ABSTRACT
Brucellosis is a major threat to public health and animal husbandry. Several in vivo vertebrate models, such as mice, guinea pigs, and nonhuman primates, have been used to study Brucella pathogenesis, bacteria-host interactions, and vaccine efficacy. However, these models have limitations whereas the invertebrate Galleria mellonella model is a cost-effective and ethical alternative. The aim of the present study was to examine the invertebrate G. mellonella as an in vivo infection model for Brucella. Infection assays were employed to validate the fitness of the larval model for Brucella infection and virulence evaluation. The protective efficacy of immune sera was evaluated by pre-incubated with a lethal dose of bacteria before infection. The consistency between the mouse model and the larval model was confirmed by assessing the protective efficacy of two Brucella vaccine strains. The results show that G. mellonella could be infected by Brucella strains, in a dose- and temperature-dependent way. Moreover, this larval model can effectively evaluate the virulence of Brucella strains in a manner consistent with that of mammalian infection models. Importantly, this model can assess the protective efficacy of vaccine immune sera within a day. Further investigation implied that haemolymph played a crucial role in the protective efficacy of immune sera. In conclusion, G. mellonella could serve as a quick, efficient, and reliable model for evaluating the virulence of Brucella strains and efficacy of immune sera in an ethical manner.
KEYWORDS: Galleria mellonella, Brucella, infection model, virulence evaluation, sera efficacy evaluation, rapid evaluation model
GRAPHICAL ABSTRACT
Introduction
Brucellosis is a zoonotic infectious disease caused by Brucella spp. It is viewed as a major threat to public health and animal husbandry, with annually about 500 thousand new reported cases worldwide, involving more than 170 countries and regions [1–5]. Vaccines are the most cost-effective means to prevent and control brucellosis. Commercial veterinary live attenuated vaccines, such as Brucella abortus S19/A19, Brucella abortus RB51, Brucella melitensis Rev.1, Brucella melitensis M5, and Brucella Suis S2, have played a substantial role in the prevention and control of brucellosis in livestock [6,7], but they still have deficiencies, such as residual virulence to the host, interfering conventional serological tests, and resulting infection in humans [8–12] (Table 1). Till now, the Brucella abortus live attenuated vaccine 104M is the only human Brucella vaccine approved by the Chinese Food and Drug Administration, but it is not suitable for mass vaccination due to scratch inoculation and residual virulence [13] (Table 1). Rodents are the most commonly used animal models for Brucella infection, virulence comparison, and vaccine evaluation, with bacterial load and histopathological changes in the spleen defined as main indicators [6,8,22,23]. Nonetheless, the evaluation process is time-consuming, costly, requires specialized animal housing, and is ethically challenging. Therefore, the establishment of a simple and time-saving alternative in vivo infection and immune model of Brucella could not only greatly reduce the use of vertebrates but also speed up the evaluation process.
Table 1.
Commercial Brucella attenuated vaccines and their virulence degree.
| Vaccine name | Origin | Degree of Virulence |
|---|---|---|
| S19/A19 | Brucella abortus | Cause abortion/orchitis [6,7] Virulent to humans [14,15] |
| RB51 | Brucella abortus | Virulent to humans [16]; |
| Rev.1 | Brucella. melitensis | More virulent than S19 [15] Cause abortion in pregnant animals [17,18]; Virulent to humans [19] |
| M5 | Brucella. melitensis | Virulent to humans [10] |
| S2 | Brucella. suis | Less virulent than S19 and Rev-1 [20,21] |
| 104M | Brucella abortus | Slightly virulent to humans [16] |
Galleria mellonella larvae, also known as wax worms, are invertebrates with a mammal-like innate immune system that consists of cellular and humoral immune responses [24,25]. In contrast to traditional models, G. mellonella larvae are an inexpensive and easily acquired resource that can be handled without difficulty, does not require feeding or specific caging, and thus could serve as a valuable tool for investigating microbial infections. Importantly, they can survive at 37°C, which is the optimum temperature for human pathogens. In addition, this model is ethically acceptable and does not require ethical approval. As a result, this model has been widely used in the evaluation of pathogen virulence, toxicity of chemicals, and rapid screening of novel antibacterial agents [24,26–30].
In this study, we described a G. mellonella larval infection and immune model for Brucella and validated its potential to evaluate the virulence of Brucella strains and the efficacy of immune sera in a fast, high-throughput, and ethical manner.
Materials and Methods
Bacterial strains and culture conditions
All strains were stored at −80°C in tryptone soya broth (TSB; Oxoid) containing 15% glycerol (SCR). Bacteria were cultured in TSB at 37°C with 220 rpm until the mid-log phase was reached. The samples were serially diluted and plated on tryptone soya agar (TSA, Oxoid) for CFU determination. B. abortus A19, A19ΔVirB12 and B. suis S2 were obtained from Tecon Biology, and B. abortus 104M was obtained from Lanzhou Institute of Biological Products. The 104M:Omp19 overexpression mutant was generated as previously described [31]. The 104MΔOmp19 deletion mutant was generated by amplification of the upstream and downstream flanking regions and the kanamycin resistance gene (kanR gene) using the primers Omp19-up-F, Omp19-up-R, Omp19-down-F, Omp19-down-R, kanR-F, and kanR-R (Additional file 1). Subsequently, the upstream, kanR and downstream regions of Omp19 were ligated by overlapping PCR, and the ligated product was inserted into pEASY-Blunt 3 vector (Transgen Biotech). The resulting vector was transformed into the genome of strain 104M by electroporation at 625 V for 10 ms and then transferred to selective TSA plates containing 100 μg/ml kanamycin..
G. mellonella larvae
Last-instar antibiotic-free G. mellonella larvae were purchased from Tianjin Huiyude Biotechnology Ltd. and were kept for one day before infection, without feeding during experimentation. Larvae with uniform colour, no markings, and high motility were deemed healthy and were used in subsequent experiments.
G. mellonella larvae infection assay and virulence evaluation assay
Brucellacultures were centrifuged for 5 min at 5 000 g. Bacteria pellets were washed once and resuspended in 1× PBS buffer (Gibco) to the desired density (5 × 104 CFU/ml to 5 × 109CFU/ml). To inactivate the bacteria, mid-log phase strains were suspended in PBS buffer (Gibco) heat inactivated at 100 °C for 10 min, or suspended in 4% paraformaldehyde (PFA, Leagene) for 1 h. Inactivity was confirmed by plating 100 μl of the suspension onto TSA plates and incubating for three days. A 25 μl suspension of bacteria (1.25 × 103 CFU/larva to 1.25 × 108 CFU/larva) was injected into G. mellonella larvae through the lower left proleg using a sterile 29 G insulin syringe (BD). Control groups were left untreated or injected with 25 μl of sterile PBS (Gibco) or pooled sera from PBS-treated groups of mice (PBS sera). After infection, larvae were kept at 25°C or 37°C in a lidded 9-cm Petri-dish in the dark, monitored, and scored daily according to the “Health Index Scoring System (HISS) (Figure 1a) [10]. ” for up to 5 days. The larvae were determined to be dead when they did not respond to stimulation. Pupae were excluded from the analysis.
Figure 1.
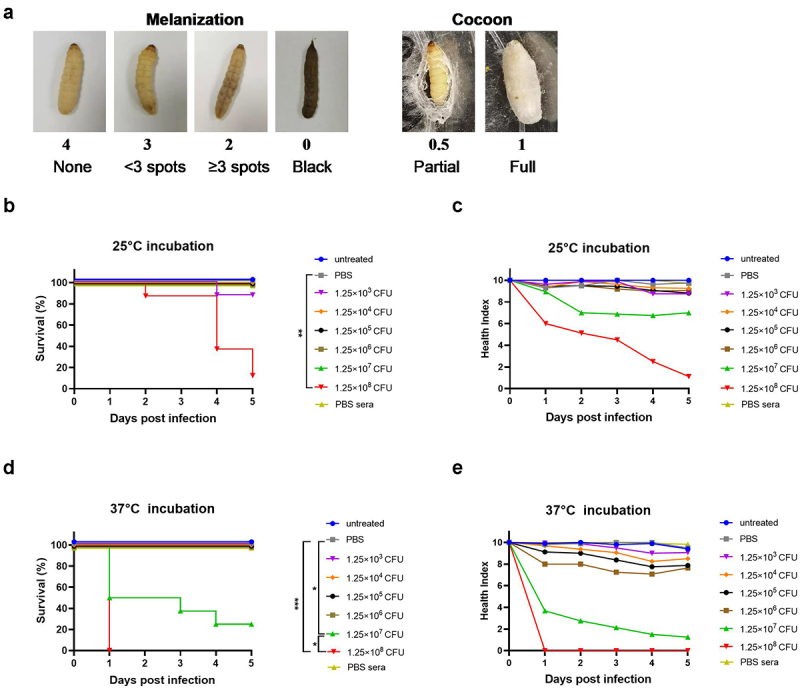
Establishment of the G. mellonella model of Brucella infection. G. mellonella larvae were injected with 25 μl bacteria suspension, incubated at 25°C or 37°C and scored daily according to the health index scoring system. a) Representative photographs of melanization scores and cocoon scores. b) Effect of different inoculum doses of Brucella strain A19 on G. mellonella larval survival at 25°C. c) Health index scores of larvae infected with different doses of Brucella strain A19 at 25°C. d) Effect of different inoculum doses of Brucella strain A19 on G. mellonella larval survival at 37°C. e) Health index scores of larvae infected with different doses of Brucella strain A19 at 37°C. Significant differences between the Kaplan-Meier curves were identified with the log-rank test (GraphPad Prism). *P< 0.05,**P < 0.01, ***P < 0.001.
Mouse immunization, sera collection and mouse infection
Six-to eight-week-old female BALB/c mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were immunized subcutaneously with 1 × 105 CFU of strain 104M, strain 104MΔOmp19 or PBS on day 0, and infected with 1 × 108 CFU of strain A19 intraperitoneally on day 28. Sera were collected before infection, aliquoted, and stored at −80°C until use. On the 7th day post infection, the mice were euthanized, and their spleens were harvested. Mouse spleens from each group were fixed in 4% PFA, or homogenized, serially diluted, and spread on TSA plates for CFU determination.
Histopathology
Larvae were fixed in 4% PFA on day 1 and day 5 post infection, and mouse spleens were fixed on the 7th day post infection. Fixed samples were embedded and sectioned, followed by dewaxing and hydration. The tissue sections were stained using the Hematoxylin and Eosin (H&E) staining kit (Beyotime) or Brucella staining kit (Leagene), according to the manufacturer’s instructions. Subsequently, dehydration, transparency, and neutral resin sealing were performed.
Evaluation of the protective efficacy of immune sera
B. abortus A19 was suspended in PBS buffer containing 4%-50% (v/v) pooled normal sera or heat-inactivated (56°C, 30 min) sera from mice immunized with strain 104M (hereafter referred to as 104M sera), strain 104MΔOmp19 (hereafter referred to as 104MΔOmp19 sera), or PBS control (hereafter abbreviated as PBS sera) and incubated at 37°C for 30, 45, or 60 min, evenly mixed every 10 min. Larvae were injected with a 25 μl mixture containing 1.25 × 108 CFU of bacteria, incubated at 37°C and scored daily according to HISS.
Depletion of G.mellonella hemocytes
Dichloromethylene-bisphosphonate (clodronate) has been reported to deplete the haemocytes of G. mellonella and Anopheles gambiae mosquitos [30,32]. Liposomes containing 5 mg/ml clodronate (CDLip) and control liposomes (Lip) in PBS were purchased from Liposoma Research and used in the assays. G. mellonella larvae were injected with 5 µL of Lip or CDLip in the last left proleg. After 24 h, the larvae were injected with 20 µL of Brucella A19 suspension or A19-sera pre-incubated mixture containing 1.25 × 108 CFU of bacteria in the last right proleg, and larval survival was monitored over a 5-day period.
Effect of priming G.mellonella with a non-lethal dose of Brucella
G. mellonella larvae were injected in the last left proleg with 5 µL of bacterial suspension containing 1.25 × 105 CFU of live or fixed Brucella strains. After 24 h, a lethal dose of 10 µL of Brucella A19 was used to infect the larvae in the last right proleg. Larval survival and health status were evaluated for 5 days following infection.
Statistical analyses
Data were analysed for statistical significance using GraphPad Prism Version 9. Significant differences between the Kaplan-Meier curves were identified with the log-rank test (GraphPad Prism). *P < 0.05, **P < 0.01, ***P < 0.001. Significant differences in the bacterial load of mouse spleens were identified by one-way ANOVA (GraphPad Prism). *P < 0.05, ***P < 0.001, ****P < 0.0001.
Results
G. mellonella larval survival was Brucella dose-dependent and temperature-dependent
First, we determined the virulence of B.abortus A19 to G. mellonella larvae at 25°C and 37°C using 37°C cultured strain A19, which were the optimum survival temperatures for larvae and bacteria, respectively. To begin with, larvae were injected with a series of doses of strain A19 at 25°C to verify whether Brucella could establish infection in G. mellonella larvae. Untreated larvae and larvae injected with PBS buffer both showed 100% survival when incubated at this temperature over a 5-day period, demonstrating that neither needle injection nor PBS buffer had any influence on larval survival. However, larvae infected with 1.25 × 108 CFU of bacteria showed significantly decreased survival (P < 0.01), with less than 20% survival on day 5; however, at lower infection doses, larvae showed almost 100% survival (Figure 1b). We also employed a health index scoring system to assess the health status of larvae, which enabled the observation of general health status and slight differences in larval activity, cocoon formation, melanization, and survival (Figure 1a) [24]. The daily health index scores of larvae infected with 1.25 × 108 CFU dropped with time, eventually falling below 2 on day 5. The daily scores of larvae infected with 1.25 × 107 CFU were always above 6, although they decreased gradually. On the contrary, the health index scores of groups treated with doses lower that 1.25 × 107 CFU were always above 8 (Figure 1c).
We subsequently conducted the larval infection assay at 37°C, as this was the optimal growth temperature for Brucella. As expected, both untreated larvae and larvae injected with PBS buffer also exhibited 100% survival at 37°C. Furthermore, larval survival was also dose-dependent at this temperature, with higher concentrations correlating with reduced survival. Larvae infected with less than 1.25 × 107 CFU of strain A19 all survived, while at higher infection doses, a concentration-dependent pattern on both survival and health index scores was observed. All larvae died within 1 day and typically scored 0 or 2 when infected with 1.25 × 108 CFU of strain A19, whereas 50% mortality was reached on day 1 with 1.25 × 107 CFU of bacteria (Figure 1d,e). Similar results were obtained using 25°C cultured strain A19 (Fig. S1).
Overall, larval survival depends on both the infection dose of Brucella and the post infection incubation temperature. Consequently, a standard inoculum of 1.25 × 108 CFU/larva and a standard temperature of 37°C were determined for all subsequent experiments, which killed all larvae consistently and reproducibly within a day.
G. mellonella developed Granuloma-like structures in response to non-lethal Brucella infection
To examine histopathological changes, larvae were infected with 1.25 × 105 CFU or 1.25 × 108 CFU of A19 and processed at 1-day and 5-day post infection for H&E staining or Koster’s staining. Larvae treated with the same volume of PBS were used as controls. Compared to the PBS control (Figure 2a), larvae infected with a non-lethal dose of A19 on day 1 and day 5 showed granuloma-like structures formed by the aggregation of haemocytes with melanin deposition, but no evidence of severe damage to the adipose body or total tissue structure (Figure 2b,c). Conversely, larvae infected with a lethal dose of A19 on day 1 showed extensive tissue destruction with severely disseminated basophils (Figure 2d).
Figure 2.
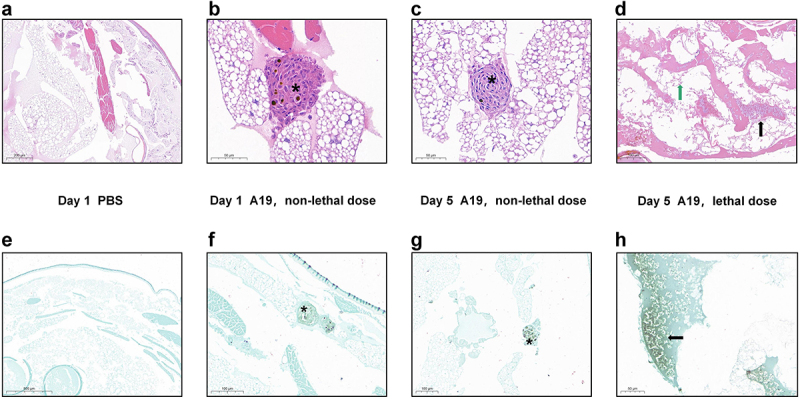
Histopathology examination of G. mellonella infected with Brucella strain A19. Larvae were infected with Brucella strain A19 (1.25 × 105 or 1.25 × 108 CFU) and histopathology were examined at 1- and 5-day post infection on fixed larval sections following H&E staining or Koster’s staining. a) PBS control, H&E staining larval section on day 1. Normal histological structures, no haemocyte reaction or melanization was observed. b) Infected, non-lethal dose (1.25 × 105 CFU), H&E staining larval section on day 1. Granuloma-like structures formed by haemolymph were observed. Asterisks represented the granuloma-like structures formed by the aggregation of haemocytes. c) Infected, non-lethal dose (1.25 × 105 CFU), H&E staining larval section on day 5. d) Infected, lethal dose (1.25 × 108 CFU), H&E staining larval section on day 1. Extensive destruction of the adipose body with scattered melanin were observed. The black arrow represented disseminated Brucella. The green arrow represented damaged adipose body. e) PBS control, Koster’s staining larval section on day 1. No bacteria were observed in the section. f) Infected, non-lethal dose (1.25 × 105 CFU), Koster’s staining larval section on day 1. Granuloma-like structure containing individual Brucella or colonies of Brucella were observed. g) Infected, non-lethal dose (1.25 × 105 CFU), Koster’s staining larval section on day 5. Brucella-positive components were observed within the nodules of haemocytes. h) Infected, lethal dose (1.25 × 108 CFU), Koster’s staining larval section on day 1. Large numbers of Brucella were distributed in the haemocoel cavity.
To further confirm the distribution of bacteria, Koster’s staining was performed. Brucella strains were stained magenta using this specific staining method, while non-Brucella strains and tissue structures were stained green. No Brucella-positive components were detected in the sections of the PBS control group (Figure 2e), whereas Brucella-positive elements were observed within the nodules of haemocytes in the sections of larvae infected with the non-lethal dose of strain A19 (Figure 2f,g). Moreover, in the sections of larvae infected with the lethal dose, large numbers of individual bacteria were stained magenta, indicating severe disseminated infection in this group (Figure 2h).
G. mellonella as a model for rapid evaluation of Brucella strain virulence
To further investigate whether killing G. mellonella requires live bacteria, G. mellonella larvae were injected with a dose of 1.25 × 108 CFU of live strain A19, live strain 104M, or with the same dose of heat-inactivated bacteria. Larvae infected with both live strains died by day 1, whereas those infected with heat-inactivated strains survived at all time points (Figure 3a), with average health score of 8.1 (A19-HI) and 8.4 (104M-HI) on day 5 (Figure 3b), indicating that only live strains had a significant effect on larval survival at this infection dose.
Figure 3.
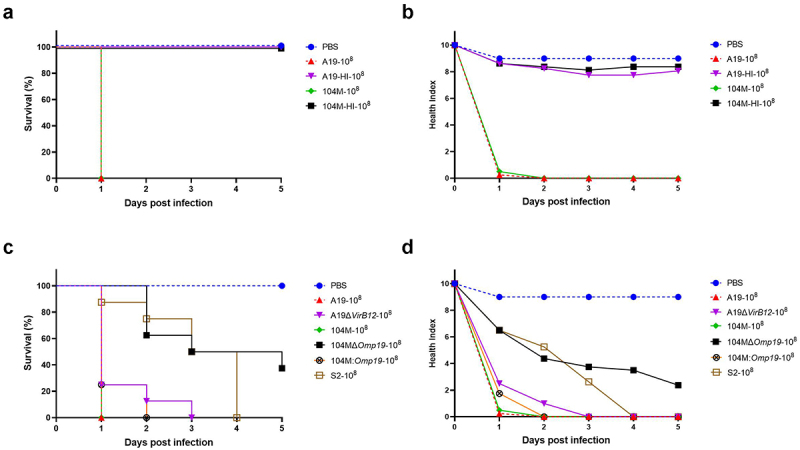
G. mellonella as a model for rapid evaluation of Brucella strain virulence. Groups of larvae were infected with 1.25 × 108 CFU of live or heat-inactivated Brucella strain A19, strain 104M, strain 104MΔOmp19 deletion mutant, strain 104M:Omp19 overexpression mutant or strain S2, incubated at 37°C and monitored for 5 days. a) Kaplan-Meier curves representing daily percentage of survived larvae infected with live or heat-inactivated Brucella strains. b) Daily health index scores of larvae infected with live or heat-inactivated Brucella strains. c) Kaplan-Meier curves representing daily percentage of survived larvae infected with Brucella strains. d) Daily health index scores of larvae infected with Brucella strains.
We subsequently evaluated the presence of virulence factors on larval survival by inoculating larvae with 1.25 × 108 CFU of different Brucella strains or their deletion/over-expression mutants. Brucella abortus A19 and A19ΔVirB12 are both commercially available live vaccines that show similar virulence in mice [33]. Also, we generated the deletion mutant and overexpression mutant of strain 104M, as recent evidence suggests that Omp19 is a crucial virulence factor of Brucella [34–36]. Consistent with a previous report in the mouse model, strain A19 and strain A19ΔVirB12 showed comparable pathogenicity in the larval model. As anticipated, larvae infected with strain 104MΔOmp19 showed prolonged survival compared to the parental strain, with all larvae surviving on day 1 (Figure 3c). However, there was no significant difference in the survival rate and health index scores between the 104M-infected group and the 104M:Omp19-infected group (Figure 3c,d), implying that overexpression of the virulence factor Omp19 did not increase the virulence of the strain. Finally, we compared the differences in virulence between Brucella species. Strain S2 showed lower virulence in the larval model than strain A19 and strain 104M, which is consistent with previous reports [37–39]. Taken together, these data indicate that the G. mellonella model is suitable for the rapid virulence evaluation of Brucella strains.
G. mellonella as a model for rapid evaluation of Brucella vaccine immune sera efficacy
G. mellonella larvae have been used for the evaluation of monoclonal antibodies [40,41]. We therefore made an effort to develop the G. mellonella larvae as a rapid evaluation model for Brucella vaccine immune sera. To begin with, we determined whether the serum components affected the survival of G. mellonella larvae. PBS sera had no effect on larval survival (Figure 4a), indicating that serum components were not toxic to G. mellonella larvae. Likewise, the health index scores of the larvae injected with PBS sera were always above 9, proving that the larvae were healthy (Figure 4b).
Figure 4.
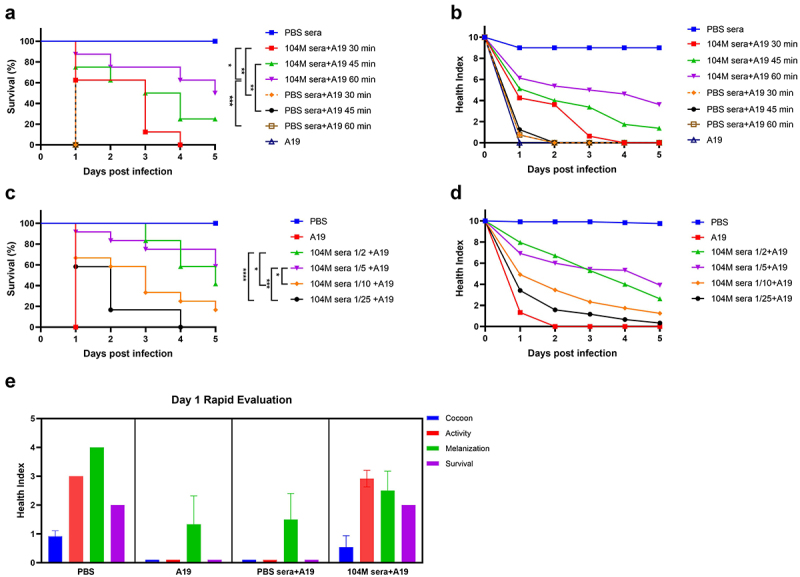
G. mellonella as a model for rapid evaluation of Brucella vaccine immune sera efficacy. a) & b) The effect of pre-incubation time of immune sera and bacteria on G. mellonella larval survival. a) Survival rate and b) health index scores of larvae infected with Brucella strain A19 pre-treated with sera for 30,45 or 60 minutes. c) & d) The effect of immune sera concentration on G. mellonella larval survival. c) Survival rate and d) health index scores of larvae infected with A19 pre-treated with different concentrations of sera. e) Cocoon formation, activity, melanization and survival of G. mellonella larvae on day 1. Significant differences between the Kaplan-Meier curves were identified with the log-rank test (GraphPad Prism). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, as a proof of concept, we applied this model to examine the efficacy of 104M sera, since 104M live attenuated vaccine has already been proven to be highly protective against brucellosis [13,42]. To study the inhibitory effect of the antibody, strain A19 was pre-treated with pooled sera before inoculation. To optimize the pre-incubation time, bacteria were suspended in PBS buffer containing 50% (v/v) 104M immune sera or PBS sera and incubated at 37°C for 30, 45, or 60 min. Larvae were then inoculated with a 25 μl mixture containing 1.25 × 108 CFU of bacteria. It is not surprising that PBS sera had no protective effect on the larvae, regardless of the pre-incubation time. Contrastingly, 104M sera showed an excellent protective effect. The survival rates of groups pre-incubated with 104M sera for 30, 45, or 60 min were 62.5%, 75%, and 87.5% on day 1, respectively (Figure 4a). The protective efficacy of the 60-minute pre-incubated group outperformed the other two groups, as reflected in both the daily survival rates and health index scores (Figure 4b). Thus, 60 min was employed as the pre-incubation time in subsequent studies.
To determine the optimal sera concentration, strain A19 was suspended in PBS containing 1/2 diluted to 1/25 diluted [4%-50% (v/v)] 104M sera and incubated at 37°C for 60 min before being inoculated into the larvae. Larval survival rates were all improved in the 104M sera pre-incubated groups by 58.3%-100% on day 1, compared with the PBS sera pre-treated group. All larvae inoculated with 1/25 diluted sera died by day 4, whereas those injected with 1/2 or 1/5 diluted sera showed more than 40% survival by day 5 (Figure 4c). Similar observations were apparent when comparing the health index scores of these different treated groups. Larvae in the 1/25 and 1/10 diluted 104M sera pre-incubated groups scored consistently lower than those in the 1/2 and 1/5 diluted 104M sera pre-incubated groups (Figure 4d). Therefore, these data indicate that 104M sera provide good protection against a lethal Brucella challenge when pre-incubated with bacteria for 60 min, with 1/2, 1/5, and 1/10 diluted sera outperforming 1/25 diluted sera.
Except for the total health index scores, individual indicator (activity, cocoon formation, melanization, and survival) scores also reflected that 104M sera had a good protective effect against A19 during the infection course. It is worth mentioning that differences in the protective efficacy of immune sera were already rather noticeable on the first day of infection (Figure 4e), suggesting that the G. mellonella model offers the possibility to rapidly evaluate the efficacy of serum antibodies.
Haemocytes play a major role in the protective effect of 104M sera
To better understand the bacteria-larvae interaction and compare the histopathological differences between the 104M sera pre-incubated group and the PBS sera pre-incubated group, transverse sections of larvae were examined following H&E or Koster’s staining. Consistent with our expectations, larvae in the PBS sera pre-incubated group showed extensive tissue destruction, with Brucella scattered within the coelom (Figure 5a,b). In contrast, in the 104M sera pre-incubated group, bacteria were confined to the granuloma-like structures, thus preventing diffuse infection of the organism on day 1 (Figure 5c,d). Additionally, an increase in the size of granulomatous-like lesions was observed on day 5 (Figure 5e,f).
Figure 5.
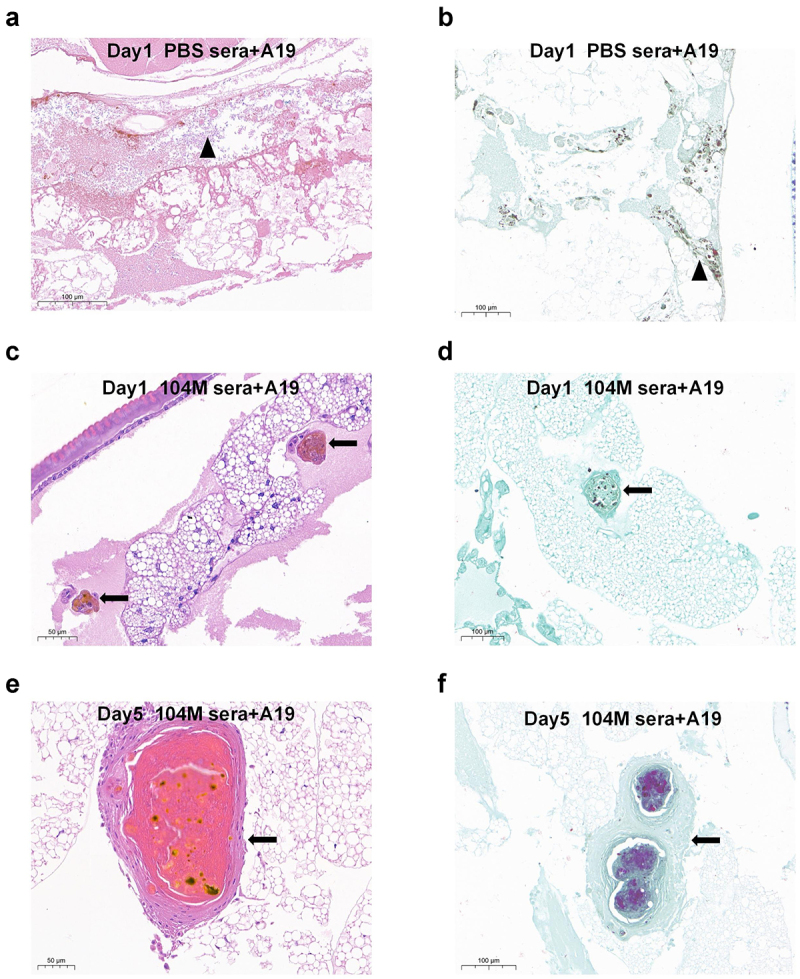
Histological analysis of G. mellonella larvae infected with sera-pre-treated Brucella strain A19. Groups of larvae were infected with 1.25 × 108 CFU of live Brucella strain A19 pre-treated with 104M sera or PBS sera for 1 hour. a) Larvae infected with PBS sera pre-treated bacteria on day 1, H&E staining. Black triangles represented disseminated Brucella. b) Larvae infected with PBS sera pre-treated bacteria on day 1, Koster’s staining. c) Larvae infected with 104M sera pre-treated bacteria on day 1, H&E staining. d) Larvae infected with 104M sera pre-treated bacteria on day 1, Koster’s staining. e) Larvae infected with 104M sera pre-treated bacteria on day 5, H&E staining. f) Larvae infected with 104M sera pre-treated bacteria on day 5, Koster’s staining. Black arrows represented the granuloma-like structures formed by the aggregation of haemocytes.
One possible explanation for our observations is that the pre-incubation of bacteria and immune sera may contribute to the recognition, phagocytosis, and encapsulation of bacteria by larval haemocytes. To test this hypothesis, we injected larvae with CDLip to deplete haemocytes, or Lip as a control; after 24 h, we exposed the larvae to the 104M sera pre-incubated bacteria, and measured larval survival over 5 days. Lip and CDLip alone showed no significant toxicity towards larvae (data not shown). In line with our speculation, larval survival was significantly reduced in the CDLip pre-treated larvae, compared with the larvae pre-treated with Lip, indicating that the protective effect of 104M sera is dependent on larval haemocytes. However, it should be noted that pretreatment with CDLip only decreased, but did not completely inhibit, the protective effect of 104M sera, suggesting that several mechanisms may play a role in the protective effect of 104M sera (Figure 6a,b).
Figure 6.
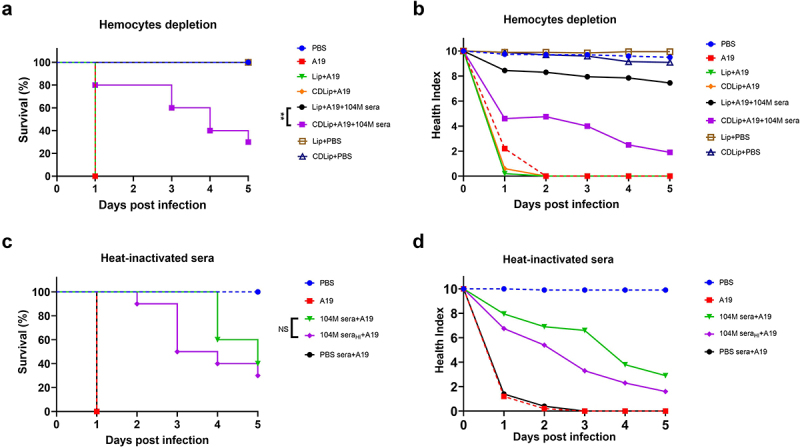
The effect of haemocytes depletion and complement inactivation on larval survival. Larvae were either injected with CDLip to deplete haemocytes or injected with Lip as a control 24 hours before infection (a&b). Sera were untreated or heat-inactivated at 56°C for 30 minutes before co-incubated with bacteria (c&d). a) The effect of haemocytes depletion on G. mellonella larval survival. b) Health index scores of haemocytes depletion larvae infected with a lethal dose of Brucella. c) The effect of complement inactivation on G. mellonella larval survival. d) Health index scores of larvae infected with a lethal dose of Brucella strain A19 pre-incubated with complement inactivated immune sera. Significant differences between the Kaplan-Meier curves were identified with the log-rank test (GraphPad Prism). **P < 0.01.
To verify whether the protective effect of 104M sera is complement-related, we further inactivated the serum complement. Immune sera were first heat-inactivated at 56°C for 30 min and then co-incubated with 1.25 × 108 CFU of strain A19. The protective effect of the 104M sera was not affected by complement inactivation, suggesting that the serum complement is not relevant to the protective effect of the 104M sera in the larval model (Figure 6c,d).
Priming with a non-lethal dose of Brucella strains protects G.mellonella larvae against a subsequent lethal challenge
Several studies have shown that priming low-dose injections of microorganisms shielded the larvae from repeated exposure to the same or other microorganisms. To verify whether pre-exposure to a non-lethal dose of Brucella strains protects the larvae from a subsequent lethal challenge, groups of larvae were first primed with 1.25 × 105 CFU of live or fixed strain A19 through the last left proleg, followed by infection with 1.25 × 108 CFU of the same strain 24 h later via the last right proleg. As expected, priming with a non-lethal dose of either live or fixed strains protected the larvae from a subsequent lethal challenge, with the live strain outperforming the fixed strain (Figure 7a).
Figure 7.
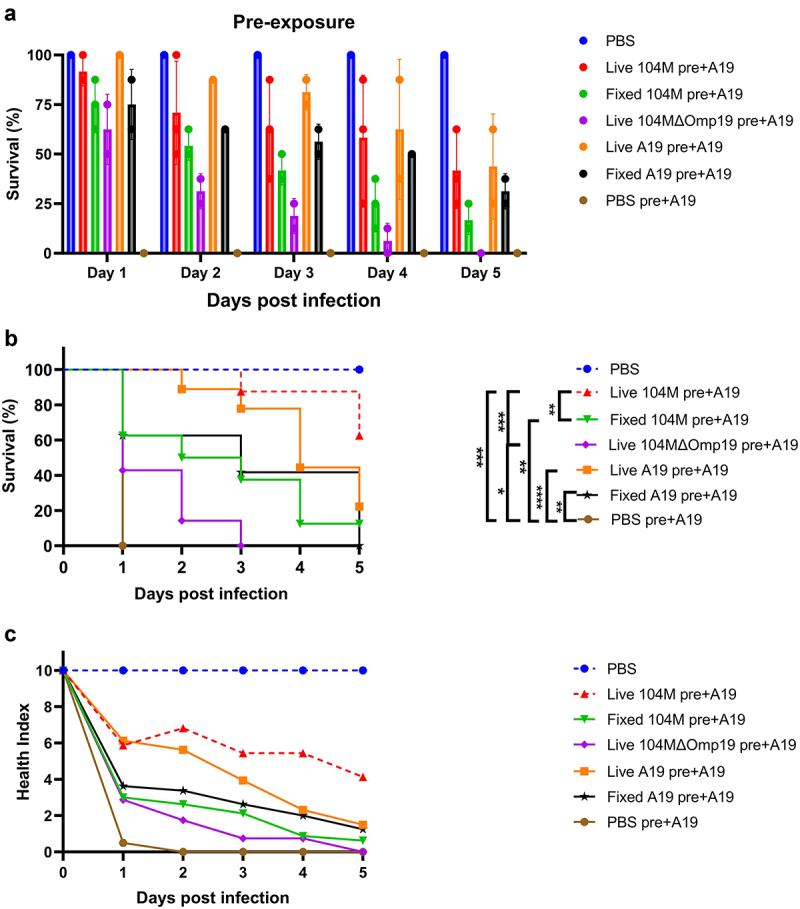
The effect of priming G. mellonella larvae with a non-lethal dose of Brucella strains. Larvae were primed with a non-lethal dose of Brucella or PBS control 24 hours before infection with a lethal dose of strain A19. a) & b) Survival rate of larvae primed with non-lethal dose of bacteria. c) Health index scores of larvae primed with non-lethal dose of bacteria. Significant differences between the Kaplan-Meier curves were identified with the log-rank test (GraphPad Prism). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We simultaneously explored whether priming with heterologous Brucella strains could also protect against a subsequent lethal challenge. To this end, larvae were primed with 1.25 × 105 CFU of strain 104M or strain 104MΔOmp19 24 h before confronted with 1.25 × 108 CFU of strain A19. The protective efficacy of the 104M primed group was not statistically different from that of the A19 primed group, while the 104MΔOmp19 strain primed group showed significantly decreased protective efficacy (Figure 7b,c), demonstrating that the presence of Omp19 is of great importance for the 104M vaccine.
G. mellonella model and the mouse model are comparable in evaluation of vaccine efficacy
To verify whether the G. mellonella and mouse models were comparable in evaluation of vaccine efficacy, strain 104M and strain 104MΔOmp19 were employed. The protective efficacy of the two strains was firstly tested in a mouse infection model. Groups of mice were immunized with 1 × 105 CFU of strain 104M, strain 104MΔOmp19, or the PBS control on day 1. Four weeks after immunization, the mice were infected intraperitoneally with 1 × 108 CFU of strain A19. The protective efficacy of 104M was far superior to that of the 104MΔOmp19 deletion mutant, with a 15-fold difference in the spleen bacterial load (P < 0.001) between these two groups. (Figure 8a).
Figure 8.
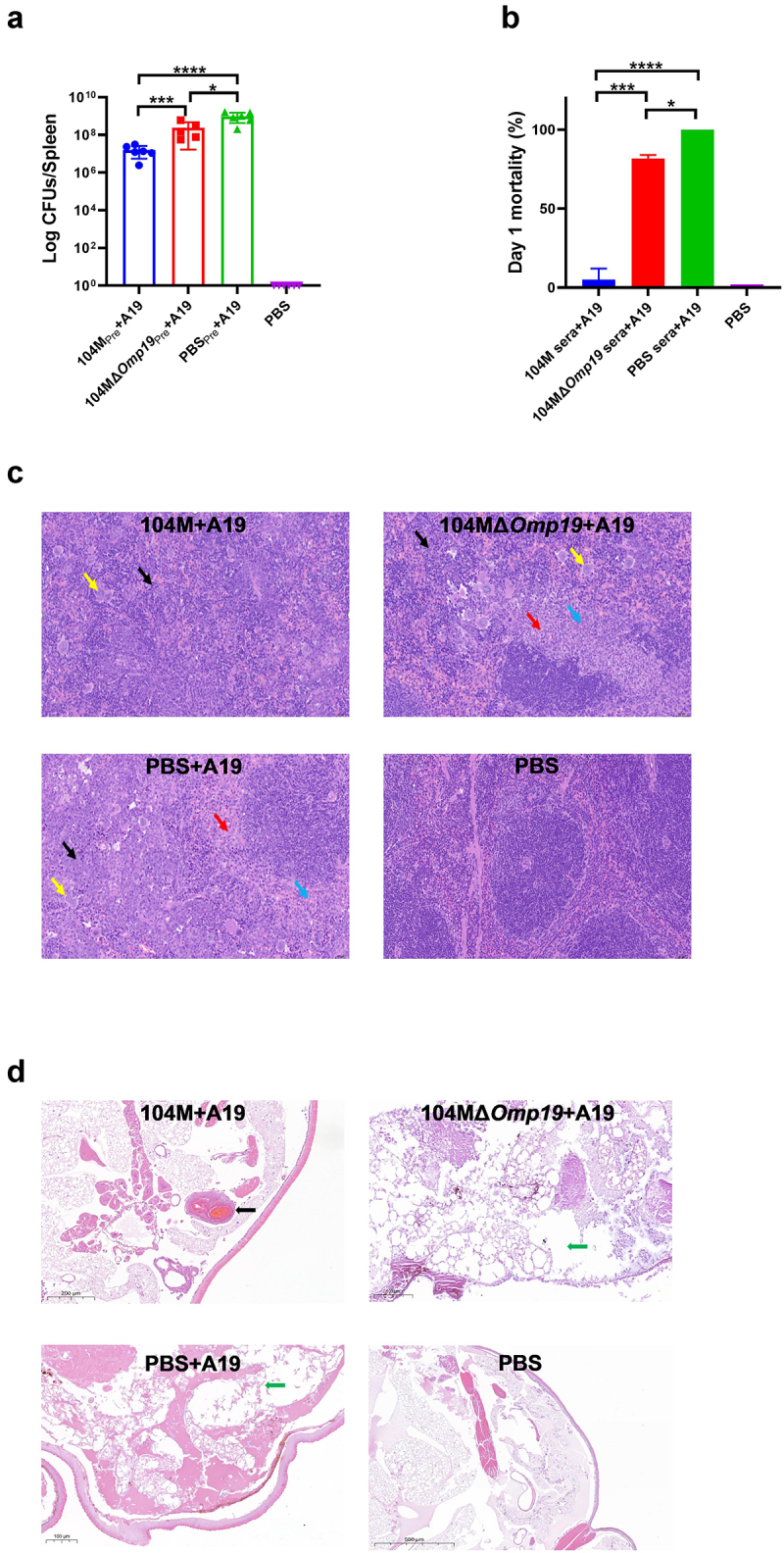
Evaluation of the consistency between the invertebrate G. mellonella model and the mouse model. Groups of mice were immunized with 1 × 105 CFU of strain 104M, strain 104MΔOmp19, or the PBS control on day 1, and 4 weeks after immunization, the mice were infected intraperitoneally with 1 × 108 CFU of strain A19. Sera were collected on day 28. a) CFU counts from 104M and 104MΔOmp19 immunized mice spleens in the challenge study. b) Day 1 mortality of larvae infected with 1.25 × 108 CFU of Brucella strain A19 pre-treated with 104M sera, 104MΔOmp19 sera or PBS sera for 1 hour. c) Representative images of H&E staining mouse spleens in different treatment groups on day 35. Red arrows represented hyperplasia of connective tissue around splenic nodules, blue arrows represented punctate neutrophil infiltration, black arrows represented large areas of extramedullary haematopoiesis in the red pulp, and yellow arrows represented multinucleated giant cells. d) Representative images of H&E staining larvae sections in different treatment groups on day 1. The black arrow represented the granuloma-like structures formed by the aggregation of haemocytes, and green arrows represented damaged adipose body. Significant differences were identified by one-way ANOVA (GraphPad Prism). *P < 0.05, ***P < 0.001, ****P < 0.0001.
We then compared the protective efficacy of 104M and 104MΔOmp19 immune sera from the two groups in the G. mellonella model. Compared with PBS sera, 104MΔOmp19 immune sera did have some protective effects, as reflected in the 20% survival rate on day 1 and prolonged survival time (Figure 8b and Additional file 2). However, this protective effect was rather limited because all larvae died on day 4 (Additional file 2). In contrast, the 104M sera pre-incubated group, consistent with the results from the mouse model, demonstrated a significant protective effect. 90% of the larvae survived on day 1 (Figure 8b) and the survival rate remained at 30% on day 5 (Additional file 2). Meanwhile, the daily health index scores of the 104M sera pre-incubated group were consistently higher than that of the 104MΔOmp19 sera pre-incubated group (Additional file 2).
Histopathological changes in mouse spleens (Figure 8c) and G. mellonella larvae (Figure 8d) were then investigated by H&Estaining sections. No abnormalities were observed in the spleen of the PBS-immunized, uninfected group, while in the PBS-immunized, challenged group and 104MΔOmp19 immunized, challenged group, the number of splenic nodules was decreased, and hyperplasia of connective tissue around splenic nodules (red arrows) was observed, with punctate neutrophil infiltration (blue arrows). Large areas of extramedullary haematopoiesis can be observed in the red pulp (black arrows), accompanied by an increase in the number of multinucleated giant cells (yellow arrows). In contrast, fewer severe lesions in the spleen section were observed in the 104M immunized group, with small extramedullary haematopoietic foci (black arrows) and a slight increase in the number of multinucleated giant cells (yellow arrows). Similar observations were discovered when comparing lesions in the larval sections. Larval sections in the PBS sera pre-treated group showed extensive disruption of the tissue structure and diffuse distribution of bacteria, while those in the 104MΔOmp19 sera pre-treated group only showed partial disruption. A portion of the bacteria was distributed in the haemocoel, while the remainder was encapsulated in the haemolymph nodes. Notably, in the 104M serum pre-treated group, almost all bacteria were encapsulated in the granuloma-like nodes formed by haemocytes. Consequently, the tissue structure was relatively intact. Taken together, these data indicate that comparable results to murine studies can be obtained using the G. mellonella model when 104M and 104MΔOmp19 are used as model vaccines. Consequently, as a bridge between the in vitro and in vivo models, G. mellonella model could be used as a simple, rapid, and high-throughput screening tool to evaluate the protective efficacy of immune sera.
Discussion
Although there is an urgent demand for new strategies and therapies to control, combat, and eradicate brucellosis, this process is extremely slow. Animal models are indispensable for studying virulence, pathogenic mechanisms, bacteria-host interactions, and vaccine development of Brucella. However, commonly used vertebrate evaluation models require feeding and housing, are time-consuming and costly. On the other hand, the in vitro evaluation methods, such as the serum bactericidal activity assay and the minimum inhibitory concentration assay, are not recommended because of the potential for creating aerosols that are hazardous to laboratory personnel. As such, the G. mellonella larvae in vivo evaluation model is of great value, as it offers a bridge between in vitro cell assays and final vertebrate studies. The use of the G. mellonella larvae model in pathogen-host interaction, virulence comparison, and antibiotic screening has been broadly investigated, but few studies have focused on this model for Brucella infection or evaluation of immune serum efficacy. In 2014, Nicolas et al. presented a protocol to analyse the virulence of select agents, including two highly virulent Brucella strains [43]. Here, we report that G. mellonella larvae could be used as a complementary in vivo model for infection and virulence comparison of Brucella strains. Also, we showed that this simple invertebrate model exhibited promise as a novel, fast, and efficient evaluation model for the efficacy of immune sera, which, to our knowledge, is a brand new finding.
To characterize the Brucella infection model, we first evaluated the virulence of A19, an attenuated strain of B. abortus, to the larvae using a range of doses at 25°C and 37°C, and determined the survival of the larvae over a 5-day infection course. Also, the infection process was monitored by the health index scores of larvae. Larval survival occurred in a temperature- and dose-dependent manner. A dose of 1.25 × 108 CFU of Brucella A19 killed almost all the larvae by the end of infection regardless of the temperature, and a delay in larval death was observed when they were maintained at a lower temperature. This difference may be related to the growth disadvantage of bacteria at lower temperatures or the expression of temperature-dependent virulence factors. Similarly, health index scores correlated well with infection doses, with lower doses leading to higher scores. As a result, the infection dose of 1.25 × 108 CFU and the incubation temperature of 37°C were used in subsequent studies. In addition, the histopathological sections showed that Brucella was restricted to granuloma-like structures formed by haemocytes during non-lethal dose of infection, whereas bacteria were scattered throughout the course of the lethal dose of infection, indicating that larval death may be related to haemocyte depletion.
Next, since the G. mellonella larval model has been reported for the virulence evaluation of bacteria and fungi [24,26,27,44,45], we compared the virulence differences between live bacteria and heat-inactivated bacteria. Live strain 104M and live strain A19 both killed the larvae completely at an inoculation dose of 1.25 × 108 CFU, indicating that there was probably little difference in virulence between these two Brucella strains in the G. mellonella model. Meanwhile, the killing activity could be annulled by heat inactivation, which agreed with the results of Dijokaite et al. [30], implying that inactivated bacterial lysate and endotoxin did not play a significant role in larval survival. Strain A19 and strain A19ΔVirB12 showed equal pathogenicity in the larval model, which is in keeping with an earlier report in the mouse model [33]. Importantly, the survival rate of larvae inoculated with the 104MΔOmp19 deletion mutant was greatly increased at the same infection dose, proving that Omp19 is indeed a key virulence factor of strain 104M. In addition, strain S2 was less virulent in the larval model than strain A19, which is consistent with the results of previous studies [37,39]. Together, the G. mellonella model could detect virulence differences of Brucella strains, and a good correlation has been established between the G. mellonella model and the murine model. Therefore, the larval model might be a good substitute for the early evaluation of potential virulence genes when using a mammalian model, which might not be morally or practically appropriate. While preparing this article, a study was published by Aitor et al., which focused on utilizing the larval model to screen for potential Brucella factors modulating innate immunity and yielded similar findings to our own [46].
After successfully established the larval model of Brucella infection and virulence evaluation, we determined the possibility of using this model to assess the efficacy of immune sera. As expected, the serum components did not affect the survival of G. mellonella, which is the prerequisite to the G. mellonella model for evaluation of immune serum efficacy. Then, the protective efficacy of 104M immune sera was determined. Unsurprisingly, 104M sera provided good protection against a Brucella lethal challenge when incubated with bacteria in advance, while PBS sera showed no effect on preventing Brucella induced larval death. This protective effect depends on both the concentration of immune sera and the co-incubation time of the bacteria and immune sera. These data highlight the potential of the G. mellonella larvae model for the evaluation of immune serum efficacy. Notably, the differences in the survival rates and health index scores between the 104M sera and PBS sera were quite clear by day 1. Accordingly, the survival rates and health index scores on day 1 could be used as predictors of the overall protective effect without monitoring the larvae for 5 days, which greatly shortens the experimental cycle.
To explore the mechanisms underlying the protective efficacy of 104M sera in the G. mellonella model, histopathological differences between the 104M sera and PBS sera pre-treated groups were analysed. Strain A19 caused severe disseminated infection and tissue destruction in the PBS sera pre-treated group, whereas in the 104M sera pre-treated group, bacteria were confined to the granulomatous structures formed by haemocytes, thus preventing disseminated infection.
We then depleted larval haemocytes to verify whether they cooperated with 104M sera. As expected, the protective effect of 104M sera decreased but did not completely disappear, indicating that multiple mechanisms may be involved in the protective effect of 104M immune sera. Additionally, we heat-inactivated the serum complement to determine whether it caused a difference in the serum protective efficacy. As the outcome demonstrates, complement is not important for understanding how 104M sera protects. Omp19 plays a significant role in bacterial adhesion, invasion, colonization, and intracellular survival; thus, it is considered a leading target for Brucella vaccines. To verify whether the protective efficacy of strain 104M decreases after deletion of Omp19, groups of larvae were primed with a non-lethal dose of Brucella strain 104M and strain 104MΔOmp19, followed by a lethal dose of strain A19 challenge. Indeed, priming with strain 104M or 104MΔOmp19 could protect larvae from the lethal challenge, with strain 104M outcompeting strain 104MΔOmp19, as reflected in the daily survival rate and health index scores of different groups. In line with our expectations, the protective effect of pre-immunization with a non-lethal dose of live vaccine was superior to that of the corresponding inactivated vaccine, demonstrating the reliability of the model.
Finally, we compared the consistency between the larval and mouse models. A prophylactic study was performed in the mouse model, and the protective efficacy of strain 104M, 104MΔOmp19 and PBS control were determined. As predicted, strain 104MΔOmp19 showed a much lower protective efficacy than strain 104M, reflecting the difference in bacterial load. Subsequently, the protective efficacy of 104M sera, 104MΔOmp19 sera, and PBS sera from the mouse study was determined using the larval model. Similarly, 104MΔOmp19 sera showed much lower protective efficacy than 104M sera, following the same pattern as the mouse model. Further analysis of the histopathological changes also showed that strain 104M provided better protection than strain 104MΔOmp19 in the mouse and G. mellonella models. Combined, these data highlight the correlation between the larval model and the mouse model, implying the potential of the larval model in immune sera evaluation.
In conclusion, in the present study, we characterized a G. mellonella larval model for Brucella infection and virulence evaluation. Moreover, we report for the first time a G. mellonella larval model for potential evaluation of the protective efficacy of immune sera, which shortens the evaluation cycle to 1 day and therefore greatly accelerates the evaluation process. Future research will concentrate on the cellular, proteomic, and metabolomic responses to Brucella infection in the G. mellonella larval model. We anticipate that the G. mellonella infection and immune model will lessen our reliance on mammalian infection models, while enabling us to gain a better understanding of Brucella virulence factors, bacteria-host interactions, and vaccine development.
Supplementary Material
Acknowledgements
We thank Dr. Shanhu Li and Dr. Chenfeng Mao of Beijing Institute of Biotechnology for their helpful discussions on topics related to this work. We would also like to thank Dr. Wei Fan for valuable suggestions on animal studies.
Funding Statement
The work was supported by the National Natural Science Foundation of China [31800770]; National Natural Science Foundation of China [32170945].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Ethics approval
The animal studies and procedures were approved by the Animal Care and Use Committee of Beijing Institute of Biotechnology under project license number IACUC-SWGCYJS-2022-004. The animal procedures followed the guidelines of the Administration of Affairs Concerning Experimental Animals.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2268496
References
- [1].Pappas G, et al. Brucellosis. N Engl J Med. 2005;352(22):2325–16. doi: 10.1056/NEJMra050570 [DOI] [PubMed] [Google Scholar]
- [2].Dean AS, Crump LGreter H, et al., Global burden of human brucellosis: a systematic review of disease frequency. PLoS Negl Trop Dis. 2012;6(10): e1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Franco MP, Mulder M, Gilman RH, et al. Human brucellosis. Lancet Infect Dis. 2007;7(12):775–786. doi: 10.1016/S1473-3099(07)70286-4 [DOI] [PubMed] [Google Scholar]
- [4].Boschiroli ML, Foulongne V, O’Callaghan D.. Brucellosis: a worldwide zoonosis. Curr Opin Microbiol. 2001;4(1):58–64. doi: 10.1016/S1369-5274(00)00165-X [DOI] [PubMed] [Google Scholar]
- [5].Pappas G, Papadimitriou P, Akritidis N, et al. The new global map of human brucellosis. Lancet Infect Dis. 2006;6(2):91–99. doi: 10.1016/S1473-3099(06)70382-6 [DOI] [PubMed] [Google Scholar]
- [6].Lalsiamthara J, Lee JH. Development and trial of vaccines against Brucella. J Vet Sci. 2017;18(S1):281–290. doi: 10.4142/jvs.2017.18.S1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cheng Z, Li Z, Yin Y, et al. Characteristics of Brucella abortus vaccine strain A19 reveals its potential mechanism of attenuated virulence. Vet Microbiol. 2021;254:109007. doi: 10.1016/j.vetmic.2021.109007 [DOI] [PubMed] [Google Scholar]
- [8].Dorneles EM, Sriranganathan N, Lage AP. Recent advances in Brucella abortus vaccines. Vet Res. 2015;46(1):76. doi: 10.1186/s13567-015-0199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Heidary M, Dashtbin S, Ghanavati R, et al. Evaluation of brucellosis vaccines: a comprehensive review. Front Vet Sci. 2022;9:925773. doi: 10.3389/fvets.2022.925773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hou H, Liu X, Peng Q. The advances in brucellosis vaccines. Vaccine. 2019;37(30):3981–3988. doi: 10.1016/j.vaccine.2019.05.084 [DOI] [PubMed] [Google Scholar]
- [11].Yang X, Skyberg JA, Cao L, et al. Progress in Brucella vaccine development. Front Biol (Beijing). 2013;8(1):60–77. doi: 10.1007/s11515-012-1196-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schurig GG, Sriranganathan N, Corbel MJ. Brucellosis vaccines: past, present and future. Vet Microbiol. 2002;90(1–4):479–496. doi: 10.1016/S0378-1135(02)00255-9 [DOI] [PubMed] [Google Scholar]
- [13].Yu D, Hui Y, Zai X, et al. Comparative genomic analysis of Brucella abortus vaccine strain 104M reveals a set of candidate genes associated with its virulence attenuation. Virulence. 2015;6(8):745–754. doi: 10.1080/21505594.2015.1038015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arenas-Gamboa AM, Ficht TA, Kahl-McDonagh MM, et al. The Brucella abortus S19 DeltavjbR live vaccine candidate is safer than S19 and confers protection against wild-type challenge in BALB/c mice when delivered in a sustained-release vehicle. Infect Immun. 2009;77(2):877–884. doi: 10.1128/IAI.01017-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spink WW, HALL JW, FINSTAD J, et al. Immunization with viable Brucella organisms. Results of a safety test in humans. Bull World Health Organ. 1962;26(3):409–419. [PMC free article] [PubMed] [Google Scholar]
- [16].Ashford DA, di Pietra J, Lingappa J, et al. Adverse events in humans associated with accidental exposure to the livestock brucellosis vaccine RB51. Vaccine. 2004;22(25–26):3435–3439. doi: 10.1016/j.vaccine.2004.02.041 [DOI] [PubMed] [Google Scholar]
- [17].López-Goñi I, García-Yoldi D, Marín CM, et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol. 2008;46(10):3484–3487. doi: 10.1128/JCM.00837-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bardenstein S, Mandelboim M, Ficht TA, et al. Identification of the Brucella melitensis vaccine strain Rev.1 in animals and humans in Israel by PCR analysis of the PstI site polymorphism of its omp2 gene. J Clin Microbiol. 2002;40(4):1475–1480C. doi: 10.1128/JCM.40.2.1475-1480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blasco JM, Díaz R. Brucella melitensis Rev-1 vaccine as a cause of human brucellosis. Lancet. 1993;342(8874):805. doi: 10.1016/0140-6736(93)91571-3 [DOI] [PubMed] [Google Scholar]
- [20].Xin X. Orally administrable brucellosis vaccine: Brucella suis strain 2 vaccine. Vaccine. 1986;4(4):212–216. doi: 10.1016/0264-410X(86)90131-3 [DOI] [PubMed] [Google Scholar]
- [21].Mustafa AA, Abusowa M. Field-oriented trial of the Chinese Brucella suis strain 2 vaccine on sheep and goats in Libya. Vet Res. 1993;24(5):422–429. [PubMed] [Google Scholar]
- [22].Silva TM, Costa EA, Paixão TA, et al. Laboratory animal models for brucellosis research. J Biomed Biotechnol. 2011;2011:518323. doi: 10.1155/2011/518323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grilló MJ, Blasco JM, Gorvel JP, et al. What have we learned from brucellosis in the mouse model? Vet Res. Vet Res. 2012;43(1):29. doi: 10.1186/1297-9716-43-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229C. doi: 10.1080/21505594.2015.1135289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sheehan G, Garvey A, Croke M, et al. Innate humoral immune defences in mammals and insects: the same, with differences ? Virulence. 2018;9(1):1625–1639. doi: 10.1080/21505594.2018.1526531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cutuli MA, Petronio Petronio G, Vergalito F, et al. Galleria mellonella as a consolidated in vivo model hosts: new developments in antibacterial strategies and novel drug testing. Virulence. 2019;10(1):527–541. doi: 10.1080/21505594.2019.1621649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Champion OL, Wagley S, Titball RW. Galleria mellonella as a model host for microbiological and toxin research. Virulence. 2016;7(7):840–845. doi: 10.1080/21505594.2016.1203486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Desbois AP, Coote PJ. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of New antimicrobial agents. Adv Appl Microbiol. 2012;78:25–53. [DOI] [PubMed] [Google Scholar]
- [29].Tsai CJ, Loh JMS, Proft T. The use of Galleria mellonella (wax moth) as an infection model for group a streptococcus. Methods Mol Biol. 2020;2136:279–286. [DOI] [PubMed] [Google Scholar]
- [30].Dijokaite A, Humbert MV, Borkowski E, et al. Establishing an invertebrate Galleria mellonella greater wax moth larval model of Neisseria gonorrhoeae infection. Virulence. 2021;12(1):1900–1920. doi: 10.1080/21505594.2021.1950269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang Q, Zai X, Yin Y, et al. Construction and immunoprotective evaluation of Brucella 104M: Omp19 overexpression mutant strain. Lett Biotechnol. 2018;29(1):6. [Google Scholar]
- [32].Kwon H, Smith RC. Chemical depletion of phagocytic immune cells in Anopheles gambiae reveals dual roles of mosquito hemocytes in anti-Plasmodium immunity. Proc Natl Acad Sci USA. 2019. Jul 9;116(28):14119–14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang J, He C, Zhang H et al. Evaluation and differential diagnosis of a genetic marked Brucella vaccine A19ΔvirB12 for cattle. Front Immunol. 2021;12:679560. doi: 10.3389/fimmu.2021.679560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zai X, Yin Y, Guo F, et al. Screening of potential vaccine candidates against pathogenic Brucella spp. using compositive reverse vaccinology. Vet Res. 2021;52(1):75. doi: 10.1186/s13567-021-00939-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pasquevich KA, Carabajal MV, Guaimas FF, et al. Omp19 enables Brucella abortus to evade the antimicrobial activity from host’s proteolytic defense System. Front Immunol. 2019;10:1436. doi: 10.3389/fimmu.2019.01436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Uslu A, Erganis O. Development of Brucella melitensis Rev.1 ΔOmp19 mutants with DIVA feature and comparison of their efficacy against three commercial vaccines in a mouse model. Mol Immunol. 2021;133:44–52C. doi: 10.1016/j.molimm.2021.02.006 [DOI] [PubMed] [Google Scholar]
- [37].Wang X, Li Z, Li B, et al. Bioluminescence imaging of colonization and clearance dynamics of Brucella suis vaccine strain S2 in mice and Guinea pigs. Mol Imaging Biol. 2016;18(4):519–526. doi: 10.1007/s11307-015-0925-6 [DOI] [PubMed] [Google Scholar]
- [38].Bosseray N, Plommet M. Brucella suis S2, Brucella melitensis Rev. 1 and Brucella abortus S19 living vaccines: residual virulence and immunity induced against three Brucella species challenge strains in mice. Vaccine. 1990;8(5):462–468. doi: 10.1016/0264-410X(90)90247-J [DOI] [PubMed] [Google Scholar]
- [39].Zhu L, Feng Y, Zhang G, et al. Brucella suis strain 2 vaccine is safe and protective against heterologous Brucella spp. infections. Vaccine. 2016;34(3):395–400. doi: 10.1016/j.vaccine.2015.09.116 [DOI] [PubMed] [Google Scholar]
- [40].Antoran A, Aparicio-Fernandez L, Pellon A, et al. The monoclonal antibody Ca37, developed against Candida albicans alcohol dehydrogenase, inhibits the yeast in vitro and in vivo. Sci Rep. 2020;10(1):9206. doi: 10.1038/s41598-020-65859-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fregonezi NF, Oliveira LT, Singulani JD, et al. Heat shock protein 60, insights to its importance in Histoplasma capsulatum: from biofilm formation to host-interaction. Front Cell Infect Microbiol. 2020;10:591950. doi: 10.3389/fcimb.2020.591950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wei C, Yang WH, Hou XX, et al. Immunization efficacy and safety of Brucella 104M against aerosol challenge in BALB/c mice. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(7):1103–1109. doi: 10.3760/cma.j.cn112338-20190911-00665 [DOI] [PubMed] [Google Scholar]
- [43].Sprynski N, Valade E, Neulat-Ripoll F. Galleria mellonella as an infection model for select agents. Methods Mol Biol. 2014;1197:3–9. [DOI] [PubMed] [Google Scholar]
- [44].Torres M, Pinzón EN, Rey FM, et al. Galleria mellonella as a novelty in vivo model of host-pathogen interaction for malassezia furfur CBS 1878 and malassezia pachydermatis CBS 1879. Front Cell Infect Microbiol. 2020;10:199. doi: 10.3389/fcimb.2020.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hernando-Ortiz A, Mateo E, Perez-Rodriguez A, et al. Virulence of candida auris from different clinical origins in Caenorhabditis elegans and Galleria mellonella host models. Virulence. 2021;12(1):1063–1075. doi: 10.1080/21505594.2021.1908765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Elizalde-Bielsa A, Aragón-Aranda B, Loperena-Barber M, et al. Development and evaluation of the Galleria mellonella (greater wax moth) infection model to study Brucella host-pathogen interaction. Microb Pathog. 2023;174:105930. doi: 10.1016/j.micpath.2022.105930 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


