Abstract
Members of the phylum Cnidaria include sea anemones, corals and jellyfish, and have successfully colonized both marine and freshwater habitats throughout the world. The understanding of how cnidarians adapt to extreme environments such as the dark, high-pressure deep-sea habitat has been hindered by the lack of genomic information. Here, we report the first chromosome-level deep-sea cnidarian genome, of the anemone Actinernus sp., which was 1.39 Gbp in length and contained 44 970 gene models including 14 806 tRNA genes and 30 164 protein-coding genes. Analyses of homeobox genes revealed the longest chromosome hosts a mega-array of Hox cluster, HoxL, NK cluster and NKL homeobox genes; until now, such an array has only been hypothesized to have existed in ancient ancestral genomes. In addition to this striking arrangement of homeobox genes, analyses of microRNAs revealed cnidarian-specific complements that are distinctive for nested clades of these animals, presumably reflecting the progressive evolution of the gene regulatory networks in which they are embedded. Also, compared with other sea anemones, circadian rhythm genes were lost in Actinernus sp., which likely reflects adaptation to living in the dark. This high-quality genome of a deep-sea cnidarian thus reveals some of the likely molecular adaptations of this ecologically important group of metazoans to the extreme deep-sea environment. It also deepens our understanding of the evolution of genome content and organization of animals in general and cnidarians in particular, specifically from the viewpoint of key developmental control genes like the homeobox-encoding genes, where we find an array of genes that until now has only been hypothesized to have existed in the ancient ancestor that pre-dated both the cnidarians and bilaterians.
Keywords: adaptations, deep-sea, cnidarian, genome, homeobox, circadian
1. Introduction
Deep-sea hydrothermal vents and seeps are characterized by darkness, high hydrostatic pressure and the presence of reducing chemicals such as hydrogen sulfide and methane that serve as energy sources to fuel chemosynthesis. These habitats provide a dramatically different ecological niche from those that rely on photosynthesis for primary production, and as such provide us with very distinctive ecological and evolutionary systems to those commonly studied. Our knowledge of metazoan genomic adaptations to these extreme environments has mainly derived from genome analyses of annelids [1], crustaceans [2], molluscs [3–7], echinoderms [8,9] and fishes [10–12]. The phylum Cnidaria contains over 10 000 species of animals including sea anemones, corals and jellyfishes, and its members play important ecological roles in both shallow-water and deep-sea habitats throughout the world. Nevertheless, there are currently only a limited number of deep-sea cnidarian genomes sequenced, including one from a deep-sea coral [13] and one from a deep-sea anemone [14], both at draft genome qualities, hindering the understanding of how cnidarians adapt to this distinctive, extreme environment.
Higher quality genome assemblies allow analyses of genome architecture and much better inferences of ancestral states and the evolutionary dynamics governing genome organization. The homeobox-encoding genes are excellent markers for analyses of genome architecture due to their abundance in animal genomes and a classification scheme that enables detection of classes, families and orthologues across wide spans of the animal kingdom [15,16]. Given the dramatic expansion in homeobox families early in animal evolution, particularly in the ANTP-class, it is assumed that the predominant mode of their origin was via tandem duplication. This led to an influential hypothesis for the origin of the ANTP-class homeobox genes via a Mega-cluster [17]. Evidence supporting this hypothetical Mega-cluster comes from comparisons of various extant animal genomes in which subsets of ANTP-class genes are still found to be linked, such as the Hox cluster and various families of Hox-linked (HoxL) genes, the NK cluster and NK-linked (NKL) genes and other families like the ParaHox and NK2 genes. However, in bilaterians there is a tendency to find the splits amongst subsets of these genes onto distinct chromosomes are in similar places [18,19]. This implies that the last common ancestor of the bilaterians already had these ANTP-class families distributed across at least four chromosomes, which raises the possibility that the ANTP-class Mega-cluster never actually existed after all. Outgroups of the bilaterians, such as cnidarians, are thus a key group of animals to further test the plausibility of the Mega-cluster hypothesis as high-quality genome assemblies become available.
Actinernus is a genus of deep-sea anemones currently represented by six species distributed in the Southern Ocean, Indian Ocean, Atlantic Ocean and Pacific Ocean [20–23]. However, molecular data in this genus is scarce. To date, little is known about the phylogeny of Actinernus as only several mitochondrial and ribosomal gene fragments are available from two species, A. robustus and A. elongatus [23,24], let alone further molecular biological information in these deep-sea species. Here, we assembled and analysed the high-quality genome of a deep-sea anemone Actinernus sp. sampled from the South China Sea to deepen our understanding on its genomic architecture. Our analyses revealed a striking finding of a mega-array of ANTP-class homeobox genes that may well reflect an ancient ancestral state for these key developmental control genes, alongside likely molecular adaptations to the deep-sea environment.
2. Results and discussion
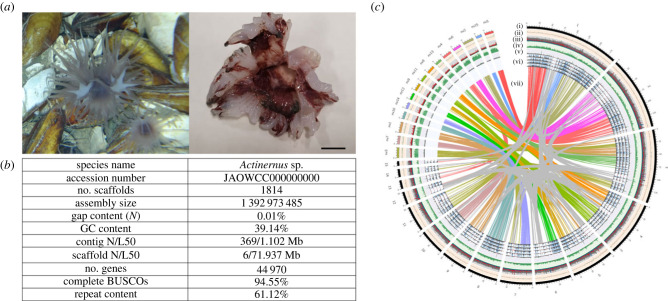
The deep-sea anemone Actinernus sp. individuals were collected from the Haima cold seep in the South China Sea at a depth of 1386 m (figure 1a), where Actinernus sp. was commonly observed on the shells of the deep-sea clam Archivesica marissinica around cold-seep mussel beds [20,25,26]. The genome of Actinernus sp. was sequenced and assembled with PacBio HiFi reads followed by further scaffolding with Omni-C reads (electronic supplementary material, S1 and S2). The genome assembly was 1.39 Gbp with a contig N50 of 1.1 Mbp and scaffold N50 of 71.9 Mbp (figure 1b). This high physical contiguity was matched by high completeness, with 94.55% complete BUSCO score (metazoa_genes version odb10) (figure 1b; electronic supplementary material, S3). The genome contained 44 970 gene models including 14 806 tRNA genes and 30 164 protein-coding genes, and the majority of the sequences assembled (approx. 80%) were anchored to 15 linkage groups (electronic supplementary material, figure S1), which we labelled according to descending size (figure 1c).
Figure 1.
(a) Photographs of the Actinernus sp. on site (left) and the sampled specimen (right, scale bar = 1 cm); (b) genome statistics of Actinernus sp.; (c) circos plot of the genome assembly of Actinernus sp. in comparison with Nematostella vectensis, with tracks (i–vii) specified as follows: (i) chromosomes; (ii) GC content percentage (greater than 41.9% in green; less than 37.7% in red); (iii) distribution of repeat elements where DNA transposons, LTRs, LINEs, SINEs, other repeats and unclassified repeats are coloured in red, green, blue, purple, pink and grey, respectively; (iv) gene density; (v) exon density; (vi) transcriptome reads coverage of trunk and tentacle samples; (vii) syntenic blocks showing synteny conservation with N. vectensis in colours and self-syntenic regions in grey. All tracks were plotted with 100k window size except tracks (iv) and (v) where 200k window size was used.
Macrosynteny analyses revealed that the genome of Actinernus sp. comprised a unique chromosome organization. For instance, the longest chromosome, chr 1, corresponded to at least three chromosomes in other animals while chr 2, 3, 4, 5 and 7 shared syntenic regions with at least two chromosomes (figure 1c; electronic supplementary material, figures S2 and S3). Furthermore, 129 self-syntenic blocks (SSBs) were identified, comprising 974 gene pairs across approximately 208 Mbp (approx. 19%) of the 15 chromosomes (electronic supplementary material, figure S4 and S4). Moreover, transposable elements (TEs) accounted for 61.12% of the genome, which was dominated by unclassified TEs (39.05%) with recent expansion activity (electronic supplementary material, figure S5). The burst of TEs was significantly correlated to the large genome size in Actinernus sp., as compared with genomes of other cnidarians, deep-sea animals and other non-bilaterian organisms (electronic supplementary material, figures S5 and S6). While the potential link between genome size of eukaryotes and TEs has been demonstrated in arthropods and chordates (e.g. insects, myriapods, larvaceans and other chordates [27–29]), the genome of Actinernus sp. demonstrates that this association is phylogenetically more widespread, now including a cnidarian.
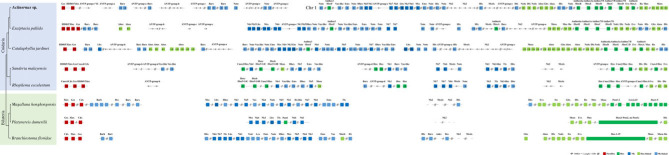
Homeobox genes are important developmental genes and can also serve as markers of large-scale genomic changes during metazoans evolution [16,30]. Previous studies suggested that a ProtoANTP gene underwent a series of tandem duplications to produce the Hox, ParaHox and NK cluster genes that at some point deep in animal ancestry existed as a ‘Mega-cluster’ of ANTP-class homeobox genes [17]. Genomic analyses have identified that cnidarians and bilaterians seemingly took different evolutionary pathways to their ANTP-class homeobox genes organization since their last common ancestor. Bilaterians tend to have a similar split of their ANTP-class gene families across approximately four distinct chromosomes, consisting of the Hox cluster genes with a handful of HoxL genes on one chromosome, the NK cluster genes and a few NKL genes on another, with the ParaHox cluster and the NK2 genes on two further distinct chromosomes [18,19,31–33]. In contrast, some cnidarians have been found to have mixtures of a small number of Hox, HoxL, NK and NKL genes dispersed around their respective genomes [34,35] (figure 2). To our surprise, we found the first case of the existence of the previously hypothetical mega-homeobox array of Hox cluster, HoxL, NK cluster and NKL genes in an extant species, on Actinernus chr 1 (figure 2; electronic supplementary material, S6 and figures S7–S10). Since chr 1 is the largest chromosome in the Actinernus genome, whether this genomic organization represents a primary linkage array directly descendant and retained from the cnidarian–bilaterian ancestor, or instead represents a secondary ‘coming together’ of these genes (e.g. via chromosome fusion events), remains to be established, for example, by further genome sequencing and linkage analyses in a wider diversity of species [16]. Nevertheless, the widespread linkage of Hox/HoxL with NK/NKL genes that is also found in other cnidarian species (figure 2) would imply that this Actinernus condition may well reflect the ancestral state.
Figure 2.
ANTP-class homeobox gene arrangement in high-quality cnidarian genomes compared with selected bilaterians. Chromosome 1 of Actinernus sp. is indicated with ‘Chr 1’. Double slash denotes genomic distance between 100 kb and 1 Mb; triple slash denotes genomic distance over 1 Mb. ParaHox, NK, NK-linked, Hox and Hox-linked genes (defined as previously described [16]) are coloured in red, blue, light blue, green and light green, respectively, while other genes are coloured in grey. Arrows inside gene boxes represent transcriptional orientation.
Further homeodomain phylogenetic tree analyses also identified an anthozoan-specific group of ANTP-class genes, termed ANTP-group-c here that was expanded in Actinernus sp. (electronic supplementary material, figure S8). Twenty-four out of 28 of these Actinernus ANTP-group-c genes resulted from tandem duplications on four chromosomes based on the comparable patterns of conserved TEs around them (electronic supplementary material, figure S11). Interestingly, this preponderance of tandem duplications leading to the expansion of the complement of homeobox genes, as shown here in these ANTP-group-c genes, is consistent with the assumptions underpinning the Mega-cluster hypothesis discussed above, whereby ancestral precursors to the ANTP-class gene families are hypothesized to most likely have originated via tandem duplications. Whether this ANTP-class homeobox gene expansion of ANTP-group-c occurred more widely in deep-sea cnidarians remains to be tested.
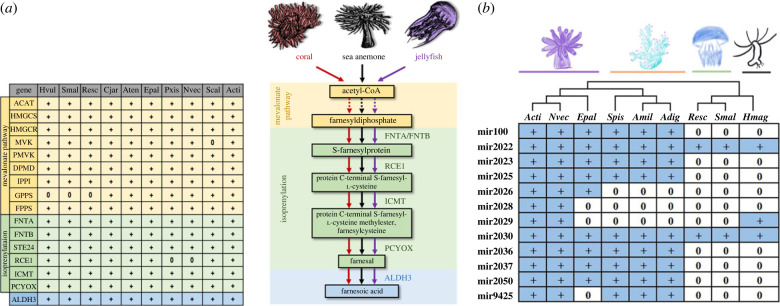
Beyond the homeobox complement, genes for sesquiterpenoid hormone production such as farnesoic acid, methyl farnesoate and juvenile hormone were long thought to be confined to regulation of development and reproduction of insects and other arthropods [29,36,37]. Recently, genes for this pathway were unexpectedly revealed in jellyfish genomes [32]. Sesquiterpenoid hormone systems are now also known in other cnidarian genomes, and farnesoic acid has also been detected in jellyfish [34,35,38]. In the Actinernus sp. genome and transcriptomes, we found that genes were expressed throughout the whole sesquiterpenoid biosynthetic pathway, presumably leading to the de novo synthesis of farnesoic acid in this species as well (figure 3a; electronic supplementary material, S7 and figures S12 and S13). The identification of these genes implies that sesquiterpenoids are of widespread importance across the cnidarians, even in the deep-sea environment, where their precise function remains to be determined.
Figure 3.
(a) Conserved genes in the sesquiterpenoid biosynthesis pathway among cnidarians and (b) conserved cnidarian microRNAs that are shared between at least two sea anemone species. ‘+’ and ‘0’ signs denote the presence and absence of genes/microRNAs in respective species. Species abbreviations: Acti, Actinernus sp.; Adig, Acropora digitifera; Amil, Acropora millepora; Aten, Actinia tenebrosa; Cjar, Catalaphyllia jardinei; Epal, Exaiptasia pallida; Hmag, Hydra magnipapillata; Hvul, Hydra vulgaris; Nvec, Nematostella vectensis; Pxis, Paraphelliactis xishaensis; Resc, Rhopilema esculentum; Scal, Scolanthus callimorphus; Smal, Sanderia malayensis; Spis, Stylophora pistillata.
MicroRNAs are 21–23 nucleotides small RNAs that play important post-transcriptional gene regulation roles in animals. Previous studies have shown that cnidarians and bilaterians only share one conserved microRNA (miR-100) between them [32,34,35,39,40]. We produced small RNA transcriptomes for Actinernus sp. and annotated a total of 28 microRNAs in its genome (figure 3b, electronic supplementary material, S8). Among these annotated microRNAs, we identified 12 conserved microRNAs across different lineages of cnidarians, including miR-100, miR-2022, miR-2023, miR-2030, miR-2025, miR-2026, miR-2028, miR-2029, miR-2036, miR-2037, miR-2050 and miR-9425 (figure 3b; electronic supplementary material, figure S14). Taking Actinernus sp. as a reference point, we see a gradually nested pattern of these conserved microRNAs at successive nodes in the cnidarian phylogeny (figure 3b). Such a pattern would be consistent with them having been incorporated into the gene regulatory networks of functions that have progressively assembled at distinct nodes that represent successive ancestors within the cnidarians. In addition, independent expansions of microRNAs also happened in individual cnidarian lineages, illustrating the converse pattern to the widely conserved genes and demonstrating the divergent evolution of these ancient cnidarian lineages. MicroRNAs thus provide a mixed evolutionary pattern ranging from widely conserved to lineage-specific genes, but in total the cnidarian microRNA complements highlight the distinct nature of these animals from bilaterians.
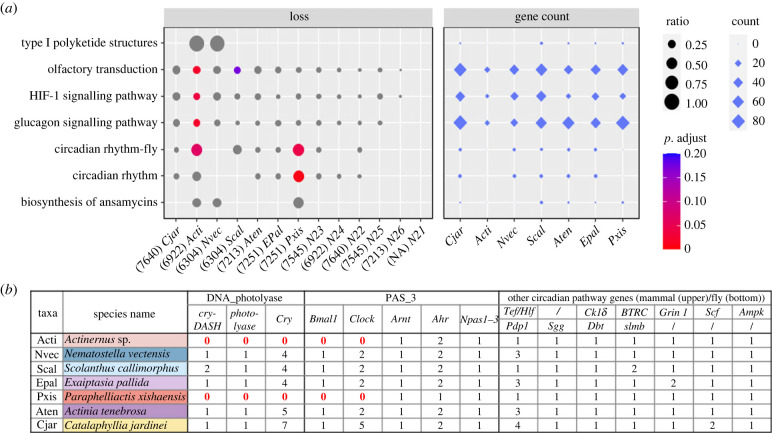
Phylogenomic analyses using 332 single-copy orthologues suggested Actinernus diverged from the last common ancestor of itself and the sea anemones Scolanthus callimorphus and Nematostella vectensis about 250 Ma (electronic supplementary material, figure S15). Gene loss has been suggested as an important force in metazoan evolution [41], and our enrichment analyses identified the loss of circadian rhythm genes and DNA photolyases in both Actinernus sp. and another deep-sea anemone, Paraphelliactis xishaensis (figure 4; electronic supplementary material, S9 and figures S16–S19). Although the evidence of gene loss in P. xishaensis could be limited by the quality of the available draft genome [14], the high quality and physical continuity of this Actinernus sp. genome enabled us to carry out microsyntenic analyses to confirm the gene losses, including cryptochrome, cryptochrome-DASH, deoxyribodipyrimidine photolyase, brain and muscle Arnt-like protein-1 (Bmal1), and circadian locomotor output cycles kaput (Clock) genes (electronic supplementary material, figure S20). Evolution of morphology and physiology in concert with gene loss has been documented in multiple examples in metazoans, including changes to the pelvis and spines in stickleback fishes [42], eye degeneration and pigment losses in cavefish [43], visual adaptation and loss of photoreceptors and olfactory receptors in deep-sea fishes [44,45] and loss of circadian genes in a sub-surface blind centipede [46]. Taken together, the adaptations to the deep-sea environment found in this anemone include gene losses associated with processes that have become redundant in the darkness (e.g. circadian clock), demonstrating the dynamic nature of gene complement evolution in relation to ecological evolutionary adaptation.
Figure 4.
(a) Enriched KEGG pathways of gene family loss (left panel) and gene count in Hexacorallia taxa. The gene ratio was defined by the number of gene loss at each taxon/internode divided by the total number of genes in its ancestral state. P-value was calculated to test significantly over-represented genes using the hypergeometric model and was adjusted using the Benjamini and Hochberg method (BH) and (b) gene count table of circadian rhythm pathway and related gene families with DNA photolyase and PAS fold (PAS_3) domains.
This study has provided high-quality genomic and transcriptomic resources of a deep-sea cnidarian, Actinernus sp. Our analyses reveal a distinctive genome architecture spread over 15 chromosomes that includes a mega-array of ANTP-class homeobox genes that until now was only hypothesized to exist in ancient animal ancestors. In addition, patterns of gene losses such as those in the circadian clock genes coincide with living in the deep-sea habitat, and appear to be generally applicable across animals inhabiting this extreme environment.
3. Methods
(a) . Sample collection and genome sequencing
Six deep-sea anemone specimens were collected from Haima cold seep in the South China Sea at 1386 m depth during a HYDZ6-202005 cruise of the research vessel R/V Haiyangliuhao in September 2020. Samples were instantly frozen in liquid nitrogen and kept frozen during delivery and storage. The whole body was sent to Novogene (Hong Kong) to perform genomic DNA extraction. PacBio Biosciences HiFi circular consensus sequencing (CCS) library was constructed and sequenced by Novogene (Hong Kong).
(b) . Dovetail Omni-C library preparation and sequencing
For each Dovetail Omni-C library, chromatin was fixed in place with formaldehyde in the nucleus and then extracted. Fixed chromatin was digested with DNAse I, chromatin ends were repaired and ligated to a biotinylated bridge adapter followed by proximity ligation of adapter-containing ends. After proximity ligation, crosslinks were reversed and the DNA purified. Purified DNA was treated to remove biotin that was not internal to ligated fragments. Sequencing libraries were generated using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. The library was sequenced on an Illumina HiSeqX platform to produce an approximately 49× sequence coverage (electronic supplementary material, S2). Then HiRise was applied for reads with MQ > 50 for scaffolding.
(c) . mRNA and small RNA transcriptomes
mRNA and small RNA were extracted from tentacle and trunk tissues of two individuals using TRIzol (Ambion) and the miRVana Isolation kit, respectively. The concentrations were measured using a NanoDrop One/One C Microvolume UV Spectrophotometer (Thermo Fisher Scientific, Fitchburg, WI, USA). Agarose gel electrophoresis was used to assess the quality. Samples were stored at −80°C and sent to Novogene (Hong Kong) for transcriptome (Novaseq, PE150 platform for strand-specific library construction) and small RNA library (Novaseq, 50SE platform for strand-specific library construction) sequencing (electronic supplementary material, S2).
(d) . Genome assembly and scaffolding with HiRise
The de novo genome assembly was carried out with Hifiasm [47], and haplotypic duplication was identified and removed with purge_dups based on the depth of HiFi reads [48]. The primary genome assembly and Dovetail Omni-C library reads were then used as input data for HiRise [49]. Dovetail Omni-C library sequences were aligned to the draft input assembly using bwa (https://github.com/lh3/bwa). The separations of Dovetail Omni-C read pairs mapped within draft scaffolds were analysed by HiRise to produce a likelihood model for genomic distance between read pairs, and the model was used to identify and break putative misjoins, to score prospective joins and make joins. The mitochondrial genome of Actinernus sp. was assembled by MitoHiFi [50] (v.2.2, https://github.com/marcelauliano/MitoHiFi) using PacBio HiFi reads and Macrodactyla doreensis (NC_066448.1) as the mitochondrial reference sequence.
(e) . Gene model prediction
Gene model annotations were processed as previously described [32]. In brief, gene models were trained and predicted using funannotate (v.1.8.9). Protein-coding genes were searched with BLASTp against the nr and swissprot databases by diamond (v.0.9.24) with parameters ‘—more-sensitive—e-value 1 × 10−3,’ and mapped by HISAT2 v.2.1.0 with transcriptome reads from the tentacle and columnar trunk tissues. Gene models with no similarities to any known protein and no messenger RNA support were removed from the final version, filtered by CGAT [51], and only the longest genes were retained for subsequent analyses. Repetitive elements were identified using the TE annotation pipeline Earl Grey [52] as described previously [53].
(f) . Macrosynteny analysis
Orthologous gene pairs were retrieved from reciprocal BLASTp hits (e-value 1 × 10−5) between Actinernus sp. and each taxon in the orthology analysis. Macrosynteny was inferred by comparing Actinernus sp. with four chromosome-level Hexacorallia genomes (S. callimorphus, N. vectensis, Exaiptasia pallida and Catalaphyllia jardinei) using MCScanX under default parameters [54] and visualized with TBtools v.1.055 [55]. DupGen_finder [56] was used to parse MCScanX results and classify duplicated genes in Actinernus sp. into self-syntenic duplications (originally whole-genome duplication), tandem duplications, proximal duplications, transposed duplications and dispersed duplications using the four chromosome-level Hexacorallia genomes as outgroups. Further, reciprocal best hits from the highest BLASTp bit-score between Acinernus sp. and the four Hexacorallia genomes and five other outgroups (Rhopilema esculentum, Sanderia malayensis, Hydra vulgaris, Ephydatia muelleri and Hormiphora californensis) were used to generate Oxford dot plots as described [29,57].
(g) . Annotation of sesquiterpenoid pathway genes
Gene sequences involved in the sesquiterpenoid biosynthetic pathway were retrieved from Nematostella and previous studies [32] and were used for searching against the gene models and genome using BLASTP and TBLASTN, respectively. Putatively identified orthologues with a threshold of e-value equal to 10−3 were tested by reciprocal searches in the NCBI nr database using BLASTP. The gene expression profile of the sesquiterpenoid biosynthetic pathway was visualized with a heatmap of trimmed mean of M-values normalized counts from the tentacle and trunk transcriptomes under log2 scale in TBtools [55].
(h) . microRNA annotation
Cnidarian microRNAs were annotated as previously described [32]. Adaptor sequences were trimmed from small RNA sequencing reads and reads with Phred quality scores less than 20 were removed. Processed reads of lengths between 18 bp and 27 bp were then mapped to the genomes using the mapper.pl module of the mirDeep2 package [58]. To identify known miRNAs, the predicted deep-sea anemone microRNA hairpins were compared against metazoan microRNA precursor sequences from miRbase [59] using BLASTN (e-value < 1 × 10−2). miRNAs with no significant sequence similarity to any of the miRNAs in miRBase were checked manually. Novel microRNAs were defined as those that fulfilled the criteria of microRNAs in MirGeneDB [60–62]. In addition, precursor sequences of microRNAs from other sea anemones in previous studies [40,63,64] were also used to carry out BLASTN searches for identification of any missed microRNA annotations. Multiple alignments of conserved miRNAs were carried out by MEGA7 [65].
(i) . Annotation of homeobox genes
Potential homeobox genes were identified by searching with homeodomain sequences from N. vectensis [66], Branchiostoma floridae, Drosophila melanogaster (retrieved from HomeoDB2 [67] in the Actinernus sp. genome and previous published high-quality cnidarian genomes [14,31,32,34,35,57,68–73]) using tBLASTN (electronic supplementary material, S4). NCBI CD-Search [74] was then used to validate the presence of homeodomains in the retrieved sequences. Identification of each putative gene was tested by comparison to sequences confirmed in the NCBI nr database and previous studies [32,67,75–86] using BLASTx and BLASTp, phylogenetic analysis and syntenic analysis (electronic supplementary material, S4). Syntenic relationships between E. pallida, N. vectensis, Acropora millepora, R. esculentum and H. vulgaris were computed using MCScanX [54] with default parameters. MG2C [87] was used to visualize the mega-array of homeodomain genes on a chromosome map of Chr 1 in Actinernus sp.
(j) . Orthology and gene family evolution analysis
The gene orthology of the longest gene transcripts from six actiniarians and nine outgroup taxa was inferred by OrthoMCL [88]. Three-hundred and thirty-two single-copy genes were aligned by MAFFT v.7.271 [89], trimmed with trimAl v.1.4.rev15 [90] and concatenated to construct a species phylogenetic tree using RAxML v.8.2.9 [91] with the PROTGAMMAILGF model and 1000 bootstrap replicates. BEAST v.2.6.2 [92] was employed to infer the divergence times with a relaxed clock rate and calibrated Yule model with gamma prior distribution. The calibration points were retrieved from the TimeTree web database (http://timetree.org): Salpingoeca rosetta–H. californensis (mean 908 Ma, sigma: 30), H. californensis–E. muelleri (mean 851 Ma, sigma: 35), Trichoplax adhaerens–H. vulgaris (mean 661, sigma: 27), H. vulgaris–R. esculentum (581 Ma, sigma: 30) and C. jardinei–Actinia tenebrosa (mean 532, sigma: 25). The first 30% of the MCMC sampling was discarded as burn-in. The resulting species tree was used as the input tree for CAFE analysis [93], where multiple lambdas were used to infer the birth–death rate for four groups, namely Choanozoa-Placozoa, Hydrozoa-Scyphozoa, Scleractinia and Actinaria.
(k) . Functional enrichment analysis
Functional annotations were processed in each analysed proteome with eggnog [94]. Orthologous groups were assigned for annotation terms from Gene Ontology (GO), EuKaryotic Orthologous Groups (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG) and KEGG Orthology (KO) when they were observed in genes within an orthologous group. Functional enrichment was tested as previously described [29,38]. Enrichment was tested with the function ‘compareCluster()’ from R package ‘clusterProfiler’ v.3.16.1 [95]. For enriched terms that resulted from gene family gain, their significance was determined with p-value cut-off of 0.05 and q-value cut-off of 0.2, according to adjustment of p-value using the Benjamini and Hochberg (BH) method. To validate gene family gains, protein family searches using HMMER [96] (v.3.3.1; cut-off e-value < 10−5) followed by confirmation with Reverse Position-Specific BLAST (RPS-BLAST) with the NCBI's Conserved Domain Database (CDD) [97] were used to generate a gene count table. For enriched terms that resulted from gene loss, as statistical significance may not be applicable, they were manually checked by sorting from high to low (ratio of 1.0 conveys complete loss of a gene family). The absence of gene family was further validated with tBLASTn and BLASTp as well as protein family (HMMER) searches [96,98] of reference protein sequences in the genome and proteome, respectively. Microsynteny of neighbouring conserved genes at a loss site was inspected by comparing with other Hexacorallia genomes using MCScanX [54], followed by visualization in the gggenomes package [99] in R v.4.1.1 [100].
Contributor Information
David E. K. Ferrier, Email: dekf@st-andrews.ac.uk.
Jianwen Qiu, Email: qiujw@hkbu.edu.hk.
Peiyuan Qian, Email: boqianpy@ust.hk.
Jerome Ho Lam Hui, Email: jeromehui@cuhk.edu.hk.
Ethics
This is an invertebrate sea anemone, which was sampled during the HYDZ6-202005 cruise and approved by Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou).
Data accessibility
The Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the accession JAOWCC000000000. The raw reads generated in this study were deposited to the NCBI database under the BioProject accessions PRJNA889077. The genome annotation files were deposited in Figshare: https://figshare.com/s/162e2b28fc843c840a65. Files can also be found here: https://datadryad.org/stash/share/8KypWmpBYJgpPXhWPE5PsO4citcscg4IJShJ61VQgI4 [101].
The data are provided in the electronic supplementary material [102].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
S.T.S.L.: data curation, formal analysis, investigation, methodology, validation, writing—original draft, writing—review and editing; Y.Y.: formal analysis, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing; W.N.: data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing—original draft, writing—review and editing; W.L.S.: formal analysis, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing; Y.L.: formal analysis, investigation, methodology, validation, visualization, writing—review and editing; T.S.: data curation, methodology, resources, validation, visualization, writing—original draft, writing—review and editing; D.E.K.F.: conceptualization, formal analysis, funding acquisition, investigation, methodology, writing—original draft, writing—review and editing; J.Q.: conceptualization, data curation, funding acquisition, investigation, methodology, resources, writing—original draft, writing—review and editing; P.Q.: conceptualization, funding acquisition, investigation, methodology, resources, writing—original draft, writing—review and editing; J.H.L.H.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This study was supported by Hong Kong Research Grant Council Collaborative Research Fund CRF (grant no. C4015-20EF), General Research Fund GRF (grant no. 14100420), CUHK Direct Grant (grant nos. 4053489 and 4053547), Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (grant nos. HJ202101, SMSEGL20SC01 and SMSEGL20SC02) and Major Project of Basic and Applied Basic Research of Guangdong Province (grant no. 2019B030302004).
References
- 1.Sun Y, et al. 2021. Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica. Mol. Biol. Evol. 38, 4116-4134. ( 10.1093/molbev/msab203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan J, et al. 2022. Genome of a giant isopod, Bathynomus jamesi, provides insights into body size evolution and adaptation to deep-sea environment. BMC Biol. 20, 113. ( 10.1186/s12915-022-01302-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayavedra L, et al. 2015. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. Elife 4, e07966. ( 10.7554/eLife.07966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat. Ecol. Evol. 1, 121. ( 10.1038/s41559-017-0121) [DOI] [PubMed] [Google Scholar]
- 5.Liu R, et al. 2020. De novo genome assembly of limpet Bathyacmaea lactea (Gastropoda: Pectinodontidae): the first reference genome of a deep-sea gastropod endemic to cold seeps. Genome Biol. Evol. 12, 905-910. ( 10.1093/gbe/evaa100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ip JC-H, et al. 2021. Host–endosymbiont genome integration in a deep-sea chemosymbiotic clam. Mol. Biol. Evol. 38, 502-518. ( 10.1093/molbev/msaa241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan Y, et al. 2021. Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis. Nat. Commun. 12, 1165. ( 10.1038/s41467-021-21450-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao G, et al. 2022. The genome of a hadal sea cucumber reveals novel adaptive strategies to deep-sea environments. iScience 25, 105545. ( 10.1016/j.isci.2022.105545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, et al. 2022. The genome of an apodid holothuroid (Chiridota heheva) provides insights into its adaptation to a deep-sea reducing environment. Commun. Biol. 5, 1-11. ( 10.1038/s42003-022-03176-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaither MR, et al. 2018. Genomics of habitat choice and adaptive evolution in a deep-sea fish. Nat. Ecol. Evol. 2, 680-687. ( 10.1038/s41559-018-0482-x) [DOI] [PubMed] [Google Scholar]
- 11.Mu Y, et al. 2021. Whole genome sequencing of a snailfish from the Yap Trench (∼7,000 m) clarifies the molecular mechanisms underlying adaptation to the deep sea. PLoS Genet. 17, e1009530. ( 10.1371/journal.pgen.1009530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bo J, Xu H, Lv W, Wang C, He S, Yang L. 2022. Molecular mechanisms of the convergent adaptation of bathypelagic and abyssopelagic fishes. Genome Biol. Evol. 14, evac109. ( 10.1093/gbe/evac109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Feng C, Pu Y, Liu J, Liu R, Zhang H. 2021. The first draft genome of a cold-water coral Trachythela sp. (Alcyonacea: Stolonifera: Clavulariidae). Genome Biol. Evol. 13, evaa265. ( 10.1093/gbe/evaa265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng C, et al. 2021. The genome of a new anemone species (Actiniaria: Hormathiidae) provides insights into deep-sea adaptation. Deep Sea Res. Part I Oceanogr. Res. Pap. 170, 103492. ( 10.1016/j.dsr.2021.103492) [DOI] [Google Scholar]
- 15.Holland PWH, Booth HAF, Bruford EA. 2007. Classification and nomenclature of all human homeobox genes. BMC Biol. 5, 47. ( 10.1186/1741-7007-5-47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrier DEK. 2016. Evolution of homeobox gene clusters in animals: the Giga-cluster and primary vs. secondary clustering. Front. Ecol. Evol. 4, 36. ( 10.3389/fevo.2016.00036) [DOI] [Google Scholar]
- 17.Pollard SL, Holland PWH. 2000. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 10, 1059-1062. ( 10.1016/S0960-9822(00)00676-X) [DOI] [PubMed] [Google Scholar]
- 18.Castro LFC, Holland PWH. 2003. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol. Dev. 5, 459-465. ( 10.1046/j.1525-142x.2003.03052.x) [DOI] [PubMed] [Google Scholar]
- 19.Hui JHL, Mcdougall C, Monteiro AS, Holland PWH, Arendt D, Balavoine G, Ferrier DEK. 2012. Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol. Biol. Evol. 29, 157-165. ( 10.1093/molbev/msr175) [DOI] [PubMed] [Google Scholar]
- 20.Dong D, et al. 2021. Report of epibenthic macrofauna found from Haima cold seeps and adjacent deep-sea habitats, South China Sea. Mar. Life Sci. Technol. 3, 1-2. ( 10.1007/s42995-020-00073-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gates A. 2016. In Deep-sea life of Tanzania, pp. 27. Southampton, UK: National Oceanography Centre. [Google Scholar]
- 22.Gusmão LC, Rodríguez E. 2021. Deep-sea anemones (Cnidaria: Anthozoa: Actiniaria) from the South Atlantic. Bull. Am. Mus. Nat. Hist. 444, 1-69. ( 10.1206/0003-0090.444.1.1) [DOI] [Google Scholar]
- 23.Izumi T, Fujii T, Yanagi K, Higashiji T, Fujita T. 2019. Redescription of Synactinernus flavus for the first time after a century and description of Synactinernus churaumi sp. nov. (Cnidaria: Anthozoa: Actiniaria). Zool. Sci. 36, 528-538. ( 10.2108/zs190040) [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez E, Barbeitos MS, Brugler MR, Crowley LM, Grajales A, Gusmão L, Häussermann V, Reft A, Daly M. 2014. Hidden among sea anemones: the first comprehensive phylogenetic reconstruction of the order Actiniaria (Cnidaria, Anthozoa, Hexacorallia) reveals a novel group of hexacorals. PLoS ONE 9, e96998. ( 10.1371/journal.pone.0096998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Guan H, Qiu JW, Xu T, Peckmann J, Chen D, Feng D. 2022. Macro-ecology of cold seeps in the South China Sea. Geosystems Geoenvironment 1, 100081. ( 10.1016/j.geogeo.2022.100081) [DOI] [Google Scholar]
- 26.He X, Xu T, Qian P. 2023. Same (sea) bed different dreams: biological community structure of the Haima seep reveals distinct biogeographic affinities. Innov. Geosci. 1, 100019. ( 10.59717/j.xinn-geo.2023.100019) [DOI] [Google Scholar]
- 27.Canapa A, Barucca M, Biscotti MA, Forconi M, Olmo E. 2016. Transposons, genome size, and evolutionary insights in animals. Cytogenet. Genome Res. 147, 217-239. ( 10.1159/000444429) [DOI] [PubMed] [Google Scholar]
- 28.Naville M, Henriet S, Warren I, Sumic S, Reeve M, Volff J-N, Chourrout D. 2019. Massive changes of genome size driven by expansions of non-autonomous transposable elements. Curr. Biol. 29, 1161-1168. ( 10.1016/j.cub.2019.01.080) [DOI] [PubMed] [Google Scholar]
- 29.So WL, et al. 2022. Myriapod genomes reveal ancestral horizontal gene transfer and hormonal gene loss in millipedes. Nat. Commun. 13, 3010. ( 10.1038/s41467-022-30690-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland PWH. 2013. Evolution of homeobox genes. Wiley Interdiscip. Rev.: Dev. Biol. 2, 31-45. ( 10.1002/wdev.78) [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Nong W, Baril T, Yip HY, Swale T, Hayward A, Ferrier DEK, Hui JHL. 2020. Reconstruction of ancient homeobox gene linkages inferred from a new high-quality assembly of the Hong Kong oyster (Magallana hongkongensis) genome. BMC Genom. 21, 713. ( 10.1186/s12864-020-07027-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nong W, et al. 2020. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat. Commun. 11, 3051. ( 10.1038/s41467-020-16801-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu Z, et al. 2020. Millipede genomes reveal unique adaptations during myriapod evolution. PLoS Biol. 18, e3000636. ( 10.1371/journal.pbio.3000636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shum CWY, et al. 2022. Genome of the sea anemone Exaiptasia pallida and transcriptome profiles during tentacle regeneration. Front. Cell Dev. Biol. 10, 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, et al. 2022. Genome of elegance coral Catalaphyllia jardinei (Euphylliidae). Front. Mar. Sci. 9, 991391. ( 10.3389/fmars.2022.991391) [DOI] [Google Scholar]
- 36.Qu Z, Kenny NJ, Lam HM, Chan TF, Chu KH, Bendena WG, Tobe SS, Hui JHL. 2015. How did arthropod sesquiterpenoids and ecdysteroids arise? Comparison of hormonal pathway genes in noninsect arthropod genomes. Genome Biol. Evol. 7, 1951-1959. ( 10.1093/gbe/evv120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu Z, et al. 2017. MicroRNAs regulate the sesquiterpenoid hormonal pathway in Drosophila and other arthropods. Proc. R. Soc. B 284, 20171827. ( 10.1098/rspb.2017.1827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.So WL, Kai Z, Qu Z, Bendena WG, Hui JHL. 2022. Rethinking sesquiterpenoids: a widespread hormone in animals. Int. J. Mol. Sci. 23, 5998. ( 10.3390/ijms23115998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Hui JHL, Marco A, Ronshaugen M. 2011. MicroRNA evolution by arm switching. EMBO Rep. 12, 172-177. ( 10.1038/embor.2010.191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praher D, et al. 2021. Conservation and turnover of miRNAs and their highly complementary targets in early branching animals. Proc. R. Soc. B 288, 20203169. ( 10.1098/rspb.2020.3169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat. Rev. Genet. 17, 379-391. ( 10.1038/nrg.2016.39) [DOI] [PubMed] [Google Scholar]
- 42.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302-305. ( 10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mcgaugh SE, et al. 2014. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307. ( 10.1038/ncomms6307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Du K, Gan X, Yang L, He S. 2019. Massive loss of olfactory receptors but not trace amine-associated receptors in the world's deepest-living fish (Pseudoliparis swirei). Genes 10, 910. ( 10.3390/genes10110910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, et al. 2019. Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat. Ecol. Evol. 3, 823-833. ( 10.1038/s41559-019-0864-8) [DOI] [PubMed] [Google Scholar]
- 46.Chipman AD, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12, e1002005. ( 10.1371/journal.pbio.1002005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170-175. ( 10.1038/s41592-020-01056-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan D, Mccarthy SA, Wood J, Howe K, Wang Y, Durbin R. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896-2898. ( 10.1093/bioinformatics/btaa025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putnam NH, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26, 342-350. ( 10.1101/gr.193474.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Resour. 20, 892-905. ( 10.1111/1755-0998.13160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims D, et al. 2014. CGAT: computational genomics analysis toolkit. Bioinformatics 30, 1290-1291. ( 10.1093/bioinformatics/btt756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baril T, Imrie RM, Hayward A. 2022. Earl Grey: a fully automated user-friendly transposable element annotation and analysis pipeline. bioRxiv 2022.06.30.498289. ( 10.1101/2022.06.30.498289) [DOI]
- 53.Nong W, et al. 2022. Genome of the ramshorn snail Biomphalaria straminea—an obligate intermediate host of schistosomiasis. GigaScience 11, giac012. (https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giac012/6528774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. ( 10.1093/nar/gkr1293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194-1202. ( 10.1016/j.molp.2020.06.009) [DOI] [PubMed] [Google Scholar]
- 56.Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH. 2019. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 38. ( 10.1186/s13059-019-1650-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simakov O, et al. 2022. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci. Adv. 8, eabi5884. ( 10.1126/sciadv.abi5884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37-52. ( 10.1093/nar/gkr688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68-D73. ( 10.1093/nar/gkt1181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang W, Bartel DP. 2015. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell 60, 131-145. ( 10.1016/j.molcel.2015.08.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartel DP. 2018. Metazoan microRNAs. Cell 173, 20-51. ( 10.1016/j.cell.2018.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fromm B, et al. 2020. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 48, D132-D141. ( 10.1093/nar/gkz885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumgarten S, Cziesielski MJ, Thomas L, Michell CT, Esherick LY, Pringle JR, Aranda M, Voolstra CR. 2018. Evidence for miRNA-mediated modulation of the host transcriptome in cnidarian–dinoflagellate symbiosis. Mol. Ecol. 27, 403-418. ( 10.1111/mec.14452) [DOI] [PubMed] [Google Scholar]
- 64.Fridrich A, Modepalli V, Lewandowska M, Aharoni R, Moran Y. 2020. Unravelling the developmental and functional significance of an ancient Argonaute duplication. Nat. Commun. 11, 6187. ( 10.1038/s41467-020-20003-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870-1874. ( 10.1093/molbev/msw054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. 2006. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 7, R64. ( 10.1186/gb-2006-7-7-r64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong Y, Holland PWH. 2011. HomeoDB2: functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evol. Dev. 13, 567-568. ( 10.1111/j.1525-142X.2011.00513.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srivastava M, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454, 955-960. ( 10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- 69.Surm JM, Stewart ZK, Papanicolaou A, Pavasovic A, Prentis PJ. 2019. The draft genome of Actinia tenebrosa reveals insights into toxin evolution. Ecol. Evol. 9, 11 314-11 328. ( 10.1002/ece3.5633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kenny NJ, et al. 2020. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat. Commun. 11, 3676. ( 10.1038/s41467-020-17397-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simakov O, et al. 2020. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820-830. ( 10.1038/s41559-020-1156-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schultz DT, Francis WR, Mcbroome JD, Christianson LM, Haddock SHD, Green RE. 2021. A chromosome-scale genome assembly and karyotype of the ctenophore Hormiphora californensis. G3 11, jkab302. ( 10.1093/g3journal/jkab302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zimmermann B, et al. 2022. Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. BioRxiv 359448. ( 10.1101/2020.10.30.359448) [DOI] [Google Scholar]
- 74.Lu S, et al. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265-D268. ( 10.1093/nar/gkz991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanze N, Spring J, Schmidli C, Schmid V. 2001. Conservation of Hox/ParaHox-related genes in the early development of a cnidarian. Dev. Biol. 236, 89-98. ( 10.1006/dbio.2001.0299) [DOI] [PubMed] [Google Scholar]
- 76.Kamm K, Schierwater B, Jakob W, Dellaporta SL, Miller DJ. 2006. Axial patterning and diversification in the cnidaria predate the Hox system. Curr. Biol. 16, 920-926. ( 10.1016/j.cub.2006.03.036) [DOI] [PubMed] [Google Scholar]
- 77.Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86-94. ( 10.1126/science.1139158) [DOI] [PubMed] [Google Scholar]
- 78.Chiori R, Jager M, Denker E, Wincker P, Silva CD, Guyader HL, Manuel M, Quéinnec E. 2009. Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (Cnidaria). PLoS ONE 4, e4231. ( 10.1371/journal.pone.0004231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dubuc TQ, Ryan JF, Shinzato C, Satoh N, Martindale MQ. 2012. Coral comparative genomics reveal expanded Hox cluster in the Cnidarian–Bilaterian ancestor. Integr. Comp. Biol. 52, 835-841. ( 10.1093/icb/ics098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zapata F, et al. 2015. Phylogenomic analyses support traditional relationships within Cnidaria. PLoS ONE 10, e0139068. ( 10.1371/journal.pone.0139068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kayal E, Bentlage B, Sabrina Pankey M, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF. 2018. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 18, 68. ( 10.1186/s12862-018-1142-0) [DOI] [Google Scholar]
- 82.Khalturin K, et al. 2019. Medusozoan genomes inform the evolution of the jellyfish body plan. Nat. Ecol. Evol. 3, 811-822. ( 10.1038/s41559-019-0853-y) [DOI] [PubMed] [Google Scholar]
- 83.Leclère L, et al. 2019. The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat. Ecol. Evol. 3, 801-810. ( 10.1038/s41559-019-0833-2) [DOI] [PubMed] [Google Scholar]
- 84.Ohdera A, et al. 2019. Box, stalked, and upside-down? Draft genomes from diverse jellyfish (Cnidaria, Acraspeda) lineages: Alatina alata (Cubozoa), Calvadosia cruxmelitensis (Staurozoa), and Cassiopea xamachana (Scyphozoa). Gigascience 8, giz069. ( 10.1093/gigascience/giz069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Debiasse MB, et al. 2022. A cnidarian phylogenomic tree fitted with hundreds of 18S leaves. bioRxiv 510641. ( 10.1101/2022.10.03.510641) [DOI]
- 86.Steinworth BM, Martindale MQ, Ryan JF. 2023. Gene loss may have shaped the cnidarian and bilaterian Hox and ParaHox complement. Genome Biol. Evol. 15, evac172. ( 10.1093/gbe/evac172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chao J, Li Z, Sun Y, Aluko OO, Wu X, Wang Q, Liu G. 2021. MG2C: a user-friendly online tool for drawing genetic maps. Mol. Hortic. 1, 1-4. ( 10.1186/s43897-021-00020-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178-2189. ( 10.1101/gr.1224503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972-1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537. ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987-1997. ( 10.1093/molbev/mst100) [DOI] [PubMed] [Google Scholar]
- 94.Huerta-Cepas J, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309-D314. ( 10.1093/nar/gky1085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene custers. OMICS J. Integr. Biol. 16, 284-287. ( 10.1089/omi.2011.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. ( 10.1371/journal.pcbi.1002195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang M, Derbyshire MK, Yamashita RA, Marchler-Bauer A. 2020. NCBI's conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinform. 69, e90. ( 10.1002/cpbi.90) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinf. 10, 421. ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hackl T, Duponchel S, Barenhoff K, Weinmann A, Fischer MG. 2021. Virophages and retrotransposons colonize the genomes of a heterotrophic flagellate. Elife 10, e72674. ( 10.7554/eLife.72674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 101.Law STS, et al. 2023. Data from: Adaptations to the deep-sea alongside presence of a mega-array of homeobox genes in the genome of the cnidarian Actinernus. Dryad Digital Repository. ( 10.5061/dryad.dbrv15f6k) [DOI]
- 102.Law STS, et al. 2023. The genome of the deep-sea anemone Actinernus sp. contains a mega-array of ANTP-class homeobox genes. Figshare. ( 10.6084/m9.figshare.c.6875449) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Law STS, et al. 2023. Data from: Adaptations to the deep-sea alongside presence of a mega-array of homeobox genes in the genome of the cnidarian Actinernus. Dryad Digital Repository. ( 10.5061/dryad.dbrv15f6k) [DOI]
- Law STS, et al. 2023. The genome of the deep-sea anemone Actinernus sp. contains a mega-array of ANTP-class homeobox genes. Figshare. ( 10.6084/m9.figshare.c.6875449) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the accession JAOWCC000000000. The raw reads generated in this study were deposited to the NCBI database under the BioProject accessions PRJNA889077. The genome annotation files were deposited in Figshare: https://figshare.com/s/162e2b28fc843c840a65. Files can also be found here: https://datadryad.org/stash/share/8KypWmpBYJgpPXhWPE5PsO4citcscg4IJShJ61VQgI4 [101].
The data are provided in the electronic supplementary material [102].