Abstract
From each of two AIDS patients with oropharyngeal candidiasis, five Candida albicans isolates from recurrent episodes of infection which became gradually resistant against fluconazole during antimycotic treatment were analyzed for molecular changes responsible for drug resistance. In both patients, a single C. albicans strain was responsible for the recurrent infections, but the CARE-2 fingerprint pattern of the isolates exhibited minor genetic alterations, indicating that microevolution of the strains took place during fluconazole therapy. In the isolates from patient 1, enhanced mRNA levels of the MDR1 gene, encoding a multiple drug resistance protein from the superfamily of major facilitators, and constitutive high expression of the ERG11 gene, coding for the drug target enzyme sterol 14α-demethylase, correlated with a stepwise development of fluconazole resistance. The resistant strains exhibited reduced accumulation of fluconazole and, for the last in the series, a slight increase in drug needed to inhibit sterol 14α-demethylation in vitro. In the isolates from patient 2, increased MDR1 mRNA levels and the change from heterozygosity to homozygosity for a mutant form of the ERG11 gene correlated with continuously decreased drug susceptibility. In this series, reduced drug accumulation and increased resistance in the target enzyme activity, sterol 14α-demethylase, were observed. These results demonstrate that different molecular mechanisms contribute to a gradual development of fluconazole resistance in C. albicans.
Candida albicans is an important opportunistic fungal pathogen of humans and the major cause of oropharyngeal candidiasis (OPC) in AIDS patients (24). The azole antifungal agent fluconazole is used widely to treat OPC. In recent years, however, there have been numerous reports of treatment failures in patients receiving prolonged fluconazole therapy, and these treatment failures have been demonstrated to be due to the emergence of fluconazole-resistant C. albicans strains (5, 26). Different mechanisms may be responsible for drug resistance. Changes in the level of the drug target enzyme, sterol 14α-demethylase, as a consequence of enhanced transcription or amplification of the ERG11 gene (previously termed ERG16), may lead to reduced susceptibility of yeasts to fluconazole (35, 37) although gene dosage effects are limited (9). Mutations in this gene which lower the affinity of the enzyme for the drug have also been detected (15, 38). Central to the mode of action of azole antifungals against C. albicans is the accumulation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol during treatment, and defects in sterol C5-desaturation prevent the diol from accumulating and also cause resistance in the clinic in isolates from AIDS patients (10–12). Another common resistance mechanism in C. albicans which has been described by several authors is enhanced expression of certain multiple drug resistance proteins, leading to increased fluconazole efflux out of the cell. In some cases, reduced accumulation of drug in cells appears to account for resistance without changes in sterol 14α-demethylase or sterol C5-desaturase (36). The highly homologous CDR1 and CDR2 genes encode proteins which belong to the superfamily of ATP-binding cassette transporters (25, 31), and the MDR1 gene (previously termed BENr) encodes a protein from the major facilitator superfamily (3). These efflux pumps have been implicated in fluconazole resistance in C. albicans because some fluconazole-resistant C. albicans isolates which accumulated less intracellular fluconazole exhibited increased mRNA levels of the corresponding genes compared to fluconazole-susceptible isolates (1, 29, 31, 37). When expressed in fluconazole-hypersusceptible Saccharomyces cerevisiae mutants lacking specific multidrug transporters, CDR1, CDR2, and MDR1 conferred fluconazole resistance upon transformants (25, 29, 31). In addition, disruption of CDR1 and CDR2 in C. albicans resulted in enhanced susceptibility to the drug (30, 31).
In order to determine which of the described mechanisms commonly lead to fluconazole resistance in clinical C. albicans strains, we undertook a molecular analysis of serial C. albicans isolates from different episodes of OPC in AIDS patients who, after successful treatment of the initial episodes, failed to respond to fluconazole therapy. This report presents a complete characterization, encompassing all the known fluconazole resistance mechanisms, for two series of matched isolates which exhibit gradually increasing resistance in vitro and which were obtained from infections resistant to fluconazole therapy in the patient.
MATERIALS AND METHODS
C. albicans strains and culture conditions.
The C. albicans isolates used in this study are listed in Table 1. The isolates were obtained from recurrent episodes of OPC in two AIDS patients. Isolates were recovered by oral washings with 0.9% NaCl solution either before (isolates F1, F2, G1, G2) or during (F3, G3, G4) treatment of the episode with 100 mg of fluconazole per day. Isolate F4 was obtained from an episode which did not respond to normal fluconazole doses but which could be cured by increasing the dose to 300 mg per day. The last isolates of each series were from episodes which did not respond to enhanced doses of fluconazole (300 mg per day for patient 1 and 400 mg per day for patient 2) and which were treated with intravenous amphotericin B desoxycholate (patient 1, isolate F5) or itraconazole capsules (patient 2, isolate G5). MIC determinations have been described in detail in a previous study (27). The isolates were kept as frozen stocks at −80°C and were subcultured on YPD agar plates (10 g of yeast extract, 20 g of peptone, 20 g of glucose, 15 g of agar per liter) at 30°C.
TABLE 1.
Characteristics of serial C. albicans isolates from two AIDS patients with OPC
| C. albicans isolate | Date of isolation (mo/day/yr) | MIC (μg/ml)a
|
Flz doseb (mg/day) | Molecular changesc | ||
|---|---|---|---|---|---|---|
| Flz | Keto | Itra | ||||
| Patient 1 | ||||||
| F1 | 07/05/90 | 3.12 | 0.09 | 0.39 | 100 | |
| F2 | 08/22/90 | 6.25 | 0.048 | 0.39 | 100 | |
| F3 | 09/14/90 | 12.5 | 0.09 | 0.78 | 100 | Enhanced MDR1 mRNA levels |
| F4 | 10/12/90 | 25 | 0.09 | 0.78 | 100 | Enhanced MDR1 mRNA levels |
| F5 | 01/14/91 | ≥50 | 0.19 | 0.78 | 300 | Enhanced MDR1 and ERG11 mRNA levels |
| Patient 2 | ||||||
| G1 | 11/27/90 | 0.39 | 0.048 | 0.19 | 100 | |
| G2 | 12/04/90 | 0.39 | 0.048 | 0.39 | 100 | |
| G3 | 04/15/91 | 6.25 | 0.048 | 0.39 | 100 | Enhanced MDR1 mRNA levels |
| G4 | 05/23/91 | 25 | 0.048 | 0.39 | 100 | Enhanced MDR1 mRNA levels |
| G5 | 07/30/91 | ≥50 | 0.19 | 0.78 | 400 | Enhanced MDR1 mRNA levels, mutation in ERG11 gene, change from ERG11 heterozygosity to homozygosity |
MICs were determined by the microdilution method as described previously (27). Flz, fluconazole; Keto, ketoconazole; Itra, itraconazole.
Fluconazole dose used to treat the respective episode.
Molecular changes as compared to the first isolate of each series.
Construction of DNA probes for Southern and Northern hybridization.
A 954-bp CARE-2 fragment was amplified by PCR from the template plasmid pRFL37 (17) by using the primers 5′-CTCTAAAACTGTGCTTGGTG-3′ and 5′-AATTTGCACTCATCGAGAGC-3′. Other probes used for Southern hybridization were obtained by PCR amplification from chromosomal DNA of C. albicans strain CAI4 (4) using primers derived from the published sequences of the ERG11, MDR1, and CDR1 genes (3, 14, 25). The following oligonucleotides were used for amplification: ERG11 probe (positions 148 to 1694 in the ERG11 gene), 5′-ATGGCTATTGTTGAAACTGTCATTG-3′ (ERG1) and 5′-GCTGGTTCAGTAGGTAAAACCACC-3′ (ERG2); MDR1 probe (positions 2697 to 4391 in the MDR1 gene), 5′-ATGCATTACAGATTTTTAAGAGAT-3′ (MDR1) and 5′-CTAATTAGCATACTTAGATCTTGC-3′ (MDR2); CDR1/2 probe (positions 1832 to 3870 in the CDR1 gene), 5′-CACATTGGTAAAGAATCCCAAATTAC-3′ (CDR3) and 5′-GGTTTGACCCATCCATCAACA-3′ (CDR4). The PCR products were cloned into the SmaI site of pUC18, yielding plasmids pERG1, pMDR1, and pCDR1/2, and were partially sequenced to confirm their identity. Sequencing demonstrated that the fragment obtained with primers CDR3 and CDR4 corresponded to the CDR2 gene. As it was recently demonstrated that a corresponding probe hybridizes to both CDR1 and CDR2 (31), we refer to this fragment as CDR1/2 probe.
For the detection of mRNAs in Northern hybridization experiments, hybrid probes containing part of the C. albicans ACT1 gene in addition to sequences from the ERG11, MDR1, or CDR2 gene were constructed to include an internal control for equal RNA loading. A 0.8-kb fragment from the ACT1 coding region (positions 1808 to 2606 [20]) was ligated together with the 1.5-kb ERG11 fragment from pERG1, the 1.7-kb MDR1 fragment from pMDR1, or with a 1.6-kb CDR2 fragment from pCDR1/2 (positions 1567 to 3135 in the CDR2 gene) and cloned into pBluescript KSII, resulting in plasmids pERG2, pMDR2, or pCDR1/2N, respectively. The hybrid fragments were gel purified and used for Northern hybridizations.
Isolation of chromosomal DNA and Southern hybridization.
Chromosomal DNA from C. albicans strains was isolated as described by Millon et al. (22). Ten micrograms of DNA was digested with EcoRI, separated on a 1% (wt/vol) agarose gel, and after ethidium bromide staining, transferred by vacuum blotting to a nylon membrane and fixed by UV cross-linking. Probe labeling, hybridization, washing, and signal detection were performed with the ECL labeling and detection kit provided by Amersham (Braunschweig, Germany), in accordance with the instructions of the manufacturer. After signal detection from one probe, the blot was kept for 1 day in the detection solution to eliminate any remaining signal, washed with 5× SSC, and rehybridized with the next probe.
Isolation of total RNA and Northern hybridization.
Total RNA from C. albicans strains grown at 30°C in YPD medium to mid-log phase was isolated by the hot acidic phenol method (2). For measuring transcript levels in the presence or absence of fluconazole, overnight cultures grown in YPD medium were diluted 1:100 in fresh YPD medium and in YPD medium containing 5 μg of fluconazole (Pfizer UK, Sandwich, United Kingdom)/ml, and grown to mid-log phase. Ten micrograms of RNA was separated on a 1.2% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane by capillary blotting, and fixed by UV cross-linking. Radioactive labeling of the hybrid probes described above was performed with a random-primer DNA labeling kit (Boehringer, Mannheim, Germany). Hybridization and washing of the blots were performed under stringent conditions using standard protocols (28), and transcripts were detected by autoradiography. Blots were used only once for each probe.
Sequencing of the ERG11 alleles.
The coding region from the start codon at position 148 to position 1694 near the stop codon of the ERG11 genes from C. albicans isolates G1 to G5 was PCR amplified by using primers ERG1 and ERG2. The PCR products were phenol extracted, ethanol precipitated, and dissolved in distilled water. Sequencing was performed with 200 ng of the PCR products as template and the thermo Sequenase fluorescence-labeled primer cycle sequencing kit with deaza dGTP (Amersham). The following IRD 41 dye-labeled oligonucleotides (MWG Biotech, Ebersberg, Germany) were used for sequencing: 5′-CCCATTAAGAATCCCTGAAACC-3′ (ERG16seq1); 5′-CAGGGTCAGGCACTTTATAACC-3′ (ERG16seq2); 5′-GAAGCAGAAGTATGTTGACCACCC-3′ (ERG16seq3); 5′-CCCCTTTACCGAAAACTGGAGTAG-3′ (ERG16seq4); 5′-CGTGGTGATATTGATCCAAATCGTG-3′ (ERG16seq5). After denaturing the template DNA for 2 min at 95°C, 30 cycles of sequencing were performed, with 15 s of denaturation at 95°C, 15 s of annealing at 58°C, and a 30-s extension at 70°C. Sequence analysis was performed on a LI-COR model 4000 automated sequencer (MWG Biotech). The sequences were analyzed visually for positions of heterozygosity in the ERG11 alleles.
Identification of sterols by GC-MS.
Samples for gas chromatography-mass spectrometry (GC-MS) were prepared from 50-ml cultures in the exponential phase of growth on RPMI 1650 medium (Sigma). Following silylation for 1 h at 60°C with BSTFA (50 μl) in 50 μl of toluene, sterols were analyzed by GC-MS (VG 12-250 [VG BIOTECH]) with sterol identification by reference to relative retention time and mass spectra as reported previously (36).
Inhibition of sterol 14α-demethylase activity.
Inhibition of sterol 14α-demethylase by azole antifungals was investigated by assessing the cell-free biosynthesis of ergosterol from mevalonic acid in accordance with the methods of previous studies (36). Following extraction of nonsaponifiable lipids (sterols and sterol precursors) by two treatments with 3 ml of heptane, samples were dried under nitrogen, applied to thin-layer chromatography plates (Merck), and developed in toluene-diethyl ether (9:1 [vol/vol]). Radioactive metabolites were located by autoradiography, the band corresponding to ergosterol was excised, and radioactivity was assessed by liquid scintillation counting.
Azole content of cells.
Cellular content of fluconazole was investigated by using 109 cells incubated in 1 × 10−5 M [14C]fluconazole in 100 mM potassium phosphate buffer (pH 7.4) at 37°C (150 rpm) as described in previous studies (36). Cells were washed three times in 10 ml of unlabeled 10−4 M fluconazole prior to collection on Whatman GF/C filters to establish a favorable fluconazole concentration gradient to reduce loss of radiolabeled fluconazole from cells and to wash off nonspecifically bound fluconazole. The samples were assayed for radioactivity on a Philips 4700 scintillation counter, and efficiency was examined by the external standard method.
RESULTS
Genomic alterations in C. albicans strains during fluconazole therapy.
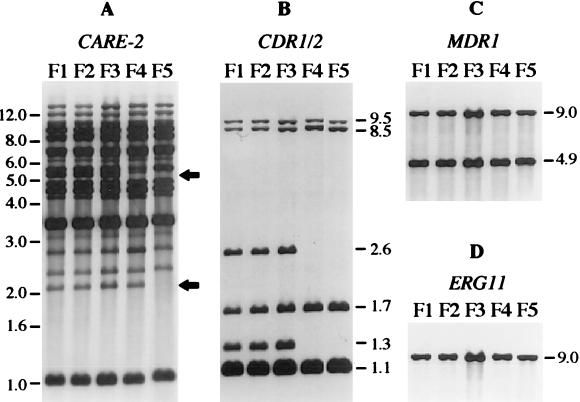
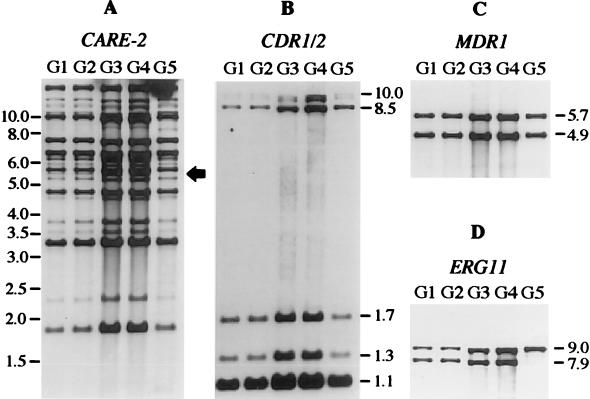
From each of two AIDS patients with recurrent OPC, five C. albicans isolates from different episodes of infection were selected for this study on the basis of their gradually increased in vitro resistance to fluconazole (Table 1). Comparison of the CARE-2 hybridization pattern of these isolates demonstrated that in both patients, a single C. albicans strain was responsible for the recurrent infections, as the fingerprint pattern of all five isolates was highly similar in each case (Fig. 1A and 2A). However, the CARE-2 hybridization pattern also revealed subtle genomic alterations in some isolates with increased MICs. In the isolates from patient 1, the disappearance of one hybridizing band was observed in isolate F4, and a second band was missing in isolate F5 (Fig. 1A, lanes 4 and 5). A similar finding was observed with the isolates from patient 2, where isolate G5 differed from the previous isolates by the absence of one band (Fig. 2A, lane 5). This indicates that during fluconazole therapy microevolution of infecting C. albicans strains takes place, a phenomenon which has also been described by others (18, 34).
FIG. 1.
Southern hybridization with different probes of EcoRI-digested chromosomal DNA of C. albicans isolates F1 to F5 from patient 1. (A) CARE-2 fingerprint pattern of the isolates. The arrows indicate the fragments missing in isolates F4 and F5. The positions of molecular size markers (in kb) are shown on the left side. (B) Hybridization with the CDR1/2 probe. (C) Hybridization with the MDR1 probe. (D) Hybridization with the ERG11 probe. The sizes of hybridizing fragments (in kb) are indicated on the right side of the blots in panels B, C, and D.
FIG. 2.
Southern hybridization with different probes of EcoRI-digested chromosomal DNA of C. albicans isolates G1 to G5 from patient 2. (A) CARE-2 fingerprint pattern of the isolates. The arrow indicates the fragment missing in isolate G5. The positions of molecular size markers (in kb) are shown on the left side. (B) Hybridization with the CDR1/2 probe. (C) Hybridization with the MDR1 probe. (D) Hybridization with the ERG11 probe. The sizes of hybridizing fragments (in kb) are indicated on the right side of the blots in panels B, C, and D.
No changes in the ergosterol content of the clinical strains was observed, thereby excluding sterol C5-desaturase defects as the basis of the observed resistance (Table 2). To investigate if the observed genomic alterations involved genes coding for multiple drug resistance proteins or the drug target enzyme, sterol 14α-demethylase, the Southern blots were rehybridized with probes specific for the CDR1/2 genes, the MDR1 gene, and the ERG11 gene. With the isolates from patient 1, a change in the CDR1/2 hybridization pattern was detected in isolates F4 and F5 (Fig. 1B, lanes 4 and 5). These isolates had lost two fragments of 1.3 and 2.6 kb which were present in the previous isolates. No change in the MDR1 or ERG11 hybridization pattern was observed in the isolates from this patient (Fig. 1C and D).
TABLE 2.
Percentage of ergosterol among total sterols, cellular concentration of fluconazole, and IC50 for inhibition of sterol 14α-demethylase among the two clinical series of C. albicans strains
| C. albicans isolate | Ergosterol level (% of total sterol) | [14C]fluconazole uptake (pmol/109 cells) | IC50 (nM) |
|---|---|---|---|
| F1 | 89.6 | 25.7 ± 2.8 | 54.2 ± 4.3 |
| F2 | 92.4 | 23.1 ± 3.9 | 51.7 ± 4.5 |
| F3 | 97.2 | 12.4 ± 2.3 | 59.4 ± 5.3 |
| F4 | 96.3 | 2.2 ± 0.8 | 69.8 ± 4.7 |
| F5 | 92.5 | 3.2 ± 1.2 | 126.6 ± 5.7 |
| G1 | 98.8 | 26.4 ± 5.9 | 53.8 ± 3.2 |
| G2 | 95.9 | 25.4 ± 4.7 | 56.6 ± 7.5 |
| G3 | 96.1 | 11.2 ± 3.1 | 59.2 ± 4.3 |
| G4 | 93.7 | 4.2 ± 1.2 | 52.6 ± 6.5 |
| G5 | 91.2 | 1.2 ± 0.9 | 625.9 ± 44.8 |
The first four isolates (G1 to G4) from patient 2 exhibited two EcoRI fragments which hybridized with the ERG11 probe (Fig. 2D). As the ERG11 gene does not contain an EcoRI site, the fragments most probably represent the two alleles of the ERG11 gene which are differentiated by a restriction site polymorphism in these isolates. The last isolate (G5), which had the highest MIC of fluconazole, had lost the smaller fragment. The most likely explanation for this observation is that isolate G5 became homozygous for one of the two ERG11 alleles, either by mitotic recombination or by a gene conversion event. No change in the CDR1/2 or MDR1 hybridization pattern was detected in the strain from patient 2 (Fig. 2B and C).
The Southern hybridizations in Fig. 1 and 2 also demonstrated that no stable amplification of any of the genes investigated had occurred during resistance development in the C. albicans strains from both patients, as there was no increase in relative signal strength and no appearance of additional hybridizing fragments.
Enhanced expression of ERG11 and MDR1 genes in fluconazole-resistant C. albicans.
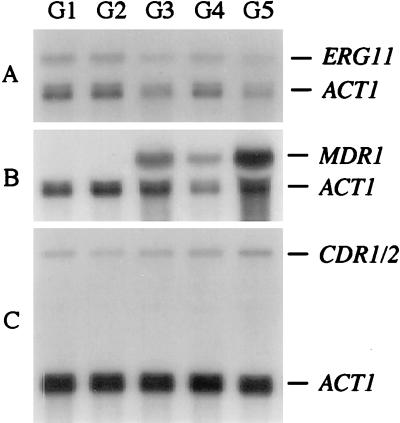
To investigate if changes in the expression of ERG11, MDR1, or CDR1/2 correlated with fluconazole resistance, the mRNA levels of these genes in the C. albicans isolates were compared after Northern hybridization of total RNA with ERG11-ACT1, MDR1-ACT1, or CDR1/2-ACT1 hybrid probes (see Materials and Methods). Isolates F1 and F2 from patient 1 did not contain detectable amounts of MDR1 mRNA (Fig. 3B, lanes 1 and 2). In contrast, isolate F3 expressed the MDR1 gene (Fig. 3B, lane 3), and there was a further 3.5-fold increase in the amount of MDR1 mRNA in isolate F4 (Fig. 3B, lane 4). This corresponded with an observed reduction in intracellular fluconazole accumulating in these strains and supports the role of MDR1 in causing resistance (Table 2). Isolate F5, which had the highest MIC (Fig. 3B, lane 5), did not contain significantly higher MDR1 mRNA levels than isolate F4, but F5 exhibited 3.5-fold greater amounts of ERG11 mRNA compared to the previous isolates F1 to F4 (Fig. 3A). This last isolate from patient 1 was the only one to show a slight increase in the amount of drug needed to inhibit the cell-free synthesis of ergosterol in vitro (Table 2). No significant alterations in the level of the CDR1/2 transcripts were observed in the isolates F1 to F5 (Fig. 3C).
FIG. 3.
Northern hybridization with different probes of total RNA of C. albicans isolates F1 to F5 from patient 1. (A) Hybridization with an ERG11-ACT1 hybrid probe. (B) Hybridization with an MDR1-ACT1 hybrid probe. (C) Hybridization with a CDR1/2-ACT1 hybrid probe. The identity of the mRNAs is indicated.
Isolates G1 to G5 from patient 2 all contained similar amounts of ERG11 mRNA (Fig. 4A). No MDR1 mRNA was detected in isolates G1 and G2 (Fig. 4B, lanes 1 and 2), but isolate G3 expressed the MDR1 gene (Fig. 4B, lane 3). MDR1 mRNA levels in G3 and G4 were comparable (Fig. 4B, lanes 3 and 4; the difference in signal strength is due to unequal loading as can be seen from the signal corresponding to the ACT1 transcript), but there was a further 1.9-fold increase in the level of MDR1 mRNA in isolate G5 compared to G4 (Fig. 4B, lane 5). Again, no significant differences in CDR1/2 transcript levels were observed in the isolates G1 to G5 (Fig. 2C). For this series the change in MDR1 transcript level also corresponded with reduced accumulation of fluconazole observed in cells from G3, G4, and G5 compared to G1 and G2.
FIG. 4.
Northern hybridization with different probes of total RNA of C. albicans isolates G1 to G5 from patient 2. (A) Hybridization with an ERG11-ACT1 hybrid probe. (B) Hybridization with an MDR1-ACT1 hybrid probe. (C) Hybridization with a CDR1/2-ACT1 hybrid probe. The identity of the mRNAs is indicated.
Mutations in the ERG11 gene correlating with fluconazole resistance.
The change from heterozygosity for ERG11 in isolates G1 to G4 to homozygosity in isolate G5 suggested that allelic differences in this gene might account for reduced fluconazole susceptibility of the enzyme, and that conversion to homozygosity could have rendered isolate G5 more resistant. To test this hypothesis, the sequences of both ERG11 alleles in isolates G1 to G5 were determined by direct sequencing of PCR-amplified DNA fragments which contained almost all of the ERG11 coding region. The results confirmed that isolates G1 to G4 indeed contained two different alleles of the ERG11 gene, whereas isolate G5 had become homozygous for one of these alleles. An illustration of this analysis is presented in Fig. 5, which shows three positions of heterozygosity in the ERG11 alleles of isolates G1 to G4. In isolate G5, only one of the two respective nucleotides was present at the corresponding positions. Overall, heterozygosity was detected at 13 positions in the ERG11 coding region in isolates G1 to G4 (Table 3). Although sequencing did not distinguish which allele was associated with which nucleotide at heterozygous positions, it is highly probable that the nucleotides detected in ERG11 from isolate G5 were contained in one of the alleles of isolates G1 to G4 and the remaining nucleotides in isolates G1 to G4 were present in the other allele. Most of the codon exchanges were silent; only two codon differences resulted in the conserved amino acid exchanges E266D and V488I. However, isolate G5 additionally contained an A at position 1537 in both alleles of the ERG11 gene whereas isolates G1 to G4 contained a G at this position in both ERG11 alleles. Therefore, the G-to-A mutation must have occurred in one ERG11 allele in an intermediate isolate which was not recovered from patient 2 before the strain became homozygous for the ERG11 gene. This mutation resulted in the substitution of serine for glycine at the corresponding position in isolate G5 and this isolate exhibited a large increase in the 50% inhibitory concentration (IC50) for the effect of fluconazole on ergosterol biosynthesis by cell-free extracts.
FIG. 5.
Detection of allelic polymorphisms in the ERG11 gene of C. albicans isolates from patient 2. The samples corresponding to the same nucleotides were in all cases run next to each other in order from isolate G1 to G5. A part of the sequencing gel where three examples of allelic polymorphism between the two ERG11 alleles in isolates G1 to G4 were detected (indicated on the right side) is shown. Isolate G5 became homozygous for one of the two alleles.
TABLE 3.
Sequence differences between the ERG11 alleles of strains G1 to G4 and G5a
| Nucleotide positionb | G1–G4 | G5 | Amino acid position | G1–G4 | G5 |
|---|---|---|---|---|---|
| 462 | T/C | C | 105 | Phe | Phe |
| 558 | C/T | T | 137 | Ser | Ser |
| 696 | T/C | C | 183 | His | His |
| 805 | C/T | T | 220 | Leu | Leu |
| 945 | A/C | C | 266 | Glu/Asp | Asp |
| 1173 | A/G | G | 342 | Lys | Lys |
| 1230 | G/A | A | 361 | Ser | Ser |
| 1431 | C/T | T | 428 | Asp | Asp |
| 1443 | C/T | T | 432 | Ala | Ala |
| 1449 | T/C | C | 434 | Ala | Ala |
| 1537 | G | A | 464 | Gly | Ser |
| 1587 | G/A | A | 480 | Leu | Leu |
| 1609 | G/A | A | 488 | Val/Ile | Ile |
| 1617 | C/T | T | 490 | Asn | Asn |
Sequence differences resulting in amino acid exchanges are highlighted in bold.
The position refers to the published sequence of the ERG11 gene (GenBank accession no. X13296).
Inducibility of ERG11 gene expression by fluconazole in susceptible and resistant C. albicans isolates.
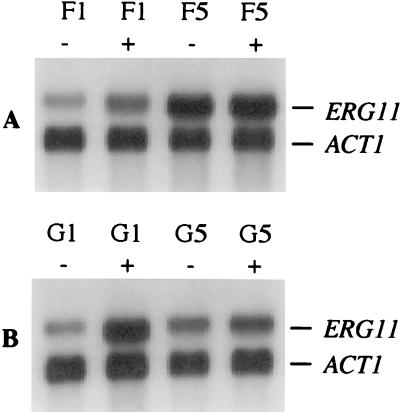
The response of fluconazole-susceptible and -resistant isolates to subinhibitory concentrations of the drug was investigated with respect to the expression of the ERG11, MDR1, and CDR1/2 genes. The growth-inhibitory effect of fluconazole strongly depends on the culture conditions (5), and preliminary experiments showed that the isolates used in this study grew well in YPD medium containing 5 μg of fluconazole/ml (data not shown). Therefore, the mRNA levels of the genes under study in the absence or presence of 5 μg of fluconazole/ml were compared in the first (susceptible) and last (resistant) selected isolate of each series. As can be seen in Fig. 6, fluconazole induced the expression of the ERG11 gene encoding the target enzyme. The susceptible C. albicans isolates from both patients exhibited enhanced ERG11 mRNA levels when grown in the presence of fluconazole as compared to the controls grown in drug-free medium (2-fold for F1 and 3.4-fold for G1). In contrast, the resistant isolates did not change the level of ERG11 mRNA in response to fluconazole. The resistant isolate F5 constitutively expressed the ERG11 gene even in the absence of fluconazole at higher levels than the susceptible isolate F1 under induced conditions, but ERG11 mRNA levels were not further increased by fluconazole (Fig. 6A). In drug-free medium, the resistant isolate G5 exhibited an amount of ERG11 mRNA similar to that of the susceptible isolate G1 but, in contrast to G1, ERG11 expression was not inducible by fluconazole in isolate G5 (Fig. 6B). The expression of neither MDR1 nor CDR1/2 was induced by fluconazole in any of the strains tested (data not shown).
FIG. 6.
ERG11 mRNA levels in susceptible and resistant C. albicans strains during growth in the absence (−) or presence (+) of 5 μg of fluconazole/ml. The ERG11 and ACT1 mRNAs are indicated on the right side. (A) Isolates F1 (susceptible) and F5 (resistant) from patient 1. (B) Isolates G1 (susceptible) and G5 (resistant) from patient 2.
DISCUSSION
The results of the present study confirm earlier observations that fluconazole resistance in previously susceptible C. albicans strains can develop during therapy. In two series of isolates from two different patients analyzed in this study, resistance developed gradually, and different molecular changes were correlated at various steps with reduced drug susceptibility of the isolates. Whereas no alteration was detected which could account for the enhanced in vitro resistance of isolate F2 compared to F1, the enhanced MIC for isolate F3 correlated with the expression of MDR1 mRNA which was not detected in F1 and F2 and the reduced accumulation of fluconazole. The further increase in MDR1 expression could also explain the enhanced resistance of isolate F4. This isolate additionally exhibited genomic alterations involving the CDR1 or CDR2 genes. The loss of two hybridizing fragments may be due to deletion of one of the genes or to mitotic recombination which could result in the loss of fragments representing heterozygous alleles. The probe used in this study could not differentiate between CDR1 and CDR2, and recently, additional CDR genes have been identified by other researchers (33). Such genes might also hybridize to the CDR1/2 probe and could have suffered from genomic rearrangements. However, the observed genomic changes did not lead to alterations in the overall expression of the CDR genes as judged by Northern hybridization. The last isolate of this series, F5, which had the highest MIC of fluconazole, constitutively expressed the ERG11 gene at high levels, and this corresponded with a slightly increased IC50 for fluconazole in cell-free studies on the susceptibility of the enzyme target. Therefore, correspondingly greater amounts of the target enzyme sterol 14α-demethylase may have contributed to the resistant phenotype of this isolate. Definitive proof will require specific antibodies to quantify sterol 14α-demethylase and cytochrome P45051, together with knowledge of the biodiversity of the cytochrome P450 superfamily in C. albicans.
Multiple mechanisms also seem to be involved in fluconazole resistance development in the isolates from patient 2. Again, expression of MDR1 presumably accounts for the elevated resistance, as reflected in reduced accumulation of fluconazole, of isolate G3 compared to that of G1 and G2. No molecular changes which could explain the differences in drug susceptibility between isolates G3 and G4 were detected. However, as G4 accumulated less intracellular fluconazole than G3, other unidentified efflux pumps may be involved. Several alterations were detected in the isolate with the highest resistance, G5. This isolate exhibited a further increase in the level of MDR1 mRNA compared to the previous isolates. In addition, G5 became homozygous for a mutated allele of the ERG11 gene. In contrast to isolate F5 from patient 1, ERG11 mRNA levels were not enhanced in isolate G5, but the differences in the amino acid sequence of the enzyme may have led to a reduced affinity for fluconazole. The G464S alteration was previously observed in a resistant isolate as were the two other polymorphisms detected in separate strains (19). As the initial strains of this series were sensitive to fluconazole that implies the E266D and V488I changes do not contribute to resistance, as an enzyme with reduced affinity for fluconazole should exert a resistance phenotype in the heterozygous state. A change from heterozygosity to homozygosity for the ERG11 gene has also recently been described to be responsible for enhanced fluconazole resistance in a clinical C. albicans strain from another matched set of isolates (38). In that case, an R467K amino acid substitution was found in both ERG11 alleles from the isolate with elevated resistance which was not present in the previous isolates and must therefore have occurred in an intermediate isolate not recovered from the patient. A similar situation was detected in the isolates from patient 2 described in our present study. The G464S mutation, which is located in the heme binding domain of the Erg11p protein near the R467K substitution described by White (38), must have been introduced into one ERG11 allele before conversion to homozygosity. Molecular modeling of azole binding to sterol 14α-demethylase of C. albicans has included the known binding of the triazole of fluconazole to the heme moiety and interactions of the N-1 substituent group with the apoprotein, particularly F233,235 (16). One obvious mechanism which might account for the effect of a G464S mutation observed in the cell-free studies is a subtle alteration in the plane or position of the heme, resulting in interference in the aromatic interactions of apoprotein and fluconazole which occurs above the plane of the heme. The contribution of the G464S mutation in sterol 14α-demethylase to fluconazole resistance was recently confirmed by Sanglard et al. (32) who, after submission of our manuscript, reported the same mutation in several drug-resistant isolates. However, the two other amino acid differences between the ERG11 alleles in isolates G1 to G4, E266D, and V488I may also have increased fluconazole resistance after strain G5 became homozygous for the allele with D266 and I488. Determination of the influence of these amino acid substitutions, individually and in combination, on drug susceptibility, however, awaits the further molecular characterization of mutant enzymes in vitro.
Our finding that multiple molecular mechanisms contribute to the development of fluconazole resistance in clinical C. albicans isolates confirm the recently published observation by White (37, 38), who demonstrated that increased levels of ERG11, CDR, and MDR1 mRNAs as well as an altered ERG11 gene all correlated with increased drug resistance of C. albicans isolates from an HIV-infected patient. In the isolates presented in our study, we could not demonstrate an involvement of CDR genes in fluconazole resistance, which was found by White (37) and also by other researchers (1, 29, 31). Remarkably, in both series of C. albicans isolates, enhanced MDR1 mRNA levels were clearly correlated with elevated fluconazole resistance. MDR1 has been reported to mediate resistance against fluconazole but not against other azoles, in contrast to CDR1 and CDR2 (29, 31). Correspondingly, overexpression of MDR1 in the strains analyzed here did not result in cross-resistance against ketoconazole and itraconazole (Table 1). In a previous study, Sanglard et al. (29) found that MDR1 was not involved in fluconazole resistance as often as CDR1, and it was also shown that an mdr1-negative derivative of C. albicans strain CAF4-2 was not hypersusceptible to fluconazole (30). Our results support the idea of a more frequent role of enhanced MDR1 expression in fluconazole resistance in C. albicans. In addition, we found that the C. albicans strain CAI4, like CAF4-2 a derivative of strain SC5314, did not detectably express the MDR1 gene during growth in YPD medium (unpublished observation). Low-level MDR1 expression has also been reported for strain CAF4-2 (30). Therefore, it is not surprising that inactivation of the MDR1 gene in this strain did not influence fluconazole susceptibility. Another mdr1-negative derivative of a strain expressing the MDR1 gene was constructed by Goldway et al. (6), but fluconazole susceptibilities of the mutant and its parent were not tested in that study. A definite genetic proof for MDR1-mediated clinical fluconazole resistance would require inactivation of the gene in resistant strains which express MDR1, like the isolates described in our study. The development of positive selection markers (13) will make the introduction of such mutations in wild-type, clinical C. albicans strains feasible.
We demonstrated that C. albicans responds to the presence of fluconazole by enhancing the expression of the ERG11 gene, a phenomenon recently also described by others (1). Although other researchers supposed that increased ERG11 expression is not a major cause of fluconazole resistance (29), increased ERG11 mRNA levels were clearly correlated with drug resistance in isolate F5 in our study. The ERG11 gene in this isolate was constitutively expressed at higher levels than in previous isolates from the same patient even after induction of the gene by fluconazole. This finding suggests that mutations, either in the ERG11 regulatory region or in an unknown regulatory protein, which increase expression of the gene above normally inducible levels, result in fluconazole resistance. Further investigation of isolate F5 is warranted to substantiate this through characterization of sterol 14α-demethylase among the multiplicity of cytochrome P450 which may be present in C. albicans.
The MDR1 and CDR genes were not inducible by fluconazole in the strains tested. Susceptible isolates did not detectably express the MDR1 gene, either in the absence or in the presence of fluconazole. In contrast, resistant isolates constitutively expressed the MDR1 gene at high levels. Promoter mutations could account for this observation. Another explanation which has also been proposed by others (29) is that mutations in a regulator(s) of MDR1 and/or other genes encoding multiple drug resistance proteins result in unrestricted MDR1 expression. Such a putative regulator would not respond to fluconazole, as the normal biological function of the efflux pump is presumably different from conferring resistance to synthetic drugs. In S. cerevisiae, Pdr1p and Pdr3p activate transcription of the genes PDR5, SNQ2, and YOR1 encoding ATP-binding cassette transporters (7, 8, 21), and very recently it was shown that Pdr1p and Pdr3p also regulate the expression of genes belonging to the major facilitator superfamily (23). Analogous proteins may be responsible for the regulation of C. albicans genes encoding multiple drug resistance proteins, like MDR1.
In conclusion, our results demonstrate that C. albicans uses different mechanisms to develop fluconazole resistance which can be combined in the same strain to generate high levels of resistance during antimycotic treatment.
ACKNOWLEDGMENTS
This study was supported by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMBF grant O1 K1 8906-0).
We thank B. Lasker for the gift of plasmid pRFL37. Gerwald Köhler and Jörg Hacker are acknowledged for critical reading of the manuscript.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Fling M E, Kopf J, Tamarkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 4.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghannoum M A, Rex J H, Galgiani J N. Susceptibility testing of fungi: current status of correlation of in vitro data with clinical outcome. J Clin Microbiol. 1996;34:489–495. doi: 10.1128/jcm.34.3.489-495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldway M, Teff D, Schmidt R, Oppenheim A B, Koltin Y. Multidrug resistance in Candida albicans: disruption of the BENr gene. Antimicrob Agents Chemother. 1995;39:422–426. doi: 10.1128/aac.39.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzmann D J, Burnett P E, Golin J, Mahé Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly S L, Arnoldi A, Kelly D E. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans. 1993;21:1034–1038. doi: 10.1042/bst0211034. [DOI] [PubMed] [Google Scholar]
- 10.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals is associated with the formation of 14α-methylergosta-8,24(28)-3β,6β-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 11.Kelly S L, Lamb D C, Kelly D E, Loeffler J, Einsele H. Resistance to fluconazole in Candida albicans from AIDS patients involving cross-resistance to amphotericin. Lancet. 1996;348:1523–1524. doi: 10.1016/S0140-6736(05)65949-1. [DOI] [PubMed] [Google Scholar]
- 12.Kelly S L, Lamb D C, Kelly D E, Manning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 13.Köhler G A, White T C, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14α-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb D C, Kelly D E, Schunck W H, Shyedehi A Z, Akhtar M, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 16.Lamb D C, Kelly D E, Baldwin B C, Gozzo F, Boscott P, Richards W G, Kelly S L. Differential inhibition of Candida albicans CYP51 by stereoisomers of azole antifungals. FEMS Microbiol Lett. 1997;149:23–30. doi: 10.1111/j.1574-6968.1997.tb10303.x. [DOI] [PubMed] [Google Scholar]
- 17.Lasker B A, Page L S, Lott T J, Kobayashi G S. Isolation, characterization, and sequencing of Candida albicans repetitive element 2. Gene. 1992;116:51–57. doi: 10.1016/0378-1119(92)90628-3. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart S R, Reed B D, Pierson C L, Soll D R. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeffler J, Kelly S L, Hebart H, Schumacher U, Lass-Floer C, Einsele H. Sequence analysis of CYP51 in nineteen fluconazole resistant and sensitive strains of Candida albicans. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 20.Losberger C, Ernst J F. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 1989;17:9488. doi: 10.1093/nar/17.22.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahé Y, Parle-McDermott A, Nourani A, Delahodde A, Lamprecht A, Kuchler K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol. 1996;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 22.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nourani A, Wesolowski-Louvel M, Delaveau T, Jacq C, Delahodde A. Multiple-drug-resistance phenomenon in the yeast Saccharomyces cerevisiae: involvement of two hexose transporters. Mol Cell Biol. 1997;17:5453–5460. doi: 10.1128/mcb.17.9.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odds F C. Candida and candidosis: a review and bibliography. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 25.Prasad R, Dewergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 26.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruhnke M, Eigler A, Tennagen I, Geiseler B, Engelmann E, Trautmann M. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J Clin Microbiol. 1994;32:2092–2098. doi: 10.1128/jcm.32.9.2092-2098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 32.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherer S, Ran Y. Candida albicans information: webpage. 1997. http://alces.med.umn.edu/Candida.html http://alces.med.umn.edu/Candida.html. . [Google Scholar]
- 34.Schröppel K, Rotman M, Galask R, Soll D R. Evolution and replacement of Candida albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J Clin Microbiol. 1994;32:2646–2654. doi: 10.1128/jcm.32.11.2646-2654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 36.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Resistance to fluconazole in Candida albicans from AIDS patients associated with decreased accumulation of drug. FEMS Microbiol Lett. 1995;131:337–341. doi: 10.1111/j.1574-6968.1995.tb07797.x. [DOI] [PubMed] [Google Scholar]
- 37.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]