. J. Clin. Endocrinol. Metab. 2001;86:4970–4973. doi: 10.1210/jcem.86.10.7957. [DOI] [PubMed] [Google Scholar]
- 4.Pinto E.M., Zambetti G.P. What 20 Years of Research Has Taught Us About the TP53 p.R337H Mutation. Cancer. 2020;126:4678–4686. doi: 10.1002/cncr.33143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto E.M., Billerbeck A.E.C., Villares M.C.B.F., Domenice S., Mendonça B.B., Latronico A.C. Founder effect for the highly prevalent R337H mutation of tumor suppressor p53 in Brazilian patients with adrenocortical tumors. Arq. Bras. Endocrinol. Metabol. 2004;48:647–650. doi: 10.1590/s0004-27302004000500009. [DOI] [PubMed] [Google Scholar]
- 6.Custódio G., Parise G.A., Kiesel Filho N., Komechen H., Sabbaga C.C., Rosati R., Grisa L., Parise I.Z.S., Pianovski M.A.D., Fiori C.M.C.M., et al. Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J. Clin. Oncol. 2013;31:2619–2626. doi: 10.1200/JCO.2012.46.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seidinger A.L., Caminha I.P., Mastellaro M.J., Gabetta C.S., Nowill A.E., Pinheiro V.R.P., Yunes J.A. TP53 p.Arg337His geographic distribution correlates with adrenocortical tumor occurrence. Mol. Genet. Genomic Med. 2020;8:e1168. doi: 10.1002/mgg3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto E.M., Figueiredo B.C., Chen W., Galvao H.C.R., Formiga M.N., Fragoso M.C.B.V., Ashton-Prolla P., Ribeiro E.M.S.F., Felix G., Costa T.E.B., et al. XAF1 as a modifier of p53 function and cancer susceptibility. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M.G., Han J., Jeong S.I., Her N.G., Lee J.H., Ha T.K., Kang M.J., Ryu B.K., Chi S.G. XAF1 directs apoptotic switch of p53 signaling through activation of HIPK2 and ZNF313. Proc. Natl. Acad. Sci. USA. 2014;111:15532–15537. doi: 10.1073/pnas.1411746111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson E.A. Identity by Descent: Variation in Meiosis, Across Genomes, and in Populations. Genetics. 2013;194:301–326. doi: 10.1534/genetics.112.148825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whibley C., Pharoah P.D.P., Hollstein M. p53 polymorphisms: cancer implications. Nat. Rev. Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 12.Levine A.J. Spontaneous and inherited TP53 genetic alterations. Oncogene. 2021;40:5975–5983. doi: 10.1038/s41388-021-01991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers J., Pinto E.M., Barnoud T., Leung J.C., Martynyuk T., Kossenkov A.V., Philips A.H., Desai H., Hausler R., Kelly G., et al. A rare TP53 mutation predominant in Ashkenazi Jews confers risk of multiple cancers. Cancer Res. 2020;80:3732–3744. doi: 10.1158/0008-5472.CAN-20-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonçalves F.T., Fridman C., Pinto E.M., Guevara-Aguirre J., Shevah O., Rosembloom A.L., Hwa V., Cassorla F., Rosenfeld R.G., Lins T.S.S., et al. The E180splice mutation in the GHR gene causing Laron syndrome: witness of a Sephardic Jewish exodus from the Iberian Peninsula to the New World? Am. J. Med. Genet. 2014;164A:1204–1208. doi: 10.1002/ajmg.a.36444. [DOI] [PubMed] [Google Scholar]
- 15.Gusmão L., Butler J.M., Carracedo A., Gill P., Kayser M., Mayr W.R., Morling N., Prinz M., Roewer L., Tyler-Smith C., et al. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci. Int. 2006;157:187–197. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Roewer L., Andersen M.M., Ballantyne J., Butler J.M., Caliebe A., Corach D., D'Amato M.E., Gusmão L., Hou Y., de Knijff P., et al. DNA Commission of the International Society of Forensic Genetics (ISFG): Recommendations on the interpretation of Y-STRs results in forensic analysis. Forensic Sci. Int. 2020;48 doi: 10.1016/j.fsigen.2020.102308. [DOI] [PubMed] [Google Scholar]
- 17.Futreal P.A., Barrett J.C., Wiseman R.W. An Alu polymorphism intragenic to the TP53 gene. Nucleic Acids Res. 1991;19:6977. doi: 10.1093/nar/19.24.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn M., Fislage R., Pingoud A. Polymorphism of the pentanucleotide repeat d(AAAAT) within intron 1 of the human tumor suppressor gene p53 (17p13.1) Hum. Genet. 1995;95:471–472. doi: 10.1007/BF00208983. [DOI] [PubMed] [Google Scholar]
- 19.Jones M.H., Nakamura Y. Detections of Loss of Heterozigosity at the Human TP53 Locus Using a Dinucleotide Repeat Polymorphism. Genes Chromosomes Cancer. 1992;5:89–90. doi: 10.1002/gcc.2870050113. [DOI] [PubMed] [Google Scholar]
- 20.Aungkawattanapong N., Techavichit P., Lauhasurayotin S., Chiengthong K., Sosothikul D., Monsereenusorn C., Supornsilchai V., Suphapeetiporn K., Teerapakpinyo C., Shuangshoti S. A study of the TP53 Germline Mutation and Clinicopathologic Features in Thai Children with Adrenocortical Carcinoma. J. Health Sci. Med. Res. 2021;39:491–502. [Google Scholar]
- 21.Macaulay S., Goodyear Q.C., Kruger M., Chen W., Essop F., Krause A. The first two confirmed sub-Saharan African families with germline TP53 mutations causing Li-Fraumeni syndrome. Fam. Cancer. 2018;17:607–613. doi: 10.1007/s10689-018-0075-5. [DOI] [PubMed] [Google Scholar]
- 22.Kratz C.P., Freycon C., Maxwell K.N., Nichols K.E., Schiffman J.D., Evans D.G., Achatz M.I., Savage S.A., Weitzel J.N., Garber J.E., et al. Analysis of the Li-Fraumeni Spectrum Based on an International Germline TP53 Variant Data Set. JAMA Oncol. 2021;7:1800–1805. doi: 10.1001/jamaoncol.2021.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer N.W., Ma Y.-H.V., Gariépy J. Emerging insights into ethnic-specific TP53 germline variants. J. Natl. Cancer Inst. 2023 doi: 10.1093/jnci/djad106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kou S.H., Li J., Tam B., Lei H., Zhao B., Xiao F., Wang S.M. TP53 germline pathogenic variants in modern humans were likely originated during recent human history. NAR Cancer. 2023;5:zcad025. doi: 10.1093/narcan/zcad025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiGiammarino E.L., Lee A.S., Cadwell C., Zhang W., Bothner B., Ribeiro R.C., Zambetti G., Kriwacki R.W. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat. Struct. Biol. 2002;9:12–16. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 26.Park J.H., Li J., Starost M.F., Liu C., Zhuang J., Chen J., Achatz M.I., Kang J.G., Wang P.Y., Savage S.A., Hwang P.M. Mouse homolog of the human TP53 R337H mutation reveals its role in tumorigenesis. Cancer Res. 2018;78:5375–5383. doi: 10.1158/0008-5472.CAN-18-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffers J.R., Pinto E.M., Rehg J.E., Clay M.R., Wang J., Neale G., Heath R.J., Lozano G., Lalli E., Figueiredo B.C., et al. The Common Germline TP53-R337H Mutation is Hypomorphic and Confers Incomplete Penetrance and Late Tumor Onset in a Mouse Model. Cancer Res. 2021;81:2442–2456. doi: 10.1158/0008-5472.CAN-20-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guha T., Malkin D. Inherited TP53 Mutations and the Li-Fraumeni Syndrome. Cold Spring Harb. Perspect. Med. 2017;7:a026187. doi: 10.1101/cshperspect.a026187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momand J., Zambetti G.P., Olson D.C., George D., Levine A.J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 30.Bergamaschi D., Samuels Y., Sullivan A., Zvelebil M., Breyssens H., Bisso A., Del Sal G., Syed N., Smith P., Gasco M., et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat. Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 31.Frank A.K., Leu J.I.J., Zhou Y., Devarajan K., Nedelko T., Klein-Szanto A., Hollstein M., Murphy M.E. The codon 72 polymorphism of p53 regulates interaction with NF-{kappa}B and transactivation of genes involved in immunity and inflammation. Mol. Cell Biol. 2011;31:1201–1213. doi: 10.1128/MCB.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurochkina N., Guha U. SH3 domains: modules of protein–protein interactions. Biophys. Rev. 2012;5:29–39. doi: 10.1007/s12551-012-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong B.S., Hu W., Belyi V., Rabadan R., Levine A.J. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. Faseb. J. 2010;24:1347–1353. doi: 10.1096/fj.09-146001. [DOI] [PubMed] [Google Scholar]
- 34.Gemignani F., Moreno V., Landi S., Moullan N., Chabrier A., Gutiérrez-Enríquez S., Hall J., Guino E., Peinado M.A., Capella G., Canzian F. A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene. 2004;23:1954–1956. doi: 10.1038/sj.onc.1207305. [DOI] [PubMed] [Google Scholar]
- 35.Sagne C., Marcel V., Amadou A., Hainaut P., Olivier M., Hall J. A meta-analysis of cancer risk associated with the TP53 intron 3 duplication polymorphism (rs17878362): geographic and tumor-specific effects. Cell Death Dis. 2013;4:e492. doi: 10.1038/cddis.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castel S.E., Cervera A., Mohammadi P., Aguet F., Reverter F., Wolman A., Guigo R., Iossifov I., Vasileva A., Lappalainen T. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat. Genet. 2018;50:1327–1334. doi: 10.1038/s41588-018-0192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alves-Silva J., da Silva Santos M., Guimarães P.E., Ferreira A.C., Bandelt H.J., Pena S.D., Prado V.F. The ancestry of Brazilian mtDNA Lineages. Am. J. Hum. Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pena S.D.J., Santos F.R., Tarazona-Santos E. Genetic Admixture in Brazil. Am. J. Hum. Genet. 2020;184:928–938. doi: 10.1002/ajmg.c.31853. [DOI] [PubMed] [Google Scholar]
- 39.Adams S.M., Bosch E., Balaresque P.L., Ballereau S.J., Lee A.C., Arroyo E., López-Parra A.M., Aler M., Grifo M.S.G., Brion M., et al. The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am. J. Hum. Genet. 2008;83:725–736. doi: 10.1016/j.ajhg.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Avila R.L., Sweetwood K. International Institute for Jewish Genealogy and Paul Jacobi Center; 2006. Sephardic Origins and Transformations in the Spanish Extremadura: A Historical and Socio-Demographic Investigation. [Google Scholar]
- 41.Riechelmann R.P., Soares D.C., Dias C., Carraro D.M., Torrezan G.T. Li-Fraumeni-associated pancreatic neuroendocrine tumour and XAF1 p.Glu134Ter risk modifier variant. Ecancermedicalscience. 2022;16:1487. doi: 10.3332/ecancer.2022.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvalho N.d.A.d., Santiago K.M., Maia J.M.L., Costa F.D., Formiga M.N., Soares D.C.d.Q., Paixão D., Mello C.A.L.d., Costa C.M.L.D., Rocha J.C.C.D., et al. Prevalence and clinical implications of germline pathogenic variants in cancer predisposing genes in young patients across sarcoma subtypes. J. Med. Genet. 2023:109269. doi: 10.1136/jmg-2023-109269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.