ABSTRACT
Ferroptosis is an important mode of regulated cell death (RCD). Its inhibition is closely related to therapeutic resistance and poor prognosis in hepatocellular carcinoma (HCC). Previous reports have demonstrated ferroptosis as a biological process highly dependent on selective autophagy, such as ferritinophagy, lipophagy, and clockophagy. Our study also revealed a role for ER-phagy-mediated ferroptosis in HCC cells treated with multi-targeted tyrosine kinase inhibitors (TKIs). In the current study, we found that the homologous circular RNA (circRNA) of the family with sequence similarity 134, member B (FAM134B), hsa_circ_0128505 (was abbreviated as circFAM134B in the present study), was identified to specifically target ER-phagy to promote lenvatinib (LV)-induced ferroptosis using reactive oxygen species (ROS), Fe2+, malondialdehyde (MDA), and western blot (WB) assays in HCC cells. RNA pull-down and mass spectrometry analyses suggested that circFAM134B and FAM134B mRNA were enriched with several common interacting proteins. Among them, poly (A) binding protein cytoplasmic 4 (PABPC4) was identified as the most enriched binding partner. It was proven to be a novel antagonist against the nonsense-mediated mRNA decay (NMD) mechanism. We then applied RNA immunoprecipitation (RIP), RNA pull-down, luciferase reporter, and NMD reporter gene assays to further explore the exact role and underlying mechanism of circFAM134B-PABPC4-FAM134B axis in HCC cells. circFAM134B was confirmed as a sponge that competitively interacted with PABPC4, thereby influencing FAM134B mRNA nonsense decay. Our results provide novel evidences and strategies for the comprehensive treatment of HCC.
KEYWORDS: Hepatocellular carcinoma, ferroptosis, lenvatinib, circFAM134B, PABPC4, nonsense-mediated mRNA decay
Introduction
Liver cancer is the sixth most common and third leading cause of cancer-related deaths worldwide [1,2]. Hepatocellular carcinoma (HCC) is the main histological type of liver cancer and the cause of 75–85% of primary liver cancer cases [3]. Most HCC patients are diagnosed at advanced stage, thereby the survival rate is poor [4,5]. Comprehensive multidisciplinary treatment is key to prolonging the overall survival time and improving the quality of life of patients with HCC [6,7]. Sorafenib (SF) is the first-line therapeutic drug in the treatment of HCC for 10 years, as it can slightly improve the survival rate of patients with advanced HCC [8]. However, therapeutic resistance is a problem, resulting in poor prognosis [9]. With the gradual broadening and deepening of the concept of regulated cell death (RCD) in recent years, targeting strategies for HCC treatment have also undergone profound changes [10–12]. Several studies have confirmed that precise regulation of ferroptosis is an extremely critical way to enhance the sensitivity of HCC cells to multi-targeted tyrosine kinase inhibitors (TKIs), such as SF, regorafenib, and lenvatinib (LV) [13–15], and further research on the exact mechanism is of great significance for clinical translation.
Ferroptosis is an iron-dependent RCD caused by cystine depletion and overproduction of lipid reactive oxygen species (ROS) [12,16]. Several recent studies have clarified the induction, execution, and regulation of ferroptosis and its association with various pathological and physiological processes [17,18]. The classical ferroptosis pathway directly or indirectly targets the core protein glutathione peroxidase 4 (GPX4) via lipid hydroperoxidation, thereby inducing ferroptosis [19]. Recently, ferroptosis was demonstrated to be closely related to selective autophagy [20,21]. Compared with macroautophagy, selective autophagy is associated with a more precise regulation of ferroptosis owing to its dependence on specific receptors. It is expected to become a novel anti-cancer strategy [22]. Under endoplasmic reticulum (ER) stress induced by starvation, hypoxia, SF or LV, loss of redox homeostasis is a common inducing factor for autophagy and ferroptosis [23,24], both of which are key mechanisms of precise quality control in cancer cells [25]. Further exploring the regulatory mechanisms of autophagy and ferroptosis in liver cancer has significant meanings.
In SF-treated HCC cells, for instance, the unfolded protein response (UPR) is activated, leading to a swollen ER, which is then specifically recognized by ER-phagy. ER-phagy is a catabolic process that counteracts the UPR when cells are trapped under ER stress conditions [26]. It is mediated by various receptor proteins, including FAM134B [27], RTN3 [28], TEX264 [29], Sec62 [30], and CCPG1 [31]. They all contain one or more LC3 domains (LC3-interacting regions [LIR]), which are the key elements for selectively recognizing and degrading ER fragments [32,33]. Among them, the FAM134B (family with sequence similarity 134, member B) protein can specifically target the ER sheet through a conserved LIR motif and recruit autophagosomes, degrading and remodeling the ER fragments in a lysosome-dependent manner [34]. Previous studies have confirmed that ER stress is important in regulating ferroptosis [35,36]. Our previous studies showed that FAM134B silencing blocked ER-phagy and activated ferroptosis in HCC cells [37].
Circular RNAs (circRNAs), a group of transcripts characterized by covalently closed continuous loops, are generated from exons or introns of parent genes [38]. According to authoritative research, circRNAs play an essential role in tumors and selective autophagy [39]. Xi Y et al. [40] found that circBCAR3 interacted with miR-27a-3p to upregulate TNPO1, thereby exerting its biological functions in esophageal cancer cells. Liu et al. [41] revealed that circEZH2 could reverse KLF5 post-transcriptional inhibition by sponging miR-217-5p which could accelerate CXCR4-induced epithelial-mesenchymal transition (EMT) of breast cancer (BC). Wang et al. [42] demonstrated that circROBO1 facilitated the carcinogenesis and liver metastasis of BC through the circROBO1/KLF5/FUS feedback loop, which inhibited the selective autophagy of afadin by suppressing the transcription of BECN1. Thus, exploring the roles of circRNAs in autophagy and cancer progression has significant meanings.
To further explore the specific regulatory mechanism of FAM134B-mediated ER-phagy in HCC cells, we evaluated the changes in the expressions of FAM134B and circFAM134B [43] at the transcriptional level and the relationship between them. FAM134B and circFAM134B were highly expressed in HCC cells treated with lenvatinib. This significant change in transcript levels and the relationship between circular and linear transcripts were explored in this study.
Materials and methods
Cell culture and cell transfection
The human HCC cell lines (HepG2 and Huh7) were obtained from the National Infrastructure of Cell Line Resource. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Shanghai BasalMedia Technologies Co., Ltd.) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific [China] Co., Ltd.) and 1% penicillin/streptomycin (Shanghai BasalMedia Technologies Co., Ltd.). The cells were grown in humidified conditions with 5% CO2 at 37°C and tested for mycoplasma contamination by PCR method twice a month.
HCC cells (2 × 105 cells/well) were seeded in 6-well plates and incubated in humidified conditions with 5% CO2 at 37°C overnight. Small interfering RNA (siRNA) and negative control (NC), overexpressing plasmid and negative control (empty pcDNA) were commercially provided by Shanghai Genepharma Co., Ltd. Sequences were shown as follows (5’-3’): si-circFAM134B (Sense: UGA CCG ACC CAG UGA AAG CTT, Antisense: GCU UUC ACU GGG UCG GUC ATT), si-FAM134B (Sense: CCA CUG AGC UCA AGA GAA ATT, Antisense: UUU CUC UUG AGC UCA GUG GTT), si-PABPC4 (Sense: GCC AAA GCC AAG GAA UUC ATT, Antisense: UGA AUU CCU UGG CUU UGG CTT), si-UPF1 (Sense: GCA AGA AGU GGU UCU GCA ATT, Antisense: UUG CAG AAC CAC UUC UUG CTT), NC (Sense: UUC UCC GAA CGU GUC ACG UTT, Antisense: ACG UGA CAC GUU CGG AGA ATT). pcDNA-PABPC4 overexpressing plasmid and empty pcDNA sequences were shown in Supplementary Material. 1. They were transfected into HCC cells using Lipofectamine 3000 (Thermo Fisher Scientific [China] Co., Ltd.) according to the manufacturer’s instructions, respectively. The above cells were harvested at 48 h for further experiments.
RNA extraction and analysis
By using the Trizol reagent (Thermo Fisher Scientific [China] Co., Ltd.), we isolated total RNAs from cancer cells and tissues. The complementary DNA (cDNA) synthesis was performed using a FastKing cDNA First Strand Synthesis Kit (Tiangen Biotech (Beijing) Co., Ltd.). The cDNA was amplified using a SuperReal Fluorescence Quantification Premix Kit (Tiangen Biotech (Beijing) Co., Ltd.). The quantitative real-time polymerase chain reaction (qRT-PCR) assay was then conducted on ABI ViiA 7 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The PCR products of cDNA (RNase R+ and RNase R- groups) and genomic DNA (gDNA) were observed with 1.5% agarose gel electrophoresis. The gene expressions were normalized to GAPDH and calculated by the 2−ΔΔCt method. The primers sequences (Sangon Biotech [Shanghai] Co., Ltd.) were listed as follows (5’-3’): circFAM134B convergent primer (F: CGA TCT TGG GAA GTT ACA TTC CTG; R: CCA GTT TCA GCA GAA CTG AC), circFAM134B divergent primer (F: ACT CCA CAG ACA GAC ACT TC; R: GGT CTT TCA TCT GGT TTG GAA TTG AT), FAM134B primer (F: GTC CTA AGA TTA GCC TCA CGG TTG C; R: GGG TCG GTC AAG ATC ATC AGA AGT G), TPI primer (F: GCT AGA TCC CAA GAT TGC TGT G; R: AGA CAT GCC TTC TCT CTC TGA GTG), GAPDH primer (F: CAG GAG GCA TTG CTG ATG AT; R: GAA GGC TGG GGC TCA TTT).
Treatment of ribonuclease R (RNase R) and actinomycin D (ActD)
The total RNAs from HCC cells were divided into two parts in this experiment. As designed, we had the total RNAs (2 μg) mixed with 2 μl 10 × Reaction Buffer and 5 U RNase R (Guangzhou Geneseed Biotechnology Co., Ltd.) for the first part, and RNase R replaced by DEPC-treated water for the second part. After digestion with RNase R at 37°C for 15 min, the enzyme was inactivated by incubation at 70°C for 10 min. We used qRT-PCR assay to help with the analyses of circFAM134B, FAM134B, and GAPDH compared with the mock group.
HCC cells were cultured in a complete medium with or without 1 μg/ml (or 2.5 μg/ml) of ActD reagent (Merck [China] Technology Co., Ltd.) for 0, 2, 4, 8, 12, 24 h. Then, we isolated total RNAs from cells. The expressions of circFAM134B, FAM134B, and GAPDH were analyzed using qRT-PCR.
Western blot (WB) analysis and protein co-localization
Protein’s isolation, quantification, and WB were performed as previously mentioned. HCC cells were washed, and proteins were extracted by radioimmunoprecipitation lysis buffer (RIPA, Shanghai Beyotime Biotechnology Co., Ltd.). Equal amount of protein samples was separated by SDS-PAGE and transferred to PVDF membranes (Merck [China] Technology Co., Ltd.). The PVDF membranes were incubated with primary antibodies FAM134B (PA5–42647, Thermo Fisher), REEP5 (ab167405, Abcam), Trap-α (ab133238, Abcam), LC3B (ab192890, Abcam), GPX4 (ab125066, Abcam), SLC7A11 (ab175186, Abcam), PABPC4 (ab241616, Abcam), UPF1 (ab109363, Abcam), and GAPDH (ab8245, Abcam) at 4°C overnight, and then incubated with anti-IgG secondary antibody (Thermo Fisher Scientific [China] Co., Ltd.). Following the membranes washed in Tris Buffered Saline with Tween 20 (TBST, Thermo Fisher Scientific [China] Co., Ltd.), ChemiDocTM Imaging System (Bio-Rad Laboratories, USA) was applied to observe the results.
FAM134B and LC3B proteins co-localization in HCC cells was evaluated using a confocal microscope (Wuhan Servicebio Technology Co., Ltd.).
Cell counting kit-8 (CCK-8) assay
Transfected HCC cells were inoculated into 96-well plates at a density of 5 × 103 cells per well. After 0.8 μM lenvatinib (Merck [China] Technology Co., Ltd.) or 10 μM erastin (Merck [China] Technology Co., Ltd.) treatment for 24 h, 10 µl of CCK-8 reagent (Wuhan Elabscience Biotechnology Co., Ltd.) was added to each well. After incubation for 2 h at 37°C, optical density (OD) values at wavelength of 450 nm were assessed using a microplate reader (BioTek, VT, USA).
Measurement of Fe2+, malondialdehyde (MDA), and glutathione (GSH) levels
An Iron Assay Kit (Beijing Applygen Technologies Co., Ltd.) was used to measure the intracellular Fe2+. The concentration of MDA was detected using an MDA Assay Kit (Shanghai Beyotime Biotechnology Co., Ltd.). The concentration of GSH was quantified using a GSH Colorimetric Assay Kit (Wuhan Elabscience Biotechnology Co., Ltd.). All these experimental procedures were performed as per the manufacturer’s guide. The optical density at 550 nm (Fe2+), 532 nm (MDA) and 405 nm (GSH) were measured by the microplate reader.
Measurement of reactive oxygen species (ROS) level
The intracellular ROS level was detected using C11 BODIPY581/591 lipid peroxidation fluorescent probes (Wuhan ABclonal Technology Co., Ltd.) and a flow cytometry (Beckman Coulter, USA). HCC cells were seeded into 6-well plates at a density of 2 × 105 cells/well and incubated with 5% CO2 at 37°C for 48 h. After 0.8 μM lenvatinib or 10 μM erastin treatment for 24 h, cells were incubated with a serum-free medium containing C11 BODIPY581/591 lipid peroxidation fluorescent probes for 1 h at 37°C. After being washed with Phosphate Buffer Solution (PBS) 3 times, analysis of C11 BODIPY581/591 fluorescence was conducted using the flow cytometry.
Tumor xenograft experiment
Male BALB/c nude mice (age: 4–5 weeks, weight: 14–16 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. They were housed under standard conditions. After acclimatization for one week, mice were randomly divided into two groups (n = 5/group): the “LV + NC group” and the “LV + sh-circFAM134B group”. HepG2 cells with si-circFAM134B (or NC) were suspended in 100 μl PBS and respectively injected into each mouse at the left forelimb. 40 μl of si-circFAM134B (or NC) was injected at the tumor site every 4 days. Lenvatinib (10 mg/kg) was dissolved in 200 μl dimethyl sulfoxide (DMSO) and injected intraperitoneally every 3 days. Tumor size (width2 × length/2) was monitored every 3 days. After 4 weeks, the mice were killed, and the tumors were resected for further examination. All animal protocols were approved by the Animal Care and Use Committee of the Shandong Provincial Qianfoshan Hospital. All animal experiments were strictly implemented in compliance with the NIH Guide for the Care and Use of Laboratory Animals.
RNA pull-down assay, silver staining experiment and mass spectrometry analysis
RNA pull-down assay was performed using a Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific [China] Co., Ltd.) and biotin labeled probes (Viagene Biotech, Changzhou, China). The experimental procedure was performed as the manufacturer’s instructions. A Rapid Silver Staining Kit (Shanghai Beyotime Biotechnology Co., Ltd.), mass spectrometry analysis (assisted by Shanghai Personalbio Technology Co., Ltd.), and WB assay were used to analyze enriched proteins. Bio-circFAM134B probe: 5’-GAC TGA TAA TGG GAC CTT CAA CCT TTC AGA A-3’; Bio-circFAM134B-unrelated probe: 5’-TGT GGC TCC TCT GTG TGC TCC TCC TGC AC-3’. Bio-FAM134B probe: 5’-AGC AGC AGG AGG AGG AAG CGC AGG AAG CTG-3’; Bio-FAM134B-unrelated probe: 5’-TGT GGC TCC TCT GGG GAC TCC TCC TGC ACC-3’.
RNA immunoprecipitation (RIP) assay
A EZ-Magna RIP Kit (Merck [China] Technology Co., Ltd.) was utilized to conduct RIP assay following the manufacturer’s instructions. Briefly, HCC cells were lysed in RIP lysis buffer and incubated with RIP buffer including magnetic beads coupled with anti-IgG, anti-PABPC4, or anti-UPF1. After incubation with Proteinase K, the immunoprecipitation was set aside for qRT-PCR analysis.
Luciferase reporter assay
Dual-luciferase reporter vectors carrying the FAM134B 5’UTR, CDS, 3’UTR, 3’UTR-1, 3’UTR-2, 3’UTR-3, 3’UTR-4, 3’UTR-1 Wild and 3’UTR-1 Mutant fragments were constructed using pGL3 vectors (Shanghai Genepharma Co., Ltd.). Lipofectamine 3000 was used to transfect the vectors into cancer cells. Then, co-transfected the pcDNA-PABPC4 overexpressing plasmid with the vectors into cells. Luciferase activities were detected after transfection for 48 h by a Dual-Luciferase Reporter Assay System (Beijing Promega Biotech Co., Ltd.).
Nonsense-mediated mRNA decay (NMD) reporter assay
TPI-SMG5, TPI-4MS2-SMG5, and TPI-4MS2-FAM134B were constructed using GV657/658 vectors (Shanghai Genechem Co., Ltd.), and sequences were shown in Supplementary Material. 2. Lipofectamine 3000 was used to transfect the vectors into cells. Then, TPI-4MS2-SMG5, or TPI-4MS2-FAM134B were co-transfected with MS2-PABPC4 (Shanghai Genechem Co., Ltd.), MS2-GST (Shanghai Genechem Co., Ltd.), pcDNA-PABPC4, or pcDNA into cells. After 48 h of transfection, total RNAs were extracted from cells and we performed the expressions of TPI-4MS2-SMG5 and TPI-4MS2-FAM134B using qRT-PCR.
Statistical analysis
The data was displayed as mean ± standard deviation (SD) from at least three independent experiments and analyzed by GraphPad Prism 9.0 (San Diego, CA, USA). The comparisons between two groups were estimated by unpaired Student’s t-test, while one-way analysis of variance (ANOVA) was applied to compare differences among multiple groups. P values less than 0.05 indicated statistical significance.
Results
circFAM134B specifically regulates FAM134B-mediated ER-phagy
According to the circBase [43], circFAM134B is generated from the 2–6 exons of FAM134B pre-mRNA with genomic location of chr5: 16477,770–16,483,581, whose spliced mature sequence length is 542 bp. Sanger sequencing was conducted to identify back-spliced junction region of circFAM134B which proved the existence of circFAM134B (Figure 1a). Convergent and divergent primers for circFAM134B were used for amplification. circFAM134B was amplified by divergent primer in cDNA but not in gDNA, confirming the presence of circularized FAM134B exons and excluding trans-splicing products. After addition of RNase R, circFAM134B was amplified only by divergent primer in cDNA (Figure 1b). We further detected the stability of circFAM134B and FAM134B with RNase R. circFAM134B was more resistant to RNase R, while FAM134B was significantly decayed (P < 0.05, Figure 1c). In addition, the results of ActD assay revealed the higher stability of circFAM134B than FAM134B mRNA (P < 0.05, Figure 1d). These results indicated the cyclic structure of circFAM134B.
Figure 1.
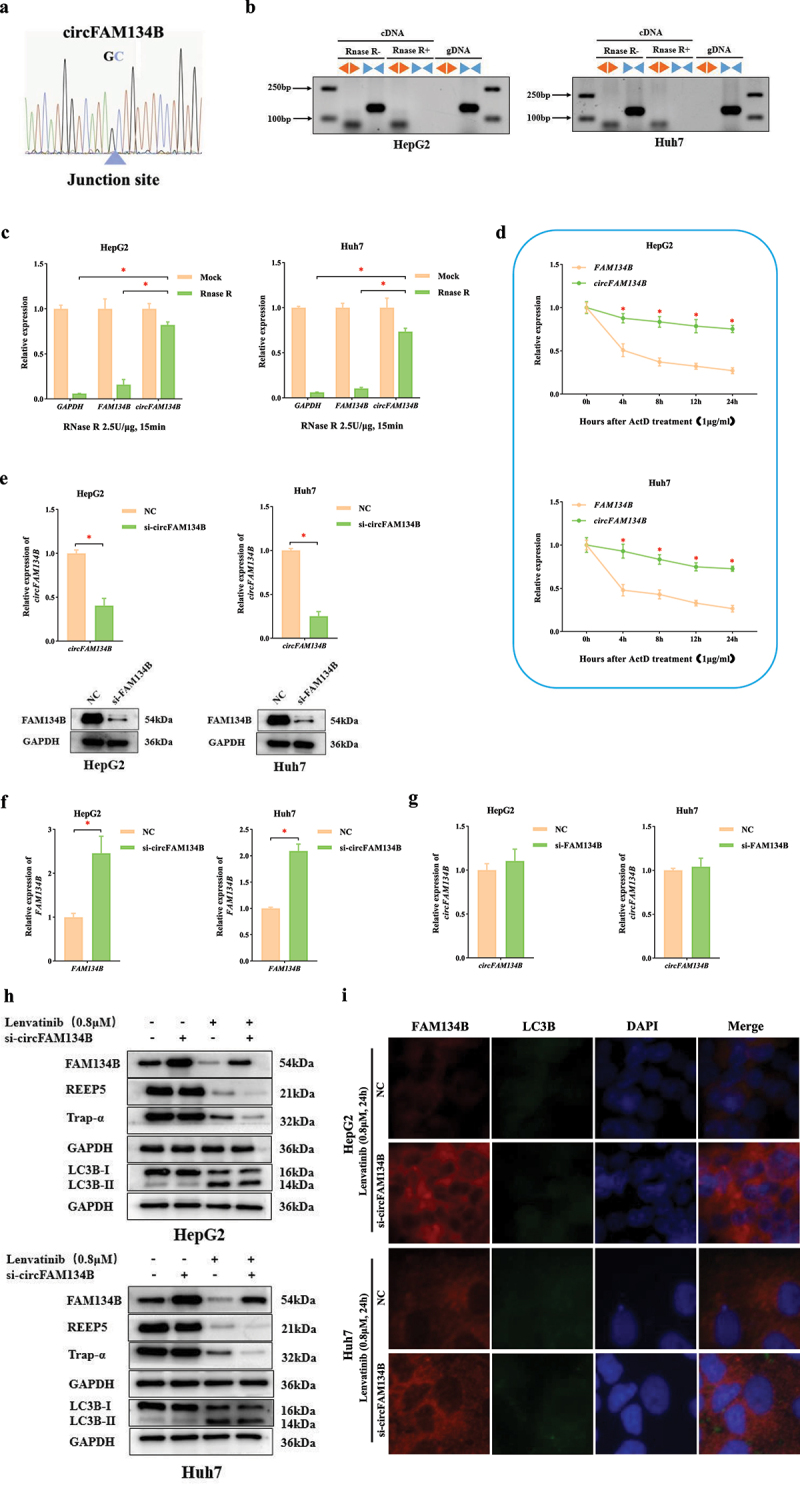
circFAM134B specifically regulates FAM134B-mediated ER-phagy.
a. The sequence around the junction site of circFAM134B was confirmed through Sanger sequencing. The blue triangle indicated the position of the junction site. b. Divergent and convergent primers were used for amplification of circFAM134B in cDNA (RNase R- group and RNase R+ group) and gDNA. Agarose gel electrophoresis visualized the products. c. Following digestion with RNase R (2.5 U/μg, 37°C, 15 min), qRT-PCR was used to measure the changes in circFAM134B and FAM134B. d. The expressions of circFAM134B and FAM134B were identified after ActD (1 μg/ml) treatment for 4 h, 8 h, 12 h, 24 h. e. qRT-PCR was used to perform the level of circFAM134B in HCC cells transfected with si-circFAM134B (or NC). WB assay revealed the protein level of FAM134B in HCC cells transfected with si-FAM134B (or NC). f. The expression of FAM134B in HCC cells transfected with si-circFAM134B (or NC) was identified by qRT-PCR. g. The expression of circFAM134B in HCC cells transfected with si-FAM134B (or NC) was identified by qRT-PCR. h. Effects of si-circFAM134B on ER-phagy levels at 24 h post-treatment with 0.8 μM lenvatinib. i. Co-localization of FAM134B and LC3B in HCC cells, red fluorescence represented FAM134B and green fluorescence represented LC3B. The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05 .
To explore the functions of circFAM134B in HCC cells, we designed siRNAs to effectively silence circFAM134B or FAM134B in HCC cells (Figure 1e). The expression of FAM134B was significantly increased in cells transfected with si-circFAM134B (P < 0.05, Figure 1f), while the si-FAM134B had no significant effects on circFAM134B expression (P > 0.05, Figure 1g). These results revealed that circFAM134B could specifically and negatively regulate the expression of FAM134B, but FAM134B could not regulate the expression of circFAM134B.
After 0.8 μM LV treatment for 24 h, we evaluated the levels of FAM134B, two ER-resident proteins (REEP5 located in the ER tubule and Trap-α located in the ER sheet), and LC3B in HCC cells by WB assay. We confirmed the downregulated expressions of FAM134B, REEP5, Trap-α, and the conversion of LC3B-I to LC3B-II (Figure 1h). Then, the knockdown of circFAM134B caused the elevation of FAM134B and further downregulated expressions of REEP5 and Trap-α, but not affected the conversion of LC3B-I to LC3B-II (Figure 1h). Similar results were obtained in HCC cells treated with erastin (a classical ferroptosis inducer), as shown in Supplementary (Figure 1a) Confocal fluorescence microscopy indicated that the levels of FAM134B and its co-localization with LC3B were significantly increased in HCC cells with si-circFAM134B transfection (Figure 1i). These experimental results illustrated that LV could induce FAM134B-mediated ER-phagy in HCC cells, and circFAM134B regulated FAM134B-mediated ER-phagy without affecting macroautophagy.
circFAM134B affects ferroptosis in HCC cells by regulating FAM134B-mediated ER-phagy
Effects of circFAM134B knockdown on ferroptosis were further explored. CCK-8 assay was used to perform that the effect of LV-induced growth inhibition was weakened after circFAM134B silencing (P < 0.05, Figure 2a). As revealed in Figure 2b,d, si-circFAM134B reduced intracellular Fe2+, MDA levels and increased GSH levels (P < 0.05). Then, we performed the ROS levels using the C11 BODIPY581/591 probe [44,45] which was a fat-soluble ratio type fluorescent probe with low fluorescence artifacts, and good separation of non-oxidized and oxidized spectra. siRNA-mediated loss of circFAM134B in HCC cells also resulted in downregulated basal levels of ROS in HCC cells (Figure 2e). These results indicated that circFAM134B was an important regulator of LV-induced ferroptosis. Meanwhile, GPX4 and SLC7A11 (the core regulators of ferroptosis) protein levels were increased by silencing of circFAM134B (Figure 2f). Similar results were obtained for erastin-treated HCC cells (Supplementary Fig. S1b – f). These results indicated that circFAM134B was a positive ferroptosis regulator in HCC cells.
Figure 2.
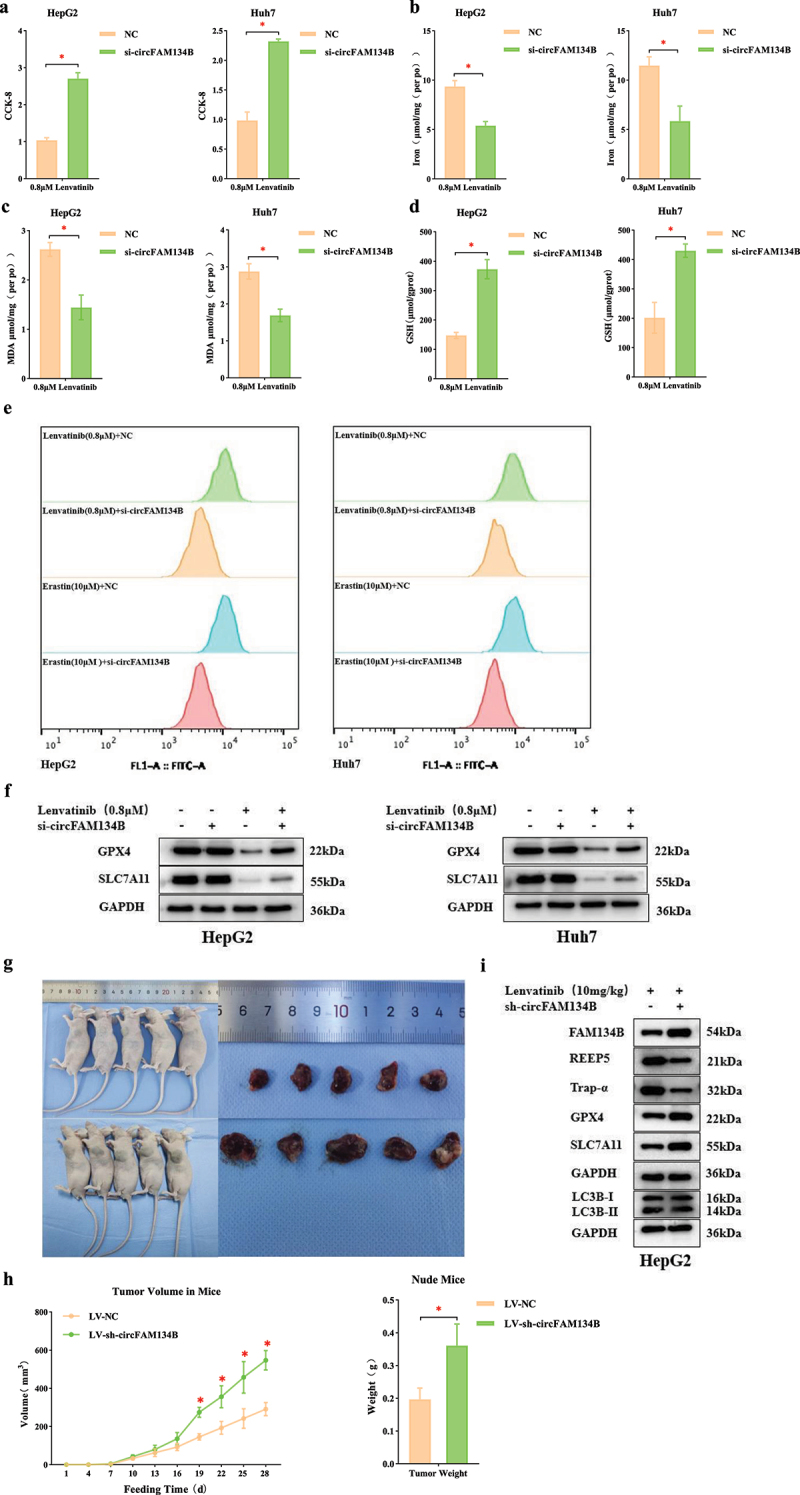
circFAM134B affects ferroptosis in HCC cells by regulating FAM134B-mediated ER-phagy.
a. CCK-8 assay. b. Intracellular Fe2+ levels in HCC cells after transfection of si-circFAM134B were detected with an Iron Assay Kit. c. MDA levels were detected with an MDA Assay Kit. d. GSH levels were measured with a GSH Colorimetric Assay Kit. e. ROS levels were assessed using C11 BODIPY581/591 probes. f. Effects of si-circFAM134B on ferroptosis levels at 24 h post-treatment with 0.8 μM lenvatinib. g. Nude mice and tumor injected with cells transfected with si-circFAM134B (or NC) and lenvatinib treatment. h. The tumor volume and tumor weight of mice in the “LV + sh-circFAM134B” and the control group were shown. i. ER-phagy and ferroptosis in nude mice injected with cells transfected with si-circFAM134B (or NC) and lenvatinib treatment. The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05.
Together, circFAM134B played a key role in FAM134B-mediated ER-phagy, and the inhibition of ER-phagy might activate ferroptosis. These results indicated that targeting circFAM134B could significantly affect FAM134B-mediated ER-phagy, thereby regulating LV-induced ferroptosis in HCC cells.
circFAM134B regulates ER-phagy and ferroptosis in vivo
To further verify the biological roles of circFAM134B in tumor growth, ER-phagy, and ferroptosis in vivo, we established liver xenograft in mice by injection of HepG2 cells which transfected with si-FAM134B (or NC). When the mice were sacrificed, the volume and weight of the tumors in “LV + sh-circFAM134B group” mice were found to be significantly increased than those in the “LV + NC group” mice (P < 0.05, Figure 2g, 2h). qRT-PCR confirmed that circFAM134B was indeed knocked down in HepG2 cells (Supplementary Fig. S1g). WB performed the protein levels of FAM134B, REEP5, Trap-α, LC3B, GPX4, SLC7A11 in the mice’s tumor tissues. In the “LV + sh-circFAM134B group” tumor tissues, FAM134B, GPX4, SLC7A11 were significantly upregulated, while REEP5, Trap-α were significantly downregulated and the conversion of LC3B-I to LC3B-II showed no change compared with those in the control (Figure 2i). These results suggested that circFAM134B knockdown promoted FAM134B-mediated ER-phagy, thereby inhibiting LV-induced ferroptosis in vivo.
Molecular mechanism of circFAM134B-specific regulation of FAM134B
Several studies have shown that homologous circRNAs and linear mRNAs competitively bind to the same proteins, which lead to changes in the expression and translation levels of linear mRNAs [46–48]. To identify the regulatory mechanism of circFAM134B and FAM134B, we designed Bio-circFAM134B probe, Bio-FAM134B probe and the corresponding Bio-unrelated probes. RNA pull-down assay was used to found that in the 70 kDa marker labeling region, Bio-circFAM134B probe and Bio-FAM134B probe could enrich much more proteins (Figure 3a). Subsequently, we performed mass spectrometry analysis of the proteins in the range of 55–100 kDa. Of them, the enrichment levels of 14 types of protein caused both by Bio-circFAM134B probe and Bio-FAM134B probe were much more (Figure 3b). Indeed, the protein that was significantly enriched and had a molecular weight of approximately 70 kDa was only poly (A) binding protein cytoplasmic 4 (PABPC4, Figure 3b).
Figure 3.
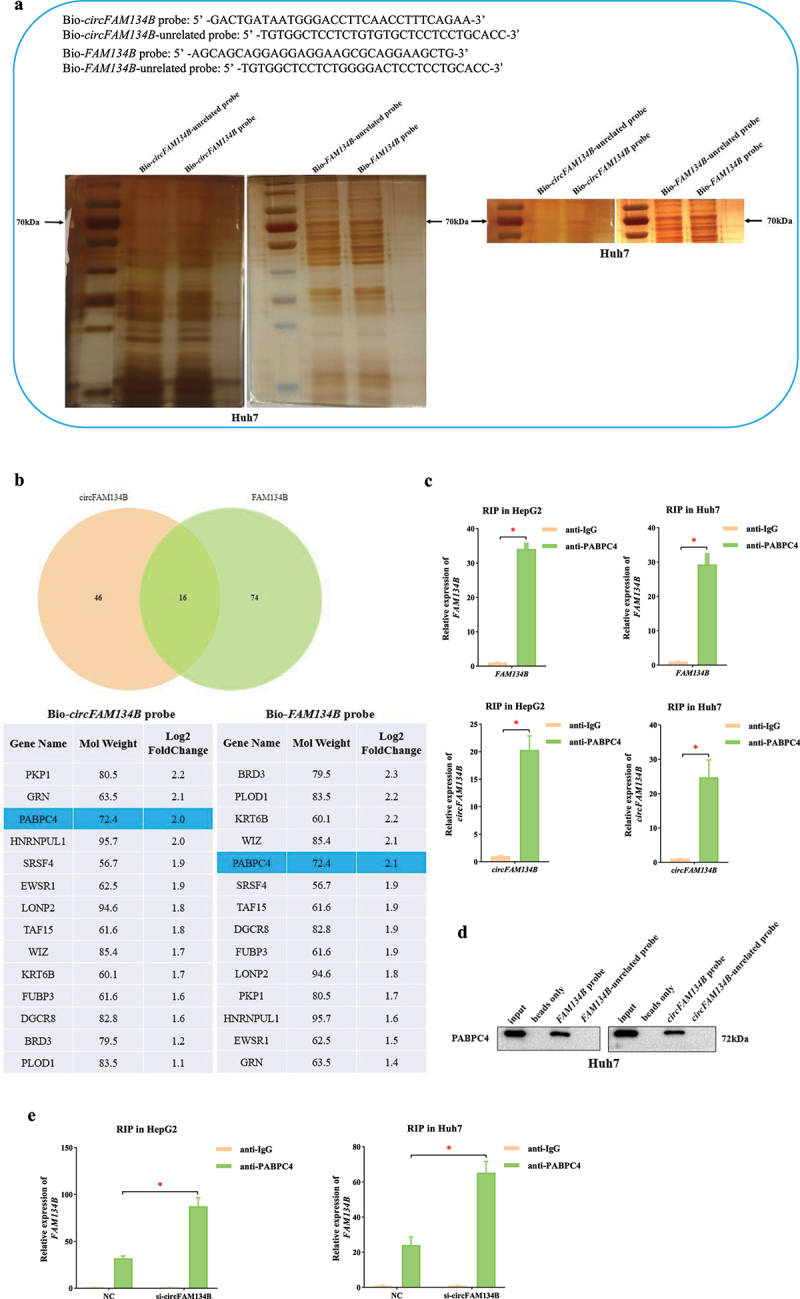
Molecular mechanism of circFAM134B-specific regulation of FAM134B a. RNA pull-down assay and silver staining experiment were performed using Bio-circFAM134B probe and Bio-FAM134B probe. b. mass spectrometry analysis of proteins in the range of 55–100 kDa. c. the RIP assay in HCC cells was used to evaluate the enrichment of circFAM134B and FAM134B mRNA in PABPC4-RNA binding complexes using anti-IgG as a negative control. d. RNA pull-down and WB assays were performed to find that both Bio-circFAM134B probe and Bio-FAM134B probe enriched PABPC4 successfully compared with their control groups. e. the RIP assay was performed that si-circFAM134B elevated the level of FAM134B mRNA enriched by anti-PABPC4.
The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05.
Then, we performed the RIP assay in HCC cells. We found higher enrichment of circFAM134B and FAM134B mRNA in the anti-PABPC4-RNA binding complex using anti-IgG as a negative control (P < 0.05, Figure 3c). Next, RNA pull-down and WB assays were performed to find that both Bio-circFAM134B probe and Bio-FAM134B probe enriched PABPC4 successfully compared with their control groups (Figure 3d). These experimental results indicated that both circFAM134B and FAM134B could effectively bind to PABPC4.
We found that si-circFAM134B elevated the level of FAM134B mRNA enriched by anti-PABPC4 (P < 0.05, Figure 3e). This result further indicated that both circFAM134B and FAM134B had significant binding relationships with PABPC4 and competitively bound to it. Based on the above results, we determined the circFAM134B-PABPC4-FAM134B axis in HCC cells.
PABPC4 is a key protein that regulates the stability of FAM134B mRNA
To further identify the role of PABPC4, we knocked down PABPC4 in HCC cells. When we decreased the expression of PABPC4, there was no significant change of circFAM134B (P > 0.05, Figure 4a), whereas the expression of FAM134B mRNA was vividly downregulated (P < 0.05, Figure 4b). This result implied that PABPC4 might regulate the expression of FAM134B mRNA. Furthermore, we explored whether PABPC4 exerted a regulatory effect on the stability of FAM134B mRNA. After cells transfected with si-PABPC4, we found that the expression of FAM134B mRNA was significantly downregulated by using the transcriptional inhibitor, ActD (P < 0.05, Figure 4c). These experimental results suggested that PABPC4 was a key RNA-binding protein (RBP) that affected the stability of FAM134B mRNA.
Figure 4.
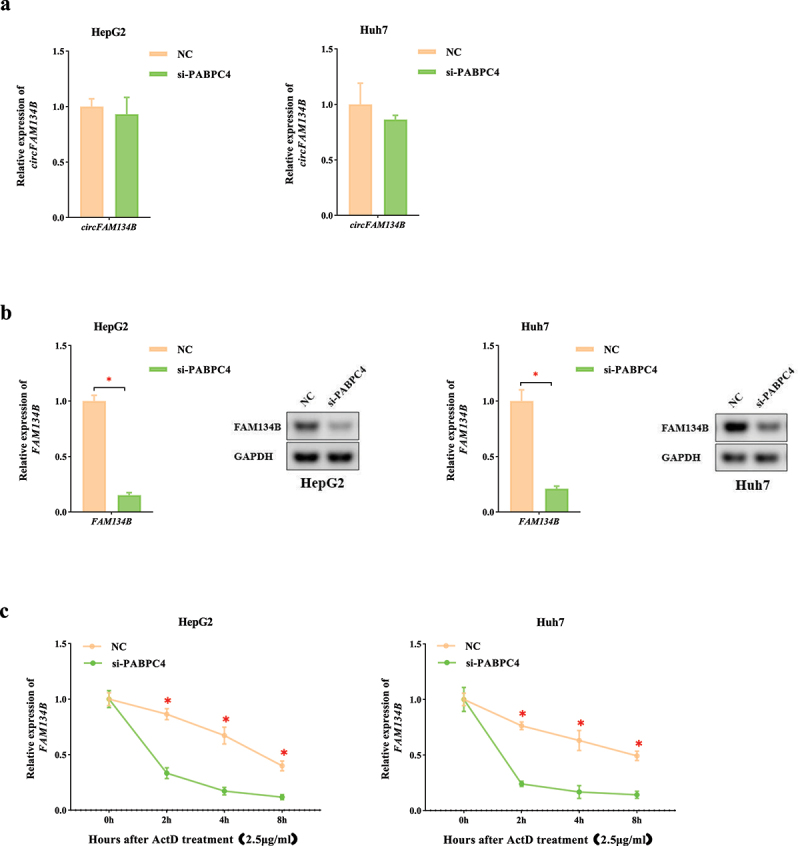
PABPC4 is a key protein that regulates the stability of FAM134B mRNA a. the expression of circFAM134B in HCC cells transfected with si-PABPC4 (or NC) was identified by qRT-PCR. b. the expression of FAM134B in HCC cells transfected with si-PABPC4 (or NC) was identified by qRT-PCR. Agarose gel electrophoresis visualized the products. c. After transfection of si-PABPC4 and ActD (2.5 μg/ml) administration, RNA decay assay was applied to evaluate the stability of FAM134B. The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05.
PABPC4 stabilizes mRNA by inhibition of NMD mechanism
PABPC1 is an important paralogous homolog of PABPC4. PABPC1 can promote mRNA circularization, facilitate ribosome recycling, and suppress nonsense-mediated mRNA decay (NMD) at normal termination codons [49]. Meanwhile, PABPC1 can use the long 3’UTR to regulate mRNA stability via NMD mechanism [50,51]. Another study revealed that mRNA decay was triggered only by removing the last PABPC-binding site [52]. These results reflected the inhibitory effect of PABPC on mRNA decay.
To explore the relationship between PABPC4 and NMD mechanism, we constructed an NMD reporter gene consisting of the triosephosphate isomerase (TPI) open reading frame (ORF) to which we added the 3’UTR of SMG5. The SMG5 mRNA had been previously shown to undergo NMD mediated by its 3’UTR and owing to the presence of the SMG5 3’ UTR, the reporter gene was degraded [53,54]. si-UPF1 (or NC) and TPI-SMG5 were co-transfected into HCC cells. This result verified the effectiveness of TPI-SMG5 in the study of NMD mechanism (P < 0.05, Figure 5a). Then, si-PABPC4 (or NC) and TPI-SMG5 were co-transfected into HCC cells. We found that the level of TPI-SMG5 was significantly lower than that in the NC group (P < 0.05, Figure 5a). This result revealed that PABPC4 could significantly change the level of TPI-SMG5 by antagonizing NMD.
Figure 5.
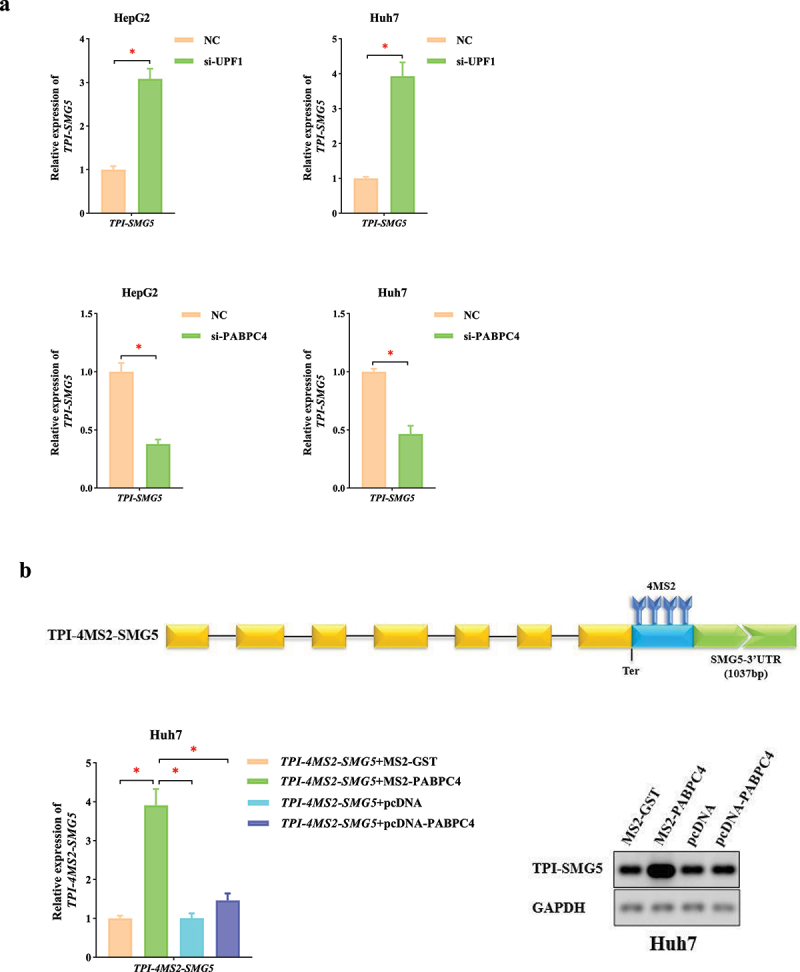
PABPC4 stabilizes mRNA by inhibition of NMD mechanism a. qRT-PCR was used to perform the TPI-SMG5 levels in HCC cells transfected with si-UPF1, si-PABPC4 (or NC). b. the expression of TPI-4MS2-TPI was identified by qRT-PCR. Agarose gel electrophoresis visualized the products. The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05.
To further elucidate the molecular mechanism by which PABPC4 affects the mRNA stability, we inserted four MS2 binding sites downstream from the termination codon of TPI-SMG5 (TPI-4MS2-SMG5), which allowed the MS2-fusion protein to be tethered to a position at the beginning of the 3’UTR. For the operability, accuracy and repeatability of the experiment, we used qRT-PCR combined with agarose gel electrophoresis to analyze the changes of mRNAs abundance. Upon co-expression of MS2-PABPC4, the level of TPI-4MS2-SMG5 significantly increased compared with that of MS2-GST, which served as NC (P < 0.05, Figure 5b). This indicated that PABPC4 was able to counteract the NMD of the reporter mRNA. Then, we co-expressed either pcDNA-PABPC4 together with TPI-4MS2-SMG5 to exclude possible trans-effects of PABPC4 expression. In this case, we observed only slight increases in level of TPI-4MS2-SMG5 when PABPC4 was not directly bound to it (P > 0.05, Figure 5b). The above experimental results showed that PABPC4 antagonized NMD when it was tethered in close proximity downstream of the termination codon.
PABPC4 stabilizes FAM134B mRNA by inhibition of NMD mechanism
To further investigate the molecular mechanism by which PABPC4 affects FAM134B mRNA stability, we preliminarily predicted the possible binding sites of them using the RBP-map database [55]. Under the condition of high Z-score (the standard set here was greater than 3), we found that there were 15 specific binding sites between PABPC4 and FAM134B mRNA. Interestingly, they were all located in FAM134B 3’UTR. Subsequently, FAM134B 5’UTR, CDS, 3’UTR were cloned into pGL3 vectors and introduced into HCC cells along with pcDNA-PABPC4 (or empty pcDNA) to perform dual-luciferase reporter assays. Overexpression of PABPC4 significantly enhanced the relative luciferase activity in HCC cells bearing the 3’UTR sequence (P < 0.05, Figure 6a). Then, according to the predicted specific binding sites using the catRAPID database [56], we divided FAM134B 3’UTR into four segments (starting from the beginning of 3’UTR, the segments were 3’UTR-1, 3’UTR-2, 3’UTR-3, and 3’UTR-4, Figure 6b). Overexpression of PABPC4 boosted the relative luciferase activity in HCC cells transfected with FAM134B 3’UTR-1 (P < 0.05, Figure 6c). Moreover, overexpression of PABPC4 further boosted luciferase activity in HCC cells transfected with the Wild FAM134B 3’UTR-1, but not the Mutant one (P < 0.05, Figure 6d). This finding suggested that PABPC4 might be sensitive to binding sites at the beginning of FAM134B 3’UTR. Based on these results, we suspected that PABPC4 might achieve the purpose of stabilizing FAM134B mRNA by antagonizing NMD.
Figure 6.
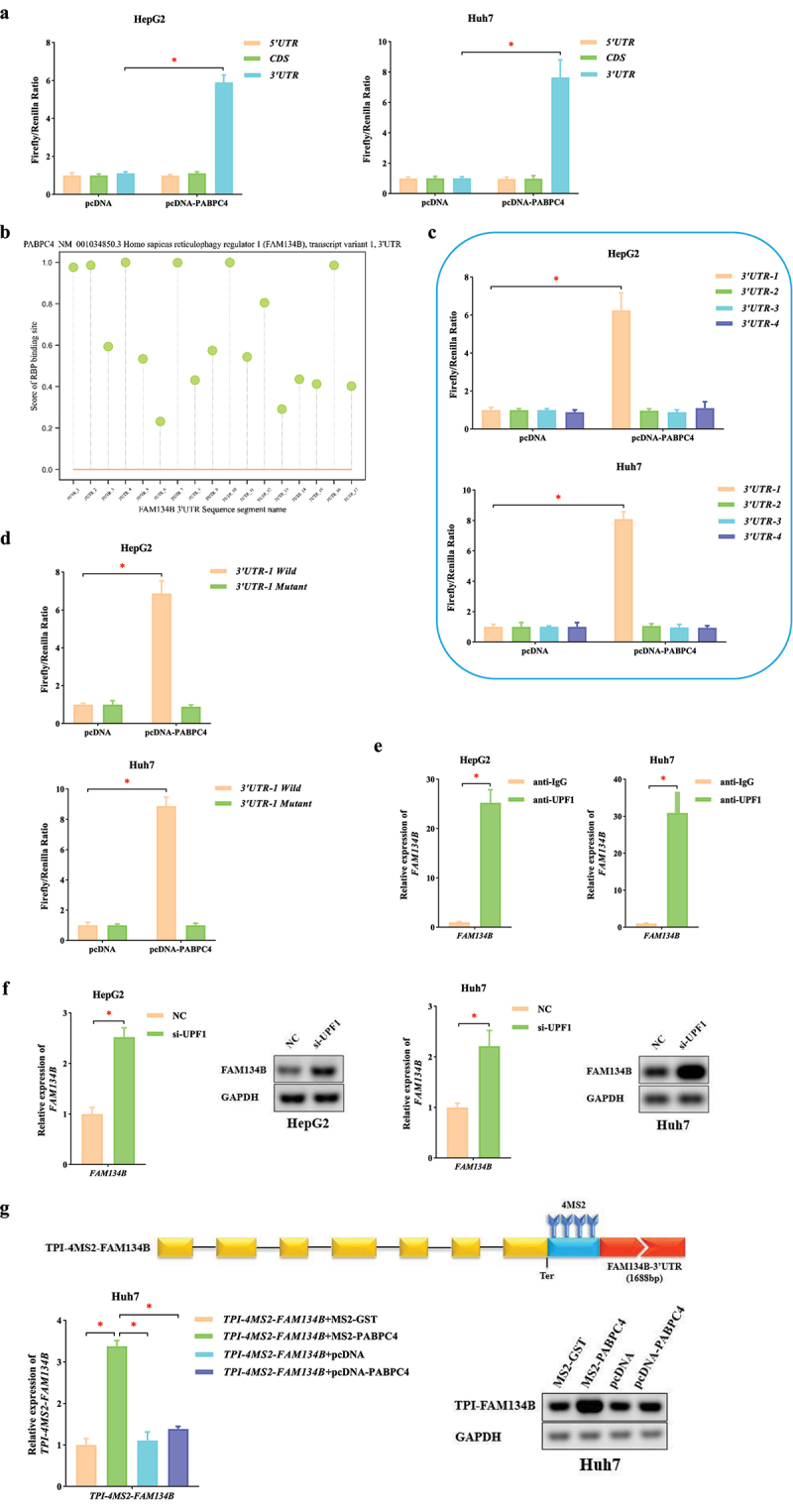
PABPC4 stabilizes FAM134B mRNA by inhibition of NMD mechanism a. the pcDNA-PABPC4 obviously enhanced luciferase activity in HCC cells transfected with FAM134B 3’UTR sequence compared with FAM134B 5’UTR, CDS sequence. b. the score of PABPC4 with binding sites of FAM134B 3’UTR segments. c. luciferase reporter vectors with FAM134B 3’UTR-1 (1 ~ 404bp), 3’UTR-2 (405 ~ 808bp), 3’UTR-3 (809 ~ 1212bp) and 3’UTR-4 (1213 ~ 1688bp) sequences were used to co-expressing with pcDNA-PABPC4 (or empty pcDNA) in HCC cells which were measured the relative luciferase activity (firefly/renilla). d. the pcDNA-PABPC4 obviously enhanced luciferase activity in HCC cells transfected with Wild FAM134B 3′ UTR-1 sequence compared with the Mutant one. e. the RIP assay was used to evaluate enrichment of FAM134B mRNA in UPF1-RNA binding complexes, using anti-IgG as a negative control. f. the expression of FAM134B in HCC cells transfected with si-UPF1 (or NC) was identified by qRT-PCR. Agarose gel electrophoresis visualized the products. g. the expression of TPI-4MS2-FAM134B was identified by qRT-PCR. Agarose gel electrophoresis visualized the products. The error bars represented the standard deviation (SD) of at least three independent experiments. *P < 0.05.
Using qRT-PCR and agarose gel electrophoresis assay, we found that the level of FAM134B was significantly elevated after UPF1 silencing (Figure 6e). Then, we performed the RIP assay and found the higher enrichment of FAM134B mRNA in the anti-UPF1-RNA binding complex using anti-IgG as a negative control (Figure 6f). These results suggested that FAM134B mRNA was significantly regulated by UPF1 (the core factor in the NMD mechanism) and could trigger NMD. Based on the above results, “PABPC4 is a key RBP that affects the stability of FAM134B mRNA” and “PABPC4 can stabilize mRNA by antagonizing NMD”, we illustrated that PABPC4 affected the level of FAM134B mRNA via NMD.
To further determine how PABPC4 regulates FAM134B mRNA via NMD, we constructed a TPI-FAM134B vector gene that replaced SMG5 3’UTR with FAM134B 3’UTR in TPI-SMG5. As mentioned above, we inserted four MS2 binding sites from the termination codon of TPI-FAM134B (TPI-4MS2-FAM134B). After co-transfecting TPI-4MS2-FAM134B with MS2-PABPC4, the level of TPI-4MS2-FAM134B significantly increased compared with NC (P < 0.05, Figure 6g). This result revealed that PABPC4 was able to antagonize NMD (triggered by FAM134B) by binding to the beginning of 3’UTR. Meanwhile, we co-expressed either pcDNA-PABPC4 together with TPI-4MS2-FAM134B to exclude possible trans-effects of PABPC4 expression. We observed only slight increases in the level of TPI-4MS2-FAM134B when PABPC4 was not bound directly to it (P > 0.05, Figure 6g). These above experimental results showed that PABPC4 could antagonize NMD (triggered by FAM134B) when it was tethered at the beginning of 3’UTR. The mechanism that caused the increase in FAM134B mRNA was NMD-specific.
Discussion
Lenvatinib is an oral multi-targeted TKI that has shown antitumor, antiangiogenic, and immunomodulatory activities across multiple cancer types [57–59]. Iseda N et al. [60] found that LV suppressed xCT expression and induced lipid ROS accumulation through FGFR4 inhibition. Accumulated lipid ROS induced ferroptosis in HCC cells. Meanwhile, LV was also revealed that it could promote autophagy in cancer cells. Fernández-Palanca P et al. [61] found that autophagy acted as a crucial mechanism involved in cell adaptation to LV treatment through NRP1 modulation, thus promoting cell resistance development. Roberts JL et al. [62] revealed that LV could interact with the novel histone deacetylase inhibitor entinostat to kill hepatoma cells, in which autophagy, ER stress and death receptor signaling all played important roles. However, there are few detailed reports on the role of LV in regulating selective autophagy and ferroptosis. In this study, we demonstrated that LV could induce FAM134B-mediated ER-phagy and ferroptosis in HCC cells. At present, the specific mechanisms of LV induced ER-phagy and ferroptosis in HCC are still not very clear, further research is necessary.
Developing therapy resistance is a general clinical challenge among the newest molecularly targeted therapies. Authoritative researches revealed that there was a close relationship between non-apoptotic RCD (autophagy, ferroptosis, etc) and therapy-resistant cancer cells [22,63–65]. And then, circRNAs are also associated with targeted drug resistance in HCC cells [66,67] and play an important regulatory role in autophagy [68,69]. These findings indicated the potential roles of circRNAs in the regulation of autophagy and promoted us to explore the roles of circRNAs in ferroptosis. In the present study, our data further demonstrated that circFAM134B knockdown exerted positive effects on FAM134B-mediated ER-phagy and negative effects on LV-induced ferroptosis of HCC in vitro and in vivo. Thus, circFAM134B is a key regulator of FAM134B-mediated ER-phagy and ferroptosis. These findings indicated the potential application of molecular therapy targeting circFAM134B in liver cancer.
Several studies have reported the interaction between circRNAs and RBPs, which offered new ideas for circRNAs research [70,71]. Liu CX et al. [46], Tsitsipatis D et al. [47] found that circRNAs and homologous linear mRNAs could bind to the same proteins competitively, leading to changes in linear mRNAs expression and translation. Li S et al. [48] revealed that circFAM120A prevented FAM120A mRNA from binding to the m6A reader and IGF2BP2 protein, which affected the translation of FAM120A required for proliferation. To explore the precise regulatory network of ER-phagy, we further investigated the specific regulatory mechanism of circFAM134B on FAM134B mRNA. Our present study identified that circFAM134B was confirmed to act as a sponge which competitively interacted with PABPC4, thereby influencing FAM134B mRNA nonsense decay.
PABPC4 belongs to the poly(A) binding protein family, which is a class of highly conserved RBPs. They are commonly found in eukaryotes and contain four RNA recognition motifs at their N-termini. PABPC4 is widely distributed in the cytoplasm and closely related to various metabolic activities of mRNAs in cells, including translation initiation [72] and mRNAs stability [73]. Our study confirmed that PABPC4 was a key RBP that affected the stability of FAM134B mRNA. An important paralogous homolog of PABPC4 is PABPC1. Studies showed that PABPC1 could use the long 3’UTR to regulate mRNA stability via the NMD mechanism and found that NMD would only be triggered by removing the last PABPC binding site [52]. The present study further identified that PABPC4 antagonized NMD when tethered downstream of the termination codon.
According to previous studies, there were several ways about the regulation of circRNAs as follows. Increased MBL leaded to reduce Mbl mRNA splicing by promoting the back-splicing of circMbl and augmented circMbl [74]. circBIRC6 contained miR-34a and miR-145 binding sites that modulated human cell pluripotency and differentiation [75], and circHIPK2 contained one miR124-2HG binding site that modulated astrocyte activation [76]. Across the BSJ site, CKAP5 mRNA bound circZNF609 to facilitate Human antigen R (HuR) binding, which increased the stability and translation of CKAP5 mRNA [77]. Hafez AK et al. [78] found that circHomer1 reduced synaptic translation of Homer1b mRNA by binding to its 3’UTR near the predicted neural Hu protein D (HuD)-binding sites. Abdelmohsen K et al. [79] demonstrated that HuR could positively regulate the translational expression of PABPN1, whereas circPABPN1 regulated the promotive effects of HuR on PABPN1 translation by suppressing HuR binding to PABPN1 mRNA. In summary, the roles of circRNAs in regulating RNA include modulation of transcription, protein sponges, miRNA sponges, translation, and signaling pathways. Interestingly, our research suggested that PABPC4 might be sensitive to binding sites at the beginning of FAM134B 3’UTR. Then, we further demonstrated that PABPC4 stabilized FAM134B mRNA by antagonizing NMD when tethered at the beginning of the 3’UTR. Among the regulatory mechanisms of circRNAs, the NMD mechanism has not been reported.
NMD (Figure 7) is an mRNA degradation pathway that removes transcripts containing premature termination codons (PTCs) [80,81]. There are two main models for NMD in eukaryotic cells. One is the “exon-junction complex (EJC) model”, in which EJC is more than 50–55 nucleotides downstream of the PTC and triggers NMD. Another model proposes that the distance of the stop codon from the poly(A) tail facilitates the targeting of NMD [82]. Amrani N et al. [83] revealed that NMD was triggered by a ribosome’s failure to terminate adjacent to a properly configured 3’UTR. Yepiskoposyan H et al. [84] demonstrated that the median 3’UTR length for the genes stabilized by UPF1 was longer than the median human 3’UTR length. And this reinforced the notion that long 3’UTRs indeed constituted an important NMD-inducing feature of endogenous mRNAs. Thus, the long 3’UTR plays an important role in NMD. Meanwhile, authoritative research showed that NMD profoundly impacted the mutagenic landscape of tumors [80]. Lindeboom et al. [85] found that the last exon and 55-nucleotide boundary rules accounted for almost half of the variation in NMD efficiency across PTCs observed in human tumors. Hu et al. [86] revealed that different tumor types depended on the tumor suppressor genes that most frequently harbored NMD-elicited mutations. Wang et al. [87] discovered that the human PC3 prostate cancer cell line repressed NMD magnitude when grown as a three-dimensional tumor in vivo than as a monolayer in vitro. These results indicate that NMD plays a key role in cancer. However, many outstanding questions remain unanswered. For example, which specific steps of tumorigenesis are affected by NMD? Does NMD promote some stages of tumor development and hinder other stages? Solutions to these problems may bring new hope for human cancer treatment. Our study discovered that PABPC4 could antagonize NMD (triggered by FAM134B) through FAM134B 3’UTR. Thus, our results revealed that targeting NMD on FAM134B might be the potential therapy options in HCC. However, the mechanism by which FAM134B mRNA is recognized as an NMD target should be further explored.
Figure 7.
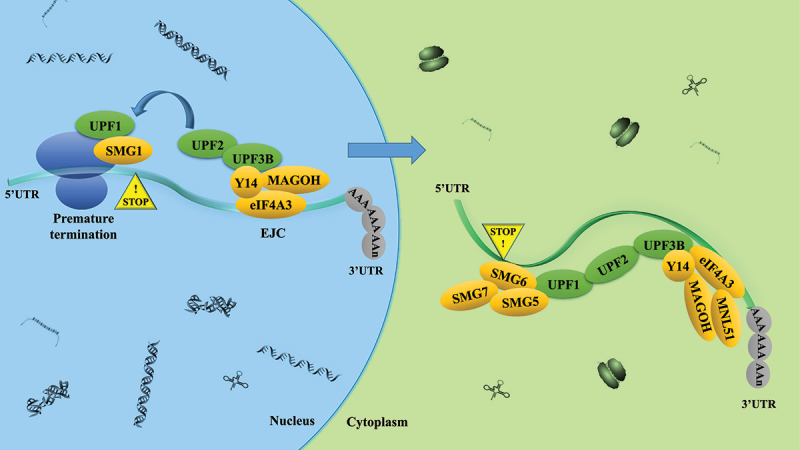
The nonsense-mediated RNA decay (NMD) mechanism UPF1 remains stably bound only to NMD target mRnas. UPF1 also binds to UPF2, that activates its RNA-helicase activity. UPF2 also binds to UPF3B, which, in turn, interacts with the exon-junction complex (EJC) that bind near exon-exon junctions in RNAs after they are spliced in the nucleus. EJC-bound RNAs are then transported to the cytoplasm, where accessory proteins, including the EJC protein MLN51 are recruited. During this chain of events, SMG1 phosphorylates UPF1, since phosphorylated UPF1 binds to the RNA endonuclease SMG6, which is known to cleave NMD target mRnas. Phosphorylated UPF1 also recruits the SMG5-SMG7 heterodimer.
In recent years, RCD-related exploratory work has broadened and deepened our knowledge of cell death patterns, where as many as 12 RCDs have been discovered and have been systematically studied [88]. Ferroptosis is one of the hotspots, and the elucidation of its regulatory pathways provides a new theoretical basis for targeted cancer treatment. However, from the macro-level analysis, the simple regulation of ferroptosis pathway is not an ideal strategy. There are complex interaction networks and linkages among various RCDs. Hence, it has become an inevitable trend to explore the interaction among different RCDs in depth while studying the molecular mechanism of a single RCD. In this research, we proposed that lenvatinib could induce ER-phagy and ferroptosis in HCC cells. circFAM134B was confirmed to competitively interacted with PABPC4, thereby influencing FAM134B mRNA nonsense decay. The results of this featured study are expected to further improve the ferroptosis sensitivity of HCC cells. Targeting circFAM134B-PABPC4-FAM134B axis can provide new ideas and strategies to address the issue of resistance to liver cancer treatment.
Supplementary Material
Funding Statement
This work was financed by grants from the National Natural Science Foundation of China (No. 82172830 and No. 81802414), and Shandong First Medical University (Shandong Academy of Medical Sciences) Youth Science Foundation Cultivation Program (No. 202201-097).
Disclosure statement
No potential conflict of interest was reported by the authors.
Authors’ contributions
Jie Li, Zhiqian Liu, Tao Bi designed this study and wrote the manuscript. Tao Bi performed the experimental work. Qianqian Lu, Xiaohong Pan provided the majority of statistical analysis as well as provided the figures and tables for the manuscript. Fenglin Dong, Yejia Hu, Zongzhen Xu, and Peng Xiu collected a large amount of data for the dataset. All authors have read and approved the final manuscript.
Author consent for publication
Consent for publication was obtained from each author.
Availability of data and materials
The datasets used during the present study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate
This study was approved by the Animal Care and Use Committee of the Shandong Provincial Qianfoshan Hospital.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2023.2249302
References
- [1].Zhu AX, Abbas AR, de Galarreta MR, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med. 2022;28(8):1599–1611. doi: 10.1038/s41591-022-01868-2 [DOI] [PubMed] [Google Scholar]
- [2].Wang H, Cao C, Wei X, et al. A comparison between drug-eluting bead-transarterial chemoembolization and conventional transarterial chemoembolization in patients with hepatocellular carcinoma: a meta-analysis of six randomized controlled trials. J Cancer Res Ther. 2020;16(2):243–249. doi: 10.4103/jcrt.JCRT_504_19 [DOI] [PubMed] [Google Scholar]
- [3].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- [4].Song R, Ma S, Xu J, et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol Cancer. 2023. [Published 2023 Jan 23];22(1):16. doi: 10.1186/s12943-023-01719-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang H, Liu Y, Shen K, et al. A comparison between radiofrequency ablation combined with transarterial chemoembolization and surgical resection in hepatic carcinoma: a meta-analysis. J Cancer Res Ther. 2019;15(7):1617–1623. doi: 10.4103/jcrt.JCRT_503_19 [DOI] [PubMed] [Google Scholar]
- [6].Benson AB, D’Angelica MI, Abbott DE, et al. Hepatobiliary cancers, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19(5):541–565. [Published 2021 May 1]: doi: 10.6004/jnccn.2021.0022 [DOI] [PubMed] [Google Scholar]
- [7].Llovet JM, Pinyol R, Kelley RK, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. doi: 10.1038/s43018-022-00357-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rimassa L, Personeni N, Czauderna C, et al. Systemic treatment of HCC in special populations. J Hepatol. 2021;74(4):931–943. doi: 10.1016/j.jhep.2020.11.026 [DOI] [PubMed] [Google Scholar]
- [9].Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151–172. doi: 10.1038/s41571-021-00573-2 [DOI] [PubMed] [Google Scholar]
- [10].Peng F, Liao M, Qin R, et al. Regulated cell death (RCD) in cancer: key pathways and targeted therapies. Signal Transduct Target Ther. 2022;7(1):286. [Published 2022 Aug 13]: doi: 10.1038/s41392-022-01110-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yao F, Deng Y, Zhao Y, et al. A targetable LIFR-NF-κB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis. Nat Commun. 2021;12(1):7333. [Published 2021 Dec 17]: doi: 10.1038/s41467-021-27452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lei G, Zhuang L, Gan B.. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22(7):381–396. doi: 10.1038/s41568-022-00459-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang M, Wu X, Hu J, et al. COMMD10 inhibits HIF1α/CP loop to enhance ferroptosis and radiosensitivity by disrupting Cu-Fe balance in hepatocellular carcinoma. J Hepatol. 2022;76(5):1138–1150. doi: 10.1016/j.jhep.2022.01.009 [DOI] [PubMed] [Google Scholar]
- [14].Gao R, Kalathur RKR, Coto-Llerena M, et al. YAP/TAZ and ATF4 drive resistance to sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol Med. 2021;13(12):e14351. doi: 10.15252/emmm.202114351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shen M, Li Y, Wang Y, et al. N6-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021;47:102151. doi: 10.1016/j.redox.2021.102151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi: 10.1038/s41571-020-00462-0 [DOI] [PubMed] [Google Scholar]
- [17].Stockwell BR. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185(14):2401–2421. doi: 10.1016/j.cell.2022.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yao X, Li W, Fang D, et al. Emerging roles of energy metabolism in ferroptosis regulation of tumor cells. Adv Sci. 2021;8(22):e2100997. doi: 10.1002/advs.202100997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen X, Kang R, Kroemer G, et al. Organelle-specific regulation of ferroptosis. Cell Death Differ. 2021;28(10):2843–2856. doi: 10.1038/s41418-021-00859-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu J, Liu Y, Wang Y, et al. TMEM164 is a new determinant of autophagy-dependent ferroptosis [published online ahead of print, 2022 Aug 22]. Autophagy. 2022;1–12. doi: 10.1080/15548627.2022.2111635 [DOI] [Google Scholar]
- [21].Chen X, Song X, Li J, et al. Identification of HPCAL1 as a specific autophagy receptor involved in ferroptosis. Autophagy. 2023;19(1):54–74. doi: 10.1080/15548627.2022.2059170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li J, Liu J, Xu Y, et al. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy. 2021;17(11):3361–3374. doi: 10.1080/15548627.2021.1872241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–388. doi: 10.1038/cdd.2014.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hou W, Xie Y, Song X, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–1428. doi: 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu J, Kuang F, Kroemer G, et al. Autophagy-dependent ferroptosis: Machinery and regulation. Cell Chem Biol. 2020;27(4):420–435. doi: 10.1016/j.chembiol.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dikic I. Open questions: why should we care about ER-phagy and ER remodelling? BMC Biol. 2018. [Published 2018 Nov 1];16(1):131. doi: 10.1186/s12915-018-0603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–358. doi: 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- [28].Grumati P, Morozzi G, Hölper S, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6:e25555. Published 2017 Jun 15. DOI: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chino H, Hatta T, Natsume T, et al. Intrinsically disordered protein tex264 mediates ER-phagy. Mol Cell. 2019;74(5):909–921.e6. doi: 10.1016/j.molcel.2019.03.033 [DOI] [PubMed] [Google Scholar]
- [30].Fumagalli F, Noack J, Bergmann TJ, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery [published correction appears in Nat Cell Biol. Nat Cell Biol. 2016;18(11):1173–1184. 19(1):76]. 2016 Dec 23. 10.1038/ncb3423 [DOI] [PubMed] [Google Scholar]
- [31].Smith MD, Harley ME, Kemp AJ, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for er-phagy and pancreatic er proteostasis. Dev Cell. 2018;44(2):217–232.e11. doi: 10.1016/j.devcel.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Grumati P, Dikic I, Stolz A. ER-phagy at a glance.J Cell Sci. 2018. [Published 2018 Sep 3];131(17):jcs217364. DOI: 10.1242/jcs.217364. [DOI] [PubMed] [Google Scholar]
- [33].Chino H, Mizushima N. ER-Phagy: quality control and turnover of endoplasmic reticulum. Trends Cell Biol. 2020;30(5):384–398. doi: 10.1016/j.tcb.2020.02.001 [DOI] [PubMed] [Google Scholar]
- [34].Zhang Z, Zhang L, Zhou L, et al. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol. 2019;25:101047. doi: 10.1016/j.redox.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen Y, Mi Y, Zhang X, et al. Dihydroartemisinin-induced unfolded protein response feedback attenuates ferroptosis via PERK/ATF4/HSPA5 pathway in glioma cells. J Exp Clin Cancer Res. 2019;38(1):402. [Published 2019 Sep 13]: doi: 10.1186/s13046-019-1413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu M, Tao J, Yang Y, et al. Ferroptosis involves in intestinal epithelial cell death in ulcerative colitis. Cell Death Dis. 2020. [Published 2020 Feb 3];11(2):86. doi: 10.1038/s41419-020-2299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu Z, Ma C, Wang Q, et al. Targeting FAM134B-mediated reticulophagy activates sorafenib-induced ferroptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2022;589:247–253. doi: 10.1016/j.bbrc.2021.12.019 [DOI] [PubMed] [Google Scholar]
- [38].Du A, Li S, Zhou Y, et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21(1):109. [Published 2022 May 6]: doi: 10.1186/s12943-022-01575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhong C, Wu K, Wang S, et al. Autophagy-related circRNA evaluation reveals hsa_circ_0001747 as a potential favorable prognostic factor for biochemical recurrence in patients with prostate cancer [published correction appears in Cell death Dis. Cell Death Dis. 2021;12(8):726. 12(8):768]. 2021 Aug 4Published 2021 Jul 22. 10.1038/s41419-021-04015-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xi Y, Shen Y, Wu D, et al. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol Cancer. 2022;21(1):145. [Published 2022 Jul 15]: doi: 10.1186/s12943-022-01615-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu P, Wang Z, Ou X, et al. The FUS/circEZH2/KLF5/feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21(1):198. [Published 2022 Oct 12]: doi: 10.1186/s12943-022-01653-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang Z, Yang L, Wu P, et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21(1):29. [Published 2022 Jan 24]: doi: 10.1186/s12943-022-01498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang F, Xiao Y, Ding JH, et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023;35(1):84–100.e8. doi: 10.1016/j.cmet.2022.09.021 [DOI] [PubMed] [Google Scholar]
- [45].Li Z, Wang C, Dai C, et al. Engineering dual catalytic nanomedicine for autophagy-augmented and ferroptosis-involved cancer nanotherapy [published online ahead of print. Biomaterials. 2022;287:121668. 2022 Jul 5]. DOI: 10.1016/j.biomaterials.2022.121668. [DOI] [PubMed] [Google Scholar]
- [46].Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications [published correction appears in Cell. 2022. [2022 Jun 23]. 185(13):2390] Cell. 185(12):2016–2034. 10.1016/j.cell.2022.04.021. [DOI] [PubMed] [Google Scholar]
- [47].Tsitsipatis D, Grammatikakis I, Driscoll RK, et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 2021;49(3):1631–1646. doi: 10.1093/nar/gkaa1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li S, Li X, Xue W, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. 2021;18(1):51–59. doi: 10.1038/s41592-020-01011-4 [DOI] [PubMed] [Google Scholar]
- [49].Peixeiro I, Inácio Â, Barbosa C, et al. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res. 2012;40(3):1160–1173. doi: 10.1093/nar/gkr820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].JM C, NK G, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12(20):3226–3235. doi: 10.1101/gad.12.20.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ivanov PV, Gehring NH, Kunz JB, et al. Interactions between UPF1, eRfs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27(5):736–747. doi: 10.1038/emboj.2008.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kühn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678(2–3):67–84. doi: 10.1016/j.bbaexp.2004.03.008 [DOI] [PubMed] [Google Scholar]
- [53].Singh G, Rebbapragada I, Lykke-Andersen J, et al. A competition between stimulators and antagonists of upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6(4):e111. doi: 10.1371/journal.pbio.0060111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fatscher T, Boehm V, Weiche B, et al. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA. 2014;20(10):1579–1592. doi: 10.1261/rna.044933.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Paz I, Kosti I, Ares MJ, et al. Rbpmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42(Web Server issue):W361–W367. doi: 10.1093/nar/gku406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Armaos A, Colantoni A, Proietti G, et al. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49(W1):W72–W79. doi: 10.1093/nar/gkab393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schlumberger M, Tahara M, Wirth LJ, et al. Lenvatinib versus placebo in radioiodine-refractory thyroid cancer. N Engl J Med. 2015;372(7):621–630. doi: 10.1056/NEJMoa1406470 [DOI] [PubMed] [Google Scholar]
- [58].Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi: 10.1016/S0140-6736(18)30207-1 [DOI] [PubMed] [Google Scholar]
- [59].Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. [Published 2021 Jan 21]: doi: 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- [60].Iseda N, Itoh S, Toshida K, et al. Ferroptosis is induced by lenvatinib through fibroblast growth factor receptor-4 inhibition in hepatocellular carcinoma. Cancer Sci. 2022;113(7):2272–2287. doi: 10.1111/cas.15378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fernández-Palanca P, Payo-Serafín T, San-Miguel B, et al. Hepatocellular carcinoma cells loss lenvatinib efficacy in vitro through autophagy and hypoxia response-derived neuropilin-1 degradation [published online ahead of print, 2022 Nov 14]. Acta Pharmacol Sin. 2022;10.1038/s41401-022-01021–2. doi: 10.1038/s41401-022-01021-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Roberts JL, Poklepovic A, Booth L, et al. The multi-kinase inhibitor lenvatinib interacts with the HDAC inhibitor entinostat to kill liver cancer cells. Cell Signal. 2020;70:109573. doi: 10.1016/j.cellsig.2020.109573 [DOI] [PubMed] [Google Scholar]
- [63].Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gao W, Wang X, Zhou Y, et al. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. 2022;7(1):196. [Published 2022 Jun 20]: doi: 10.1038/s41392-022-01046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shen LD, Qi WH, Bai JJ, et al. Resibufogenin inhibited colorectal cancer cell growth and tumorigenesis through triggering ferroptosis and ROS production mediated by GPX4 inactivation [retracted in. Anat Rec. 2021;304(2):313–322. Anat Rec (Hoboken). 2021 Dec;304 (12):2892]. doi: 10.1002/ar.24378 [DOI] [PubMed] [Google Scholar]
- [66].Xu J, Ji L, Liang Y, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5(1):298. [Published 2020 Dec 26]: doi: 10.1038/s41392-020-00375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang Y, Nguyen TM, Zhang XO, et al. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRnas. Genome Biol. 2021;22(1):41. [Published 2021 Jan 21]: doi: 10.1186/s13059-021-02263-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang Y, Mo Y, Peng M, et al. The influence of circular RNAs on autophagy and disease progression. Autophagy. 2022;18(2):240–253. doi: 10.1080/15548627.2021.1917131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhang Z, Mou Z, Xu C, et al. Autophagy-associated circular RNA hsa_circ_0007813 modulates human bladder cancer progression via hsa-miR-361-3p/IGF2R regulation. Cell Death Dis. 2021;12(8):778. [Published 2021 Aug 7]: doi: 10.1038/s41419-021-04053-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29(3):481–491. doi: 10.1038/s41418-022-00948-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xue C, Li G, Lu J, et al. Crosstalk between circRnas and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct Target Ther. 2021;6(1):400. [Published 2021 Nov 24]: doi: 10.1038/s41392-021-00788-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cragle CE, MacNicol MC, Byrum SD, et al. Musashi interaction with poly(A)-binding protein is required for activation of target mRNA translation. J Biol Chem. 2019;294(28):10969–10986. doi: 10.1074/jbc.RA119.007220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jiang X, Wang G, Liu Y, et al. A novel long non-coding RNA RP11-286H15.1 represses hepatocellular carcinoma progression by promoting ubiquitination of PABPC4. Cancer Lett. 2021;499:109–121. doi: 10.1016/j.canlet.2020.11.038 [DOI] [PubMed] [Google Scholar]
- [74].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- [75].Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149. [Published 2017 Oct 27]: doi: 10.1038/s41467-017-01216-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Huang R, Zhang Y, Han B, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG [published correction appears in autophagy. Autophagy. 2017;13(10):1722–1741. 2022 Jan;18(1):234] [published correction appears in Autophagy. 2020 Aug;16(8):1553] [published correction appears in Autophagy 2020.Nov 16(11):[2117-2118]. doi: 10.1080/15548627.2017.1356975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rossi F, Beltran M, Damizia M, et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol Cell. 2022;82(1):75–89.e9. doi: 10.1016/j.molcel.2021.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hafez AK, Zimmerman AJ, Papageorgiou G, et al. A bidirectional competitive interaction between circHomer1 and Homer1b within the orbitofrontal cortex regulates reversal learning. Cell Rep. 2022;38(3):110282. doi: 10.1016/j.celrep.2021.110282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tan K, Stupack DG, Wilkinson MF. Nonsense-mediated RNA decay: an emerging modulator of malignancy. Nat Rev Cancer. 2022;22(8):437–451. doi: 10.1038/s41568-022-00481-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cho H, Abshire ET, Popp MW, et al. AKT constitutes a signal-promoted alternative exon-junction complex that regulates nonsense-mediated mRNA decay. Mol Cell. 2022;82(15):2779–2796.e10. doi: 10.1016/j.molcel.2022.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sato H, Singer RH. Cellular variability of nonsense-mediated mRNA decay. Nat Commun. 2021. [Published 2021 Dec 10];12(1):7203. doi: 10.1038/s41467-021-27423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Amrani N, Ganesan R, Kervestin S, et al. A faux 3’-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432(7013):112–118. doi: 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- [84].Yepiskoposyan H, Aeschimann F, Nilsson D, et al. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17(12):2108–2118. doi: 10.1261/rna.030247.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48(10):1112–1118. doi: 10.1038/ng.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hu Z, Yau C, Ahmed AA. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay.Nat Commun. 2017. [Published 2017 Jun 26];8(1):15943. DOI: 10.1038/ncomms15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang D, Zavadil J, Martin L, et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31(17):3670–3680. doi: 10.1128/MCB.05704-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347–364. doi: 10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding authors on reasonable request.


