Abstract
Two extended-spectrum mutants of the class D β-lactamase OXA-10 (PSE-2) from Pseudomonas aeruginosa isolates obtained in Ankara, Turkey, were described previously and were designated OXA-11 and -14. P. aeruginosa 906 and 961, isolated at the same hospital, were highly resistant to ceftazidime (MIC ≥ 128 μg/ml) and produced a β-lactamase with a pI of 6.2. The MICs of ceftriaxone, cefoperazone, cefsulodin, and cefepime were 4- to 16-fold above the typical values for P. aeruginosa, whereas the MICs of penicillins and cefotaxime were raised only marginally. Ceftazidime MICs were not significantly reduced by clavulanate or tazobactam at 4 μg/ml. Ceftazidime resistance did not transfer conjugatively but was mobilized to P. aeruginosa PU21 by plasmid pUZ8. Both isolates gave similar DNA restriction patterns, suggesting that they were replicates; moreover, they yielded identically sized BamHI fragments that hybridized with a blaOXA-10 probe. DNA sequencing revealed that both isolates had the same new β-lactamase, designated OXA-16, which differed from OXA-10 in having threonine instead of alanine at position 124 and aspartate instead of glycine at position 157. The latter change is also present in OXA-11 and -14 and seems critical to ceftazidime resistance. Kinetic parameters showed that OXA-16 enzyme was very active against penicillins, cephaloridine, cefotaxime, and ceftriaxone, but hydrolysis of ceftazidime was not detected despite the ability of the enzyme to confer resistance.
Resistance to oxyimino cephalosporins in enterobacteria is often associated with extended-spectrum β-lactamases (ESBLs), most of which are mutants of molecular class A β-lactamases, specifically TEM-1 and SHV-1 (2, 19). In Pseudomonas aeruginosa, on the other hand, the most frequent mechanisms of resistance to the oxyimino cephalosporins are derepression of the AmpC chromosomal enzyme and up-regulation of multi-drug efflux (3, 4), and only one extended-spectrum TEM mutant (TEM-42) has been reported (24). Nevertheless, P. aeruginosa has been a major source of unusual ESBLs. Examples include IMP-I, the first plasmidic zinc β-lactamase (32); PER-1, a class A enzyme now widespread in Turkey, though not elsewhere (6, 26, 27, 30); OXA-15, an ESBL mutant of OXA-2 (7); and OXA-11 and -14, which are ESBL mutants of OXA-10 enzyme (5, 11). OXA-11 β-lactamase has two mutations compared with OXA-10, whereas OXA-14 has only one of them. OXA-11 and -14 were produced by isolates from Hacettepe University Hospital in Ankara, Turkey. Total-DNA restriction profiles of these two isolates were identical, and both carried plasmids that gave identical restriction fragments on digestion with EcoRI (5). It is likely that the producer isolates differed only in the presence or absence of the second mutation in the β-lactamase gene (5).
In the present study, we describe the discovery and characterization of a further ESBL mutant of OXA-10, also from P. aeruginosa isolates collected from Hacettepe University Hospital.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. aeruginosa 906 and 961 were isolated in June 1993 from patients treated for burns at Hacettepe University Hospital and were retained because of their considerable resistance to ceftazidime (MIC, 128 μg/ml). P. aeruginosa PU21 ilv leu Strr Rifr was used as a recipient in transconjugation (14). P. aeruginosa ABD and 455, with plasmids pMLH52 and pMLH53, encoding OXA-11 and -14 β-lactamases, respectively (5, 11), and their P. aeruginosa PU21 transconjugants were used as controls, together with P. aeruginosa PU21(pMLH51) as a reference producer of OXA-10 β-lactamase (11, 20). Gene sequencing has confirmed that the OXA-10 enzyme encoded by pMLH51 is identical to that encoded by plasmid R151, the prototype host of the OXA-10 gene (13). Plasmid pUZ8 (IncP-1), determining resistance to kanamycin, tetracycline, and mercuric chloride, was used to mobilize resistance (12). Escherichia coli NCTC 50192 (31), with plasmids of 154, 66, 38, and 7 kb, and P. aeruginosa PU21 with plasmid pMG2 (450 kb) served as controls in plasmid-sizing studies (29).
Antibiotics.
Antimicrobials tested were aztreonam and cefepime (Bristol-Myers Squibb, Syracuse, N.Y.); cefsulodin (Novartis, Basel, Switzerland); ceftazidime (Glaxo-Wellcome, Stevenage, Hertfordshire, United Kingdom); piperacillin sodium, tazobactam, and tetracycline (Wyeth-Lederle, Taplow, Berkshire, United Kingdom); cephalothin, moxalactam, and tobramycin (Lilly, Basingstoke, Hampshire, United Kingdom); imipenem (Merck Sharp and Dohme, Hoddesdon, Hertfordshire, United Kingdom); ceftriaxone (Roche, Welwyn Garden City, Hertfordshire, United Kingdom); cefotaxime and cefpirome (Roussel, Uxbridge, Middlesex, United Kingdom); benzylpenicillin, cephaloridine, cloxacillin, gentamicin, kanamycin, oxacillin, and rifampin (Sigma, St. Louis, Mo.); ampicillin sodium, carbenicillin disodium, and clavulanate lithium (SmithKline Beecham, Brentford, Middlesex, United Kingdom); and meropenem (Zeneca, Macclesfield, Cheshire, United Kingdom).
Susceptibility tests.
MICs were determined on DST agar (Oxoid, Basingstoke, Hampshire, United Kingdom) with inocula of 104 CFU per spot, as described previously (11).
Plasmid transfer to P. aeruginosa PU21.
Logarithmic-phase cells of P. aeruginosa isolates 906 and 961 were mated with similar cultures of P. aeruginosa PU21 on DST agar (11). After overnight incubation, transconjugants were selected on the DST agar containing ceftazidime at 25 or 50 μg/ml plus rifampin at 100 μg/ml. Where appropriate, the mobilizing plasmid pUZ8 was first transferred from P. aeruginosa PU21(pUZ8) to the isolates, using the same plate-mating method but with selection on DST agar containing ceftazidime at 25 μg/ml and kanamycin at 1,000 μg/ml. To confirm their presence and to estimate their sizes, the plasmids were extracted by the method of Kado and Liu (15) and electrophoresed at 100 V for 4 h in 0.7% agarose gels at 4°C.
Detection of β-lactamases and their genes.
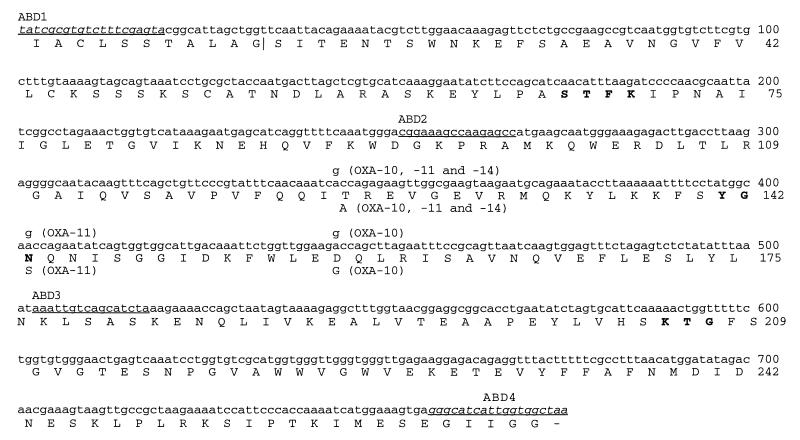
β-Lactamases were characterized by isoelectric focusing of ultrasonic extracts prepared from overnight nutrient agar cultures (Oxoid) (22). For gene probing, plasmids or total DNA were extracted and digested with BamHI and then electrophoresed on agarose, Southern blotted to nylon membrane, and hybridized with probes, exactly as described previously for strain ABD (11). The probe for blaOXA-10 was made by PCR amplification of a DNA fragment corresponding to the coding region from pMLH51, using primers ABD1 and ABD4 (Fig. 1), and was labeled with digoxigenin (DIG DNA labelling and detection kit; Boehringer, Lewes, East Sussex, United Kingdom).
FIG. 1.
Gene and protein sequences of OXA-16 β-lactamase in comparison to those of OXA-10, -11, and -14 enzymes. The nucleotide sequence of blaOXA-10 described by Huovinen et al. (13) runs from bases 57 to 957 and contains the OXA-10 coding region. Nucleotides, shown above the OXA-16 sequences, and the deduced amino acid changes, shown below the OXA-16 sequences, indicate differences in OXA-10, -11, and -14 β-lactamase. The signal peptide extends from amino acid residues 1 to 20, and the proposed cleavage site is indicated by a vertical line. The underlined nucleotide sequences represent the primers used for sequencing and PCR amplification. Sequences corresponding to the amplification primers ABD-1 and -4, shown in italics, have not been independently determined for OXA-16. Boldface letters represent amino acids around the active site. The nucleotide sequence shown corresponds to that of the OXA-16 gene, determined in this study.
Sequencing of the β-lactamase gene.
OXA-10-related genes from clinical isolates were amplified by PCR with primers ABD1 and 5′ biotin-labelled ABD4 (Fig. 1), using the methods described previously (5). The two DNA strands of the product were separated by using paramagnetic beads conjugated with streptavidin (Dynabeads M-280 Streptavidin; Dynal, New Ferry, Wirral, United Kingdom) and were used as templates for sequencing by chain termination (5), with ABD1, ABD2, and ABD3 as primers (Fig. 1).
β-Lactamase purification.
Cultures were grown overnight with shaking in 0.6 liters of antibiotic no. 3 broth (Oxoid) at 37°C and then diluted into 12 liters of identical fresh medium and incubated for 5 h to yield late-logarithmic-phase cells. The bacteria were harvested at 5,000 × g for 15 min at 37°C, washed once in 20 mM triethanolamine buffer (pH 7.6) (buffer A), and then resuspended in the same buffer and frozen and thawed twice. Debris was removed by ultracentrifugation at 100,000 × g for 45 min at 4°C, and the supernatants were loaded onto columns (2.6-cm diameter by 40 cm) of DEAE Sephadex A-50 equilibrated with buffer A. Elution was done with a gradient of 0 to 0.5 M K2SO4 in buffer A. Fractions with β-lactamase activity were dialyzed against 50 mM malonic acid (pH 7.0) overnight and then adjusted to pH 4.9 (OXA-10 enzyme) or 5.0 (enzyme from isolate 906) with 0.5 M H2SO4. These solutions were loaded on to S-Sepharose high-performance columns (1.6-cm diameter by 10 cm) (Pharmacia LKB, Milton Keynes, Buckinghamshire, United Kingdom), which were equilibrated with 50 mM malonic acid, pH 4.9 (OXA-10) or 5.0 (enzyme from isolate 906), and eluted with a gradient of 0 to 0.5 M K2SO4 in the same buffer. The fractions with β-lactamase activity were dialyzed against 4 liters of 50 mM bis-Tris–H2SO4, pH 7.0. Their purity was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by the system of Lugtenberg et al. (21), and the protein concentration was determined by the microbicinchoninic acid method (Pierce, Rockford, Ill).
Spectrophotometric β-lactamase assays.
β-Lactamase activity was assayed by UV spectrophotometry at 37°C in 0.1 M phosphate buffer at pH 7.0. The following wavelengths were used: ampicillin and penicillin G, 235 nm; cephaloridine, 295 nm; cephalothin, 262 nm; cefotaxime and ceftazidime, 257 nm; cloxacillin and oxacillin, 263 nm; and imipenem and meropenem, 297 nm. Biphasic kinetics were analyzed by the program of De Meester et al. (10), kindly provided by J.-M. Frère. Kinetic parameters were determined by using the Enzfitter program (16).
Bioassays of β-lactamase activity.
Cultures of the isolates and transconjugants were grown overnight as lawns on nutrient agar plates and then resuspended in 2 ml of sterile 10 mM phosphate buffer, pH 7.0, chilled on ice, and sonicated for two or three 10-s bursts at an amplitude of 15 μm. Debris was removed by centrifugation at 16,000 × g for 15 min, and 200 μl of the supernatants was mixed with 200 μl of a 40-μg/ml solution of ceftazidime in 10 mM phosphate buffer, pH 7.0. As a control, ceftazidime solutions (40 μg/ml) were incubated with similar extracts of P. aeruginosa PU21 containing or not containing plasmid pMLH51, pMLH52, or pMLH53. After incubation for 1 h at 37°C, samples of the mixtures were transferred to wells (0.7-cm diameter) in a 23- by 23-cm bioassay plate containing 120 ml of Mueller-Hinton agar seeded with 0.6 ml of a 500-fold dilution of an overnight culture of E. coli NCTC 10418. The plate was incubated for 18 h at 37°C, and inhibition zones were compared to those for the untreated drug.
Nucleotide sequence accession number.
The GenBank database accession number for the sequence shown in Fig. 1 is AF043100.
RESULTS
Susceptibilities, β-lactamases, and plasmids of isolates 906 and 961.
Isolates 906 and 961 exhibited similar antibiograms. Both were highly resistant to ceftazidime (MICs, 128 to 256 μg/ml) and were 4- to 16-fold less susceptible than is typical for the species (3) to cefoperazone, ceftriaxone, cefepime, cefpirome, and cefsulodin (Table 1). On the other hand, the MICs recorded for aztreonam (2 to 4 μg/ml), carbapenems (1 to 4 μg/ml), piperacillin (8 μg/ml), and carbenicillin (64 μg/ml) were no higher than is typical for P. aeruginosa. Ceftazidime resistance was only reduced twofold by clavulanate and was not affected by tazobactam.
TABLE 1.
MICs for P. aeruginosa isolates, transconjugants, and reference comparators
| Organism | MIC (μg/ml)c
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pip | Pip-Clav | Pip-Taz | Carb | Carb-Clav | Carb-Taz | Ctaz | Ctaz-Clav | Ctaz-Taz | Ctax | Ctriax | Csul | Cpim | Cpir | Cpz | Moxa | Azt | Mero | Imp | Gent | |
| Isolates | ||||||||||||||||||||
| 961 | 8 | 16 | 8 | 64 | 64 | 64 | 128 | 64 | 128 | 32 | 64 | 32 | 32 | 32 | 32 | 32 | 4 | 1 | 4 | 128 |
| 906 | 8 | 8 | 4 | 64 | 64 | 32 | 256 | 64 | 128 | 16 | 64 | 32 | 16 | 64 | 32 | 32 | 2 | 1 | 4 | 128 |
| Transconjugants | ||||||||||||||||||||
| PU21/pUZ8-906 | 32 | 8 | 16 | 128 | 64 | 128 | 512 | 128 | 256 | 64 | >128 | 64 | 32 | 128 | 128 | 64 | 16 | -d | 1 | 128 |
| PU21/pUZ8-961 | 32 | 4 | 8 | 128 | 64 | 128 | 256 | 64 | 256 | 32 | 128 | 32 | 32 | 32 | 64 | 64 | 8 | - | 1 | 128 |
| Reference isolates, transconjugants, and recipient | ||||||||||||||||||||
| 455/OXA-14a | 64 | 64 | 64 | 512 | 512 | 512 | 512 | 1,024 | 512 | 32 | 64 | 32 | 128 | 128 | 128 | 16 | 32 | 2 | 2 | >128 |
| ABD/OXA-11b | 32 | 32 | 32 | 512 | 512 | 512 | 512 | 512 | 512 | 32 | 128 | 32 | 32 | 64 | 64 | 64 | 32 | 2 | 8 | >128 |
| PU21(pMLH53)/OXA-14a | 64 | 32 | 32 | 512 | 256 | 256 | 512 | 256 | 256 | 32 | 128 | 32 | 64 | 128 | 512 | 16 | 64 | 2 | 4 | >128 |
| PU21(pMLH52)/OXA-11b | 64 | 32 | 64 | 512 | 512 | 512 | 1,024 | 1,024 | 1,024 | 64 | 128 | 128 | 128 | 128 | 128 | 32 | 128 | 2 | 2 | >128 |
| PU21(pMLH51)/OXA-10 | 64 | 32 | 64 | 512 | 256 | 512 | 4 | 4 | 4 | 16 | 64 | 32 | 2 | 8 | 128 | 16 | 16 | 2 | 2 | >128 |
| PU21(pUZ8) | 2 | 2 | 2 | 64 | 64 | 64 | 2 | 1 | 2 | 16 | 8 | 2 | 1 | 2 | 8 | 8 | 4 | 1 | 1 | 8 |
| PU21 | 2 | 4 | 4 | 64 | 64 | 64 | 4 | 2 | 2 | 16 | 16 | 2 | 2 | 8 | 8 | 8 | 4 | 1 | 2 | 8 |
Hall et al. (11).
Danel et al. (5).
Azt, aztreonam; Carb, carbenicillin; Clav, clavulanate at 4 mg/liter; Cpim, cefepime; Cpir, Cefpirome; Cpz, cefoperazone; Csul, cefsulodin; Ctax, cefotaxime; Ctaz, ceftazidime; Ctriax, ceftriaxone; Imp, imipenem; Mero, meropenem; Moxa, moxalactam (latamoxef); Pip, piperacillin; Taz, tazobactam at 4 mg/liter; Gent, gentamicin.
-, not done.
Isoelectric focusing of crude extracts showed a single pI-6.2 β-lactamase in both isolates, and electrophoresis revealed that each isolate carried a 325-kb plasmid.
Transfer of ceftazidime resistance to P. aeruginosa PU21.
Attempts to transfer ceftazidime resistance conjugatively from isolates 906 and 961 to P. aeruginosa PU21 were unsuccessful, but transfer was achieved after plasmid pUZ8 was inserted into the isolates. The transconjugant of strain 906 acquired a 90-kb plasmid, and that of 961 acquired a 60-kb plasmid, compared to 40 kb for native pUZ8. Both transconjugants expressed the pI-6.2 β-lactamase. Both also showed similar resistance profiles and were particularly resistant to ceftazidime (MIC, > 256 μg/ml, compared with 4 μg/ml for P. aeruginosa PU21). The MICs of other cephalosporins and piperacillin were raised 4- to 16-fold by acquisition of the plasmids. The transconjugants remained as susceptible as strain PU21 to imipenem and meropenem and were not significantly (i.e., more than twofold) more resistant to carbenicillin. MICs of piperacillin, carbenicillin, and ceftazidime were not reduced by more than twofold by clavulanate or tazobactam at 4 μg/ml. Similar resistance patterns—though with carbenicillin resistance—were seen for P. aeruginosa PU21(pMLH52) and PU21(pMLH53) with OXA-11 and -14 enzymes, respectively, whereas P. aeruginosa PU21(pMLH51), with classical OXA-10 enzyme, was resistant only to piperacillin, carbenicillin, and cefoperazone.
Restriction patterns of DNA digests and hybridization with gene probe.
Total DNA was extracted from isolates 906, 961, ABD, 455 and, as a negative control, P. aeruginosa PU21. After digestion with BamHI, the fragments were separated by agarose gel electrophoresis. Isolates 906 and 961 gave identical restriction patterns, suggesting that they represented a single strain, whereas a different pattern was seen for isolates 455 and ABD. Following blotting to nylon membranes, DNA from isolates 906, 961, ABD, and 455 hybridized with a blaOXA-10 probe whereas DNA from PU21 did not do so. Isolates 906 and 961 carried the gene corresponding to blaOXA-10 on a 3.5-kb BamHI fragment indistinguishable from that yielded by isolates ABD and 455. Additionally, it was found that the 325-kb plasmid from isolates 906 and 961 and the recombinant plasmids from their PU21 transconjugants hybridized with the blaOXA-10 probe.
Gene sequencing.
Sequence analysis was performed on the genes encoding the OXA-10-related β-lactamases in isolates 906 and 961. The section sequenced corresponded to nucleotide positions 156 to 931 of blaOXA-10 (13) and excluded only the regions coding for the first 16 amino acids (which are part of the signal peptide) and the last 5 residues (Fig. 1). Both isolates had the same new OXA-10-derived β-lactamase, with glycine replaced by asparagine at position 157 and alanine changed to threonine at position 124. This enzyme was named OXA-16, and its 325-kb encoding plasmid was called pMLH57.
Purification of OXA-10 and -16 β-lactamases.
Identical methods were used to purify OXA-10 and -16 β-lactamases, except that the pH for the cation exchange was increased from 4.9 to 5.0 for OXA-16. Concentrations of salt needed to elute OXA-16 and OXA-10 enzymes were 91 and 81 mM, respectively, in the anion exchange and 160 and 200 mM, respectively, in the cation exchange. Both β-lactamases eluted from the cation exchanger after all the contaminant proteins, and the purity of each exceeded 95%, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Twelve liters of culture of isolate 906 yielded 0.4 mg of OXA-16 β-lactamase, and 12 liters of P. aeruginosa PU21(pMLH51) culture yielded 3.75 mg of OXA-10 β-lactamase.
Hydrolysis of β-lactams by OXA-10 and -16 β-lactamases.
For many substrates, the time course for hydrolysis showed biphasic behavior, complicating analysis. Hydrolysis curves were characterized as biphasic if the rate within the first minute was at least 20% higher than the steady-state rate immediately afterward. Smaller differences were not analyzed, since they could not be accurately measured during the short period while the rate was changing, but they were observed frequently.
In the case of OXA-10 β-lactamase, the hydrolysis curves of penicillin G and ceftriaxone were linear, whereas biphasic kinetics were seen for ampicillin, carbenicillin, oxacillin, cloxacillin, and cephaloridine. The initial rate/steady-state rate ratios varied with the substrate concentration, but for ampicillin they were between 1.2 and 1.7 and for carbenicillin they ranged from 3.4 to 3.0, while the highest ratio was 16, for cloxacillin.
In the case of OXA-16 β-lactamase, the hydrolysis curves were linear for cephalothin and ceftriaxone and biphasic for penicillin G, ampicillin, carbenicillin, oxacillin, cloxacillin, cephaloridine, and cefotaxime. The initial rate/steady-state rate ratios were similar to those for OXA-10 enzyme.
OXA-10 enzyme showed its lowest steady-state Km values (40 to 60 μM) for penicillin G, cephalothin, and ceftriaxone, whereas the highest values (>2,000 μM) were for cloxacillin and cephaloridine (Table 2). Between these extremes were ampicillin, carbenicillin, and oxacillin, with Km values around 200 μM. The turnover number (kcat) was less than 10 s−1 for cephalothin, cefotaxime, and ceftriaxone and between 30 and 90 s−1 for carbenicillin, penicillin G, and cephaloridine but exceeded 500 s−1 for ampicillin, oxacillin, and cloxacillin. The best substrates in terms of linear or steady-state efficiency (kcat/Km) were oxacillin and ampicillin, followed by penicillin G, whereas kcat/Km values were at least 10-fold lower for the other substrates. For substrates showing biphasic kinetics, higher affinity was seen in the initial phase, except with carbenicillin; thus, the ratios of Km values in the steady-state and initial phases (KmSS/Kmo) were 2.3 and 2.4 for oxacillin and cloxacillin, respectively, 5.9 for cephaloridine, and 0.53 for carbenicillin. The ratios of initial and steady-state kcat values (kcato/kcatSS) were 2 for cephaloridine, 4 to 5 for carbenicillin and cloxacillin, and 10 for ampicillin and oxacillin, but the two parameters were similar for oxacillin. The efficiency (kcat/Km) in the initial phase was two- to threefold greater than in the steady-state phase except for cloxacillin, where a 12-fold differential was observed.
TABLE 2.
Kinetic parameters (± SD) for the initial (Vo) and steady-state (Vss) phases of hydrolysis of β-lactams by OXA-10 and -16 β-lactamase in 0.1 M phosphate buffer (pH 7.0) at 37°C
| Substrate | OXA-10
|
||||||
|---|---|---|---|---|---|---|---|
|
Vss
|
Vo
|
Initial β-lactamase concnb (μM) | |||||
| Km (μM) | kcat (s−1) | kcat/Kma | Km (μM) | kcat (s−1) | Kcat/Km | ||
| Penicillin G | 63 ± 6c | 89 ± 10 | 1,410 | -e | - | - | 0.61 |
| Ampicillin | 235 ± 30 | 5,590 ± 30 | 2,500 | 77 ± 9 | 531 ± 12 | 6,900 | 0.3 |
| Carbenicillin | 195 ± 13 | 31 ± 1 | 159 | 370 ± 44 | 126 ± 5 | 340 | 0.61 |
| Oxacillin | 222 ± 16 | 608 ± 10 | 2,740 | 96 ± 8 | 660 ± 10 | 6,870 | 0.61 |
| Cloxacillin | 2,640 ± 323 | 530 ± 36 | 196 | 1,110 ± 100 | 2,680 ± 150 | 2,420 | 1.21 |
| Cephaloridine | 2,340 ± 300 | 79 ± 8 | 33 | 395 ± 83 | 39 ± 3 | 98 | 0.61 |
| Cephalothin | 38 ± 2 | 6 ± 0.1 | 158 | - | - | - | 3.03 |
| Cefotaxime | 346 ± 19 | 9 ± 0.2 | 26 | - | - | - | 2.43 |
| Ceftriaxone | 55 ± 2 | 3 ± 0.3 | 54 | - | - | - | 12.13 |
| Ceftazidime | NDd | ND | ND | - | - | - | 12.13 |
| OXA-16
| |||||||
|---|---|---|---|---|---|---|---|
|
Vxx
|
Vo
|
Initial β-lactamase concnb (μM) | |||||
| Km (μM) | kcat (s−1) | kcat/Kma | Km (μM) | kcat (s−1) | kcat/Kma | ||
| 65 ± 1 | 48 ± 1 | 738 | <20 | >60 | - | 1.74 | |
| 205 ± 21 | 97 ± 4 | 473 | 142 ± 31 | 163 ± 8 | 1,150 | 0.58 | |
| 129 ± 32 | 17 ± 1 | 132 | 150 ± 26 | 69 ± 4 | 460 | 0.82 | |
| 960 ± 159 | 411 ± 36 | 428 | 660 ± 120 | 455 ± 35 | 758 | 0.44 | |
| 2,080 ± 333 | 264 ± 25 | 127 | 1,120 ± 261 | 1,200 ± 137 | 1,070 | 0.82 | |
| 424 ± 40 | 21 ± 1 | 50 | 379 ± 61 | 30 ± 2 | 79 | 0.82 | |
| 32 ± 3 | 3 ± 1 | 94 | - | - | - | 1.74 | |
| 346 ± 43 | 6 ± 0.5 | 17 | 157 ± 40 | 10 ± 0.5 | 64 | 0.82 | |
| 36 ± 2 | 1.4 ± 0.5 | 39 | - | - | - | 1.74 | |
| ND | ND | ND | ND | ND | ND | 1.74 | |
(kcat/Km) × 1,000, in micromolar−1 second−1.
Concentration of the stock enzyme from which samples were taken to start the assays before dilution with substrate.
Cases where Vo and Vss were significant are indicated in boldface.
ND, hydrolysis not detected.
-, linear kinetics, so Vo corresponds to Vss.
Turning to OXA-16 enzyme, the lowest Km values among linear substrates, or nonlinear ones at steady state, were for penicillin G, cephalothin, and ceftriaxone (30 to 70 μM), and the highest was for cloxacillin (>2,000 μM) (Table 2). Between these extremes were ampicillin, carbenicillin, cephaloridine, and oxacillin, with Km values between 130 and 940 μM (Table 2). kcatSS was less than 10 s−1 for cephalothin, cefotaxime, and ceftriaxone; between 20 and 100 s−1 for carbenicillin, ampicillin, and penicillin G; and exceeded 200 s−1 for oxacillin and cloxacillin. The best substrate in terms of kcatSS/KmSS values was penicillin G, followed by ampicillin and oxacillin. Km values for nonlinear substrates were 1.4- to 2.2-fold lower in the initial phase than at steady state, except with carbenicillin and cephaloridine, where the two parameters were similar. kcato was 1.5- to 4.5-fold higher than kcatSS for ampicillin, carbenicillin, cloxacillin, cephaloridine, and cefotaxime, but the two parameters were little different for oxacillin. Hydrolysis was eightfold more efficient in terms of kcat/Km values in the initial phase than at steady state for cloxacillin and 1.6- to 3.8-fold more efficient for carbenicillin, ampicillin, oxacillin, cephaloridine, and cefotaxime.
No hydrolysis of ceftazidime by OXA-16 enzyme was detected by spectrophotometry (Table 2); nevertheless, the enzyme gave considerable ceftazidime resistance (Table 1). Therefore, bioassay with crude extracts was performed and likewise failed to detect inactivation of ceftazidime by either OXA-10 or -16 enzyme, whereas this assay confirmed inactivation hydrolysis of ceftazidime by OXA-11 and -14 β-lactamases, as had been detected previously by spectrophotometry (5, 11).
DISCUSSION
OXA-16 is the third extended-spectrum mutant of OXA-10 (PSE-2) β-lactamase to be described, following OXA-11 (11) and OXA-14 (5). All these mutants have asparagine replacing glycine at position 157; in addition OXA-16 had threonine replacing alanine at position 124 and OXA-11 has serine replacing asparagine at residue 143. Mutation at position 157 thus seems critical to ceftazidime resistance, and it was also noted in a ceftazidime-selected laboratory mutant of OXA-13 β-lactamase, which is a more distant relative of OXA-10 (23). A further ESBL mutant of OXA-10, OXA-17, lacks the substitution at position 157 and does not confer significant ceftazidime resistance, although it does compromise other oxyimino aminothiazolyl cephalosporins (8).
Acquisition of OXA-16 enzyme by P. aeruginosa PU21 conferred resistance to ceftazidime (especially), ceftriaxone, cefepime, cefpirome, cefoperazone, latamoxef, and piperacillin, as did acquisition of OXA-11 or -14 enzymes (Table 1) (5, 11). However, production of OXA-16 enzyme caused only a twofold rise in the MIC of carbenicillin for strain PU21, whereas OXA-11 and -14 enzymes gave greater protection, as does classical OXA-10 enzyme. Neither OXA-16 β-lactamase nor any other OXA-10 mutant conferred resistance to carbapenems. A final general feature was that the resistance caused by OXA-10-related enzymes, including OXA-16, was reduced only two- to fourfold by clavulanate at 4 μg/ml and not at all by tazobactam at 4 μg/ml (Table 2).
The original OXA-16 producers—P. aeruginosa 906 and 961—were less resistant than their PU21 transconjugants to penicillins and cephalosporins and retained susceptibility to carbenicillin, piperacillin, and aztreonam, both relative to National Committee for Clinical Laboratory Standards breakpoints (25) and with regard to typical MICs for P. aeruginosa isolates without acquired resistance (3). This apparent susceptibility may have reflected lower β-lactamase production in the clinical isolates than in the transconjugants, due to a difference in the plasmid copy number, or a combination of greater permeability and weaker efflux function in the isolates. These aspects were not investigated.
Both OXA-16 producers were isolated from burn patients at Hacettepe University Hospital in Ankara, Turkey, which is the same establishment where the first producers of OXA-11 and -14 enzymes—P. aeruginosa ABD and 455, respectively—were found. Isolates 906 and 961 appeared to be replicates, based on the similarity of the BamHI restriction profiles of their DNAs and on the fact that each carried the OXA-16 gene on a nonconjugative 325-kb plasmid. They were less closely related to isolates ABD and 455, which themselves appear to be replicates, apart from the point mutation that distinguishes their β-lactamases (5, 11). These latter organisms yielded a BamHI restriction profile different from that of isolates 906 and 961 and carried their blaOXA genes on 475-kb plasmids, which were conjugatively self-transmissible to P. aeruginosa PU21. However, a common ancestor is suggested by the fact the OXA-10-related genes were carried by 3.5-kb BamHI fragments in all these OXA-11, -14, and -16 producers.
The OXA-16-encoding gene of the present isolates appeared to be located within a transposon, as judged by its ability to transfer to plasmid pUZ8. The gene encoding classical OXA-10 β-lactamase was previously shown to be transposon associated (1), whereas attempts to achieve transposition of blaOXA-11 and blaOXA-14 were consistently unsuccessful (5, 11). Curiously, the OXA-16-encoding pUZ8 recombinant plasmid obtained from isolate 961 was smaller than that from isolate 906 (60 kb compared with 90 kb). It is possible that another transposon besides that carrying blaOXA-16 had inserted into pUZ8-906, whereas only the OXA-16 transposon had inserted into pUZ8-961; alternatively, the pUZ8 plasmid may have acquired two copies of the OXA-16 transposon, since the size of its insert was within experimental error of twice the size of the insert in pUZ8-961. Yet another possibility is that the two isolates had different blaOXA-16-coding transposons, but this seems unlikely, both because of the general similarity of the organisms and because it would imply an exceptionally large size (50 kb) for the transposon from isolate 906.
Kinetic studies were undertaken on purified OXA-16 enzyme, with OXA-10 as a comparator. The ability of OXA-16 to give increased cephalosporin resistance could not be correlated with increased hydrolytic activity in vitro, regardless of whether kcat or kcat/Km was taken as a measure of activity. This contrasts with ESBL mutants of TEM and SHV β-lactamases, which consistently exhibit greatly increased in vitro activity against the cephalosporins to which they confer resistance (2). Most strikingly, attempts to explain the ability of OXA-16 enzyme to confer ceftazidime resistance were unsuccessful. In the case of OXA-11 enzyme, ceftazidime resistance was associated with an increased kcat/Km ratio, owing to a reduced km compared to that of OXA-10 enzyme (11), but no such association was found for OXA-16; indeed, it proved impossible to detect ceftazidime hydrolysis by OXA-16, even by a very sensitive bioassay. A possible explanation for these anomalies may lie in the biphasic kinetics often seen for OXA-10-related β-lactamases. Such kinetics were reported for several substrates with OXA-10 itself (17) and with OXA-16 (this study) (Table 2) and for all substrates with OXA-14 (5, 9). The general pattern is for the enzyme to swiftly convert from a more active to a less active form, and data for OXA-14 suggests that this may reflect a dilution-dependent monomer-dimer interconversion (9). Given that the β-lactamase in the cell periplasm is more concentrated than in any practicable assay (18), it may be that OXA-10 family enzymes are in a more active form in the cell than in the assay.
In summary, this paper has described OXA-16, the third ESBL mutant of OXA-10 enzyme. Like the previously described OXA-11 and -14 mutants, OXA-16 had glycine replaced by aspartate at position 157 and had an increased ability to confer resistance to ceftazidime and, to a lesser extent, to other oxyimino aminothiazolyl cephalosporins. All three mutants have been recorded from P. aeruginosa isolates collected at a single Turkish hospital, though we are now also aware of such mutants at a second hospital in the same city (28). Their evolution parallels that of the TEM and SHV ESBL mutants, though the relationships between resistance and hydrolytic activity are much less clear. The emergence of OXA ESBLs in P. aeruginosa is disturbing, since it narrows the range of therapeutic options. A particular concern is that, unlike with TEM, SHV, and PER ESBLs, resistance could not be overcome with clavulanic acid or with penicillanic acid sulfone β-lactamase inhibitors.
REFERENCES
- 1.Bissonnette L, Roy P H. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1992;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H Y, Yuan M, Ibrahim-Elmagboul I B, Livermore D M. National survey of susceptibility to antimicrobials amongst clinical isolates of Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:521–534. doi: 10.1093/jac/35.4.521. [DOI] [PubMed] [Google Scholar]
- 4.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 5.Danel F, Hall L M C, Gur D, Livermore D M. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danel F, Hall L M C, Gur D, Akalin H E, Livermore D M. Transferable production of PER-1 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:281–294. doi: 10.1093/jac/35.2.281. [DOI] [PubMed] [Google Scholar]
- 7.Danel F, Hall L M C, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danel F, Hall L M C, Gur D, Livermore D M. Abstracts of the European Congress of Chemotherapy. 1996. Multiple secondary β-lactamase, including a new OXA-10 mutant, in two Turkish P. aeruginosa isolates, abstr. T119. [Google Scholar]
- 9.Danel F. Extended-spectrum β-lactamases from Pseudomonas aeruginosa isolates collected in Turkey. Ph.D. thesis. London, United Kingdom: University of London; 1997. [Google Scholar]
- 10.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère J M, Waley S G. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem Pharmacol. 1987;36:2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- 11.Hall L M C, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges R W, Matthew M. Acquisition by Escherichia coli of plasmid-borne β-lactamases normally confined to Pseudomonas spp. Plasmid. 1979;2:269–278. doi: 10.1016/0147-619x(79)90045-3. [DOI] [PubMed] [Google Scholar]
- 13.Huovinen P, Huovinen S, Jacoby G A. Sequence of PSE-2 β-lactamase. Antimicrob Agents Chemother. 1988;32:134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby G A. Properties of R-plasmids determining gentamicin-resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974;6:239–252. doi: 10.1128/aac.6.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leatherbarrow R J. Enzfitter: a non-linear regression data analysis program for the IBM PC/P52. Cambridge, United Kingdom: Elsevier Biosoft; 1987. [Google Scholar]
- 17.Ledent P, Raquet X, Joris B, Van-Beemen J, Frère J M. A comparative study of Class D β-lactamases. Biochem J. 1993;292:555–562. doi: 10.1042/bj2920555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore D M, Davy K W, Williams J D. Cephalosporin resistance in Pseudomonas aeruginosa, with special reference to the proposed trapping of antibiotics by beta-lactamase. Chemioterapia. 1985;4:28–35. [PubMed] [Google Scholar]
- 19.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livermore D M, Maskell J P, Williams J D. Detection of PSE-2 β-lactamase in enterobacteria. Antimicrob Agents Chemother. 1984;25:268–272. doi: 10.1128/aac.25.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugtenberg B, Meijers J, Peters R, Van der Hoek P, Van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 22.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–175. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 23.Mugnier P, Podglajen I, Gutmann L, Collatz E. Program and abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1994. Cloning and nucleotide sequence analysis of two Pseudomonas aeruginosa genes encoding the novel OXA-type β-lactamase variants OXA-12 and OXA-13, susceptible to inhibition by imipenem, abstr. C98; p. 139. [Google Scholar]
- 24.Mugnier P, Dubroust P, Casin I, Arlet G, Collatz E. A TEM-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2488–2493. doi: 10.1128/aac.40.11.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Methods for determining bactericidal activity of antimicrobial agents. M36-P. 7, no. 2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1987. [Google Scholar]
- 26.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:962–969. doi: 10.1128/aac.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozen Y, Arman D, Livermore D M. Abstracts of the X Mediterranean Congress on Chemotherapy. 1996. Extended-spectrum β-lactamases in P. aeruginosa isolates from Ankara University Hospital, abstr. 403; p. 188. [Google Scholar]
- 29.Summers A O, Jacoby G A. Plasmid-determined resistance to tellurium compounds. J Bacteriol. 1977;129:276–281. doi: 10.1128/jb.129.1.276-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivian A. Plasmid expansion? Microbiology. 1994;140:213–214. doi: 10.1099/13500872-140-2-213-a. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]