Abstract
Objective
Tissue crosstalk mediated by secreted hormones underlies the integrative control of metabolism. We previously showed that CTRP13/C1QL3, a secreted protein of the C1q family, can improve glucose metabolism and insulin action in vitro and reduce food intake and body weight in mice when centrally delivered. A role for CTRP13 in regulating insulin secretion in isolated islets has also been demonstrated. It remains unclear, however, whether the effects of CTRP13 on cultured cells and in mice reflect the physiological function of the protein. Here, we use a loss-of-function mouse model to address whether CTRP13 is required for metabolic homeostasis.
Methods
WT and Ctrp13 knockout (KO) mice fed a standard chow or a high-fat diet were subjected to comprehensive metabolic phenotyping. Transcriptomic analyses were carried out on visceral and subcutaneous fat, liver, and skeletal muscle to identify pathways altered by CTRP13 deficiency. RNA-seq data was further integrated with the Metabolic Syndrome in Man (METSIM) cohort data. Adjusted regression analysis was used to demonstrate that genetic variation of CTRP13 expression accounts for a significant proportion of variance between differentially expressed genes (DEGs) in adipose tissue and metabolic traits in humans.
Results
Contrary to expectation, chow-fed Ctrp13-KO male mice had elevated physical activity, lower body weight, and improved lipid handling. On a high-fat diet (HFD), Ctrp13-KO mice of either sex were consistently more active and leaner. Loss of CTRP13 reduced hepatic glucose output and improved glucose tolerance, insulin sensitivity, and triglyceride clearance, though with notable sex differences. Consistent with the lean phenotype, transcriptomic analyses revealed a lower inflammatory profile in visceral fat and liver. Reduced hepatic steatosis was correlated with the suppression of lipid synthesis and enhanced lipid catabolism gene expression. Visceral fat had the largest number of DEGs and mediation analyses on the human orthologs of the DEGs suggested the potential causal contribution of CTRP13 to human metabolic syndrome.
Conclusions
Our results suggest that CTRP13 is a negative metabolic regulator, and its deficiency improves systemic metabolic profiles. Our data also suggest the reduction in circulating human CTRP13 levels seen in obesity and diabetes may reflect a compensatory physiologic response to counteract insulin resistance.
Keywords: Secreted hormone, Obesity, Diabetes, Dyslipidemia, Insulin sensitivity, Metabolic syndrome
Highlights
-
•
Chow-fed mice lacking CTRP13 have elevated physical activity, lower body weight, and improved lipid handling.
-
•
CTRP13 deficiency attenuates diet-induced obesity.
-
•
Loss of CTRP13 improved glucose and pyruvate tolerance, insulin sensitivity, and triglyceride clearance.
-
•
CTRP13 deficiency reduces hepatic steatosis, and adipose and liver inflammation.
-
•
Mediation analysis suggests the potential contribution of CTRP13 to human metabolic syndrome.
1. Introduction
Systemic metabolic homeostasis is maintained by both tissue autonomous and non-autonomous mechanisms. The integrative control of metabolism is dependent on tissue crosstalk facilitated by secreted endocrine factors [1]. Some of the secreted metabolic regulators (e.g., adiponectin, leptin, FGF-21, GDF15) are produced by only select cell types and tissues while others are expressed more broadly [1]. Dysregulated production and activity of these endocrine factors are frequently associated with and linked to metabolic disorders [2,3].
As part of an ongoing effort to decode mechanisms underlying inter-organ communication, we have described and systematically characterized the metabolic functions of a family of secreted proteins, the C1q/TNF-related proteins (CTRP1-15) [[4], [5], [6], [7], [8], [9], [10], [11], [12]]. All CTRPs possess the signature C1q globular domain in the C-terminus, and are part of the extended C1q family [13]. Gain- and loss-of-function mouse models have helped establish non-redundant metabolic functions for CTRP family members [7,[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]], and provided insights into their remarkable conservation across all vertebrate lineages. CTRPs control various aspects of glucose and lipid metabolism by either directly acting on metabolic tissues (e.g., liver, adipose tissue, skeletal muscle) or indirectly via their modulatory roles on immune cells and inflammation [15,30]. Additional functions of CTRPs have also been established in the cardiovascular system [[31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]], kidney [43,44], gut [45], eye [46,47], musculoskeletal system [[48], [49], [50]], and brain [[51], [52], [53]].
While we have described the metabolic phenotypes of most CTRP knockout (KO) mice [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]], the impact of CTRP13 deficiency on systemic metabolism has yet to be determined. We initially identified CTRP13 (also known as C1QL3 and C1QTNF13) on the basis of shared sequence homology with other CTRP family members and adiponectin [8]. As with all C1q family proteins, secreted CTRP13 forms trimers and higher-order oligomeric structures [54]. We previously showed that CTRP13 can promote glucose uptake in cultured adipocyte, hepatocyte, and myocyte cell lines, as well as ameliorate lipid-induced insulin resistance in hepatocytes [8]. We also showed that central delivery of CTRP13 suppresses food intake and reduces body weight in mice [55]. Circulating levels of CTRP13 are reduced in obese and diabetic adult humans [[56], [57], [58]] and in obese children [59]. Therefore, a reduction in plasma CTRP13 levels is thought to contribute to dysregulated glucose and lipid metabolism and insulin resistance in humans.
In the context of atherosclerosis, circulating levels of CTRP13 are reduced in patients with coronary heart disease and in ApoE-deficient mice [60]; infusion of human CTRP13 reduces atherosclerotic lesions in ApoE-deficient mice by suppressing macrophage lipid uptake and foam cell formation [60]. Additional protective roles of CTRP13, based on recombinant protein supplementation, have also been demonstrated in the contexts of vascular calcification and aortic aneurysm formation [61,62]. In contrast to its protective role in the vasculature, CTRP13 appears to be a negative regulator of insulin secretion in pancreatic β-cells. In both β-cell lines and isolated mouse islets, CTRP13 inhibits insulin secretion in response to extendin-4, elevated cAMP, or high glucose [63,64]. Although the requirement of CTRP13 for CNS function has been extensively documented based on constitutive and conditional KO mouse models [[65], [66], [67], [68]], it is unclear whether the metabolic and pharmacologic effects of CTRP13 on cultured cells, isolated pancreatic islets, and in mice reflect the physiological function of the protein.
The aim of the study was to use a genetic loss-of-function approach to establish whether CTRP13 is required for metabolic homeostasis. Based on previous findings [8,[55], [56], [57], [58],63], we speculated that CTRP13 deficiency would impact insulin secretion and systemic metabolism. Indeed, loss of CTRP13 reduced body weight gain and improved systemic glucose and lipid metabolic profiles, although the direction of change was contrary to what we had initially expected. Loss of CTRP13 did not affect basal serum insulin levels in chow- or high-fat diet (HFD)-fed mice but resulted in a modest decrease of insulin during refeeding in female mice. Consistent with the lean phenotype, Ctrp13-KO mice had lower adiposity, smaller adipocytes, and reduced adipose inflammation in response to high-fat feeding. CTRP13 deficiency also reduced hepatic inflammation and steatosis. By integrating our RNA-seq data with the Metabolic Syndrome in Man (METSIM) cohort data, and applying an adjusted regression approach (a form of mediation analysis), we further showed that CTRP13/C1QL3 expression is relevant to and potentially contributes causally to metabolic syndrome in humans. Collectively, our data provide physiologic evidence that CTRP13 is a negative metabolic regulator, in that its deficiency improves systemic metabolic profiles.
2. Materials and methods
2.1. Mouse model
Eight-week-old mouse tissues (gonadal white adipose tissue, inguinal white adipose tissue, brown adipose tissue, liver, heart, skeletal muscle, kidney, pancreas, cerebellum, cortex, hypothalamus, hindbrain, and hippocampus) from C57BL/6J male mice (The Jackson Laboratory, Bar Harbor, ME) were collected from fasted and refed experiments as we have described previously [69]. In brief, for the fasted group, food was removed for 16 h (beginning 10 h into the light cycle), and mice were euthanized 2 h into the light cycle. For the refed group, mice were fasted for 16 h and refed with chow pellets for 2 h before being euthanized. A separate cohort of mouse tissues (gonadal white adipose tissue, inguinal white adipose tissue, brown adipose tissue, liver, heart, skeletal muscle, kidney, pancreas, whole brain, spleen, colon, and small intestine) from C57BL/6J male mice fed a control low-fat diet (LFD) or a high-fat diet (HFD) for 12 weeks were also collected as we have described previously [69]. Ctrp13/C1ql3 knockout (KO) mouse strain [66] was obtained from the Jackson Laboratory (strain # 029672). The mice were backcrossed to C57BL/6J genetic background for more than 5 generations. To generate a constitutive whole-body KO mouse model, C1ql3flox/+ mice were first crossed to a CMV-Cre driver mouse line (The Jackson Laboratory mouse strain # 006054) to generate C1ql3 heterozygous (+/−) mice. Homozygous KO (−/−) mice were generated by intercrossing Ctrp13 heterozygous (+/−) mice. Deletion of exon 2 (encoding amino acid 1–199) removed 77% of the protein coding region of the gene, thus ensuring a complete null allele. Genotyping primers for the WT allele were forward (m13-WT-F) 5′-TTCAC ATAGGTGCGTGCTAG-3′ and reverse (m13-WT-R) 5′-GTCAGCATAAATAATAAATCCAGAG-3′. The size of the WT band was 186 bp. Genotyping primers for the Ctrp13 KO allele were forward (mVenus-F) 5′-CAACATCCTGGGGCACAAGCTGGAG-3′ and reverse (mVenus-R) 5′-CAGCTTGGACTGGTAGCTCAG-3′. The size of the KO band was 220 bp. The genotyping PCR parameters were as follows: 94 °C for 5 min, followed by 10 cycles of (94 °C for 10 s, 65 °C for 15 s, 72 °C for 30 s), then 25 cycles of (94 °C for 10 s, 55 °C for 15 s, 72 °C for 30 s), and lastly 72 °C for 5 min. Due to the presence of GC rich sequences, 8% DMSO was included in the PCR genotyping reaction. Ctrp13 KO (−/−) and WT (+/+) littermate controls were housed in polycarbonate cages on a 12-h light–dark photocycle with ad libitum access to water and food. Mice were fed either a standard chow (Envigo; 2018SX) or a high-fat diet (HFD; 60% kcal derived from fat; #D12492, Research Diets, New Brunswick, NJ). HFD was provided for the duration of the study, beginning at 6 weeks of age. At termination of study, mice were fasted for 2 h and euthanized. Tissues were collected, snap-frozen in liquid nitrogen, and kept at −80 °C until analysis. All mouse protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. All animal experiments were conducted in accordance with the National Institute of Health guidelines and followed the standards established by the Animal Welfare Acts.
2.2. Body composition analysis
Body composition analyses for total fat, lean mass, and water content were determined using a quantitative magnetic resonance instrument (Echo-MRI-100, Echo Medical Systems, Waco, TX) at the Mouse Phenotyping Core facility at Johns Hopkins University School of Medicine.
2.3. Indirect calorimetry
Chow or HFD-fed WT and Ctrp13 KO male and female mice were used for simultaneous assessments of daily body weight change, food intake (corrected for spillage), physical activity, and whole-body metabolic profile in an open flow indirect calorimeter (Comprehensive Laboratory Animal Monitoring System, CLAMS; Columbus Instruments, Columbus, OH) as described previously [20]. In brief, data were collected for three days to confirm mice were acclimatized to the calorimetry chambers (indicated by stable body weights, food intakes, and diurnal metabolic patterns), and data were analyzed from the subsequent three days, including one day of fasting and one day of refeeding. Rates of oxygen consumption (; mL kg−1 h−1) and carbon dioxide production (; mL kg−1 h−1) in each chamber were measured every 24 min throughout the studies. Respiratory exchange ratio (RER = /) was calculated by CLAMS software (version 5.66) to estimate relative oxidation of carbohydrates (RER = 1.0) versus fats (RER = 0.7), not accounting for protein oxidation. Energy expenditure (EE) was calculated as EE = × [3.815 + (1.232 × RER)] and normalized to lean mass. Because normalizing to lean mass can potentially lead to overestimation of EE, we also performed ANCOVA analysis on EE using body weight as a covariate [70]. Physical activities were measured by infrared beam breaks in the metabolic chamber.
2.4. Glucose, insulin, pyruvate, and lipid tolerance tests
All tolerance tests were conducted as described previously [16,19,24]. For glucose tolerance tests (GTTs), mice were fasted for 6 h before glucose injection. Glucose (Sigma, St. Louis, MO) was reconstituted in saline (0.9 g NaCl/L), sterile-filtered, and injected intraperitoneally (i.p.) at 1 mg/g body weight (i.e., 10 μL/g body weight). Blood glucose was measured at 0, 15, 30, 60, and 120 min after glucose injection using a glucometer (NovaMax Plus, Billerica, MA). Blood was collected at 0, 15, and 30 min time points for serum isolation followed by insulin ELISA. For insulin tolerance tests (ITTs), food was removed 2 h before insulin injection. 8.62 μL of insulin stock (4 mg/mL; Gibco) was diluted in 10 mL of saline, sterile-filtered, and injected i.p. at 1.0 U/kg body weight (i.e., 10 μL/g body weight). Blood glucose was measured at 0, 15, 30, 60, and 90 min after insulin injection using a glucometer (NovaMax Plus). For pyruvate tolerance tests (PTTs), mice were fasted overnight for 16 h before sodium pyruvate injection. Sodium pyruvate (Sigma) was reconstituted in saline to a final concentration of 1 g/10 mL and injected i.p. into mice at a dose of 1 g/kg body weight (i.e., 10 μL/g body weight). Blood glucose was measured at the indicated time points using a glucometer. For lipid tolerance tests (LTTs), mice were fasted for 12 h and then injected i.p. with 20% emulsified Intralipid (soybean oil; Sigma; 10 μL/g of body weight). Sera were collected via tail bleed using a Microvette® CB 300 (Sarstedt) at 0, 1, 2, 3, and 4 h post-injection. Serum triglyceride levels were quantified using kits from Infinity Triglycerides (Thermo Fisher Scientific).
2.5. Fasting–refeeding metabolite and insulin response
Mice were fasted overnight (∼16 h) then reintroduced to food as described previously [21]. Blood glucose was monitored at the 16 h fast time point (time = 0 h refed) and at 1, 2, and 3 h into the refeeding process. Serum was collected at the 16 h fast and 2 h refed time points for insulin ELISA, as well as for the quantification of triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate levels.
2.6. Blood and tissue chemistry analysis
Tail vein blood samples were allowed to clot on ice and then centrifuged for 10 min at 10,000 × g. Serum samples were stored at −80 °C until analyzed. Serum triglycerides (TG) and cholesterol were measured according to manufacturer's instructions using an Infinity kit (Thermo Fisher Scientific, Middletown, VA). Non-esterified free fatty acids (NEFA) were measured using a Wako kit (Wako Chemicals, Richmond, VA). Serum β-hydroxybutyrate (ketone) concentrations were measured with a StanBio Liquicolor kit (StanBio Laboratory, Boerne, TX). Serum insulin levels were measured by ELISA according to manufacturer's instructions (Crystal Chem, Elk Grove Village, IL; cat # 90080).
2.7. Serum triglyceride and cholesterol analysis by FPLC
Food was removed for 2 h (in the light cycle) prior to blood collection. Sera collected from mice were pooled (n = 6–10/group) and sent to the Mouse Metabolism Core at Baylor College of Medicine for analysis. Serum sample was first fractionated by fast protein liquid chromatography (FPLC). A total of 45 fractions were collected. TG and cholesterol level in each fraction were quantified.
2.8. Histology and quantification
Inguinal (subcutaneous) white adipose tissue (iWAT), gonadal (visceral) white adipose tissue (gWAT), and liver, were dissected and fixed in formalin. Paraffin embedding, tissue sectioning, and staining with hematoxylin and eosin were performed at the Pathology Core facility at Johns Hopkins University School of Medicine. Images were captured with a Keyence BZ-X700 All-in-One fluorescence microscope (Keyence Corp., Itasca, IL). Adipocyte (gWAT and iWAT) cross-sectional area (CSA), as well as total area covered by lipid in hepatocytes were measured on hematoxylin and eosin-stained slides using ImageJ software [71]. For adipose CSA and hepatocyte lipid area measurements, all cells in one field of view at 100× magnification per tissue section per mouse were analyzed. For iWAT and gWAT adipocyte CSA, an average of 600 and 450 cells were quantified per mouse, respectively. Image capturing and quantifications were carried out blinded to genotype.
2.9. Respirometry of frozen tissue samples
Respirometry was conducted on frozen tissue samples to assay for mitochondrial activity as described previously [72]. Briefly, all tissues were dissected, snap frozen in liquid nitrogen, and stored at −80 °C for later analysis. Samples were thawed in MAS buffer (70 mM sucrose, 220 mM mannitol, 5 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA, 2 mM HEPES pH 7.4), finely minced with scissors, and then homogenized with a glass Dounce homogenizer. The resulting homogenate was spun at 1000 g for 10 min at 4 °C. The supernatant was collected and immediately used for protein quantification by BCA assay (Thermo Scientific, cat # 23225). Each well of the Seahorse microplate was loaded with 8 or 15 μg of homogenate protein for liver and gastrocnemius or iWAT and gWAT, respectively. Each biological replicate is comprised of three technical replicates. Samples were run on a Seahorse XFe96 Analyzer (Agilent Technologies). Samples from all tissues were treated separately with NADH (1 mM) as a complex I substrate or succinate (a complex II substrate, 5 mM) in the presence of rotenone (a complex I inhibitor, 2 μM), then with the inhibitors rotenone (2 μM) and Antimycin A (4 μM), followed by TMPD (0.45 mM) and ascorbate (1 mM) to activate complex IV, and finally treated with azide (40 mM) to assess non-mitochondrial respiration.
Mitochondrial content of homogenates used for respirometry was quantified using MitoTracker Deep Red FM (MTDR, Invitrogen, M22426) as described previously [72]. Briefly, lysates were incubated with MTDR (1 μM) for 10 min at 37 °C, then centrifuged at 2000 g for 5 min at 4 °C. The supernatant was carefully removed and replaced with 1× MAS solution and fluorescence was read with excitation and emission wavelengths of 581 nm and 644 nm, respectively. To minimize non-specific background signal contribution, control wells were loaded with MTDR and 1× MAS and subtracted from all sample values.
2.10. RNA sequencing
Bulk RNA sequencing of liver, skeletal muscle (gastrocnemius), gonadal white adipose tissue (gWAT), and inguinal white adipose tissue (iWAT) of Ctrp13-KO mice and WT controls were performed by Novogene (Sacramento, California, USA) on an Illumina platform (NovaSeq 6000) and pair-end reads were generated. Sample size: 6 WT and 6 KO for liver and skeletal muscle; 4 WT and 6 KO for gWAT; 5 WT and 6 KO for iWAT. Novogene provided bioinformatics analysis using their standard pipeline. In brief, data analysis was performed using a combination of programs, including Hisat2, FeatureCounts, DEseq2, CluterProfiler, Gsea,and Gatk. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were implemented by ClusterProfiler.
2.11. RNA-seq analysis and integration with the METSIM cohort
All processed datasets used and R scripts to reproduce analyses are freely available at: https://github.com/Darington/CTRP13KO-and-integration. Before integrating the RNA-seq data with the METSIM data, we performed independent analysis of the RNA-seq data generated by Novogene. Raw reads were aligned to the mouse genome (GRCm38 mm10) using standard settings on STAR aligner [73] and PCR duplicates removed via gatk. Briefly, counts matrices were filtered for genes which showed >50 counts across animals and tissues. Next, differential expression was performed using limma [74]. Resulting Ctrp13-KO differentially expressed genes (DEGs) were considered significant at a logistic regression p-value of 0.005, where human orthologues were used for downstream analyses. Here, human orthologues for mouse genes were identified from the Mouse Genome Informatics database and intersected with genes measured in the METSIM cohort [75]. Within the METSIM cohort, adipose tissue gene expression data is available for 770 individuals [75,76]. We leveraged this dataset for our data integration analysis. Given the direction of effect of Ctrp13 ablation (KO mice showed favorable systemic metabolic profiles), focal genes for the analyses were used for genes upregulated in Ctrp13-KO and negatively correlated (bicor p-value < 0.01) with insulin resistance traits in METSIM or Ctrp13-KO down-regulated genes that showed negative correlations with the same traits. These were used to visualize correlation structure of human orthologues in METSIM. From these, TNFRSF1B and HIF3A were used for regression adjustments. Specifically, empirical regressions were performed between either gene and indicated metabolic traits (biweight mid-correlation coefficient [77] where the regression p-values were compared to the same analyses, except performed using residuals of indicated gene with CTRP13/C1QL3 expression and indicated trait as previously described [[78], [79], [80]]).
2.12. Quantitative real-time PCR
Total RNA was isolated from tissues using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. Purified RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-rad). Real-time quantitative PCR analysis was performed on a CFX Connect Real-Time System (Bio-rad) using iTaq™ Universal SYBR Green Supermix (Bio-rad) per manufacturer's instructions. Data were normalized to the stable housekeeping gene β-actin or 36B4 (encoding the acidic ribosomal phosphoprotein P0) and expressed as relative mRNA levels using the ΔΔCt method [81]. Real-time qPCR primers used to assess Ctrp13 expression across mouse tissues were: forward 5′-AACGCAAGATAAGCAGATGTGTG-3′ and reverse 5′-AAGGAGTATTTGCTTTGGCGG-3′. qPCR primers used to confirm the absence of Ctrp13/C1ql3 in KO mice were: forward (m13-F), 5′-CTACTTCTTCACCT ACCACGTC-3′ and reverse (m13-R), 5′-GAAGGACCACACTGTTACTGG-3′.
2.13. Statistical analyses
Sample size for each assay and test is indicated in the figure and/or figure legend. All results are expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed with Prism 10 software (GraphPad Software, San Diego, CA). Data were analyzed with two-tailed Student's t-tests or by repeated measures ANOVA. Normality tests were performed in GraphPad Prism 10 to ensure the data points are normally distributed prior to statistical analysis. All tolerance tests, body weights over time, and fasting–refeeding responses were analyzed via 2-way ANOVA with Sidak's multiple comparisons test. p < 0.05 was considered statistically significant.
3. Results
3.1. Expression of CTRP13 in peripheral tissues is modulated by diet and metabolic state
We previously showed that Ctrp13 is expressed in the brain and adipose tissue of mice [8]. We extend the finding here and showed that Ctrp13 is also expressed in other metabolically relevant peripheral tissues, including liver, skeletal muscle, and pancreas (Figure 1A). Consistent with the mouse data, human CTRP13/C1QL3 is most highly expressed in the brain, with lower expression seen in peripheral tissues (Figure 1B). Expression of Ctrp13 varies depending on the metabolic state of the animal. Refeeding after an overnight food withdrawal suppressed the expression of Ctrp13 in visceral (gonadal) white adipose tissue (gWAT), brown adipose tissue (BAT), liver, skeletal muscle, kidney, and cerebellum (Figure 1C). Fasting and refeeding, however, has no effect on the expression of Ctrp13 in subcutaneous (inguinal) white adipose tissue (iWAT) and other brains regions such as the hypothalamus, hindbrain, and hippocampus (Figure 1C). Relative to lean mice fed a control low-fat diet (LFD), obese mice fed a high-fat diet (HFD) had a modest upregulated expression of Ctrp13 in the heart, iWAT, and small intestine, and downregulated expression in the kidney (Figure 1D). These data indicate that Ctrp13 expression in peripheral tissue is responsive to nutritional cues and changes in metabolic state, suggestive of a metabolic role.
Figure 1.
Expression of Ctrp13 is responsive to nutritional and metabolic cues. (A) Expression of Ctrp13 across different mouse tissues (n = 10). Expression levels were first normalized to β-actin, then to that of the small intestine (with the lowest expression) (B) Expression of CTRP13/C1QL3 across human tissues derived from the GTEx Portal [94]. (C) Ctrp13 expression in mouse tissues in response to overnight (16 h) fast and 2 h refeeding (after an overnight fast). Expression levels were normalized to β-actin. (D) Ctrp13 expression in mouse tissue in response to a 12-week period of control low-fat diet (LFD) or a high-fat diet (HFD). Expression levels were normalized to β-actin. All data are presented as mean ± S.E.M. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
3.2. Chow-fed Ctrp13-KO mice have lower body weight and higher physical activity
We used a whole-body knockout (KO) mouse model [66] to address the requirement of CTRP13 for metabolic homeostasis. As expected, Ctrp13-KO mice lacked Ctrp13 transcript (Figure 2A–B). At 16 weeks of age, male, but not female, Ctrp13-KO mice weighed less compared to wild-type (WT) littermates (Figure 2C), and this modest difference in body weight was due to slightly lower lean mass (Figure 2D). Mice were housed in indirect calorimetry cages to assess food intake, physical activity, and energy expenditure. Food intake across the circadian cycle (light and dark) and metabolic states (ad libitum fed, fasted, refed) was not significantly different between genotypes of either sex (Figure 2E,I). Ambulatory activity levels, however, were elevated in Ctrp13-KO mice of both sexes (Figure 2F,J). Despite higher physical activity levels, increased energy expenditure (when normalized to lean mass) was only observed in Ctrp13-KO mice of either sex during the fasting period (Figure 2G,J). It is known that energy expenditure when normalized to body weight or lean mass can potentially lead to overestimation of energy expenditure [70]. We therefore performed ANCOVA analyses on energy expenditure using body weight as a covariate [70]. This analysis also indicated male KO mice had significantly higher energy expenditure (p = 0.0055) during fasting (Figure 2H,L). Together, these data show that higher physical activity likely contributes to lower body weight in Ctrp13-KO mice.
Figure 2.
Chow-fed Ctrp13-KO mice have lower body weight and higher physical activity and energy expenditure. (A) Confirmation of Ctrp13 KO mice by PCR. (B) Expression of Ctrp13 transcript is completely abolished in the frontal cortex of Ctrp13-KO mice (n = 6). Expression levels were normalized to 36B4 gene. (C) Body weights of Ctrp13-KO mice and WT littermates at 16 weeks of age (male: WT, n = 9; KO, n = 14; female: WT, n = 12; KO, n = 12). (D) Body composition analysis of fat mass, lean mass, % fat mass (relative to body weight), and % lean mass of WT and KO male mice. (E–G) Food intake, ambulatory activity, and energy expenditure in male mice across the circadian cycle (light and dark) and metabolic states (ad libitum fed, fasted, refed) (WT, n = 8; KO, n = 13). (H) ANCOVA analysis of energy expenditure using body weight as a covariate in ad libitum fed, fasted, and refed male mice. (I–K) Food intake, ambulatory activity, and energy expenditure in female mice (WT, n = 11; KO, n = 11). (L) ANCOVA analysis of energy expenditure using body weight as a covariate in ad libitum fed, fasted, and refed female mice. All data are presented as mean ± S.E.M. ∗P < 0.05.
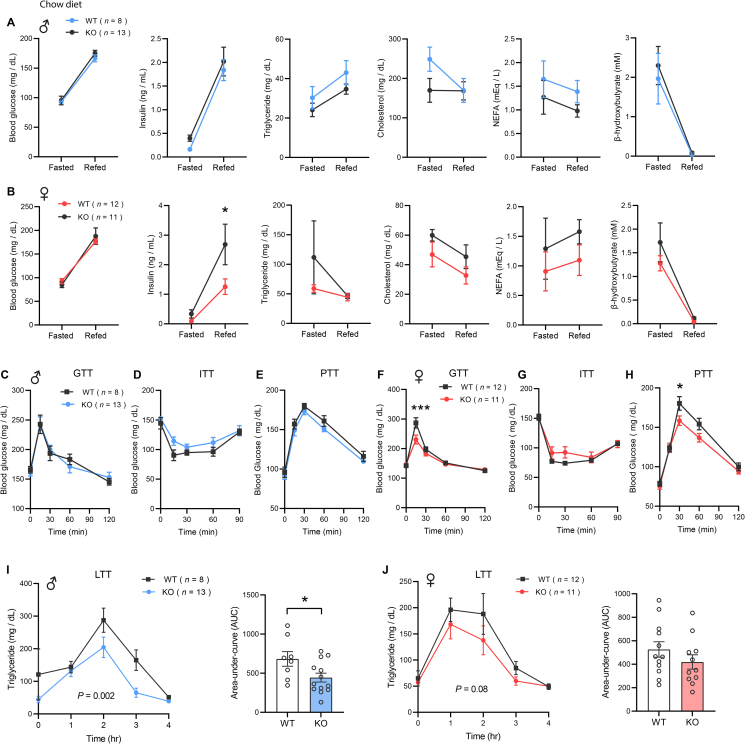
3.3. Chow-fed Ctrp13-KO mice have lower refed insulin level and a modest improvement in triglyceride clearance
Because Ctrp13 expression is regulated by nutritional state, we assessed the consequence of CTRP13 deficiency on serum metabolites in response to fasting and refeeding. Blood glucose, serum insulin, triglyceride, cholesterol, non-esterified free fatty acid (NEFA), and β-hydroxybutyrate levels in overnight fasted and refed states were not significantly different between genotypes in male mice (Figure 3A). In contrast, serum insulin levels in the refed state were significantly lower in Ctrp13-KO female mice relative to WT controls, even though refed blood glucose levels were virtually identical between genotypes (Figure 3B), suggesting an improvement in insulin action as less insulin was needed to handle the rise in blood glucose upon refeeding. Although Ctrp13 expression is modulated by fasting and refeeding in WT mice, its absence only affected insulin levels in the refed state of females, but had no impact on other serum lipid metabolite levels.
Figure 3.
Chow-fed Ctrp13-KO mice have lower refed insulin level and a modest improvement in triglyceride clearance. (A–B) Overnight fasted and refed blood glucose, serum insulin, triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate levels in male (A) and female (B) mice. (C–E) Blood glucose levels during glucose tolerance tests (GTT; C), insulin tolerance tests (ITT; D), and pyruvate tolerance tests (PTT; E) in WT and KO male mice. (F–H) Blood glucose levels during GTT (F), ITT (G), and PTT (H) in WT and KO female mice. (I–J) Serum triglyceride levels over the course of lipid tolerance tests (LTT) in male (I) and female (J) WT and KO mice. The bar graphs indicate area-under-curve of the LTT data. All data are presented as mean ± S.E.M. ∗P < 0.05.
We next determined whether CTRP13 deficiency affects glucose uptake in peripheral tissues in response to acute glucose challenge. Ctrp13-KO female, but not male, mice showed enhanced glucose tolerance, most notably at the 15 min time point (Figure 3C,F). Direct assessment of insulin action by insulin tolerance test did not reveal any differences in insulin sensitivity between genotypes of either sex (Figure 3D,G). In the fasted state, pyruvate is converted to glucose in the liver via gluconeogenesis. Because insulin suppresses gluconeogenesis, insulin sensitivity in liver affects hepatic glucose output. Assessment of hepatic glucose output and insulin sensitivity by pyruvate tolerance test revealed an improvement in Ctrp13-KO female, but not male, mice (Figure 3E,H). Lipid tolerance tests were performed to assess whether CTRP13 deficiency affects the capacity of KO mice to handle an acute fat loading. Loss of CTRP13 significantly improved the rate of triglyceride clearance in male mice (Figure 3I), and with a trend toward faster triglyceride clearance in female mice (Figure 3J). Together, these data indicate that CTRP13 deficiency improves glucose handling and hepatic insulin sensitivity in female mice and enhances lipid handling in male mice.
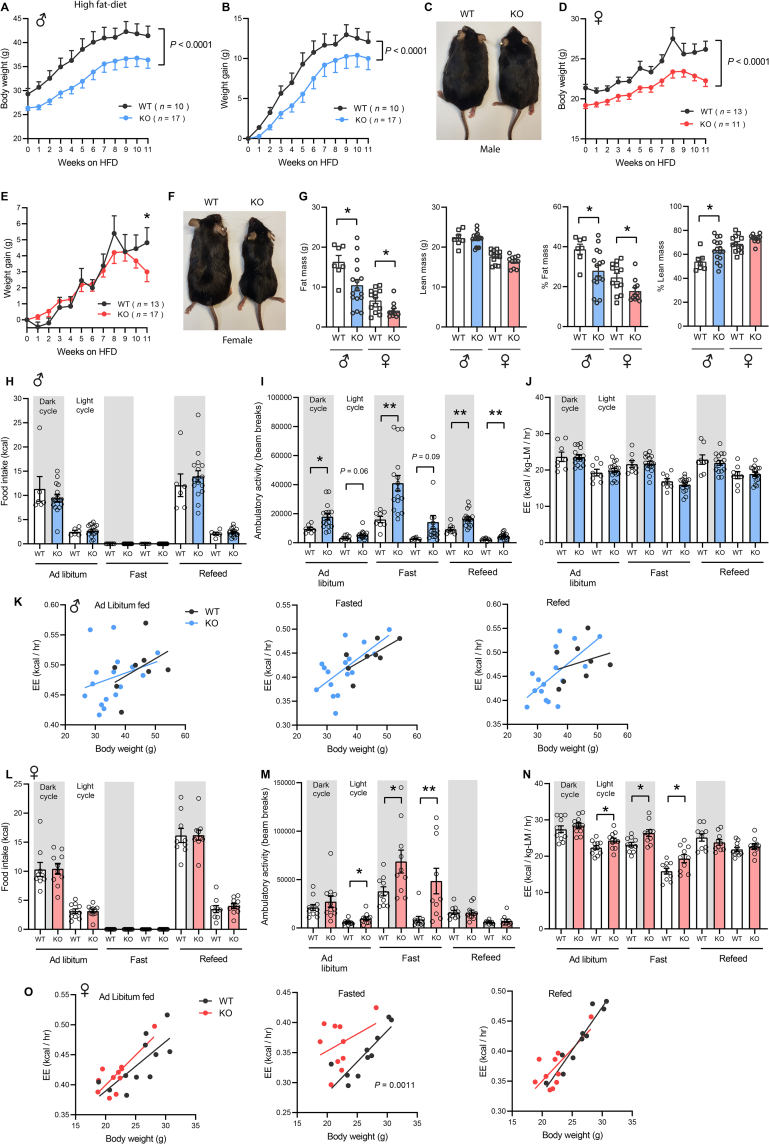
3.4. Loss of CTRP13 reduces weight gain in response to an obesogenic diet
Given the modest phenotypes seen in chow-fed Ctrp13-KO mice, we next determined whether loss of CTRP13 affects body weight gain in response to an obesogenic diet. Male KO mice had consistently lower body weight over the course of high-fat diet (HFD) (Figure 4A), and also gained significantly less weight over the same time frame (Figure 4B–C). Similar to male mice, female KO mice also had lower body weight over the course of HFD (Figure 4D). However, female mice gained similar amount of weight in the first 9 weeks on HFD, but the weight gain started to diverge due to KO female mice losing weight from 9 weeks onward (Figure 4E), resulting in lower body weight at the end of 11 weeks of HFD (Figure 4F). Lower body weight was due to reduced fat mass and not lean mass in KO male and female mice (Figure 4G). Reduced weight gain in response to high-fat feeding was not due to altered caloric intake. Food intake measurements did not reveal any differences between genotypes of either sex across the circadian cycle (light and dark) and metabolic states (ad libitum fed, fasted, refed) (Figure 4H,L). Similar to the chow-fed mice, Ctrp13-KO mice on HFD maintained elevated ambulatory activity levels (Figure 4I,M). A modest increase in energy expenditure (when normalized to lean mass) was only observed in female mice (Figure 4J,N). When body weight was used as a covariate in ANCOVA analyses, energy expenditure was shown to be significantly higher in KO female mice during fasting (p = 0.0011) (Figure 4K,O). Together, these data indicate that increased physical activity levels likely contribute to lower adiposity and body weight in Ctrp13-KO mice fed an obesogenic diet.
Figure 4.
Loss of CTRP13 reduces weight gain in response to an obesogenic diet. Body weight (A) and weight gained (B) over time of WT and KO male mice fed a high-fat diet (HFD). (C) Representative image of WT and KO male mice after 11 weeks on HFD. Body weight (D) and weight gained (E) over time of WT and KO female mice fed a high-fat diet. (F) Representative image of WT and KO female mice after 11 weeks on HFD. (G) Fat mass, lean mass, % fat mass (normalized to body weight), and % lean mass of WT and KO mice on HFD for ∼12 weeks (Male: WT, n = 7; KO, n = 16; female: WT, n = 13; KO, n = 11). (H–J) Food intake, ambulatory activity, and energy expenditure in male mice across the circadian cycle (light and dark) and metabolic states (ad libitum fed, fasted, refed) (WT, n = 8; KO, n = 16). (K) ANCOVA analysis of energy expenditure using body weight as a covariate in ad libitum fed, fasted, and refed male mice. (L–N) Food intake, ambulatory activity, and energy expenditure in female mice (WT, n = 10; KO, n = 10). (O) ANCOVA analysis of energy expenditure using body weight as a covariate in ad libitum fed, fasted, and refed female mice. All data are presented as mean ± S.E.M. ∗P < 0.05; ∗∗P < 0.01.
3.5. CTRP13 deficiency improves glucose, pyruvate, and lipid tolerance and insulin sensitivity in mice fed a high-fat diet
We again assessed whether serum metabolite response to fasting and refeeding would be different in the context of a diet-induced obese state. Blood glucose, serum insulin, triglyceride, cholesterol, NEFA, and β-hydroxybutyrate levels in the overnight fasted and refed states were not significantly different between genotypes in male mice (Figure 5A). In KO female mice, however, fasting insulin levels trended (p = 0.08) higher, and serum triglyceride and NEFA levels were lower in the refed state relative to WT controls (Figure 5B). We performed FPLC fractionation on pooled sera to assess lipoprotein-associated triglyceride and cholesterol. The profiles of very low-density lipoprotein-triglyceride (VLDL-TG) and high-density lipoprotein-cholesterol (HDL-C) were not different between genotypes in male mice (Figure 5C) but was lower in female KO mice (Figure 5D).
Figure 5.
CTRP13 deficiency improves glucose and pyruvate tolerance and insulin sensitivity in mice fed a high-fat diet. (A–B) Overnight fasted and refed blood glucose, serum insulin, triglyceride, cholesterol, non-esterified free fatty acids (NEFA), and β-hydroxybutyrate levels in male (A) and female (B) mice on HFD. (C–D) FPLC analysis of triglyceride and cholesterol levels in VLDL, LDL, and HDL of male (C) and female (D) mice. (E–G) Blood glucose levels during glucose tolerance tests (GTT; E), insulin tolerance tests (ITT; F), and pyruvate tolerance tests (PTT; G) in male and female WT and KO mice. (H) Serum triglyceride levels over the course of lipid tolerance tests (LTT) in male and female WT and KO mice. All data are presented as mean ± S.E.M. ∗P < 0.05. A 2-way ANOVA analysis was performed on all tolerance tests.
We next determined the impact of CTRP13 deficiency on glucose tolerance and insulin sensitivity in HFD-fed mice. The rate of glucose clearance in response to an acute glucose or insulin challenge was significantly higher in male, but not female, KO mice relative to WT controls (Figure 5E,F). Pyruvate tolerance tests also indicated a significant reduction in hepatic gluconeogenesis, suggesting an improvement in hepatic insulin sensitivity in KO mice, though the effects were much more pronounced in male compared to female KO mice (Figure 5G). Because we observed an improvement in triglyceride clearance in the chow-fed KO mice (Figure 3I,J), we again performed fat loading tests on the HFD-fed mice. Female, but not male, KO mice showed a faster rate of triglyceride clearance relative to WT controls (Figure 5H). Altogether, these data indicate that CTRP13 deficiency improves insulin sensitivity and systemic glucose and lipid profiles in the context of diet-induced obesity.
3.6. Loss of CTRP13 has minimal impact on mitochondrial respiration in adipose, liver, and skeletal muscle
Next, we determined whether the lean phenotype of Ctrp13-KO mice is related to increased mitochondrial activity in adipose tissue, liver, or skeletal muscle. Maximal mitochondrial respiration through complex I (CI), CII, and CIV were not significantly different between genotypes across the four major tissues examined when oxygen consumption rate (OCR) was normalized to input protein level (Figure 6A–D, panels 1–3). Mitochondrial content was quantified using the mitoTracker Deep Red (MTDR). Interestingly, Ctrp13-KO mice had lower liver mitochondrial content (Figure 6C, panel 4). When OCR was normalized to mitochondrial content, respiration through CIV was higher in the iWAT of KO animals (Figure 6B, panel 5). Altogether, the data indicate that loss of CTRP13 had minimal impact on maximal mitochondrial respiration across four major metabolic tissues.
Figure 6.
Loss of CTRP13 minimally affects mitochondrial respiration in adipose tissue, liver, and skeletal muscle. A–D) Mitochondrial respiration in the gonadal white adipose tissue (gWAT; A), inguinal white adipose tissue (iWAT; B), liver (C), and gastrocnemius skeletal muscle (D) of Ctrp13 WT (n = 4–6) and KO (n = 9) male mice fed a high-fat diet. For each tissue column, the following panels (from top to bottom) are shown: Panel 1: maximal oxygen consumption rate of mitochondrial complex CI and CIV. Panel 2: maximal oxygen consumption rate of mitochondrial complex CII and CIV. Panel 3: Quantification of maximal oxygen consumption rate of mitochondrial complex CI, CII, and CIV. Panel 4: Quantification of mitochondrial content by MitoTracker Deep Red. Panel 5: Maximal oxygen consumption rate of mitochondrial complex CI, CII, and CIV after normalization to mitochondrial content. All data are presented as mean ± SEM. ∗P < 0.05.
3.7. CTRP13 deficiency reduces adipose tissue inflammation
Consistent with lower adiposity, histological analyses of visceral (gonadal) and subcutaneous (inguinal) adipose tissues showed that male Ctrp13-KO mice had smaller adipocytes (Figure 7A–B). RNA sequencing was performed on these two major fat depots. The top biological processes downregulated in gonadal white adipose tissue (gWAT) were related to immune response (Figure 7C). In accordance, the expression of many pro-inflammatory macrophage marker genes (Tnf, Il6, Adgre1, Ccl2, Itgax, Nos2), as well as many chemokine genes, were significantly lower in Ctrp13-KO mice relative to WT controls (Figure 7D). Interestingly, we did not observe similar reduction in pro-inflammatory gene signatures in inguinal white adipose tissue (iWAT). Together, these data suggest that reduced adiposity and visceral fat inflammation likely contribute to an overall better systemic metabolic profiles in Ctrp13-KO male mice.
Figure 7.
Reduced adipose tissue inflammation and hepatic steatosis in Ctrp13 KO male mice fed a high-fat diet. (A–B) Representative H&E stained histological sections of visceral (gonadal) white adipose tissue (gWAT; A) and subcutaneous (inguinal) white adipose tissue (iWAT; B) of WT (n = 6) and KO (n = 10) male mice on HFD. The quantifications of adipocyte cell size (CSA, cross sectional area) in gWAT and iWAT are indicated by bar graphs. Data are presented as mean ± SEM. ∗∗P < 0.01; ∗∗∗P < 0.001. (C) The top biological processes in Gene Ontology that were downregulated in gWAT. The number next to each bar indicates the number of genes significantly downregulated. (D) A heatmap showing some of the differentially expressed genes (DEGs) from gWAT that affect inflammation in adipose tissue. Color gradient reflects row Z-score values. (E) Representative H&E stained liver sections of WT (n = 6) and KO (n = 10) male mice on HFD. The quantifications of lipid area are indicated by the bar graph. Data are presented as mean ± SEM. ∗∗P < 0.01. (F) Some of the top biological processes in Gene Ontology that were downregulated in liver. The number next to each bar indicates the number of genes significantly downregulated. (G) A heatmap showing some of the differentially expressed genes (DEGs) from liver that affect lipid synthesis and catabolism, as well as signaling. Color gradient reflects row Z-score values. (H) A heatmap showing some of the DEGs that affect inflammation in liver. Color gradient reflects row Z-score values.
3.8. Loss of CTRP13 reduces hepatic inflammation and steatosis
Reduced weight gain in response to an obesogenic diet generally results in reduced lipid accumulation in liver. Consistent with the lean phenotype, histological analyses and quantification showed that Ctrp13-KO male mice had reduced hepatic steatosis (Figure 7E). Based on RNA sequencing, the top biological processes downregulated in the liver of KO male mice were related to immune response and lipid synthesis (Figure 7F). The expression of many fatty acid and cholesterol synthesis and lipid droplet genes (Fasn, Scd2, Elovl3, Elovl5, Dgat1, Mgat3, Hmgcr, Hmgcs1, Sqle, Pparg, Acss3, Plin2, Plin4, Plin5) were significantly suppressed in KO mice relative to WT controls (Figure 7G). Concomitantly, the expression of a number of lipid catabolism genes (Cpt1a, Ppara, Ppargc1b, Acad11, Acad12, Pla2g15, Pla2g6) were significantly upregulated in KO mice relative to WT controls (Figure 7G). In addition, transcripts encoding proteins involved in insulin signaling (Insr, Irs2), PI3K signaling (Pik3c2a, Pik3c2g, Pik3cb), and AMPK signaling (Prkaa2, Prkacb) were also elevated in KO male mice (Figure 7G). Like the visceral fat depot, many pro-inflammatory genes were downregulated in the liver of KO male mice relative to WT controls (Figure 7H). Together, these transcriptomics changes likely contribute to reduced lipid accumulation and enhanced insulin sensitivity (as indicated by the pyruvate tolerance test) in Ctrp13-KO mouse liver.
3.9. The relevance and contribution of human CTRP13/C1QL3 to metabolic syndrome
Lastly, we addressed whether CTRP13-mediated changes are conserved in humans and relevant to metabolic traits important in metabolic disorder. To do so, we integrated our Pan-tissue sequencing data with the Metabolic Syndrome in Man (METSIM) cohort data [75]. Of the gene expression changes seen in gWAT, iWAT, liver, and gastrocnemius, the strongest degree of change was observed in gWAT (Supplementary Figure 1A–B). Substantially more differentially expressed genes (DEGs) were observed in gWAT compared to all other tissues across 3 statistical thresholds, where little overlap was observed between genes across tissues (Supplementary Figure 2).
We analyzed adipose expression array data from 770 phenotyped men from the METSIM cohort [75]. We focused our analyses on human orthologs of the mouse DEGs which were directionally consistent with associated metabolic traits. Interestingly, while human CTRP13/C1QL3 was not significantly correlated with metabolic traits itself, nearly all of the top DEGs derived from the Ctrp13-KO model showed highly significant correlations (Figure 8A). Of note, these genes clustered strongly by whether they were up- or down-regulated within the gWAT of Ctrp13 KO mice (Figure 8A). To test whether the relationship between CTRP13/C1QL3 and its top DEG orthologs could be relevant for explaining the variation with metabolic traits, we adopted an adjusted regression approach [[78], [79], [80]]. We compared the p-value of regressions between a given DEG and trait from both the raw correlations and those adjusted for the expression of C1QL3. For example, despite C1QL3 itself showing no significant correlation to fasting insulin and fat mass (Figure 8A), adjusting the correlation of TNFRSF1B (Figure 8B) or HIF3A (Figure 8C) with nearly all metabolic traits abolished the statistical significance. These data suggest that genetic variation of C1QL3 expression accounts for a significant proportion of variance which explains the relationship between adipose DEGs and relevant metabolic traits.
Figure 8.
DEG integration highlights the adipose-specific impacts of CTRP13/C1QL3, where gene expression changes strongly track with cardiometabolic outcomes in humans. (A) Human orthologs of the top-ranked differentially expressed genes (DEGs) in gonadal white adipose tissue (gWAT) were correlated with select cardiometabolic traits in the Metabolic Syndrome in Man (METSIM) population, where gene names are colored by whether they were shown to increase (blue) or decrease (brown) in Ctrp13-KO vs WT controls. Color indicates direction of correlation via biweight midcorrelation coefficient (bicor) and those which meet a regression significance <0.05 contain a “–”. (B–C) Adjusted regressions were applied to test for statistical mediation of the variance explained by relationships between TNFRSF1B (B) or HIF3A (C) and cardiometabolic traits, as well as how these shift when adjusting for C1QL3/CTRP13. Adjustments for C1QL3 notably reduced the significance of nearly every correlation between gene and trait. N = 770 individuals.
4. Discussion
This is the first study to establish the requirement of CTRP13 for metabolic homeostasis. While previous studies employing gain-of-function approaches in cell model systems [8,63,64] and in mice [55,[60], [61], [62]] have yielded valuable insights concerning CTRP13's biological functions in normal and disease settings, the physiological and metabolic impact of CTRP13 loss-of-function in peripheral tissues was unknown. Using a constitutive KO mouse model, we established that CTRP13 acts as a negative metabolic regulator and its deficiency promotes leanness and improves systemic glucose and lipid metabolism, with sex differences noted. The phenotypes of the Ctrp13-KO mice were, however, contrary of what we had initially expected based on previous in vitro and in vivo studies using recombinant protein supplementation or adenoviral-mediated overexpression [8,55,63,64]. One of the major caveats of gain-of-function studies is the difficulty of assessing whether the outcomes reflect physiological or pharmacological effects of overexpression or the recombinant protein used. Thus, our current study using a KO mouse model underscores the value and importance of using a complementary loss-of-function approach to independently validate the function of CTRP13 in a physiological context.
Contrary to our initial expectation, loss of CTRP13 did not affect food intake in chow-fed or HFD-fed mice. Despite similar caloric intake across the circadian cycle (light and dark) and metabolic states (ad libitum fed, fasted-refed), CTRP13-deficient mice were consistently leaner than the WT littermates. While the body weight differences between KO and WT male mice were very modest in chow-fed mice, the differences in body weight became much more apparent when mice were fed an HFD. Male KO mice were leaner and gained significantly less weight on HFD. Female KO mice also had lower body weight than WT controls but gained the same amount of weight in the first 9 weeks on HFD. Thereafter, Ctrp13-KO female mice started to lose weight while the WT controls continued to gain weight. This resulted in a significant difference in weight gain by week 11 on HFD. Relative to WT mice, HFD-fed KO mice of either sex had higher physical activity levels across the circadian cycles. Despite elevated physical activity, we did not observe a consistent and corresponding increase in energy expenditure. In fact, energy expenditure was only significantly higher in female, but not male, KO mice during the light cycle in ad libitum fed mice and during fasting. Thus, an increased in physical activity levels can, at best, partially account for the lean phenotype seen in Ctrp13-KO mice. We could not, however, rule out potential changes in intestinal nutrient absorption that contribute to the differences in body weight between genotypes.
Although we do not presently know how CTRP13 modulates physical activity, we presume that it likely acts in the central nervous system given its relatively broad expression patterns in the brain, with high expression seen in the cortex and hindbrain [66,82,83]. It has been shown that melanocortin-4 receptor (MC4R) signaling in the ventrolateral ventromedial hypothalamic nucleus (VMHvl)—that projects to the arousal centers in the hippocampus and hindbrain—controls physical activity level [84]. It remains to be determine whether CTRP13 acts on the VMHvl or other neuronal subtypes to affect locomotor activity.
Consistent with the lower body weight and adiposity, Ctrp13-KO male mice on HFD had significantly improved glucose, pyruvate, and insulin tolerance, indicative of enhanced insulin sensitivity. As with the chow-fed KO male mice that showed a greater capacity for lipid clearance in the fat loading tests, HFD-fed female mice also had a higher rate of triglyceride clearance upon lipid loading and lower VLDL-TG levels. The sex differences in metabolic phenotypes observed are not uncommon, as sex is considered an important biological variable affecting phenotypic outcomes [[85], [86], [87]]. The inclusion of both sexes in our metabolic analyses provides a more nuanced appreciation of the sexually dimorphic requirement for CTRP13 in maintaining systemic glucose and lipid metabolism.
Since the metabolic phenotypes of Ctrp13-KO male mice were more pronounced than the female mice, we focused our transcriptomics analyses across different tissues in males. The visceral fat depot of HFD-fed KO male mice had a transcriptomic profile indicative of reduced inflammation. Since low-grade chronic inflammation in the adipose tissue is known to disrupt local and systemic metabolism and insulin sensitivity [2,[88], [89], [90]], the significantly less inflamed visceral fat depot likely contributes, in part, to the improvements seen in systemic glucose metabolism and insulin sensitivity. Reduced weight gain in HFD-fed male KO mice also resulted in significantly less lipid accumulation in liver. Our transcriptomic analyses suggest that reduced hepatic steatosis might be the result of reduced lipid synthesis and increased lipid catabolism. Similar to the visceral fat, male KO liver also showed a reduced inflammatory profile. Combined, these changes in visceral fat and liver contribute to a healthier metabolic profile in HFD-fed Ctrp13-KO male mice.
Although the CTRP13 sequence is highly conserved between mouse and humans (differing by only one amino acid between the full-length proteins), whether they have the same function in the context of metabolism is unclear. To address whether human CTRP13/C1QL3 has any relevant role in metabolism, we integrated the human orthologs of the differentially expressed genes (DEGs) in visceral fat of Ctrp13-KO male mice with the data derived from the METSIM cohort [75]. We employed a form of mediation approach that involved an adjusted regression analysis. By comparing the p-value of regressions between a given DEG and trait from both the raw correlations and those adjusted for the expression of C1QL3/CTRP13, we could show that the statistical significance of the correlation to metabolic traits is abolished after adjusting for C1QL3 expression. This analysis suggests that genetic variation of C1QL3 expression explains a significant proportion of variance seen between adipose DEGs and relevant metabolic traits, thus highlighting the potential causal role and contribution of CTRP13 to human metabolic syndrome. Because the METSIM cohort consisted of only male samples [75], we do not know whether the same holds true in females.
Consistent with CTRP13 having a potential role in human metabolic disorders, circulating serum CTRP13 levels were previously shown to be reduced in obese and diabetic adult humans [[56], [57], [58]], as well as in obese children [59]. Without knowing the metabolic function of CTRP13, it can be a challenge to decipher whether altered circulating CTRP13 levels are causally contributing to dysregulated glucose and lipid metabolism and insulin sensitivity in humans. Our genetic loss-of-function studies suggest that CTRP13 is, in fact, a negative regulator of glucose and lipid metabolism. Based on this, we speculate that reduced serum CTRP13 levels seen in obese and diabetic humans may represent a compensatory physiologic response to counteract insulin resistance and metabolic deterioration in the face of increased adiposity.
Several limitations are noted in the present study. First, all our data were obtained from constitutive KO of CTRP13. We cannot rule out whether the metabolic phenotypes would be different or more severe if Ctrp13 gene is conditionally disrupted in adult mice. This question can be addressed in future studies by crossing the Ctrp13 floxed allele to the tamoxifen-regulated Cre driver line (CMV-CreERT2). Second, the CNS function for CTRP13 is well documented [[65], [66], [67], [68]]. Although food intake (largely controlled by the hypothalamus and hindbrain) was not different between WT and Ctrp13-KO mice, we could not rule out the potential CNS involvement in mediating some of the metabolic phenotypes we observed in peripheral tissues of the KO mice. Future studies using brain-specific KO of Ctrp13 will help resolve this issue. Third, CTRP13/C1QL3 has been shown to bind to the adhesion GPCR (Bai3/ADGRB3) and the postsynaptic GluK2 and GluK4 subunits of the kainite-type glutamate receptors (KARs) [63,67,68,91,92]. Bai3 is a promiscuous adhesion GPCR that binds to four closely related CTRP family members (i.e., C1QL1-4) [49,51,52,91,93]. Thus, it remains to be determined whether Bai3 mediates the biological function of CTRP13 in peripheral tissues. Because Ctrp13-KO mice had significant metabolic phenotypes, our data thus suggest that other closely related CTRP family members are unlikely to compensate for the loss of CTRP13.
In summary, we have provided comprehensive metabolic analyses of mice lacking CTRP13. Our results suggest that CTRP13 functions as a negative metabolic regulator and its deficiency improves systemic metabolic profiles. Using an adjusted regression approach, we further demonstrate the relevance and potential contribution of CTRP13 to human metabolic syndrome. Our data thus provide an important context to help interpret ongoing human studies looking at changes in circulating CTRP13 levels in different disease states.
Author contributions
FC and GWW: Conceptualization; GWW, FC, DCS, SA, MZ, MMS: Formal analysis; GWW: Funding acquisition; FC, DCS, MS, SA, GWW: Investigation; MZ, MMS: Methodology; GWW: Project administration; GWW, MMS: Supervision; FC, DCS, GWW, MZ, MMS: Visualization; GWW: Roles/Writing – original draft; FC, DCS, SA, MMS, GWW: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Institutes of Health (DK084171 to GWW; HL138193 and DK130640 to MMS). D.C.S. is supported by an NIH T32 training grant (HL007534). The FPLC/serum analyses were performed at the Mouse Metabolism and Phenotyping Core (MMPC) at the Baylor College of Medicine, supported by NIH grants (DK114356 and UM1HG006348).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101824.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1. RNA-seq of adipose tissue, liver, and skeletal muscle of WT and Ctrp13-KO male mice fed a high-fat diet. (A) A volcano plot showing the −log10(p-value) of significance (y-axis) and fold-change (x-axis) for all genes across the 4 metabolic tissues (gWAT, iWAT, liver, gastrocnemius) analyzed in RNA-Seq. (B) The proportion of genes that changes (y-axis) at a logistic regression threshold (limma) at P < 1e-3 are plotted against the tissue in which they are expressed (x-axis), where gWAT shows by-far the largest changes. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue
Supplementary Figure 2. Visceral (gonadal) fat has the largest number of differentially expressed genes. gWAT has substantially more differentially expressed genes (DEGs) compared to all other tissues (iWAT, liver, gastrocnemius) across 3 statistical thresholds, where little overlap was observed between genes across tissues. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue
Data availability
Data will be made available on request.
References
- 1.Priest C., Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nat Metab. 2019;1:1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 2.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman J.M., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 4.Wong G.W., Wang J., Hug C., Tsao T.S., Lodish H.F. A family of Acrp30/adiponectin structural and functional paralogs. Proc Natl Acad Sci U S A. 2004;101:10302–10307. doi: 10.1073/pnas.0403760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong G.W., Krawczyk S.A., Kitidis-Mitrokostas C., Ge G., Spooner E., Hug C., et al. Identification and characterization of CTRP9, a novel secreted glycoprotein, from adipose tissue that reduces serum glucose in mice and forms heterotrimers with adiponectin. FASEB J. 2009;23:241–258. doi: 10.1096/fj.08-114991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong G.W., Krawczyk S.A., Kitidis-Mitrokostas C., Revett T., Gimeno R., Lodish H.F. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J. 2008;416:161–177. doi: 10.1042/BJ20081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Z., Peterson J.M., Lei X., Cebotaru L., Wolfgang M.J., Baldeviano G.C., et al. C1q/TNF-related protein-12 (CTRP12), a novel adipokine that improves insulin sensitivity and glycemic control in mouse models of obesity and diabetes. J Biol Chem. 2012;287:10301–10315. doi: 10.1074/jbc.M111.303651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Z., Peterson J.M., Wong G.W. Metabolic regulation by C1q/TNF-related protein-13 (CTRP13): activation of AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J Biol Chem. 2011;286:15652–15665. doi: 10.1074/jbc.M110.201087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Z., Seldin M.M., Natarajan N., Djemal D.C., Peterson J.M., Wong G.W. C1q/tumor necrosis factor-related protein 11 (CTRP11), a novel adipose stroma-derived regulator of adipogenesis. J Biol Chem. 2013;288:10214–10229. doi: 10.1074/jbc.M113.458711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seldin M.M., Peterson J.M., Byerly M.S., Wei Z., Wong G.W. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem. 2012;287:11968–11980. doi: 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byerly M.S., Petersen P.S., Ramamurthy S., Seldin M.M., Lei X., Provost E., et al. C1q/TNF-related protein 4 (CTRP4) is a unique secreted protein with two tandem C1q domains that functions in the hypothalamus to modulate food intake and body weight. J Biol Chem. 2014;289:4055–4069. doi: 10.1074/jbc.M113.506956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson J.M., Wei Z., Wong G.W. C1q/TNF-related protein-3 (CTRP3), a novel adipokine that regulates hepatic glucose output. J Biol Chem. 2010;285:39691–39701. doi: 10.1074/jbc.M110.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seldin M.M., Tan S.Y., Wong G.W. Metabolic function of the CTRP family of hormones. Rev Endocr Metab Disord. 2014;15:111–123. doi: 10.1007/s11154-013-9255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei X., Rodriguez S., Petersen P.S., Seldin M.M., Bowman C.E., Wolfgang M.J., et al. Loss of CTRP5 improves insulin action and hepatic steatosis. Am J Physiol Endocrinol Metab. 2016;310:E1036–E1052. doi: 10.1152/ajpendo.00010.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei X., Seldin M.M., Little H.C., Choy N., Klonisch T., Wong G.W. C1q/TNF-related protein 6 (CTRP6) links obesity to adipose tissue inflammation and insulin resistance. J Biol Chem. 2017;292:14836–14850. doi: 10.1074/jbc.M116.766808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei X., Wong G.W. C1q/TNF-related protein 2 (CTRP2) deletion promotes adipose tissue lipolysis and hepatic triglyceride secretion. J Biol Chem. 2019;294:15638–15649. doi: 10.1074/jbc.RA119.009230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little H.C., Rodriguez S., Lei X., Tan S.Y., Stewart A.N., Sahagun A., et al. Myonectin deletion promotes adipose fat storage and reduces liver steatosis. FASEB J. 2019;33:8666–8687. doi: 10.1096/fj.201900520R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen P.S., Lei X., Wolf R.M., Rodriguez S., Tan S.Y., Little H.C., et al. CTRP7 deletion attenuates obesity-linked glucose intolerance, adipose tissue inflammation, and hepatic stress. Am J Physiol Endocrinol Metab. 2017;312:E309–E325. doi: 10.1152/ajpendo.00344.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez S., Lei X., Petersen P.S., Tan S.Y., Little H.C., Wong G.W. Loss of CTRP1 disrupts glucose and lipid homeostasis. Am J Physiol Endocrinol Metab. 2016;311:E678–E697. doi: 10.1152/ajpendo.00087.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarver D.C., Stewart A.N., Rodriguez S., Little H.C., Aja S., Wong G.W. Loss of CTRP4 alters adiposity and food intake behaviors in obese mice. Am J Physiol Endocrinol Metab. 2020;319:E1084–E1100. doi: 10.1152/ajpendo.00448.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarver D.C., Xu C., Aja S., Wong G.W. CTRP14 inactivation alters physical activity and food intake response to fasting and refeeding. Am J Physiol Endocrinol Metab. 2022;322:E480–E493. doi: 10.1152/ajpendo.00002.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarver D.C., Xu C., Carreno D., Arking A., Terrillion C.E., Aja S., et al. CTRP11 contributes modestly to systemic metabolism and energy balance. FASEB J. 2022;36 doi: 10.1096/fj.202200189RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan S.Y., Lei X., Little H.C., Rodriguez S., Sarver D.C., Cao X., et al. CTRP12 ablation differentially affects energy expenditure, body weight, and insulin sensitivity in male and female mice. Am J Physiol Endocrinol Metab. 2020;319:E146–E162. doi: 10.1152/ajpendo.00533.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan S.Y., Little H.C., Lei X., Li S., Rodriguez S., Wong G.W. Partial deficiency of CTRP12 alters hepatic lipid metabolism. Physiol Genomics. 2016;48:936–949. doi: 10.1152/physiolgenomics.00111.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Z., Lei X., Petersen P.S., Aja S., Wong G.W. Targeted deletion of C1q/TNF-related protein 9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice. Am J Physiol Endocrinol Metab. 2014;306:E779–E790. doi: 10.1152/ajpendo.00593.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf R.M., Lei X., Yang Z.C., Nyandjo M., Tan S.Y., Wong G.W. CTRP3 deficiency reduces liver size and alters IL-6 and TGFbeta levels in obese mice. Am J Physiol Endocrinol Metab. 2016;310:E332–E345. doi: 10.1152/ajpendo.00248.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson J.M., Seldin M.M., Tan S.Y., Wong G.W. CTRP2 overexpression improves insulin and lipid tolerance in diet-induced obese mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson J.M., Seldin M.M., Wei Z., Aja S., Wong G.W. CTRP3 attenuates diet-induced hepatic steatosis by regulating triglyceride metabolism. Am J Physiol Gastrointest Liver Physiol. 2013;305:G214–G224. doi: 10.1152/ajpgi.00102.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson J.M., Wei Z., Seldin M.M., Byerly M.S., Aja S., Wong G.W. CTRP9 transgenic mice are protected from diet-induced obesity and metabolic dysfunction. Am J Physiol Regul Integr Comp Physiol. 2013;305:R522–R533. doi: 10.1152/ajpregu.00110.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahav R., Haim Y., Bhandarkar N.S., Levin L., Chalifa-Caspi V., Sarver D., et al. CTRP6 rapidly responds to acute nutritional changes, regulating adipose tissue expansion and inflammation in mice. Am J Physiol Endocrinol Metab. 2021;321:E702–E713. doi: 10.1152/ajpendo.00299.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appari M., Breitbart A., Brandes F., Szaroszyk M., Froese N., Korf-Klingebiel M., et al. C1q-TNF-related protein-9 promotes cardiac hypertrophy and failure. Circ Res. 2017;120:66–77. doi: 10.1161/CIRCRESAHA.116.309398. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Q., Yuan Y., Yi W., Lau W.B., Wang Y., Wang X., et al. C1q/TNF-related proteins, A family of novel adipokines, induce vascular relaxation through the adiponectin receptor-1/AMPK/eNOS/nitric oxide signaling pathway. Arterioscler Thromb Vasc Biol. 2011;31:2616–2623. doi: 10.1161/ATVBAHA.111.231050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kambara T., Ohashi K., Shibata R., Ogura Y., Maruyama S., Enomoto T., et al. CTRP9 protein protects against myocardial injury following ischemia-reperfusion through AMP-activated protein kinase (AMPK)-dependent mechanism. J Biol Chem. 2012;287:18965–18973. doi: 10.1074/jbc.M112.357939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kambara T., Shibata R., Ohashi K., Matsuo K., Hiramatsu-Ito M., Enomoto T., et al. C1q/tumor necrosis factor-related protein 9 protects against acute myocardial injury through an adiponectin receptor I-AMPK-dependent mechanism. Mol Cell Biol. 2015;35:2173–2185. doi: 10.1128/MCB.01518-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanemura N., Shibata R., Ohashi K., Ogawa H., Hiramatsu-Ito M., Enomoto T., et al. C1q/TNF-related protein 1 prevents neointimal formation after arterial injury. Atherosclerosis. 2017;257:138–145. doi: 10.1016/j.atherosclerosis.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa H., Ohashi K., Ito M., Shibata R., Kanemura N., Yuasa D., et al. Adipolin/CTRP12 protects against pathological vascular remodeling through suppression of smooth muscle cell growth and macrophage inflammatory response. Cardiovasc Res. 2020;116:237–249. doi: 10.1093/cvr/cvz074. [DOI] [PubMed] [Google Scholar]
- 37.Otaka N., Shibata R., Ohashi K., Uemura Y., Kambara T., Enomoto T., et al. Myonectin is an exercise-induced myokine that protects the heart from ischemia-reperfusion injury. Circ Res. 2018;123:1326–1338. doi: 10.1161/CIRCRESAHA.118.313777. [DOI] [PubMed] [Google Scholar]
- 38.Uemura Y., Shibata R., Ohashi K., Enomoto T., Kambara T., Yamamoto T., et al. Adipose-derived factor CTRP9 attenuates vascular smooth muscle cell proliferation and neointimal formation. FASEB J. 2013;27:25–33. doi: 10.1096/fj.12-213744. [DOI] [PubMed] [Google Scholar]
- 39.Yuasa D., Ohashi K., Shibata R., Mizutani N., Kataoka Y., Kambara T., et al. C1q/TNF-related protein-1 functions to protect against acute ischemic injury in the heart. FASEB J. 2016;30:1065–1075. doi: 10.1096/fj.15-279885. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y., Yi W., Yuan Y., Lau W.B., Yi D., Wang X., et al. C1q/tumor necrosis factor-related protein-9, a novel adipocyte-derived cytokine, attenuates adverse remodeling in the ischemic mouse heart via protein kinase A activation. Circulation. 2013;128:S113–S120. doi: 10.1161/CIRCULATIONAHA.112.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi W., Sun Y., Yuan Y., Lau W.B., Zheng Q., Wang X., et al. C1q/tumor necrosis factor-related protein-3, a newly identified adipokine, is a novel antiapoptotic, proangiogenic, and cardioprotective molecule in the ischemic mouse heart. Circulation. 2012;125:3159–3169. doi: 10.1161/CIRCULATIONAHA.112.099937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han S., Jeong A.L., Lee S., Park J.S., Buyanravjikh S., Kang W., et al. C1q/TNF-alpha—related protein 1 (CTRP1) maintains blood pressure under dehydration conditions. Circ Res. 2018;123:e5–e19. doi: 10.1161/CIRCRESAHA.118.312871. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez S., Little H.C., Daneshpajouhnejad P., Fenaroli P., Tan S.Y., Sarver D.C., et al. Aging and chronic high-fat feeding negatively affect kidney size, function, and gene expression in CTRP1-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2021;320:R19–R35. doi: 10.1152/ajpregu.00139.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez S., Little H.C., Daneshpajouhnejad P., Shepard B.D., Tan S.Y., Wolfe A., et al. Late-onset renal hypertrophy and dysfunction in mice lacking CTRP1. FASEB J. 2020;34:2657–2676. doi: 10.1096/fj.201900558RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Y., Wu X., Ma Z., Tan W., Wang L., Na D., et al. Expression of the novel adipokine C1qTNF-related protein 4 (CTRP4) suppresses colitis and colitis-associated colorectal cancer in mice. Cell Mol Immunol. 2016;13:688–699. doi: 10.1038/cmi.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayyagari R., Mandal M.N., Karoukis A.J., Chen L., McLaren N.C., Lichter M., et al. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci. 2005;46:3363–3371. doi: 10.1167/iovs.05-0159. [DOI] [PubMed] [Google Scholar]
- 47.Hayward C., Shu X., Cideciyan A.V., Lennon A., Barran P., Zareparsi S., et al. Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet. 2003;12:2657–2667. doi: 10.1093/hmg/ddg289. [DOI] [PubMed] [Google Scholar]
- 48.Youngstrom D.W., Zondervan R.L., Doucet N.R., Acevedo P.K., Sexton H.E., Gardner E.A., et al. CTRP3 regulates endochondral ossification and bone remodeling during fracture healing. J Orthop Res. 2020;38:996–1006. doi: 10.1002/jor.24553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamoud N., Tran V., Aimi T., Kakegawa W., Lahaie S., Thibault M.P., et al. Spatiotemporal regulation of the GPCR activity of Bai3 by C1qL4 and Stabilin-2 controls myoblast fusion. Nat Commun. 2018;9:4470. doi: 10.1038/s41467-018-06897-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho Y., Kim H.S., Kang D., Kim H., Lee N., Yun J., et al. CTRP3 exacerbates tendinopathy by dysregulating tendon stem cell differentiation and altering extracellular matrix composition. Sci Adv. 2021;7 doi: 10.1126/sciadv.abg6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakegawa W., Mitakidis N., Miura E., Abe M., Matsuda K., Takeo Y.H., et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron. 2015;85:316–329. doi: 10.1016/j.neuron.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 52.Sigoillot S.M., Iyer K., Binda F., Gonzalez-Calvo I., Talleur M., Vodjdani G., et al. The secreted protein C1QL1 and its receptor Bai3 control the synaptic connectivity of excitatory inputs converging on cerebellar purkinje cells. Cell Rep. 2015;10:820–832. doi: 10.1016/j.celrep.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 53.Sarver D.C., Xu C., Cheng Y., Terrillion C.E., Wong G.W. CTRP4 ablation impairs associative learning and memory. FASEB J. 2021;35 doi: 10.1096/fj.202100733RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ressl S., Vu B.K., Vivona S., Martinelli D.C., Sudhof T.C., Brunger A.T. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure. 2015;23:688–699. doi: 10.1016/j.str.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 55.Byerly M.S., Swanson R., Wei Z., Seldin M.M., McCulloh P.S., Wong G.W. A central role for C1q/TNF-related protein 13 (CTRP13) in modulating food intake and body weight. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shanaki M., Fadaei R., Moradi N., Emamgholipour S., Poustchi H. The circulating CTRP13 in type 2 diabetes and non-alcoholic fatty liver patients. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afrookhteh A., Emamgholipour S., Alipoor B., Moradi N., Meshkani R., Nasli-Esfahani E., et al. The circulating levels of complement-C1q/TNF-related protein 13 (CTRP13) in patients with type 2 diabetes and its association with insulin resistance. Clin Lab. 2017;63:327–333. doi: 10.7754/Clin.Lab.2016.160609. [DOI] [PubMed] [Google Scholar]
- 58.Fadaei R., Moradi N., Baratchian M., Aghajani H., Malek M., Fazaeli A.A., et al. Association of C1q/TNF-related protein-3 (CTRP3) and CTRP13 serum levels with coronary artery disease in subjects with and without type 2 diabetes mellitus. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erbas I.M., Paketci A., Turan S., Sisman A.R., Demir K., Bober E., et al. Low complement C1q/TNF-related protein-13 levels are associated with childhood obesity but not binge eating disorder. J Clin Res Pediatr Endocrinol. 2022;14:179–187. doi: 10.4274/jcrpe.galenos.2021.2021-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang C., Xu W., Liang M., Huang D., Huang K. CTRP13 inhibits atherosclerosis via autophagy-lysosome-dependent degradation of CD36. FASEB J. 2019;33:2290–2300. doi: 10.1096/fj.201801267RR. [DOI] [PubMed] [Google Scholar]
- 61.Li Y., Wang W., Chao Y., Zhang F., Wang C. CTRP13 attenuates vascular calcification by regulating Runx2. FASEB J. 2019;33:9627–9637. doi: 10.1096/fj.201900293RRR. [DOI] [PubMed] [Google Scholar]
- 62.Xu W., Chao Y., Liang M., Huang K., Wang C. CTRP13 mitigates abdominal aortic aneurysm formation via NAMPT1. Mol Ther. 2021;29:324–337. doi: 10.1016/j.ymthe.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta R., Nguyen D.C., Schaid M.D., Lei X., Balamurugan A.N., Wong G.W., et al. Complement 1q-like-3 protein inhibits insulin secretion from pancreatic beta-cells via the cell adhesion G protein-coupled receptor Bai3. J Biol Chem. 2018;293:18086–18098. doi: 10.1074/jbc.RA118.005403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koltes J.E., Arora I., Gupta R., Nguyen D.C., Schaid M., Kim J.A., et al. A gene expression network analysis of the pancreatic islets from lean and obese mice identifies complement 1q like-3 secreted protein as a regulator of beta-cell function. Sci Rep. 2019;9:10119. doi: 10.1038/s41598-019-46219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chew K.S., Fernandez D.C., Hattar S., Sudhof T.C., Martinelli D.C. Anatomical and behavioral investigation of C1ql3 in the mouse suprachiasmatic nucleus. J Biol Rhythms. 2017;32:222–236. doi: 10.1177/0748730417704766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinelli D.C., Chew K.S., Rohlmann A., Lum M.Y., Ressl S., Hattar S., et al. Expression of C1ql3 in discrete neuronal populations controls efferent synapse numbers and diverse behaviors. Neuron. 2016;91:1034–1051. doi: 10.1016/j.neuron.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C.Y., Liu Z., Ng Y.H., Sudhof T.C. A synaptic circuit required for acquisition but not recall of social transmission of food preference. Neuron. 2020;107:144–157.e144. doi: 10.1016/j.neuron.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuda K., Budisantoso T., Mitakidis N., Sugaya Y., Miura E., Kakegawa W., et al. Transsynaptic modulation of kainate receptor functions by C1q-like proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez S., Stewart A.N., Lei X., Cao X., Little H.C., Fong V., et al. PRADC1: a novel metabolic-responsive secretory protein that modulates physical activity and adiposity. FASEB J. 2019;33:14748–14759. doi: 10.1096/fj.201901279R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tschop M.H., Speakman J.R., Arch J.R., Auwerx J., Bruning J.C., Chan L., et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Acin-Perez R., Benador I.Y., Petcherski A., Veliova M., Benavides G.A., Lagarrigue S., et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 2020;39 doi: 10.15252/embj.2019104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stancakova A., Javorsky M., Kuulasmaa T., Haffner S.M., Kuusisto J., Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A., et al. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am J Hum Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velez L.M., Van C., Moore T., Zhou Z., Johnson C., Hevener A.L., et al. Genetic variation of putative myokine signaling is dominated by biological sex and sex hormones. eLife. 2022;11 doi: 10.7554/eLife.76887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yokota T., McCourt J., Ma F., Ren S., Li S., Kim T.H., et al. Type V collagen in scar tissue regulates the size of scar after heart injury. Cell. 2020;182:545–562 e523. doi: 10.1016/j.cell.2020.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fathzadeh M., Li J., Rao A., Cook N., Chennamsetty I., Seldin M., et al. FAM13A affects body fat distribution and adipocyte function. Nat Commun. 2020;11:1465. doi: 10.1038/s41467-020-15291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 82.Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014 e1022. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iijima T., Miura E., Watanabe M., Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 84.Krause W.C., Rodriguez R., Gegenhuber B., Matharu N., Rodriguez A.N., Padilla-Roger A.M., et al. Oestrogen engages brain MC4R signalling to drive physical activity in female mice. Nature. 2021;599:131–135. doi: 10.1038/s41586-021-04010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goossens G.H., Jocken J.W.E., Blaak E.E. Sexual dimorphism in cardiometabolic health: the role of adipose tissue, muscle and liver. Nat Rev Endocrinol. 2021;17:47–66. doi: 10.1038/s41574-020-00431-8. [DOI] [PubMed] [Google Scholar]
- 86.Mauvais-Jarvis F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol Sex Differ. 2015;6:14. doi: 10.1186/s13293-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mauvais-Jarvis F., Bairey Merz N., Barnes P.J., Brinton R.D., Carrero J.J., DeMeo D.L., et al. Sex and gender: modifiers of health, disease, and medicine. Lancet. 2020;396:565–582. doi: 10.1016/S0140-6736(20)31561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 89.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Investig. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Russo L., Lumeng C.N. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018;155:407–417. doi: 10.1111/imm.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolliger M.F., Martinelli D.C., Sudhof T.C. The cell-adhesion G protein-coupled receptor Bai3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci U S A. 2011;108:2534–2539. doi: 10.1073/pnas.1019577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sticco M.J., Pena Palomino P.A., Lukacsovich D., Thompson B.L., Foldy C., Ressl S., et al. C1QL3 promotes cell-cell adhesion by mediating complex formation between ADGRB3/Bai3 and neuronal pentraxins. FASEB J. 2021;35 doi: 10.1096/fj.202000351RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan A., Ke S., Chen Y., Chen L., Lu X., Ding F., et al. Expression patterns of C1ql4 and its cell-adhesion GPCR Bai3 in the murine testis and functional roles in steroidogenesis. FASEB J. 2019;33:4893–4906. doi: 10.1096/fj.201801620RR. [DOI] [PubMed] [Google Scholar]
- 94.Consortium G.T. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. RNA-seq of adipose tissue, liver, and skeletal muscle of WT and Ctrp13-KO male mice fed a high-fat diet. (A) A volcano plot showing the −log10(p-value) of significance (y-axis) and fold-change (x-axis) for all genes across the 4 metabolic tissues (gWAT, iWAT, liver, gastrocnemius) analyzed in RNA-Seq. (B) The proportion of genes that changes (y-axis) at a logistic regression threshold (limma) at P < 1e-3 are plotted against the tissue in which they are expressed (x-axis), where gWAT shows by-far the largest changes. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue
Supplementary Figure 2. Visceral (gonadal) fat has the largest number of differentially expressed genes. gWAT has substantially more differentially expressed genes (DEGs) compared to all other tissues (iWAT, liver, gastrocnemius) across 3 statistical thresholds, where little overlap was observed between genes across tissues. gWAT, gonadal white adipose tissue; iWAT, inguinal white adipose tissue
Data Availability Statement
Data will be made available on request.