Abstract
Motivation
Coincidence of Convergent Amino Acid Substitutions (CAAS) with phenotypic convergences allow pinpointing genes and even individual mutations that are likely to be associated with trait variation within their phylogenetic context. Such findings can provide useful insights into the genetic architecture of complex phenotypes.
Results
Here we introduce CAAStools, a set of bioinformatics tools to identify and validate CAAS in orthologous protein alignments for predefined groups of species representing the phenotypic values targeted by the user.
Availability and implementation
CAAStools source code is available at http://github.com/linudz/caastools, along with documentation and examples.
1 Introduction
Convergent Amino Acid Substitutions (CAAS) provide important insights into the genetic changes underlying phenotypic variation (Zhang and Kumar 1997, Rey et al. 2019). Recent examples include the identification of genes potentially involved in marine adaptation in mammals (Foote et al. 2015) and the convergent evolution of mitochondrial genes in deep-sea fish species (Shen et al. 2019). Notably, in 2018, Muntané et al. identified a set of 25 genes involved in longevity in primates (Muntané et al. 2018). A few years later, a similar analysis for a wider phylogeny retrieved 996 genes associated with lifespan determination in mammals (Farré et al. 2021). While these analyses often need to be tailored for each particular phenotype and phylogeny, all CAAS detection and validation strategies reported in the literature share some common steps (Rey et al. 2019). First, researchers select the species to compare for CAAS analysis and split them into two or more groups according to the phenotype of interest. The criteria to select these groups can be quite diverse: for instance, groups can be formed by species having diverging values of a given continuous trait, or by species sharing different adaptations, like terrestrial and marine mammals (Foote et al. 2015). The second step consists in linking amino acid substitutions with each group. Here, different approaches can be used, such as identifying identical substitutions for the same amino acid (Besnard et al. 2009, Chabrol et al. 2018), detecting topological incongruencies (Li et al. 2008), variations in amino acid profiles (Rodrigue et al. 2010, Rey et al. 2018), or relying on consistent patterns of groups of amino acids in different groups of species (Zhang et al. 2014, Muntané et al. 2018, Farré et al. 2021). The third step consists in testing the significance of the results. Molecular convergence is a noisy process because spurious CAAS may occur at random in the absence of relationships with phenotypes or selective forces (Xu et al. 2017). To overcome this, researchers have adopted different strategies, mostly based on the idea that adaptive CAAS tend to exceed convergent noise. The delta Site-Specific log-Likelihood Score (ΔSSLS), for instance, is a method that consists in comparing the CAAS likelihood for different phylogenetic topologies (Castoe et al. 2009, Parker et al. 2013, Wang et al. 2013). Another approach uses bootstrap resampling tests to evaluate whether the number of detected CAAS is larger than expected by chance (Muntané et al. 2018, Farré et al. 2021). Alternatively, some authors have adopted a strategy that consists in quantifying the convergent noise and focus on the detection of Convergence on Conservative Sites (Xu et al. 2017, He et al. 2020). In spite of all these contributions, there is still no consensus approach. Some authors question whether phenotypic convergence matches genome-wide molecular convergence (Zou and Zhang 2015b), or whether adaptive substitutions outnumber random CAAS (Thomas and Hahn, 2015; Zou and Zhang 2015a). Access to free software tools that are specifically designed to retrieve CAAS will allow the wider research community to compare and validate different strategies, boosting future methodological developments in the field of phylogenetic analysis.
Here we present CAAStools, a toolbox to identify and validate CAAS in a phylogenetic context. CAAStools is based on the strategy applied in our previous studies (Muntané et al. 2018, Farré et al. 2021) and implements different testing strategies through bootstrap analysis. CAAStools is designed to be included in parallel workflows and is optimized to allow scalability at proteome level.
2 Implementation
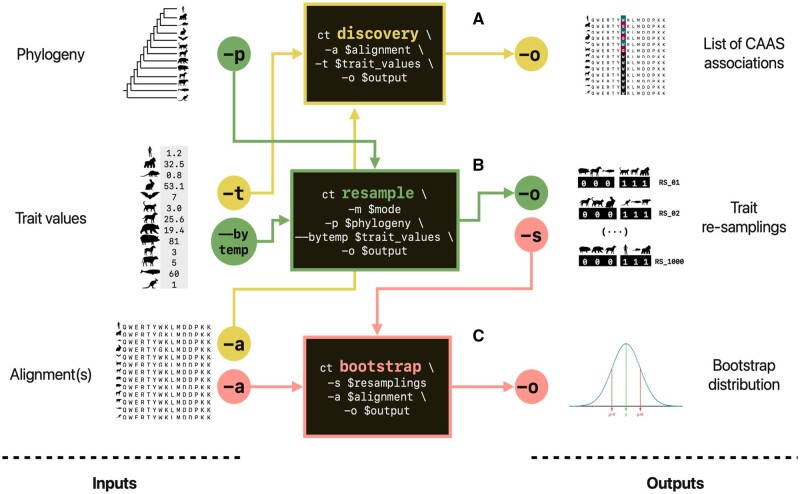
CAAStools is a multi-modular python application organized into three tools. The outline of the suite is presented in Fig. 1. The discovery tool is based on the protocol described in Muntané et al. (2018) and Farré et al. (2021). This approach identifies CAAS between two groups of species in an amino-acid Multiple Sequence Alignment (MSA) of orthologous proteins. These groups are named Foreground Group (FG) and Background Group (BG). Collectively, the two groups are called Discovery Groups (DG), as they represent the base for CAAS discovery. The CAAS identification algorithm scans each MSA and returns those positions that meet the following conditions: First, the FG and the BG species must share no amino acids in that position. Second, all the species in at least one of the two discovery groups (FG or BG) must share the same amino acid. The combination of these two conditions determines a set of different mutation patterns that the tool identifies as CAAS. Details on these patterns are provided in Supplementary Table S1.
Figure 1.
CAAStools layout. The three tools of the CAAStools suite rely on three pieces of information; a phylogenetic tree, the trait information, and an amino acid MSA. The discovery tool (A) detects the CAAS between two groups of species that are defined by the user on the basis of trait values. The resample tool (B) performs n trait resamplings in different modalities, on the bases of the phylogeny and the trait value distributions. The output of this resampling is processed by the bootstrap tool (C) that elaborates a bootstrap distribution from the MSA. All the tools can be executed independently.
Finally, CAAStools calculates the probability of obtaining a CAAS in a given position compared to randomized DGs, corresponding to the empirical P-value of the predicted CAAS in that position. This P-value represents a quantification of the convergent noise (Shahoua et al. 2017) that is associated with a specific position. The details of this calculation are presented in Supplementary Section S3. The Resample tool sorts species into n virtual DGs (resamplings) for bootstrap analysis according to different combination strategies. This tool enables bootstrap analyses based on CAAS excess or likelihood (Castoe et al. 2009, Muntané et al. 2018, Farré et al. 2021). In a Naive modality, the probability of every species being included in a DG is considered identical and independent. This feature allows for bootstrap analyses aimed at quantifying convergent noise. However, species are phylogenetically related, biasing their probability of sharing a phenotype or amino acid. To address these phylogenetic dependencies CAAStools includes two other testing strategies. In the Phylogeny-restricted modality, the randomization can be restricted to some taxonomic orders or defined clades. These clades will match the ones of the species included in the DGs. In the Brownian motion modality, resampling is based on Brownian Motion simulations. The latter builds on the “permulation” strategy for trait randomization (Saputra et al. 2021) and its implementation relies on the simpervec() function from the RERconverge package (Kowalczyk et al. 2019). Finally, the bootstrap tool determines the iterations returning a CAAS for each position in a MSA to establish the corresponding empirical P-value for the detection of a CAAS in that position. Both the discovery and the bootstrap tools are designed to be launched on single MSAs, in order to allow the user to parallelize the workflow for large protein sets.
3. Usage and testing
CAAStools users should take special care when designing the analysis and interpreting the results. The comparison should be made between species with diverging values of a convergent phenotype. Each DG should include species with comparable phenotype values from different lineages. The values between the two DGs must diverge, ideally representing the extreme top and bottom values in a continuous distribution or different binary conditions. The resulting output will consist of a list of positions where at least one DG shares the same amino acid, which differs from those found in the other DG. Depending on the DGs selected (often limited by the available phenotypic and genetic information), this outcome may be influenced by various uninformative sources of sequence variability, such as convergent noise and identity-by-descent. Therefore, it is advisable to complement the CAAS analysis with other approaches that have different limitations, such as ancestral state reconstruction (Royer-Carenzi and Didier, 2016), selection studies (Kosakovsky Pond et al., 2020), or dN/dS analysis (Yang, 1997). For e.g., we tested CAAStools on the dataset from Farré et al., (2021). The details of this test are reported in Supplementary 3. The full dataset is available in the /test folder within the CAAStools repository.
Supplementary Material
Acknowledgements
We would like to thank our collaborators at the Institute of Evolutionary Biology of Pompeu Fabra University for supporting us and sharing their ideas in scientific discussions, with particular mention of Dr Tomas Marques Bonet and members of his group.
Contributor Information
Fabio Barteri, IBE, Institute of Evolutionary Biology (UPF-CSIC), Department of Medicine and Life Sciences, Universitat Pompeu Fabra. C. Doctor Aiguader 88, Barcelona 08003, Spain; BarcelonaBeta Brain Research Center, Pasqual Maragall Foundation, C/ Wellington 30, Barcelona 08006, Spain.
Alejandro Valenzuela, IBE, Institute of Evolutionary Biology (UPF-CSIC), Department of Medicine and Life Sciences, Universitat Pompeu Fabra. C. Doctor Aiguader 88, Barcelona 08003, Spain; BarcelonaBeta Brain Research Center, Pasqual Maragall Foundation, C/ Wellington 30, Barcelona 08006, Spain.
Xavier Farré, Genomes for Life-GCAT Lab, GermanTrias i Pujol Research Institute (IGTP), Camí de les Escoles, s/n, Badalona 08916, Spain.
David de Juan, IBE, Institute of Evolutionary Biology (UPF-CSIC), Department of Medicine and Life Sciences, Universitat Pompeu Fabra. C. Doctor Aiguader 88, Barcelona 08003, Spain.
Gerard Muntané, IBE, Institute of Evolutionary Biology (UPF-CSIC), Department of Medicine and Life Sciences, Universitat Pompeu Fabra. C. Doctor Aiguader 88, Barcelona 08003, Spain; Institut d’Investigació Sanitària Pere Virgili (IISPV), Hospital Universitari Institut Pere Mata, Universitat Rovira i Virgili. Avda. Josep Laporte, 2 – Planta 0 – E2 color taronja, Reus 43204, Spain; Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Av. Monforte de Lemos, 3-5. Pabellón 11. Planta 0. Madrid 28029, Spain.
Borja Esteve-Altava, European Molecular Biology Laboratory, Meyerhofstraße 1, Heidelberg 69117, Germany.
Arcadi Navarro, IBE, Institute of Evolutionary Biology (UPF-CSIC), Department of Medicine and Life Sciences, Universitat Pompeu Fabra. C. Doctor Aiguader 88, Barcelona 08003, Spain; BarcelonaBeta Brain Research Center, Pasqual Maragall Foundation, C/ Wellington 30, Barcelona 08006, Spain; Institució Catalana de Recerca i Estudis Avançats (ICREA) and Universitat Pompeu Fabra, Pg. Lluís Companys 23, Barcelona 08010, Spain; Center for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, C. Doctor Aiguader N88, Barcelona 08003, Spain.
Author contributions
F.B. has been in charge of the development of CAAStools. A.V. carried on the beta-testing and code debugging. F.B. and A.V. have contributed equally to this manuscript. X.F.), G.M., and A.N. designed the CAAS identification and validation protocol. X.F. wrote a set of scripts to identify CAAS that served as template for CAAStools implementation. A.N. and G.M. conceptualized the method. A.N., D.J., and B.E.-A.—along with G.M.—participated in the scientific discussion and supervision of this project. D.J.’s contribution was particularly helpful for the optimization of CAAStools code.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by the I+D+i project PID2021-127792NB-I00 funded by MCIN/AEI/10.13039/501100011033 (FEDER Una manera de hacer Europa); and “Unidad de Excelencia María de Maeztu,” funded by the AEI [CEX2018-000792-M] and Departament de Recerca i Universitats de la Generalitat de Catalunya [GRC 2021 SGR 0467].
References
- Besnard G, Muasya AM, Russier F et al. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol Biol Evol 2009;26:1909–19. [DOI] [PubMed] [Google Scholar]
- Castoe TA, Jason de Koning AP, Kim H-M et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci USA 2009;106:8986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabrol O, Royer-Carenzi M, Pontarotti P et al. Detecting the molecular basis of phenotypic convergence. Methods Ecol Evol 2018;9:2170–80. [Google Scholar]
- Farré X, Molina R, Barteri F et al. Comparative analysis of mammal genomes unveils key genomic variability for human life span. Mol Biol Evol 2021;38:4948–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Liu Y, Thomas GWC et al. Convergent evolution of the genomes of marine mammals. Nat Genet 2015;47:272–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Xu S, Zhang Z et al. ; The International Mangrove Consortium. Convergent adaptation of the genomes of woody plants at the land–sea interface. Natl Sci Rev 2020;7:978–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Poon AFY, Velazquez R et al. HyPhy 2.5—a customizable platform for evolutionary hypothesis testing using phylogenies. Mol Biol Evol 2020;37:295–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A, Meyer WK, Partha R et al. RERconverge: an R package for associating evolutionary rates with convergent traits. Bioinformatics 2019;35:4815–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang J, Rossiter SJ et al. The hearing gene Prestin reunites echolocating bats. Proc Natl Acad Sci USA 2008;105:13959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntané G, Farré X, Rodríguez JA et al. Biological processes modulating longevity across primates: a phylogenetic genome-phenome analysis. Mol Biol Evol 2018;35:1990–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 2013;502:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey C, Guéguen L, Sémon M et al. Accurate detection of convergent amino-acid evolution with PCOC. Mol Biol Evol 2018;35:2296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey C, Lanore V, Veber P et al. Detecting adaptive convergent amino acid evolution. Phil Trans R Soc B Biol Sci 2019;374:20180234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N, Philippe H, Lartillot YN. Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc Natl Acad Sci USA 2010;107:4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer-Carenzi M, Didier YG. A comparison of ancestral state reconstruction methods for quantitative characters. J Theor Biol 2016;404:126–42. [DOI] [PubMed] [Google Scholar]
- Saputra E, Kowalczyk A, Cusick L et al. Phylogenetic permulations: a statistically rigorous approach to measure confidence in associations in a phylogenetic context. Mol Biol Evol 2021;38:3004–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Pu Z, Chen X et al. Convergent evolution of mitochondrial genes in deep-sea fishes. Front Genet 2019;10:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, , Hahn MW. Determining the null model for detecting adaptive convergence from genomic data: A case study using echolocating mammals. Mol Biol Evol 2015;32:1232–6. 10.1093/molbev/msv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-C, Susko E, Roger YAJ. The site-wise log-likelihood score is a good predictor of genes under positive selection. J Mol Evol 2013;76:280–94. [DOI] [PubMed] [Google Scholar]
- Xu S, He Z, Guo Z et al. Genome-Wide convergence during evolution of mangroves from woody plants. Mol Biol Evol 2017;34:1008–15. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 1997;13:555–6. [DOI] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q et al. ; Avian Genome Consortium. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014;346:1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kumar YS. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol 1997;14:527–36. [DOI] [PubMed] [Google Scholar]
- Zou Z, Zhang YJ. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol Biol Evol 2015a;32:2085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zhang YJ. No genome-wide protein sequence convergence for echolocation. Mol Biol Evol 2015b;32:1237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.