Abstract
Background
Early life adversity and psychiatric disorders are associated with earlier declines in neurocognitive abilities during adulthood. These declines may be preceded by changes in biological aging, specifically epigenetic age acceleration, providing an opportunity to uncover genome-wide biomarkers that identify individuals most likely to benefit from early screening and prevention.
Methods
Five unique epigenetic age acceleration clocks derived from peripheral blood were examined in relation to latent variables of general and speeded cognitive abilities across two independent cohorts: 1) the Female Growth and Development Study (FGDS; n = 86), a 30-year prospective cohort study of substantiated child sexual abuse and non-abused controls, and 2) the Biological Classification of Mental Disorders study (BeCOME; n = 313), an adult community cohort established based on psychiatric disorders.
Results
A faster pace of biological aging (DunedinPoAm) was associated with lower general cognitive abilities in both cohorts and slower speeded abilities in the BeCOME cohort. Acceleration in the Horvath clock was significantly associated with slower speeded abilities in the BeCOME cohort but not the FGDS. Acceleration in the Hannum clock and the GrimAge clock were not significantly associated with either cognitive ability. Accelerated PhenoAge was associated with slower speeded abilities in the FGDS but not the BeCOME cohort.
Conclusions
The present results suggest that epigenetic age acceleration has the potential to serve as a biomarker for neurocognitive decline in adults with a history of early life adversity or psychiatric disorders. Estimates of epigenetic aging may identify adults at risk of cognitive decline that could benefit from early neurocognitive screening.
Keywords: Epigenetic age acceleration, Cognitive abilities, Childhood maltreatment, Early life adversity, General cognitive, Speeded performance
1. Introduction
Models of normative cognitive aging identify early- and mid-adulthood as developmental periods when several neurocognitive abilities peak before gradually declining into later life (Deary et al., 2009; Hartshorne and Germine, 2015; Salthouse, 2009; Schaie, 2005). Such age-related declines are observed across multiple neurocognitive domains including memory, reasoning, and executive function but the earliest and most prominent changes are in processing speed (Lipnicki et al., 2017; Salthouse, 2019). Deviations from normative cognitive aging processes may be partly attributable to early life experiences, including one's opportunity for educational attainment (Chan et al., 2019; Livingston et al., 2020; Oveisgharan et al., 2020), experience of adversity (Anda et al., 2006; Korten et al., 2014; Ritchie et al., 2011; Roberts et al., 2020; Trickett et al., 2011) and is also observed across multiple psychiatric disorders (Rock et al., 2014; Castaneda et al., 2008; McIntyre et al., 2013; Bhattarai et al., 2019; Millan et al., 2012; East-Richard et al., 2020). Deficits in neurocognitive function appearing earlier or with greater severity than expected may signal premature cognitive aging, a risk factor for impaired well-being (Allerhand et al., 2014; Llewellyn et al., 2008) and later-life cognitive impairment (Gustavson et al., 2020; Knopman et al., 2018). The biological and neuropathological processes underlying later-life cognitive impairment commence decades prior to the emergence of clinical symptoms (Katsuno et al., 2018; Gandal et al., 2018), indicating an extended preclinical period that may be detectable earlier in life. This prolonged preclinical period thus provides an opportunity to identify biomarkers related to advanced cognitive decline earlier in adulthood that have the potential to inform interventions to delay, reverse, or prevent later-life impairment.
Changes in biological and neuropathological processes that typically covary with chronological age, known as biological age (BA), may also contribute to accelerated rates of cognitive decline into later-life (Harrington et al., 2021; Hohman et al., 2017; Yu et al., 2015; Wilson et al., 2020). Along with other measures of BA, epigenetic clocks quantify predictable epigenetic changes in DNA methylation (DNAm) of specific sites in the genome, which index multiple molecular processes that contribute to BA (Kabacik et al., 2022; Raj and Horvath, 2020; Yang et al., 2023). DNAm BA estimates that deviate from chronological age are referred to as epigenetic age acceleration. So far, several well-established estimates of epigenetic age acceleration in the general population have been associated with neurocognitive performance across several domains, including lower IQs and general cognitive abilities in adolescence and later-life (PhenoAge clock) (Stevenson et al., 2019), reduced attention (Horvath clock) (Shiau et al., 2021), and processing speed, working memory, and faster cognitive declines in reasoning, and processing speed (DunedinPoAm clock) (Belsky et al., 2020). However, less is known about the utility of epigenetic age acceleration estimates as a cognitive impairment biomarker among populations who have experienced early life adversity and may be at risk for psychiatric disorders (Schaefer et al., 2022).
Although some research has found a link between epigenetic age acceleration and neurocognitive function in maltreated and psychiatric populations (Yusupov et al., 2023), other findings were null (Vaccarino et al., 2021; Marioni et al., 2015; Starnawska et al., 2017; Harvanek et al., 2023). These inconsistent findings may be due to the reliance on examining a small number of epigenetic clock estimates, a lack of controlling for early life adversity and psychiatric burden and relying on individual neuropsychological tests. Using two distinct cohorts - the Female Growth and Development Study (FGDS) (Trickett et al., 2011) from the United States and the Biological Classification of Mental Disorders (BeCOME) (Brückl et al., 2020) study from Germany, we examined associations between epigenetic age acceleration and a broad range of neurocognitive abilities in adulthood. We used structural equation modeling (SEM) of our comprehensive neurocognitive batteries to distinguish general from speeded cognitive abilities. Speeded cognitive abilities tend to show the earliest and most prominent change across the lifespan (Lipnicki et al., 2017; Salthouse, 2019), and as such may be more sensitive to differences in epigenetic age acceleration. We tested associations between epigenetic age acceleration and the two neurocognitive abilities in six models: 1) first-generation clocks (i.e., Horvath (2013) and Hannum (Hannum et al., 2013)), 2) second-generation clocks (i.e., GrimAge (Lu et al., 2019) and PhenoAge (Levine et al., 2018)), and 3) pace of aging predictor (DunedinPoAm (Belsky et al., 2020)). The models tested the epigenetic clocks of the same generation in the same models to determine whether age-related or morbidity/mortality clocks were more relevant for detecting neurocognitive impairment. The primary aim of this study was to test whether epigenetic age acceleration could serve as a biomarker of poorer performance of neurocognitive abilities in adulthood and provide evidence for its transdiagnostic potential.
2. Methods and materials
2.1. Cohorts
2.1.1. Female Growth and Development Study (FGDS)
The FGDS (N = 172) began in 1987 using an accelerated cross-sequential cohort design to study the long-term consequences of child sexual abuse (CSA) (Trickett et al., 2011). FGDS data used in the present study were obtained from the most recent wave of data collection (T7) when females were entering mid-life (Mage = 39.91, Range: 29–45 years of age). Eighty-six females with (n = 37) and without (n = 49) substantiated CSA completed a neurocognitive battery and consented to biospecimen sampling for determining estimates of epigenetic age acceleration at T7. Earlier assessments indicate that around 25% of participants were at least mildly depressed for the two weeks prior to study assessment (Shenk et al., 2010). Full study protocol for the FGDS has been published elsewhere (Trickett et al., 2011; Shenk et al.). See Table 1 for descriptive statistics of the FGDS cohort.
Table 1.
Descriptive statistics.
| FGDS (N = 86) | BeCOME (N = 313) | |||
|---|---|---|---|---|
| Female (n) | 86 | 200 | ||
| Age (M/SD) | 36.91/3.79 | 35.40/12.10 | ||
| Race/Ethnicity (n) | ||||
| NH White | 45 | 277 | ||
| NH Black | 36 | 1 | ||
| Latinx/Hispanic | 4 | 1 | ||
| Other | 1 | 34 | ||
| Education Level (n) | ||||
| At Least High School | 38 | 233 | ||
| College or Graduate Degree | 29 | 48 | ||
| Maltreated (n) | 37 | 73 | ||
| Lifetime Stress (M/SD) |
7.29/4.25 |
19.80/12.80 |
||
| % Missing |
% Missing |
|||
| Outcomes (M/SD) | ||||
| PPVT-R | 155.27/13.58 | 1 | – | |
| WCJ-R PV | 18.75/1.70 | 1 | – | |
| WCJ-R CF | 34.46/5.17 | 3 | – | |
| WCJ-R DR | 18.67/13.87 | 1 | – | |
| CCS Strict Score | 22.13/13.03 | 12 | – | |
| Fluency F | 12.42/4.46 | 10 | – | |
| Fluency S | 14.40/4.44 | 10 | – | |
| Fluency Animal | 22.36/5.60 | 13 | – | |
| Fluency Fruit/Vegetable | 20.91/4.83 | 14 | – | |
| Symbol Search RT | 3302.04/704.32 | 10 | – | |
| Flanker Incongruent RT | 165.05/262.50 | 0 | – | |
| Choice Vocabulary | – | 29.90/3.34 | 25.2 | |
| Delayed Recall | – | 10.20/3.65 | 25.2 | |
| Fluency S | – | 32.70/9.01 | 25.2 | |
| Cognitive Flexibility RT | – | 667.00/226.00 | 25.6 | |
| Go No-Go RT | – | 402.00/71.3 | 25.9 | |
| Sustained Attention | – | 162.00/36.10 | 24.9 |
Notes: PPVT-R is the Peabody Picture Vocabulary Test Revised; WCJ-R is the Woodcock Johnson-Revised test; WCJ-R PV is the Picture Vocabulary Test; WCJ-R CF is the Concept Formation Test; WCJ-R DR is the Delayed Recall test; CCS is the complex counting span; Fluency F and S are fluency for F- and S-words; Fluency Animal and Fruit/Vegetable are fluency for Animal and Fruit/Vegetable words.
The Biological Classification of Mental Disorders (BeCOME). Data from the BeCOME cohort (N = 313, nfemale = 200) included patients between 18 and 66 years old (Mage = 35.40) with psychiatric disorders and self-reported healthy controls who consented for the Max Planck Institute of Psychiatry (MPIP) and were recruited in Munich, Germany to participate in the ongoing BeCOME study (registered on ClinicalTrials.gov, TRN: NCT03984084) (Brückl et al., 2020). Self-reported demographic data were available for age, sex, ethnicity and, school education as stated in the study protocol (Brückl et al., 2020) (see Table 1). Full study protocol for the BeCOME has been published elsewhere (Brückl et al., 2020).
2.2. Measures
2.2.1. Neurocognitive measures
A neurocognitive battery that assessed multiple cognitive domains was administered to assess performance in the FGDS and BeCOME cohorts. The T7 FGDS protocol administered the following battery: a) The Peabody Picture Vocabulary Test-Revised (PPVT-R), a test of receptive language abilities; b) three tests from the Woodcock-Johnson Revised including the Picture Vocabulary for receptive abilities, Concept Formation for fluid reasoning, and Visual Auditory Learning-Delayed Recall for long-term memory retrieval; c) Complex Counting Span Task to assess working memory; d) Symbol Search Task to assess processing speed; e) word fluency task to assess; and f) Flanker Attention Task to assess attention inhibition (Supplementary Table 1 for details on each neurocognitive measure).
In the BeCOME cohort, the neurocognitive battery was conducted on the first study day. The following tests were administered in the battery: a) Multiple-choice vocabulary intelligence test (MWT-B), Choice Vocabulary Test of receptive language abilities; b) two subscales from the Materialien und Normwerte für die neuropsychologische Diagnostik (MNND) neuropsychological test battery including the Delayed-Recall Task for episodic memory and the Word Fluency Task for phenomic and semantic fluency; c) two subscales from the Test for Attentional Performance (TAP) including the Cognitive Flexibility Task for the ability to switch between different tasks rules and the Go No-Go task for inhibitory control; and d) the d2 Test of Sustained Attention (Supplementary Table 1 for details on each neurocognitive measure).
2.3. Genomic and epigenomic analyses
2.3.1. DNA methylation
FGDS Cohort. Whole blood samples were collected and randomized across plates during the T7 assessment and genomic DNA extracted from whole blood using a semi-automated approach (Qiasymphony, Qiagen) and purity assessed using a nanophotometer (ImplenP300, Implen). Genomic DNA (1 μg) from whole blood was treated with sodium bisulfite using Zymo EZ-96 DNA Methylation KIT™ (Zymo Research, Orange, CA, USA) with 200 ng of bisulfite-treated DNA amplified, fragmented, and hybridized on the EPIC array. The Infinium MethylationEPIC Beadchip (EPIC array, Illumina, San Diego, CA, USA) was used for epigenome-wide DNAm analysis. Raw intensity values were directly loaded into R for quality control and normalization using the minfi R package (Aryee et al., 2014). Standard quality control was conducted with meffil R package (Suderman et al., 2020), and poorly performing samples were removed (n = 1, due to low signal intensity). Predicted biological sex from DNAm data matched the reported sex in all samples. Likewise, a selection of single-nucleotide polymorphisms (SNPs) shared between the EPIC array and genotyping showed 100% within-person concordance. Normalization was carried out using Noob (Triche et al., 2013). The final sample included DNAm from 86 females who consented to long term storage of their DNA.
BeCOME Cohort. Whole blood samples were randomized with regards to sex, age, childhood maltreatment, and self-reported case-control status using the omixer R package (Sinke et al., 2021) in a 96-well format before DNA extraction. Bisulfite-conversion of 400 ng DNA was performed with the EZ-96 DNA Methylation kit (Zymo Research, Irvine, CA, USA). Illumina Infinium Methylation EPIC BeadChip (Illumina, San Diego, CA, USA) was used for epigenome-wide methylation analysis of samples according to manufacturer protocols. Preprocessing of DNAm data was performed using a standard pipeline (Maksimovic et al., 2016) with the minfi R package (Aryee et al., 2014). After loading raw intensity values directly into R version 4.0.4 (R Core Team, 2018) and transforming them into beta-values, a quality control was performed. Samples with a mean detection p-value >0.05 (n = 7), samples presenting with distribution artefacts in raw beta-values (n = 0) or sex mismatches between estimated sex from DNAm data and reported sex (n = 1) were excluded. Normalization was performed using stratified quantile normalization (Touleimat and Tost, 2012) and subsequently beta-mixture quantile normalization (BMIQ) (Teschendorff et al., 2013). After transforming beta-values into M values, we performed principal components analysis (PCA). One outlier deviating more than three standard deviations from the mean of the first two principal components was excluded. Next, we checked which batches had the strongest associations with the principal components and corrected batch effects of plate, array, and row sequentially with ComBat of the sva R package (Leek et al., 2012). Batch corrected M values were transformed into beta-values and MixupMapper (Westra et al., 2011) confirmed that no sample mix-ups had taken place during the experiment. The final sample included DNAm data from 320 individuals.
2.3.2. Epigenetic age acceleration
Four measures of epigenetic age acceleration were generated using a publicly available tool (https://dnamage.genetics.ucla.edu/home) (Horvath, 2013): 1) Horvath, 2) Hannum, 3) PhenoAge, 4) GrimAge, with a fifth measure 5) Dunedin Pace of Aging methylation (DunedinPoAm) calculated in R according to standard procedure (Belsky et al., 2020). The current analyses focus on measures of epigenetic age acceleration, that is, residualized scores of epigenetic age determined by DNAm after accounting for each person's chronological age at the time of the biological sample collection. DunedinPoAm provides an index of the pace of epigenetic aging adjusted for chronological age. Although these five epigenetic age estimates are moderately correlated (rs = 0.17–0.45) (Lu et al., 2019), they were derived from DNAm at largely non-overlapping sites across the genome (Field et al., 2018). For each measure, positive values of epigenetic age acceleration indicate faster aging (i.e., acceleration) and negative values indicate slower aging (i.e., deceleration). Cell-type heterogeneity across samples were deconvolved using a well-established reference-based approach (Houseman et al., 2012), with proportion of cell-types included in models where appropriate.
2.3.3. Control variables
Polygenic Score for Educational Attainment. Polygenic scores that quantify the genetic contributions accounting for a statistically significant portion of variability in educational attainment (11–13%) were constructed from SNPs identified from population-level genome-wide association studies (Lee et al., 2018). The p-value threshold for SNPs was set to 1.00 in both cohorts. Imputed genotypes were used to derive the polygenic score for educational attainment using summary statistics as per standard protocol (Lee et al., 2018). See Supplemental Table 2 for further details on genotyping used in the FGDS and BeCOME cohorts.
Childhood Maltreatment Status. CSA and non-CSA statuses in the FGDS cohort was substantiated by Child Protective Services (CPS) when females. 6–16 years of age, were originally enrolled in the study (Trickett et al., 2011). Child maltreatment (CM) in the BeCOME cohort was determined by the widely used and reliable self-reported short version of the Childhood Trauma Questionnaire (CTQ) (Bernstein et al., 2003). A participant was defined as abused if a moderate or severe exposure was present in any of the subscales for emotional, sexual, or physical abuse.
Lifetime Trauma. In the FGDS cohort, lifetime trauma was measured using the non-child maltreatment items of the Comprehensive Trauma Interview (CTI) (Shenk et al., 2016), which included 22 items about different potentially traumatic events across the lifespan. A total score was calculated by summing the number of items endorsed. In the BeCOME cohort, lifetime trauma was assessed with a reduced version of the Munich Event-Questionnaire (MEL), which consisted of 27 items covering potentially traumatic events from different areas of life and their frequencies (Friis et al., 2002). A total score was calculated using the number of events endorsed, weighted by their frequencies.
Psychiatric Burden. In the BeCOME cohort, the amount of psychiatric burden was measured using the weighted score (i.e., two-points for full diagnosis and one-point for sub-threshold) of a number of diagnoses from the modified Munich-Composite International Diagnostic Interview (M-CIDI) (DIA-X/M-CIDI) conducted by trained study assistants (full details in BeCOME study protocol (Brückl et al., 2020)) to assess current (last four weeks) or past lifetime DSM-IV diagnosis (DSM-IV, 1994).
2.4. Statistical analyses
Confirmatory factor analyses (CFAs) (Furr et al., 2014) were used to determine whether the neurocognitive batteries administered in each cohort could be reduced to two correlated domains of cognitive function, General Cognitive Abilities and Speeded Cognitive Abilities. All neurocognitive measures were first z-scored. In the FGDS cohort, an item parcel was created from the two picture vocabulary measures (from the Peabody and Woodcock-Johnson Revised) by first z-scoring each and then averaging. Adequate model fit was determined from several model fit indices including a χ2 likelihood-ratio test with a p-value >0.05, a comparative fit index (CFI) above 0.90, and a root mean square error of approximation (RMSEA) with a 95% confidence interval (CI95%) that covered or had an upper bound below 0.05 (Hooper et al., 2008), (Barrett, 2007). Meaningful indicators of their corresponding domain of neurocognitive function were identified as those with standardized factor loadings > |0.30| with associated p-values <0.05. CFAs were estimated in the lavaan R package using the cfa function (Rosseel, 2012) with a robust full-information maximum-likelihood estimator and fixing the first indicator of each latent variable to 1.00 for model identification.
The associations between epigenetic age acceleration and neurocognitive function, specifically General Cognitive Abilities and Speeded Cognitive Abilities, were estimated in the structural equation modeling (SEM) framework (Kline, 2010) using the sem function of the lavaan R package (Rosseel, 2012). SEMs were estimated using a full-information MLR estimator for each generation of epigenetic age acceleration estimate in each cohort, for a total of six models. All predictors were first z-scored. The measurement models were specified the same as in the CFAs. Additional covariates were added to the models to adjust for participant ancestry from genome wide genotype data (first three principal components for all models) and cell-type counts in (GrimAge, PhenoAge, and DunedinPoAm models). In the BeCOME cohort, self-reported gender and the amount of psychiatric burden was also included as covariates. Covariates were regressed onto the latent variables for cognitive function onto indicators of the latent variables in the BeCOME cohort to preserve factor loading patters observed in the CFA and keep the meaning of these constructs intact). Specification of the SEMs were otherwise similar between cohorts. Model results were presented as standardized estimates (β) using p-values based on α < 0.05.
3. Results
3.1. Cognitive abilities
In both cohorts, CFAs revealed that the 2-factor solution, reflecting General Cognitive Abilities and Speeded Cognitive Abilities, fit the data well, χ2 (19)FGDS = 13.235, p = 0.826, χ2 (8)BeCOME = 6.589, p = 0.382, CFIs = 1.00, RMSEAs = 0.00. C.I.FGDS [0.00, 0.05], C.I.BeCOME [0.00, 0.07]. In both cohorts, all factor loadings were statistically significant (ps < 0.01) and the magnitudes of the standardized factor loadings were > |0.367|. Moreover, in both cohorts, the magnitude and the direction of the correlation between General Cognitive Abilities and Speeded Cognitive Abilities was similar, r = 0.580 (p < 0.001) in the FGDS cohort and r = 0.590 (p = 0.010) in the BeCOME cohort, providing evidence that the latent variables in each cohort reflect similar constructs despite using different neurocognitive batteries. See Supplementary Table 3 for full results of parameter estimates in both cohorts.
3.2. First-generation clocks (horvath and hannum) and cognitive abilities
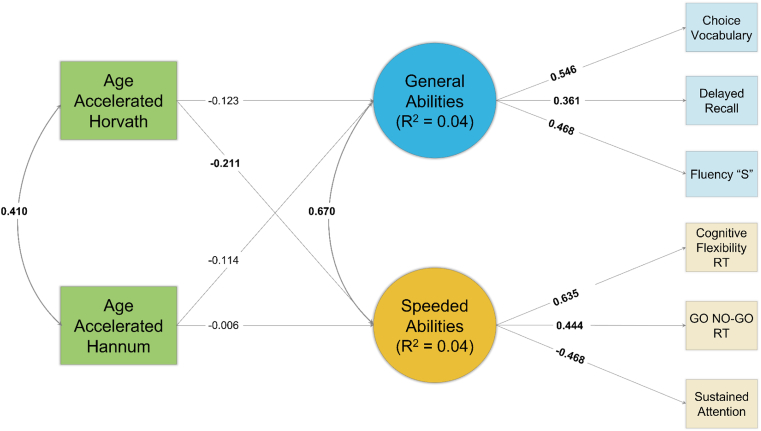
Path diagrams of results are in Fig. 1 for FGDS and Fig. 2 for BeCOME. Epigenetic acceleration as measured by the Horvath or Hannum clocks were not significantly associated with the General Cognitive Abilities in either cohort (ps > 0.185). However, greater epigenetic acceleration as measured by the Horvath clock was significantly associated with slower Speeded Cognitive Abilities in the BeCOME cohort (β = −0.211, p = 0.012), and although in the same direction, the association was not statistically significant in the FGDS cohort (β = −0.113, p = 0.166).
Fig. 1.
Path Diagram of First-Generation Clocks Results – FGDS
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Squares reflect measured variables and circles reflect latent variables.
Fig. 2.
Path Diagram of First-Generation Clocks Results – BeCOME
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Squares reflect measured variables and circles reflect latent variables. “RT” = response time.
3.3. Second-generation clocks (GrimAge and PhenoAge) and cognitive abilities
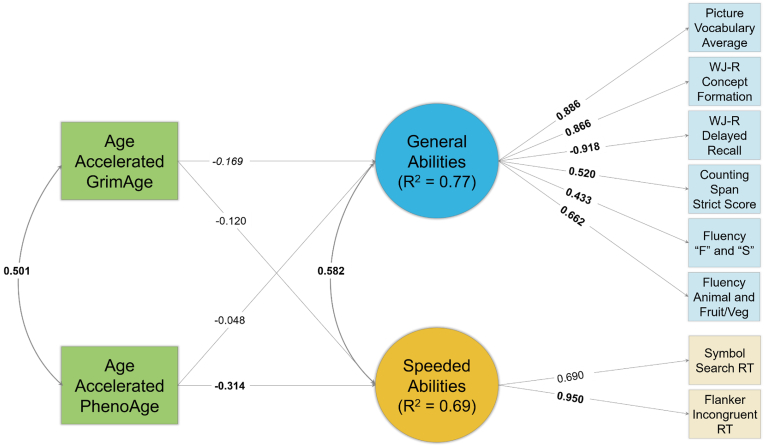
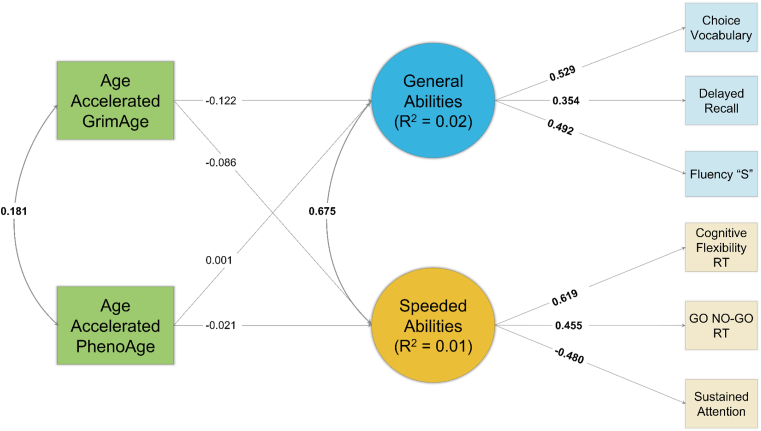
Path diagrams of results from the FGDS and BeCOME cohorts are in Fig. 3, Fig. 4, respectively. Acceleration of epigenetic age as measured by the GrimAge clock was nearly significantly associated with lower General Cognitive Abilities in the FGDS (β = −0.169, p = 0.062), but not in the BeCOME (β = −0.122, p = 0.279) cohort. Acceleration of epigenetic age measured by the GrimAge clock was not associated with Speeded Cognitive Abilities in either cohort (ps > 0.206). Acceleration in epigenetic age measured by the PhenoAge clock was associated with slower Speeded Cognitive Abilities in the FGDS cohort (β = −0.314, p = 0.044), but not in the BeCOME cohort (β = −0.021, p = 0.804). Acceleration in the PhenoAge clock was not significantly associated with General Cognitive Abilities in either cohort (ps > 0.613).
Fig. 3.
Path Diagram of Second-Generation Clocks Results – FGDS
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Italicized coefficients indicate a near statistically significant (p < 0.10) association. Squares reflect measured variables and circles reflect latent variables. “RT” = response time.
Fig. 4.
Path Diagram of Second-Generation Clocks Results – BeCOME
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Squares reflect measured variables and circles reflect latent variables. “RT” = response time.
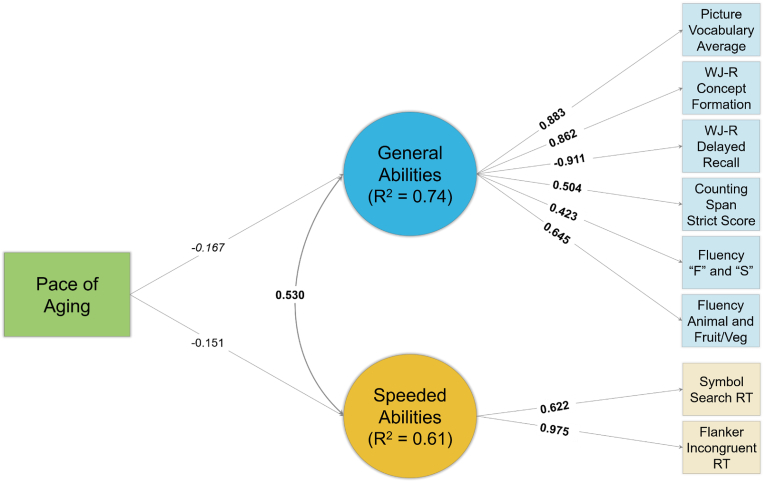
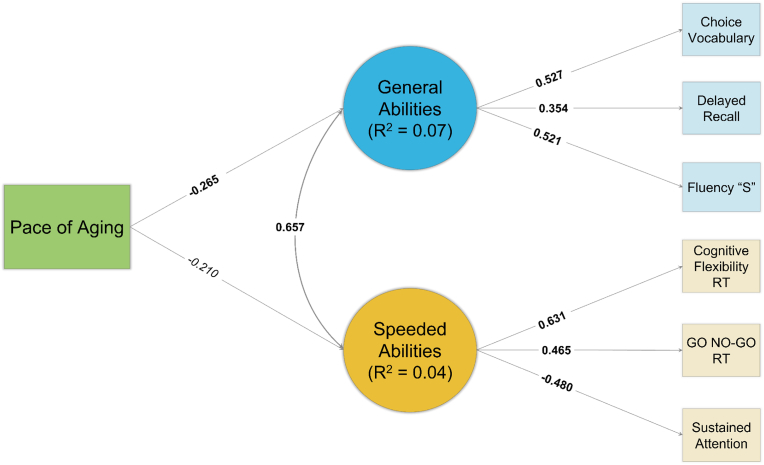
3.4. Pace of aging clock (DunedinPoAm) and cognitive abilities
See Fig. 5, Fig. 6 for path diagrams from the FGDS and BeCOME cohorts, respectively. Acceleration of epigenetic age as measured by the DunedinPoAm clock was nearly significantly associated with lower General Cognitive Abilities in the FGDS cohort (β = −0.167, p = 0.061) and significantly in the BeCOME cohort (β = −0.265, p = 0.017). Although acceleration in the DunedinPoAm was not significantly associated with slower Speeded Cognitive Abilities in the FGDS cohort (β = −0.151, p = 0.172), it was nearly significant in the BeCOME cohort (β = −0.210, p = 0.058). See Table 2 for the general pattern of results across clocks and between cohorts.
Fig. 5.
Path Diagram of DunedinPoAm Results – FGDS
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Italicized coefficients indicate a near statistically significant (p < 0.10) association. Squares reflect measured variables and circles reflect latent variables. “RT” = response time.
Fig. 6.
Path Diagram of DunedinPoAm Results – BeCOME
Note: Standardized coefficients depicted. Bolded coefficients indicate a statistically significant (p < 0.05) association. Italicized coefficients indicate a near statistically significant (p < 0.10) association. Squares reflect measured variables and circles reflect latent variables. “RT” = response time.
Table 2.
General pattern of results across cohorts.
| FGDS | BeCome | |||
|---|---|---|---|---|
| General Cognitive | Speeded Cognitive | General Cognitive | Speeded Cognitive | |
| Horvath | No | No | No | Yes |
| Hannum | No | No | No | No |
| GrimAge | Close | Close | No | No |
| PhenoAge | Close | Yes | No | No |
| DunedinmPoA | Close | Close | Yes | Close |
Note: No means not statistically significant; Yes mean statistically significant; Close means it was nearly statistically significant (p > 0.05 and p < 0.10.
4. Discussion
The current study leveraged data from two independent, international cohorts to examine whether epigenetic age acceleration was associated with neurocognitive function in samples with histories of child maltreatment or psychiatric disorders. We took a novel approach to the investigation of epigenetic age acceleration and neurocognitive function by using SEM to test whether epigenetic age acceleration was associated with general and speeded cognitive abilities. Leveraging the possibility to examine the general cognitive and speeded cognitive ability factors underlying differences in individuals neurocognitive function, we reduce the potential for test specific effects and measurement error (Kline, 2010), potentially increasing the sensitivity to detect the effects of BA acceleration (Deary et al., 2009; Hartshorne and Germine, 2015; Salthouse, 2009, 2019; Schaie, 2005; Lipnicki et al., 2017). This study provided a robust assessment of whether epigenetic age acceleration was associated with neurocognitive functioning in a sample with independent substantiation of child sexual abuse and in a sample recruited for psychiatric disorder status. In all six analyses, we controlled for the most prominent risk factors for cognitive impairment and variables that explain extraneous variation in epigenetic age acceleration estimates. We found evidence that epigenetic age acceleration was associated with neurocognitive functioning, although these associations depended upon which clock was investigated in each cohort (see Table 2 for the side-by-side comparison of the findings). Based on the strengths of this study and corresponding results, there are several important directions for future research with patients treated for child maltreatment or psychiatric disorders.
First, we found evidence that accelerated epigenetic age using DNAm-driven biomarkers was associated with general and speeded cognitive abilities differentially in each cohort. Specifically, associations between acceleration in the Horvath clock (a first-generation clock) was associated with slower speeded cognitive abilities in the BeCOME cohort, but not in the FGDS cohort. Additionally, associations between acceleration in the second-generation clocks were only significant with neurocognitive abilities in the FGDS cohort. Namely, acceleration in the GrimAge clock was associated with lower general cognitive abilities and acceleration in the PhenoAge clock was associated with slower speeded cognitive abilities. These discrepant findings may be due to differences in how these samples were recruited. The FGDS cohort was 6–16 years of age at the time of enrollment and were between the ages of 29 and 45 when the neurocognitive battery and epigenetic age acceleration estimates were collected (Trickett et al., 2011; Shenk et al.; Felt et al., 2022). Patients from the BeCOME cohort were recruited to evaluate psychiatric diagnoses, excluded if they had any evidence of physical impairment, neurocognitive degeneration, or substance abuse, and all were between the age of 18 and 6543. The relatively restricted age range in the FGDS could contribute to why associations were not found in that cohort as the first generation clocks (Horvath, Hannum) were explicitly trained on chronological age (Horvath, 2013; Hannum et al., 2013). It is possible that associations are only detectable for these clocks in samples with a wider age range such as in the BeCOME cohort. Likewise, the BeCOME cohort was recruited for evaluation of psychiatric diagnoses and patients were excluded if they had any evidence of neurodegenerative disease or severe physical health ailments, two phenotypes of adult aging that the second-generation (i.e., GrimAge (Lu et al., 2019) and PhenoAge (Levine et al., 2018)) clocks were trained on. As recruitment for the FGDS cohort occurred when females were in childhood, these common phenotypes of aging-related morbidity could not be excluded upon enrollment. It is possible that associations with the second-generation clocks can only be detected in samples that were not restricted for aging-related morbidities, such as in the FGDS. Additionally, there were significant racial and ethnic differences between the cohorts, with the FGDS including a relatively diverse sample and the BeCOME cohort comprising a primarily white-European sample, which could explain differences in findings. However, some work has found similar findings in multicultural samples and primarily white-European samples. Future work with larger, multicultural samples is needed to investigate potential differences between racial and ethnic groups in associations between epigenetic age acceleration and cognitive functioning.
Interestingly, significant (or near significant) associations between the DNAm-derived pace of BA and neurocognitive function were found in both cohorts. In the FGDS cohort, a faster pace of BA as measured by the DunedinPoAm was marginally associated with lower general cognitive abilities but not with speeded cognitive abilities. In the BeCOME cohort, acceleration in the DunedinPoAm was significantly associated with lower general cognitive abilities and marginally with slower speeded cognitive abilities. Although results for speeded cognitive abilities did not reach a statistically significant threshold in either cohort, associations were in the same direction and of a similar magnitude, suggesting that there may not have been enough power to detect the associations. These mostly parallel findings of the DunedinPoAm between the cohorts suggest that this measure of DNAm-derived BA may be sensitive enough to detect cognitive impairment in a wider-range of samples because it was derived from a longitudinal cohort to estimate the pace of BA, rather than a point-estimate of a specific age (Belsky et al., 2020). The DunedinPoAm clock was explicitly trained for early detection of cardiovascular, metabolic, renal, hepatic, pulmonary, periodontal, and immune system dysfunction (Belsky et al., 2020). Although these aging phenotypes may not have been clinically present in these samples yet, the biological processes underlying these may already be detectable and covary with cognitive decline (Harrington et al., 2021; Hohman et al., 2017; Yu et al., 2015; Wilson et al., 2020).
Impaired neurocognitive functioning is a common and prominent disabling factor in several psychiatric disorders (McIntyre et al., 2013; Millan et al., 2012; East-Richard et al., 2020), underscoring the clinical relevance of these findings for psychiatric patients. Our findings linking epigenetic age acceleration with neurocognitive function is consistent with previous work suggesting that impaired cognition was a transdiagnostic phenomenon in psychiatric disorders (Millan et al., 2012; McTeague et al., 2016; Abramovitch et al., 2021; Chavez-Baldini et al., 2021). The burden of poor cognitive performance in psychiatric disorders can be devastating and may mediate functional impairments in both personal and professional life, and ultimately quality of life (McIntyre et al., 2013; Woo et al., 2016; Brissos et al., 2008). Furthermore, current pharmacological therapies do not lead to a sufficient treatment of cognitive deficits in psychiatric disorders (Millan et al., 2012). Not only are cognitive symptoms poorly controlled during the course of disease, but they also remain long after the improvement of the affective state of patients, which have responded to current medical treatment (e.g. in 55 of 75 cognitive variables in recent MDD meta-analysis (Semkovska et al., 2019)). Therefore, identifying blood-derived epigenetic biomarkers, which covary with cognitive function, could be used to identify subpopulations for future exploration of pharmacological and non-pharmacological treatments. However, given the mixed findings in the literature (Harvanek et al., 2023) and in this study, more future work is needed before epigenetic clocks may be useful in clinical settings with patients.
Our study has several limitations to consider when interpreting these findings. First, peripheral blood, and not brain tissue, was used for the detection of epigenetic age. Currently, the relation between epigenetic mechanisms in different tissues is still unclear (Bakulski et al., 2016) but similarities of age-related DNAm across tissues and cell types have been observed in previous work (Horvath, 2013; Horvath et al., 2012). Associations examined in this study were cross-sectional and directionality of effects are difficult to determine. Future work should consider measuring epigenetic age acceleration and neurocognitive abilities longitudinally in maltreated and psychiatric populations to elucidate when epigenetic age acceleration might be used to detect earlier impairments associated with later-life neurocognitive degeneration. With the exception of the PhenoAge clock, none of the clocks used to estimate epigenetic age were explicitly trained on biomarkers of neurocognitive function, which may explain some variation in the associations across cohorts. However, these findings suggest that epigenetic clocks can be sensitive to individual differences in neurocognitive functioning despite not being explicitly trained for these purposes. Future work developing new epigenetic clocks maty consider optimizing the clocks for biomarkers of neurocognitive function to further enhance their diagnostic utility, particularly in psychiatric and maltreated samples. Finally, the different pattern of results observed in this study may be partially a function of the different characteristics between the cohorts, specifically with respect to how participants were sampled (prospective cohort study for FGDS and cross-sectional study for BeCOME), different measures used, and cultural differences between the U.S. and Germany (Linberg et al., 2019). Future research should look into cohorts that use the same measures but vary on the populations sampled to rule out the impact different measures have on the findings. However, many potential limitations of this study were mitigated from the use of the two distinct cohorts with complementary strengths and weaknesses. For instance, retrospective self-report measures of child maltreatment in the BeCOME cohort are complemented by prospective substantiated measures of child maltreatment in the FGDS cohort and the relatively small sample size in the FGDS cohort (N = 86) is complemented by the larger sample size in the BeCOME cohort (N = 313). Additionally, the use of two independent cohorts provided side-by-side elucidates potential reasons of conflicting findings across studies internationally and may provide insight into future study designs. A final limitation is that we did not correct for multiple tests. However, this was done to maintain power given the relatively low sample sizes and goal to discover associations that have not been consistent in the extant literature. Future work should consider recruiting larger, more heterogenous samples and control for the false discovery rate (Benjamini and Hochberg, 1995).
Despite these limitations, the current study provides evidence, above and beyond established risk factors for neurocognitive impairment, that epigenetic age acceleration may be a novel biomarker covarying with impaired neurocognitive function in patients with psychiatric disorders and histories of child maltreatment. Future work is still needed to establish clinical relevance of this blood-based biomarker that covaries with cognitive function across adulthood.
Funding
Data collection and analysis from the FGDS cohort was supported by the National Institutes of Health under award numbers R01AG059682 (Shenk), R01HD072468 (Noll); R01AG04879 (Noll), and P50HD089922 (Noll). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Zhenyu Zheng was funded through a T32 funded through the NICHD under award number T32HD101390 (Jackson & Noll). Karra Harrington was funded by a National Institute on Aging T32 grant under award number T32AG049676. Kieran O'Donnell is a Pfeil Investigator (Brain and Behavior Research Foundation) and received support from the Chamandy Foundation, Healthy Brains for Healthy Lives and NHLBI (R01HL157787-02S1). The work of Natan Yusupov was funded by an Else-Kroener-Fresenius scholarship (EKFS). The BeCOME study received funding from University of Tübingen, Grant/Award Number: 2453-0-0.
CRediT authorship contribution statement
John M. Felt: Conceptualization, Methodology, Software, Data curation, Formal analysis, Supervision, Validation, Writing – original draft, Writing – review & editing, Visualization. Natan Yusupov: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Validation. Karra D. Harrington: Conceptualization, Validation, Writing – review & editing. Julia Fietz: Conceptualization, Validation, Writing – review & editing. Zhenyu “Zach” Zhang: Writing – review & editing. Martin J. Sliwinski: Investigation, Resources, Writing – review & editing, Funding acquisition. Nilam Ram: Funding acquisition, Supervision, Validation, Writing – review & editing. Kieran J. O'Donnell: Methodology, Investigation, Resources, Data curation, Writing – review & editing, Funding acquisition. BeCOME Working Group Resources; Data curation; Funding. Michael J. Meaney: Investigation, Writing – review & editing, Funding acquisition. Frank W. Putnam: Investigation, Methodology, Resources, Project administration, Writing – review & editing, Funding acquisition. Jennie G. Nol: Investigation, Methodology, Resources, Project administration, Writing – review & editing, Funding acquisition. Elisabeth B. Binder: Conceptualization, Investigation, Resources, Writing – review & editing, Funding acquisition. Chad E. Shenk: Conceptualization, Methodology, Validation, Investigation, Resources, Data curation, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the BeCOME study team for their efforts: Victor I. Spoormaker, Angelika Erhardt, Susanne Lucae, Philipp G. Saemann, Norma C. Grandi, Tamara Namendorf, Michael Czisch, Immanuel Elbau, Laura Leuchs, Anna Katharine Brem, Leonhard Schilbach, Sanja Ilić-Ćoćić, Julius Ziebula, Iven-Alex von Mücke-Heim, Yeho Kim and Julius Pape.
The authors would like also to thank Stephanie Alam, Julia-Carolin Albrecht, Anastasia Bauer, Anja Betz, Miriam El-Mahdi, Gertrud Ernst-Jansen, Carolin Haas, Karin Hofer, Lisa Kammholz, Elisabeth Kappelmann, Sophia Koch, Alexandra Kocsis, Anna Lorenz, Rebecca Meissner, Jessie Osterhaus, Liisbeth Pirn and Linda Schuster for their help with data collection, study management, the recruitment, and screening of BeCOME participants, and Alexandra Bayer, Ines Eidner, Anna Hetzel, Elke Frank-Havemann, Viktoria Messerschmidt, and Ursula Ritter-Bohnensack for assisting with MRI scanning. We also thank Karina Van Bogart for feedback on earlier drafts of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2023.100577.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
The data that has been used is confidential.
References
- Abramovitch A., Short T., Schweiger A. The C Factor: cognitive dysfunction as a transdiagnostic dimension in psychopathology. Clin. Psychol. Rev. 2021;86 doi: 10.1016/j.cpr.2021.102007. [DOI] [PubMed] [Google Scholar]
- Allerhand M., Gale C.R., Deary I.J. The dynamic relationship between cognitive function and positive well-being in older people: a prospective study using the English Longitudinal Study of Aging. Psychol. Aging. 2014;29(2):306. doi: 10.1037/a0036551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anda R.F., Felitti V.J., Bremner J.D., et al. The enduring effects of abuse and related adverse experiences in childhood. Eur. Arch. Psychiatr. Clin. Neurosci. 2006;256(3):174–186. doi: 10.1007/s00406-005-0624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski K.M., Halladay A., Hu V.W., Mill J., Fallin M.D. Epigenetic research in neuropsychiatric disorders: the “tissue issue.”. Curr. Behav. Neurosci. Rep. 2016;3(3):264–274. doi: 10.1007/s40473-016-0083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett P. Structural equation modelling: adjudging model fit. Indiv. Differ. 2007;42(5):815–824. doi: 10.1016/j.paid.2006.09.018. [DOI] [Google Scholar]
- Belsky D.W., Caspi A., Arseneault L., et al. In: Hagg S., Tyler J.K., Hagg S., Justice J., Suderman M., editors. vol. 9. 2020. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. (eLife). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. http://www.jstor.org/stable/2346101 (Published online) [Google Scholar]
- Bernstein D.P., Stein J.A., Newcomb M.D., et al. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl. 2003;27(2):169–190. doi: 10.1016/s0145-2134(02)00541-0. [DOI] [PubMed] [Google Scholar]
- Bhattarai J., Oehlert M.E., Multon K.D., Sumerall S.W. Dementia and cognitive impairment among US veterans with a history of MDD or PTSD: a retrospective cohort study based on sex and race. J. Aging Health. 2019;31(8):1398–1422. doi: 10.1177/0898264318781131. [DOI] [PubMed] [Google Scholar]
- Brissos S., Dias V.V., Kapczinski F. Cognitive performance and quality of life in bipolar disorder. Can. J. Psychiatr. 2008;53(8):517–524. doi: 10.1177/070674370805300806. [DOI] [PubMed] [Google Scholar]
- Brückl T.M., Spoormaker V.I., Sämann P.G., et al. The biological classification of mental disorders (BeCOME) study: a protocol for an observational deep-phenotyping study for the identification of biological subtypes. BMC Psychiatr. 2020;20(1):213. doi: 10.1186/s12888-020-02541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda A.E., Tuulio-Henriksson A., Marttunen M., Suvisaari J., Lönnqvist J. A review on cognitive impairments in depressive and anxiety disorders with a focus on young adults. J. Affect. Disord. 2008;106(1–2):1–27. doi: 10.1016/j.jad.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Chan T., Parisi J.M., Moored K.D., Carlson M.C. Variety of enriching early-life activities linked to late-life cognitive functioning in urban community-dwelling African Americans. J. Gerontol. Ser. B. 2019;74(8):1345–1355. doi: 10.1093/geronb/gby056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Baldini U., Nieman D.H., Keestra A., et al. The relationship between cognitive functioning and psychopathology in patients with psychiatric disorders: a transdiagnostic network analysis. Psychol. Med. 2021:1–10. doi: 10.1017/S0033291721001781. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary I.J., Corley J., Gow A.J., et al. Age-associated cognitive decline. Br. Med. Bull. 2009;92(1):135–152. doi: 10.1093/bmb/ldp033. [DOI] [PubMed] [Google Scholar]
- DSM-IV . American Psychiatric Association; 1994. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- East-Richard C., R-Mercier A., Nadeau D., Cellard C. Transdiagnostic neurocognitive deficits in psychiatry: a review of meta-analyses. Can. Psychol. 2020;61(3):190–214. doi: 10.1037/cap0000196. [DOI] [Google Scholar]
- Felt J.M., Harrington K.D., Ram N., et al. Receptive Language abilities for females exposed to early life adversity: modification by epigenetic age acceleration at midlife in a 30-year prospective cohort study. J. Gerontol. Ser. B. 2022 doi: 10.1093/geronb/gbac158. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A.E., Robertson N.A., Wang T., Havas A., Ideker T., Adams P.D. DNA methylation clocks in aging: categories, causes, and consequences. Mol. Cell. 2018;71(6):882–895. doi: 10.1016/j.molcel.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis R.H., Wittchen H.U., Pfister H., Lieb R. Life events and changes in the course of depression in young adults. Eur. Psychiatr. 2002;17(5):241–253. doi: 10.1016/s0924-9338(02)00682-x. [DOI] [PubMed] [Google Scholar]
- Furr R.M., Bacharach V.R., Furr R.M., Bacharach V.R. Psychometrics: an Introduction. Sage; 2014. Confirmatory factor analysis; pp. 331–353. [Google Scholar]
- Gandal M.J., Haney J.R., Parikshak N.N., et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359(6376):693–697. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavson D.E., Elman J.A., Panizzon M.S., et al. Association of baseline semantic fluency and progression to mild cognitive impairment in middle-aged men. Neurology. 2020;95(8):e973–e983. doi: 10.1212/WNL.0000000000010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G., Guinney J., Zhao L., et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington K.D., Aschenbrenner A.J., Maruff P., et al. Undetected neurodegenerative disease biases estimates of cognitive change in older adults. Psychol. Sci. 2021;32(6):849–860. doi: 10.1177/0956797620985518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne J.K., Germine L.T. When does cognitive functioning peak? The asynchronous rise and fall of different cognitive abilities across the life span. Psychol. Sci. 2015;26(4):433–443. doi: 10.1177/0956797614567339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvanek Z.M., Boks M.P., Vinkers C.H., Higgins-Chen A.T. The cutting edge of epigenetic clocks: in Search of mechanisms linking aging and mental health. Biol. Psychiatr. 2023 doi: 10.1016/j.biopsych.2023.02.001. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohman T.J., Dumitrescu L., Oksol A., et al. APOE allele frequencies in suspected non-amyloid pathophysiology (SNAP) and the prodromal stages of Alzheimer's Disease. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0188501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D., Coughlan J., Mullen M. 2008. Structural Equation Modelling: Guidelines for Determining Model Fit.http://arrow.dit.ie/cgi/viewcontent.cgi?article=1001&context=buschmanart Articles. Published online. [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Zhang Y., Langfelder P., et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):1–18. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E.A., Accomando W.P., Koestler D.C., et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabacik S., Lowe D., Fransen L., et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging. 2022;2(6):484–493. doi: 10.1038/s43587-022-00220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno M., Sahashi K., Iguchi Y., Hashizume A. Preclinical progression of neurodegenerative diseases. Nagoya J. Med. Sci. 2018;80(3):289. doi: 10.18999/nagjms.80.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline R.B. third ed. The Guilford Press; 2010. Principles and Practice of Structural Equation Modeling. Third Edition. [Google Scholar]
- Knopman D.S., Gottesman R.F., Sharrett A.R., et al. Midlife vascular risk factors and midlife cognitive status in relation to prevalence of mild cognitive impairment and dementia in later life: the Atherosclerosis Risk in Communities Study. Alzheimers Dement. 2018;14(11):1406–1415. doi: 10.1016/j.jalz.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korten N.C., Penninx B.W., Pot A.M., Deeg D.J., Comijs H.C. Adverse childhood and recent negative life events: contrasting associations with cognitive decline in older persons. J. Geriatr. Psychiatr. Neurol. 2014;27(2):128–138. doi: 10.1177/0891988714522696. [DOI] [PubMed] [Google Scholar]
- Lee J.J., Wedow R., Okbay A., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50(8):1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M.E., Lu A.T., Quach A., et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linberg T., Schneider T., Waldfogel J., Wang Y. Socioeconomic status gaps in child cognitive development in Germany and the United States. Soc. Sci. Res. 2019;79:1–31. doi: 10.1016/j.ssresearch.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Lipnicki D.M., Crawford J.D., Dutta R., et al. Age-related cognitive decline and associations with sex, education and apolipoprotein E genotype across ethnocultural groups and geographic regions: a collaborative cohort study. PLoS Med. 2017;14(3) doi: 10.1371/journal.pmed.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston G., Huntley J., Sommerlad A., et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet. 2020;396(10248):413–446. doi: 10.1016/S0140-6736(20)30367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn D.J., Lang I.A., Langa K.M., Huppert F.A. Cognitive function and psychological well-being: findings from a population-based cohort. Age Ageing. 2008;37(6):685–689. doi: 10.1093/ageing/afn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A.T., Quach A., Wilson J.G., et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic J., Phipson B., Oshlack A. A cross-package Bioconductor workflow for analysing methylation array data. F1000Research. 2016;5 doi: 10.12688/f1000research.8839.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni R.E., Shah S., McRae A.F., et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre R.S., Cha D.S., Soczynska J.K., et al. Cognitive deficits and functional outcomes in major depressive disorder: determinants, substrates, and treatment interventions. Depress. Anxiety. 2013;30(6):515–527. doi: 10.1002/da.22063. [DOI] [PubMed] [Google Scholar]
- McTeague L.M., Goodkind M.S., Etkin A. Transdiagnostic impairment of cognitive control in mental illness. J. Psychiatr. Res. 2016;83:37–46. doi: 10.1016/j.jpsychires.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan M.J., Agid Y., Brüne M., et al. Cognitive dysfunction in psychiatric disorders: characteristics, causes and the quest for improved therapy. Nat. Rev. Drug Discov. 2012;11(2):141–168. doi: 10.1038/nrd3628. [DOI] [PubMed] [Google Scholar]
- Oveisgharan S., Wilson R.S., Yu L., Schneider J.A., Bennett D.A. Association of early-life cognitive enrichment with Alzheimer disease pathological changes and cognitive decline. JAMA Neurol. 2020;77(10):1217–1224. doi: 10.1001/jamaneurol.2020.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statsitical Computing; 2018. R: A Language and Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- Raj K., Horvath S. Current perspectives on the cellular and molecular features of epigenetic ageing. Exp. Biol. Med. 2020;245(17):1532–1542. doi: 10.1177/1535370220918329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K., Jaussent I., Stewart R., et al. Adverse childhood environment and late-life cognitive functioning. Int. J. Geriatr. Psychiatr. 2011;26(5):503–510. doi: 10.1002/gps.2553. [DOI] [PubMed] [Google Scholar]
- Roberts A.L., Sumner J.A., Koenen K.C., et al. Childhood abuse and cognitive function in a large cohort of middle-aged women. Child. Maltreat. 2020 doi: 10.1177/1077559520970647. Published online November 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock P.L., Roiser J.P., Riedel W.J., Blackwell A. Cognitive impairment in depression: a systematic review and meta-analysis. Psychol. Med. 2014;44(10):2029–2040. doi: 10.1017/S0033291713002535. [DOI] [PubMed] [Google Scholar]
- Rosseel Y. Lavaan: an R package for structural equation modeling. J. Stat. Software. 2012;48(2):1–36. [Google Scholar]
- Salthouse T.A. Decomposing age correlations on neuropsychological and cognitive variables. J. Int. Neuropsychol. Soc. JINS. 2009;15(5):650–661. doi: 10.1017/S1355617709990385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse T.A. Trajectories of normal cognitive aging. Psychol. Aging. 2019;34(1):17. doi: 10.1037/pag0000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J.D., Cheng T.W., Dunn E.C. Sensitive periods in development and risk for psychiatric disorders and related endpoints: a systematic review of child maltreatment findings. Lancet Psychiatr. 2022;9(12):978–991. doi: 10.1016/S2215-0366(22)00362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaie K.W. What can we learn from longitudinal studies of adult development? Res. Hum. Dev. 2005;2(3):133–158. doi: 10.1207/s15427617rhd0203_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semkovska M., Quinlivan L., O'Grady T., et al. Cognitive function following a major depressive episode: a systematic review and meta-analysis. Lancet Psychiatr. 2019;6(10):851–861. doi: 10.1016/S2215-0366(19)30291-3. [DOI] [PubMed] [Google Scholar]
- Shenk C, Felt J, Ram N, et al. Cortisol Trajectories Measured Prospectively across Thirty Years of Female Development Following Exposure to Childhood Sexual Abuse: Moderation by Epigenetic Age Acceleration at Midlife. Published online (in press).. [DOI] [PMC free article] [PubMed]
- Shenk C.E., Noll J.G., Putnam F.W., Trickett P.K. A prospective examination of the role of childhood sexual abuse and physiological asymmetry in the development of psychopathology. Child Abuse Negl. 2010;34(10):752–761. doi: 10.1016/j.chiabu.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk C.E., Noll J.G., Griffin A.M., et al. Psychometric evaluation of the comprehensive trauma Interview PTSD symptoms scale following exposure to child maltreatment. Child. Maltreat. 2016;21(4):343–352. doi: 10.1177/1077559516669253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau S., Arpadi S.M., Shen Y., et al. Epigenetic aging biomarkers associated with cognitive impairment in older african American adults with human immunodeficiency virus (HIV) Clin. Infect. Dis. 2021;73(11):1982–1991. doi: 10.1093/cid/ciab563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinke L., Cats D., Heijmans B.T. Omixer: multivariate and reproducible sample randomization to proactively counter batch effects in omics studies. Bioinformatics. 2021;37(18):3051–3052. doi: 10.1093/bioinformatics/btab159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnawska A., Tan Q., McGue M., et al. Epigenome-wide association study of cognitive functioning in middle-aged monozygotic twins. Front. Aging Neurosci. 2017;9:413. doi: 10.3389/fnagi.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A.J., McCartney D.L., Hillary R.F., et al. Childhood intelligence attenuates the association between biological ageing and health outcomes in later life. Transl. Psychiatry. 2019;9(1):1–8. doi: 10.1038/s41398-019-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M., Hemani G., Min J. 2020. Meffil: Efficient Algorithms for DNA Methylation. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., Marabita F., Lechner M., et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touleimat N., Tost J. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- Triche T.J., Jr., Weisenberger D.J., Van Den Berg D., Laird P.W., Siegmund K.D. Low-level processing of Illumina infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickett P.K., Noll J.G., Putnam F.W. The impact of sexual abuse on female development: lessons from a multigenerational, longitudinal research study. Dev. Psychopathol. 2011;23(2):453–476. doi: 10.1017/S0954579411000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino V., Huang M., Wang Z., et al. Epigenetic age acceleration and cognitive decline: a twin study. J. Gerontol. Ser. A. 2021;76(10):1854–1863. doi: 10.1093/gerona/glab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H.J., Jansen R.C., Fehrmann R.S., et al. MixupMapper: correcting sample mix-ups in genome-wide datasets increases power to detect small genetic effects. Bioinformatics. 2011;27(15):2104–2111. doi: 10.1093/bioinformatics/btr323. [DOI] [PubMed] [Google Scholar]
- Wilson R.S., Wang T., Yu L., Bennett D.A., Boyle P.A. Normative cognitive decline in old age. Ann. Neurol. 2020;87(6):816–829. doi: 10.1002/ana.25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo Y.S., Rosenblat J.D., Kakar R., Bahk W.M., McIntyre R.S. Cognitive deficits as a mediator of poor occupational function in remitted major depressive disorder patients. Clin. Psychopharmacol. Neurosci. 2016 doi: 10.9758/cpn.2016.14.1.1. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.H., Hayano M., Griffin P.T., et al. Loss of epigenetic information as a cause of mammalian aging. Cell. 2023 doi: 10.1016/j.cell.2022.12.027. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Boyle P.A., Segawa E., et al. Residual decline in cognition after adjustment for common neuropathologic conditions. Neuropsychology. 2015;29(3):335. doi: 10.1037/neu0000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov N., Dieckmann L., Erhart M., et al. Transdiagnostic evaluation of epigenetic age acceleration and burden of psychiatric disorders. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2023;48(9):1409–1417. doi: 10.1038/s41386-023-01579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.