Abstract
Faced with intense level of chloroquine (CQ) resistance in Plasmodium falciparum malaria, Rwanda replaced CQ with amodiaquine (AQ) + sulfadoxine-pyrimethamine (SP) in 2001, and subsequently with artemether–lumefantrine (AL) in 2006, as first-line treatment for uncomplicated malaria. Following years of discontinuation of CQ use, re-emergence of CQ-susceptible parasites has been reported in countries including Malawi, Kenya and Tanzania. In contrast, high levels of SP resistant mutant parasites continue to be reported even in countries of presumed reduced SP drug selection pressure. The prevalence and distributions of genetic polymorphisms linked with CQ and SP resistance at two sites of different malaria transmission intensities are described here to better understand drug-related genomic adaptations over time and exposure to varying drug pressures in Rwanda. Using filter paper blood isolates collected from P. falciparum infected patients, DNA was extracted and a nested PCR performed to identify resistance-mediating polymorphisms in the pfcrt, pfmdr1, pfdhps and pfdhfr genes. Amplicons from a total of 399 genotyped samples were analysed by ligase detection reaction fluorescent microsphere assay. CQ susceptible pfcrt 76K and pfmdr1 86N wild-type parasites were found in about 50% and 81% of isolates, respectively. Concurrently, SP susceptible pfdhps double (437G-540E), pfdhfr triple (108N-51I-59R), the quintuple pfdhps 437G-540E / pfdhfr 51I-59R-108N and sextuple haplotypes were found in about 84%, 85%, 74% and 18% of isolates, respectively. High-level SP resistance associated pfdhfr 164L and pfdhps 581G mutants were noted to decline. Mutations pfcrt 76T, pfdhfr 59R and pfdhfr 164L were found differentially distributed between the two study sites with the pfdhfr 164L mutants found restricted in at Ruhuha site, eastern Rwanda. Overall, sustained intense levels of SP resistance mutations and a recovery of CQ susceptible parasites were found in this study following 7 years and 14 years of the drug withdrawal from use, respectively. Most likely, the high prevalence of resistant parasites selected by the continued use of DHFR/DHPS inhibitors like trimethoprim-sulfamethoxazole (TS) for the treatment of and prophylaxis against bacterial infections among HIV infected individuals as well as the continued use of IPTp-SP within the East and Central African regions for malaria prevention among pregnant women may partly account for the observed sustained SP resistant parasite prevalent. With regard to CQ, the slow recovery of CQ susceptible parasites may have been caused partly by the continued use of CQ and/or CQ mimicking antimalarial drugs like AQ in spite of policies to withdraw it from Rwanda and the neighbouring countries of Uganda and Tanzania. Continued surveillance of P. falciparum CQ and SP associated polymorphisms is recommended for guiding future rational drug policy-making and mitigation of future risk of anti-malaria drug resistance development.
Keywords: Plasmodium falciparum, chloroquine, sulfadoxine-pyrimethamine, resistance, polymorphisms
Graphical abstract
Background
Globally, malaria accounts for about 214 million cases and over 438,000 deaths annually (World Health Organization, 2015). A major hindrance to malaria control is the development of resistance in malaria parasites to available antimalarial therapies. Currently, artemisinin-based combination therapies (ACTs) (that consist of a combination of a fast-acting artemisinin component (artesunate, artemether, or dihydroartemisinin) and a longer-acting partner drug (lumefantrine, amodiaquine, piperaquine, or mefloquine) are widely used for treatment of P. falciparum malaria Although not a problem in Sub-Saharan Africa, ACTs clinical failure has been confirmed in Southeast Asia raising concerns of a possible lack of malaria treatment in the near future at a time when anti-malarial therapy options are limited (Dondorp et al., 2009; Ashley et al., 2015).
Prior to introduction of ACTs, widespread resistance to two more affordable and safe anti-malarial drugs; chloroquine (CQ) - a highly effective first line anti-malarial monotherapy used for about 50 years, and anti-folate drug sulfadoxine - pyrimethamine (SP), led to their withdrawal from primary use in many malaria-endemic settings (Young & Moore. 1961; Harinasuta et al., 1965; Enosse et al., 2008; Hastings et al., 2002; Kublin et al., 2003; Pearce et al., 2009, White. 1999). Although completely withdrawn from use in Rwanda in 2008, SP continues to be used for intermittent preventive treatment of malaria during pregnancy (IPTp-SP), infancy (IPTi) and for seasonal malaria chemoprevention (SMC) strategies in countries surrounding Rwanda. This practice poses a spillover risk of continued impact on SP resistance levels in Rwanda. Resistance to SP has been associated with polymorphisms in the P. falciparum dihydrofolate reductase (pfdhfr) and dihydropteroate synthase (pfdhps) genes while polymorphisms in the P. falciparum CQ resistance transporter (pfcrt) gene is the major mediator of resistance to CQ and amodiaquine (AQ) (Su et al., 1997; Fidock et al., 2000; Nzila-Mounda et al., 1998; Triglia et al., 1997). In addition, the P. falciparum multidrug resistance (pfmdr1) gene polymorphisms are associated with increased sensitivity to lumefantrine, mefloquine, and dihydroartemisinin, and to decreased sensitivity to CQ and AQ (Rosenthal, 2013; Koenderink et al., 2010).
Faced with emerging resistance in P. falciparum in Rwanda, CQ was replaced with AQ + SP in 2001 and the later subsequently replaced with artemether–lumefantrine (AL) in 2006, as first line antimalarial therapies for uncomplicated malaria (Zeile et al., 2012). For SP however, its use in intermittent preventive treatment of malaria in pregnancy (IPTp) continued until 2008 when it was withdrawn due to increasing anti-folate resistance (Karema et al., 2012). Elsewhere, after periods of complete CQ withdrawal, re-emergence of CQ-sensitive parasite strains, albeit at varying rates over time and across different geographic settings, has been reported suggesting that CQ-sensitive parasites may have a fitness advantage over resistant parasites in the absence of CQ drug selection pressure (Ndiaye et al., 2012; Mwai et al., 2009; Laufer et al., 2006). This is further evidenced by the notably lower prevalence of mutant pfcrt 76T and pfmdr1 86Y alleles in low malaria transmission settings where drug pressure is presumably less (Ord et.al, 2007). Similar to the CQ experience, use of SP has been associated with ever increasing levels of resistance in P. falciparum in malaria endemic countries including Rwanda (Matondo et al., 2014; Karema et al., 2010). However, four years after cessation of SP use, high-level SP resistance was still observed in Rwanda (Karema et al., 2010).
Data on anti-malarial drug resistance is needed for rational drug policy-making, effective malaria management and designing strategies to mitigate risk and burden of drug resistance. For Rwanda, there is paucity of these data. This study measured the prevalence and distributions of P. falciparum molecular markers of resistance to CQ and SP, 14 and 7 years after a policy change involving withdrawal of these two drugs from use, respectively, at two sites of different transmission intensities.
Materials and methods
Study area and design
Rwanda is broadly divided into four malaria ecologic zones based on altitude, climate, level of transmission, and disease vector prevalence (President’s Malaria Initiative, 2015). Topographically, malaria transmission is considered meso-endemic in the plain regions of eastern and southern provinces while being epidemic-prone in the high plateau and hill settings of northern and western provinces, respectively. Ruhuha sector, Bugesera district, eastern province is located within the high malaria transmission zone whilst Mubuga sector, Kalongi district, western province is located in the low transmission zone (President’s Malaria Initiative, 2015) (Figure 1). P. falciparum infected isolates were collected from malaria confirmed cases seen at two rural health facilities located in the two sectors in a cross-sectional survey carried out between January and February 2015.
Figure 1.
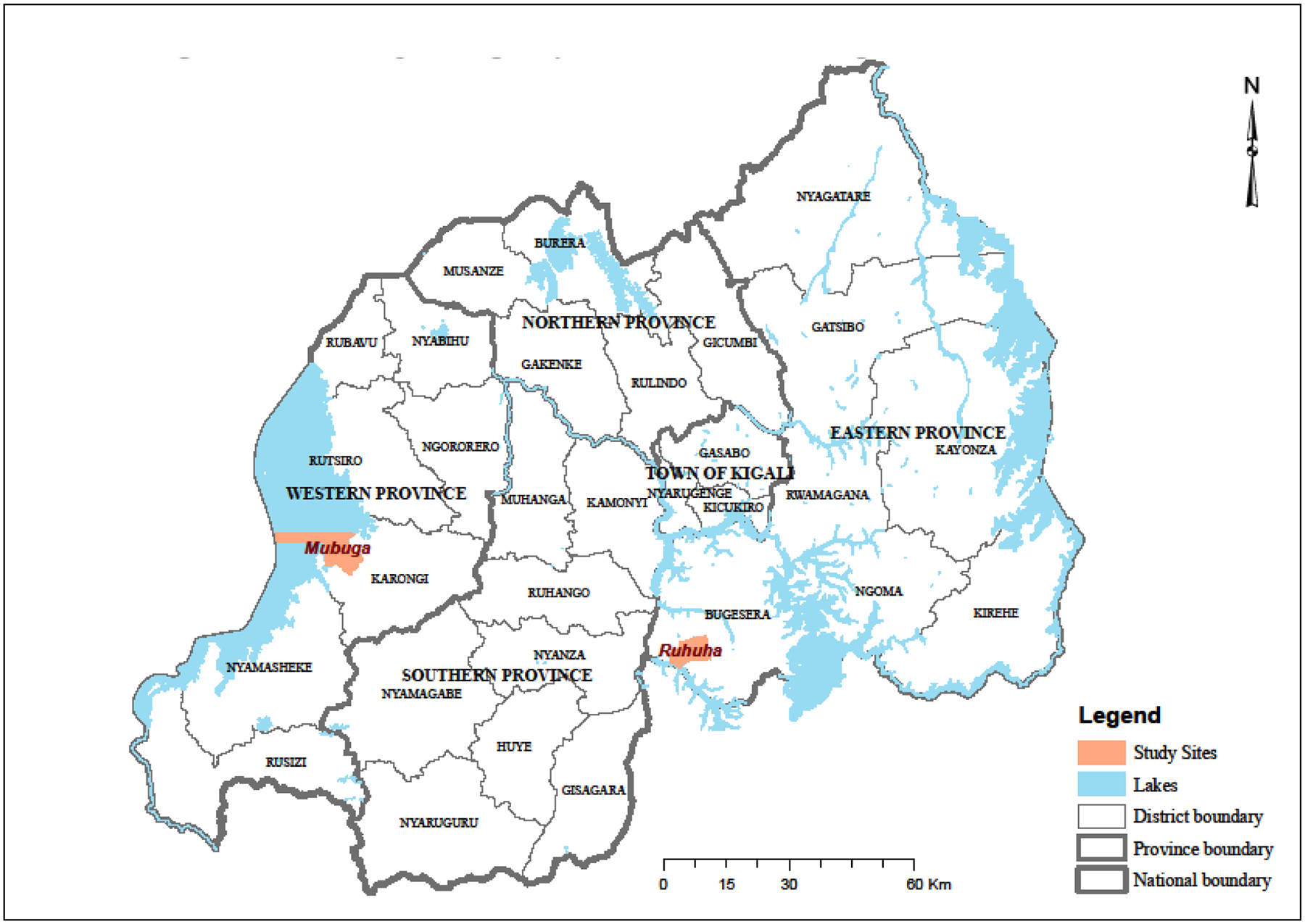
Location map showing study sites of Ruhuha and Mubuga in Rwanda.
Study participant enrolment and assessments
Study inclusion was limited to health-facility area residents who were microscopically confirmed with P. falciparum infections and who were aged ≥ 6 months. Upon provision of a written informed consent, finger-prick blood samples were then collected and used for preparation of thick and thin blood film for microscopy and for blotting on to filter papers.
Ligase Detection Reaction-Fluorescent Microsphere (LDR-FM) Assay
DNA was extracted from filter paper bloodspots using Chelex® (Bio-Rad, Germany) as described elsewhere (Kain et al., 1991). Genomic DNA was used to analyse single nucleotide polymorphisms (SNPs) in pfcrt, pfmdr1 and pfdhfr and pfdhps genes by nested PCR as previously described (LeClair et al., 2013). The ligase detection reaction-fluorescent microsphere assay was used to analyse all SNPs of interest (Nankoberanyi et al., 2014). SNPs were categorized as wild type (WT), mutant and mixed alleles against the comparator control reference strain DNA.
Statistical analysis
All statistical analyses were done using STATA version 13.1 (STATA Corp Inc., TX, USA). Differences in characteristics distribution of the study population for the different sites were tested by analysis of variance (ANOVA). Prevalence of SNPs was calculated for WT or mixed infections or pure mutants. In the final analysis, all pure mutant and mixed infections were summed up to generate the number of mutant genotypes per codon. Genotype proportions between the two study sites were compared using Pearson’s chi square test. A p value of < 0.05 was considered statistically significant.
Ethical clearance
All adults and caregivers of children < 18 years were informed of the study purpose and procedures; recruitment was done only after obtaining informed written consent. The study was reviewed and approved by the National Health Research Committee (NHRC) and the Rwanda National Ethics Committee (No. 020/RNEC/2015), Kigali, Rwanda.
Results
Patient characteristics and variable distributions
Four hundred and two (402) patients aged 6 months to 73 years were enrolled. Of these, 399 patients whose isolates provided at least one genotype result were included in the current study. Table 1 describes study participant baseline data.
Table 1.
Demographic characteristics at enrolment for 399 study participants from Ruhuha and Mubuga sites, Rwanda. *
| Variable | Variable sub-group | Mubuga n =205 | Ruhuha n=194 | Overall N=399 |
|---|---|---|---|---|
| Age (Mean ± SD) | ‐ | 17.7 ± 14.0* | 13.1 ± 12.7 | 15.5 ± 13.5) |
| Age Group | 0 – 5 years | 26 (12.7) | 51 (26.3) | 77 (19.3) |
| 6 – 15 years | 96 (46.8) | 94 (48.4) | 190 (47.6) | |
| > 16 Years | 83 (40.5) | 49 (25.3) | 132 (33.1) | |
| Sex | Male | 100 (48.8) | 79 (40.7) | 179 (44.9) |
| Female | 105 (51.2) | 115 (59.3) | 220 (55.1) | |
| Geometric Mean Parasite / μl blood | ‐ | 599.5 (95% CI#: 457.2 – 786.0) | 2190.7 (95% CI#: 1649.1 – 2910.2) | 1125.7 (95% CI#: 916.7 – 1382.4) |
Shows Mean + standard deviation (SD);
shows 95% Confidence Interval (CI
Genotyping efficacy at each codon
Per codon, typing was achieved in 97.2% of samples for pfcrt 76 (Figure 2); 95.7% for codon 86, 86.7% for codon 184, 94.7% for codon 1034, 97.5% for codon 1042 and 97.0% for codon 1246 at pfmdr1(Table 2); 94.2% for codon 51, 95.5% for codon 59, 95.0% for codon108 and 94.5% for codon 164 in pfdhfr ; 91.2% for codon 437, 91.0% for codon 540, 91.0% for codon 581, and 362 for codon 613 in pfdhps (Table 3). Among the typed polymorphisms, susceptible WT infections were observed for alleles pfmdr1 1042N and 1034S and pfdhps 613A, while mixed type infections were identified for pfcrt 76 (14%), Pfmdr1 86 (17%), 184 (32%) and 1246 (16%), pfdhfr 51 (1%) and 59 (17%) and for pfdhps 437 (8%), 540 (0.5%) and 581 (9%), respectively. Saturation (100%) levels for mutant strain were only identified in pfdhfr codon 108.
Figure 2.
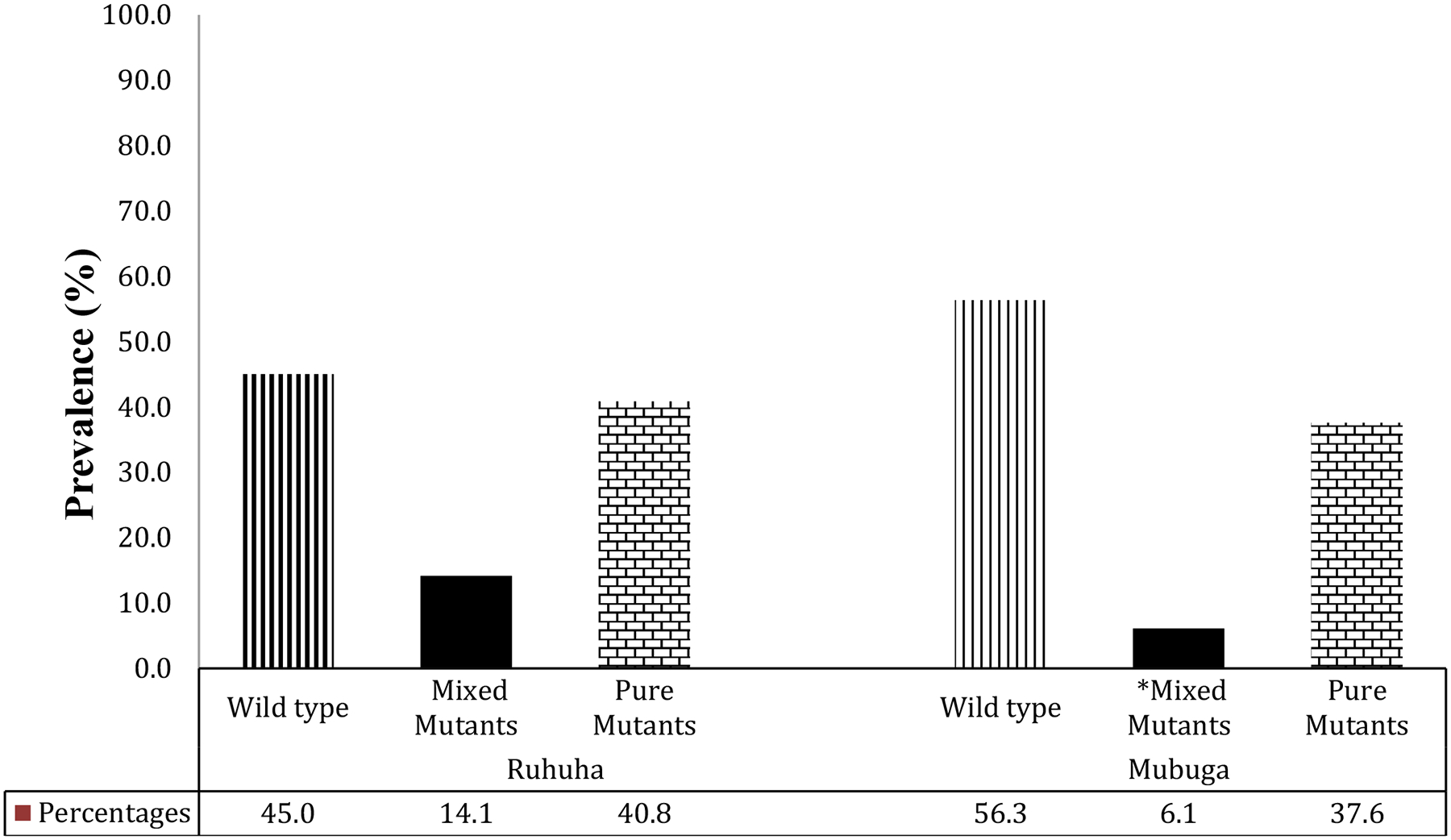
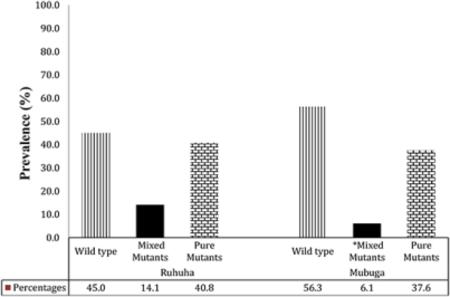
Prevalence of pfcrt K76T genotypes among isolates collected at Ruhuha vs. Mubuga sites, Rwanda.
*Mixed infections denote isolates in which both WT and mutant genotypes were detected in the same individual.
Table 2.
A comparison of pfmdr1 genotype proportions by study site Mubuga and. Ruhuha
| Ruhuha | Mubuga | All sites | |||||
|---|---|---|---|---|---|---|---|
| pfmdr1 genotypes | Wild Type n (%) | Mixed* n (%) | Mutant n (%) | Wild type n (%) | Mixed n (%) | Mutant n (%) | Total Mutants (Mixed + pure mutants) n (%) |
| N86Y | 146 (78.5) | 31 (16.7) | 9 (4.8) | 163 (83.2) | 28 (14.3) | 5 (2.6) | 73 (19.1) |
| Y184F | 64 (37.4) | 54 (31.6) | 53 (31.0) | 75 (42.9) | 58 (33.1) | 42 (24.0) | 207 (59.8) |
| N1042C | 188 (100) | 0 (0) | 0 (0) | 201 (100) | 0 (0) | 0 (0) | 0 (0) |
| S1034C | 188 (100) | 0 (0) | 0 (0) | 190 (100) | 0 (0) | 0 (0) | 0 (0) |
| D1246Y | 141 (79.2) | 29 (16.3) | 8 (4.5) | 162 (82.2) | 19 (9.6) | 16 (8.1) | 72 (19.2) |
| N86Y, D1246Y | 177 (91.2) | 17 (8.8) | 190 (92.7) | 15 (7.3) | 32 (8.0) | ||
Mixed infection denotes an isolate in which both WT and mutant genotypes were detected.
Table 3.
Prevalence of pfdhrf and pfdhps genotypes by study site Mubuga and Ruhuha
| Ruhuha | Mubuga | All sites | |||||
|---|---|---|---|---|---|---|---|
| pfdhfr alleles | Wild Type n (%) | Mixed* n (%) | Mutant n (%) | Wild type n (%) | Mixed n (%) | Mutant n (%) | All Mutants (Mixed + pure mutants) (%) |
| N51I | 0 (0) | 2 (1.1) | 188 (98.9) | 1 (0.5) | 0 (0) | 185 (99.5) | 375 (99.7) |
| C59R | 30 (15.8) | 33 (17.4) | 127 (66.8) | 7 (3.7) | 24 (12.6) | 160 (83.8) | 344 (90.3) |
| S108N | 0 (0) | 0 (0) | 190 (100) | 0 (0) | 0 (0) | 189 (100) | 379 (100.0) |
| I164L | 168 (89.8) | 9 (4.8) | 10 (5.3) | 190 (100) | 0 (0) | 0 (0) | 19 (5.0) |
| N51I, C59R, S108N | 34 (17.5) | 160 (82.5) | 26 (12.7.) | 179 (87.3) | 339 (90.2) | ||
| N51I, C59R, S108N, I164L | 194 (100) | 0 (0) | 187 (91.2) | 18 (8.8) | 18 (4.5) | ||
| pfdhps alleles | |||||||
| A437G | 15 (8.2) | 15 (8.2) | 154 (83.7) | 11 (6.1) | 28 (15.6) | 141 (78.3) | 338 (92.9) |
| K540E | 10 (5.5) | 1 (0.5) | 172 (94.0) | 10 (5.6) | 1 (0.6) | 169 (93.9) | 343 (94.5) |
| A613S | 100.0 (100) | 0.0 (0) | 0.0 (0) | 100.0 (100) | 0.0 (0) | 0.0 (0) | 0 (0.0) |
| A581G | 134 (73.2) | 16 (8.7) | 33 (18.0) | 141 (78.3) | 14 (7.8) | 25 (13.9) | 88 (24.2) |
| Double pfdhfr (A437G-K540E) | 39 (19.0) | 166 (81.0) | 26 (13.4) | 168 (86.6) | 334 (83.7) | ||
| Triple pfdhfr (A437G-K540E-A581G) | 34 (17.5) | 160 (82.5) | 29 (12.7) | 179 (87.3) | 339 (85.0) | ||
| Quintuple pfdhfr E540-G437 / pfdhfr 51I-59R-108N) | 51 (26.3) | 143 (73.7) | 52 (25.4) | 153 (74.6) | 296 (74.2) | ||
| Sextuple pfdhfr E540-G437 / pfdhfr 51I-59R-108N + pfdhps A581) | 152 (78.4) | 42 (21.6) | 174 (84.9) | 31 (15.1) | 73 (18.3) | ||
Mixed infection denotes an isolate in which both WT and mutant genotypes were detected.
Age related association with in prevalence of mutations.
For each allele (pfcrt 76, pfmdr1 86, Pfdhfr (51, 59, 108) and Pfdhps (437, 540, 581), no statistically significant difference in mean number of mutant strains between age groups 0–5 years versus 6–15 years versus >15 years and age groups 0–5 years versus > 5 years was found (data not shown).
pfcrt gene
Overall, for the 388 total isolates typed, 50.8% carried the WT pfcrt 76K allele while 10.1% were pure mutants pfcrt 76T and 39.2% mixed infections. Stratified by site, WT, mixed and mutant pfcrt 76T genotype prevalence were 45.1%, 14.1%, 40.8%, respectively at Ruhuha and 56.3%, 6.1%, and 37.6%, respectively at Mubuga (Figure 2). pfcrt 76T mutant (pure and mixed mutants combined) distribution varied significantly (p = 0.026) with higher proportions seen at Ruhuha (55%) compared to Mubuga (44%).
pfmdr1 gene
pfdmr1 WT 86N alleles were found in 80.9% isolates while 15.4% and 3.7% of isolates carried mixed and mutant type infection, respectively. WT alleles 184Y and 1246D were found in 40% and 80% isolates, respectively (Table 2). The distribution of all mutant alleles at typed pfdmr1 codons were comparable for isolates from the two sites (Table 4).
Table 4.
Comparisons in proportional distributions of Chloroquine and Sulphadoxine – Pyrimethamine polymorphisms by study sites. C
| Allele | Polymorphism | Number (%) of mutant alleles | Pearson’s X2 test | P value* | |
|---|---|---|---|---|---|
| Ruhuha - n (%) | Mubuga - n (%) | ||||
| pfcrt | 76T | 105 (55.0) | 86 (43.7) | 4.971 | 0.026 |
| pfmdrl | 86Y | 40 (21.5) | 33 (16.8) | 1.346 | 0.246 |
| 184Y | 107 (62.6) | 100 (57.1) | 1.0611 | 0.303 | |
| 1042C | 0 | 0 | - | - | |
| 1034C | 0 | 0 | - | - | |
| 1246Y | 37 (20.8) | 35 (17.8) | 0.5497 | 0.458 | |
| pfdhps | 613S | 0 | 0 | ||
| 581G | 49 (26.8) | 39 (21.7) | |||
| 437G | 169 (91.9) | 169 (93.9) | 0.572 | 0.450 | |
| 540E | 173 (94.5) | 170 (94.4) | 0.001 | 0.970 | |
| pfdhfr | 51I | 190 (100) | 185 (99.5) | 2.982 | 0.225 |
| 59R | 160 (84.2) | 184 (96.3) | 19.510 | < 0.0001 | |
| 108N | 190 (100) | 189 (100) | - | - | |
| 164L | 19 (10.2) | 0 (0) | 20.329 | < 0.0001 | |
| Grouped alleles | Double pfdhps (540E-437G) | 168 (91.8) | 166 (92.2) | 2.311 | 0.129 |
| Triple pfdhfr (51I-59R-108N) | 160 (84.2) | 179 (96.2) | 1.830 | 0.176 | |
| Quadruple (51I-59R-108N-164L) | 18 (8.8) | 0 (0.0) | 17.839 | < 0.0001 | |
| Quintuple pfdhfr 540E-G437G /pfdhfr 51I-59R-108N) | 143 (73.7) | 153 (74.6) | 0.044 | 0.833 | |
| Sextuple pfdhfr E540-G437–581G /pfdhfr 51I-59R-108N) | 42 (21.7) | 31 (15.1) | 2.8411 | 0.092 | |
P value comparing proportions of mutant parasites were based on a 2-sample t-test.
Significant values are in boldface.
pfdhps gene
High-level prevalence for pfdhps mutant (pure and mixed mutants combined) alleles 437G and 540E of 92.9% and 94.5%, respectively, was seen. At 581G and 613S codons, mutant prevalence was 24.2% and 0%, respectively. The distributions of 437G, 540E and 581G mutant alleles were comparable across the two study sites.
pfdhfr gene
pfdhfr mutant (pure and mixed mutants combined) allele prevalence at codons 51I, 59R, 108N and 164L were 99.7%, 90.3%, 100% and 5%, respectively, whilst the prevalence of the pfdhfr triple (108N-51I-59R) haplotype was 85% (Table 3). The distribution for each of pfdhfr 164L and 59R mutants varied significantly (p < 0.0001) by study sites. Notably, all 19 164L mutants were seen at the Ruhuha site of higher malaria endemicity. For the 59R mutants, a higher prevalence was observed at the lower malaria endemic Mubuga site (96.3%) compared to the Ruhuha site (84.2%).
Combination haplotypes
Of the 374 samples typed only 46 (12.2%) carried both pfcrt 76K and pfdmr1 86N WT alleles. Notably, the proportion of double pfcrt 76T and pfdmr1 86Y mutants was 2-fold higher at Ruhuha compared to Mubuga (p = 0.018). The prevalence of the pfdhps double (437G-540E), pfdhfr triple (108N-51I-59R), and pfdhps/pfdhfr quintuple haplotypes were 83.7%, 85.0% and 73.7%, respectively (Table 3). The proportions of the pfdhps double, pfdhfr triple (108N-51I-59R) and the pfdhps / pfdhfr quintuple polymorphisms were comparable across the two study sites (Table 4). In contrast, a borderline significant (p = 0.06) higher proportion of triple pfdhps 437G, 540E and 581G haplotype was seen at Ruhuha (25.3%) compared to the Mubuga (17.6%) site (Table 4). In total, about 18.3% (73 isolates) carried the sextuple (51I, 59R, 108N, 540E, 437G, 581G) mutant, with its occurrence being restricted to the Ruhuha site (p = 0.005).
Discussion
In Rwanda, CQ was replaced with AQ+SP in 2001 and the later combination was then used for only five years. No study or report on anti-malarial drug resistance neither before CQ withdrawal (2001) nor after AQ+SP withdrawal (2006) exists and thus the impact of AQ on CQ resistance has never been estimated.
In a study conducted in 2010 among under five-year old children in high malaria endemic southern Rwanda, a 74% pfcrt 76T mutant prevalence was reported (Zeile et al., 2012). In this study, conducted 5 years later, pfcrt 76T prevalence was 49%. Presuming a similar pfcrt 76T prevalence in the comparably high malaria transmission eastern and southern regions of Rwanda, ~ 25% recovery of WT pfcrt 76K strains was observed. Recovery of CQ-susceptibility after years of CQ withdrawal has shown a mixed pattern, both within and between countries. Whilst high CQ recovery rates (>85%) have been reported in Tanzania (Mohammed et al., 2013; Malmberg et al., 2013) and Malawi (Kublin et al., 2003), slower recovery rates have been reported elsewhere including Kenya (Mwai et al., 2009) and Uganda (Nsobya et al., 2010; Kamugisha et al., 2012). In our study, CQ recovery occurred in spite of the large-scale use of AL (which has been shown to select for the CQ-susceptible pfcrt 76K allele) similar other findings (Malmberg et al., 2013; Sisowath et al., 2009). Possible reasons for the observed slow CQ recovery of susceptibility include the continued use of CQ in spite of CQ withdraw policies as reported in Uganda, Rwanda and Tanzania (Karema et al., 2010; Frosch et al., 2011; Eriksen et al., 2005) and the continued use of CQ related antimalarial drugs like AQ. AQ use has been associated with limited recovery of CQ susceptibility after years of CQ withdrawal previously and has been shown to strongly select for the resistance conferring pfcrt 76T allele (Frank et al., 2011; Djimde et al., 2008). Other determinants of CQ recovery rates include time since actual CQ drug withdrawal from use, time since policy to withdraw CQ from use, baseline CQ resistance levels and malaria transmission intensities.
Polymorphisms in the P. falciparum pfmdr1 gene show mixed sensitivity responses for different anti-malarial drugs (Rosenthal, 2013). Our study showed a >80% prevalence for pfmdr1 WT 86N and 1246D alleles and a 75% prevalence for the pfmdr1 86N/1246D/184Y CQ susceptible triple haplotype. However, only 40% of isolates carried the WT 184Y alleles. Compared to a previous study in southern Rwanda where WT allelic prevalence of 61%, 88% and 48% for pfmdr1 86N, 1246D and 184Y, respectively, with ~ 60% prevalence for the pfmdr1 86N/184Y/1246D wild-type haplotype were reported, our study showed a slow recovery of WT 86N allele and the triple pfmdr1 (86N/184Y/1246D) haplotype but not the 1246Y and 184F mutants whose levels remained relatively stable (Gahutu et al., 2011). A comparably high 66% prevalence for the pfmdr1 86Y/184F/1246Y mutant haplotype was reported in Kenya (Okombo et al., 2014). This mixed selective pressure for CQ among alleles at this locus, with recovery reported for alleles 86N and 1246D but not 184Y, has been reported to be partly associated with scale-up in use of AL (Okombo et al., 2014). Our study findings are similar to those from Zanzibar, Burkina Faso, Tanzania where findings of increased prevalence of pfmdr1 86Y (Sisowath et al., 2005; Dokomajilar et al., 2006; Humphreys et al., 2007) and pfmdr1 184F have been noted to come under selection in settings of AL resistance (Vinayak et al., 2010). Analysis of P. falciparum infected samples in Mozambique showed a mixed temporal trend in the prevalence of WT 86N, 184Y and 1246D alleles. Between the 2003–2005 and 2010–2012 periods, pfmdr1 86N prevalence rose from 19.5% to 73.2%, while 184Y WT allelic prevalence remained stable (from 19.6% to 22.7%), and the WT 1246D alleles showed marginal increase from 74.4% to 96.7%, in tandem with ACT use in the 2010–2012 period (Dokomajilar et al., 2006; Lobo et al., 2014). Thus, recovery to CQ susceptible pfmdr1 alleles shows a variable temporal trend, with AL being a major influence. Findings of pfmdr1 86N, 184F, and 1246D allelic selection by treatment with AL raise concerns of a possible alteration to AL drug sensitivity by these alleles (Baliraine & Rosenthal, 2011). The relative contribution of CQ cessation and ACT scale-up on pfmdr1 epidemiology requires further exploration.
Similar to previous studies in Southern Rwanda (Zeile et al., 2012), Tanzania (Matondo et al., 2014), Kenya (Shah et al., 2015, Iriemenam et al., 2012) and Uganda (Mbogo et al., 2014), our studies also detected high levels (>92%) of pfdhps 437G, 540E and pfdhps double (437G-540E) mutants. In contrast, declines in pfdhfr and pfdhps resistance imparting polymorphisms after SP withdrawal have been reported in studies from Ethiopia (Hailemeskel et al, 2013; Tessema et al., 2015), Tanzania (Gesase et al., 2009; Matondo et al., 2014) and Mozambique (Raman et al., 2008). Overall, as seen at 6 Tanzanian sites, SP resistance continuing to increase, with emergence and dispersal of “super resistant” mutants in east Africa (Baraka et al., 2015). The sustained high prevalence of pfdhps mutants in spite of the reduced or absent SP selection pressure in Rwanda may be due to: (1) high malaria endemicity in the study areas, (ii) the continued use of PfDHFR/PfDHPS inhibitors like trimethoprim-sulfamethoxazole (TS) for the treatment of and prophylaxis against bacterial infections among HIV infected individuals, and (iii) the continued use of IPTp-SP especially in the East and Central African regions.
In our study, intense levels of 85% pfdhfr triple mutants were seen, and other studies have likewise shown increased pfdhfr triple mutant prevalence in other sites in southern, eastern and western Rwanda, in spite of the presumed 7 years absence of SP drug pressure (Zeile et al., 2012, Karema et al., 2010). Similar to the high levels of pfdhp mutants seen in this study, a possible source of pfdhfr mutants may be the use of TS. Indeed, in vitro P. falciparum culture studies have demonstrated a TS cross-resistance with SP that may lead to the development of pfdhfr and pfdhps mutants (Khalil et al., 2003; Iyer et al., 2001). Additionally, our studies also detected high prevalence of quintuple (pfdhfr triple and pfdhps double) mutants similar to those reported in Kenya and Uganda even >10 years of SP drug pressure (Iriemenam et al., 2012; Mbogo et al., 2014). High levels of the quintuple mutants have been associated with reduced efficacy of SP-IPTp (Allen et al., 2009), and the fact that IPT-Sp continues to be is use in Kenya and Uganda, unlike in Rwanda, may account for high prevalence of quintuple mutants in Kenya and Uganda.
High polymorphism at pfdhfr 164L and pfdhps 581G have been associated with increased therapeutic failure of SP (Karema et al., 2010; Lynch et al., 2008; Gesase et al., 2009; Gasasira et al., 2010; Spalding et al., 2010), however in our studies the prevalence was ~24%. A possible reason for high pfdhps 581G levels may be the continued IPTp-SP use (Harrington et al. 2009) and resulting reduced effectiveness of IPT—Sp has been attributed to significant reduction in birth weight of newborns (Minja et al., 2013). In our study, 581G levels varied by study area with a 26.8% declined at Ruhuha, eastern Rwanda vs. 60% decline reported at Rukara, eastern Rwanda (Karema et al., 2010) in the period 2005–2006 to 2016. In contrast, 581G allelic prevalence remained comparable for the Mubuga (29%) and Masheshe (21.7%) sites in western Rwanda in the same periods, respectively (Karema et al., 2010). Differences in malaria transmission intensity between the eastern and western regions may partly account for the differential 581G allelic temporal effects noted. Isolates collected from Ruhuha, eastern Rwanda revealed 5% prevalence of pfdhfr 164L mutant similar to (Karema et al., 2010), and these were found only in association with the pfdhps double (100%) but with ~95% (18/19) of alleles concurrently seen alongside the triple pfdhfr mutant. The 164L mutants are preferentially concentrated in the Kenya and Uganda eastern Africa, albeit at variable prevalence (Lobo et al., 2014; Spalding et al., 2010).
We also identified additional site-specific genotype differences. The proportion of pfcrt 76T mutant infection was significantly higher at the lower-endemic Mubuga site compared to the higher-endemic Ruhuha site. A number of factors may account for this variability including differences in access to AL external sources of pfcrt 76T mutants introduced by individuals from neighbouring countries due to high frequency of cross border movements. On the other hand the pfdhfr 164L mutants were found exclusively in the high malaria endemic Ruhuha site, while the same has been reported to vary between low (Braun et al., 2015; Alifrangis et al., 2009) and concentrated local hotspots in Rwanda and southwest Uganda (Karema et al., 2010; Lynch et al., 2008). The other difference was higher prevalence of the pfdhfr 59R mutants observed at Mubuga, western Rwanda compared to Ruhuha, eastern Rwanda. The western Rwanda borders the Democratic Republic of Congo and Burundi and hence may be more influenced by drug resistance pressure from across the border due to highly dynamic human populations, relative to the other sites. Finally, samples analysed in this study were collected from two sites located in the low and high malaria intensities zones where as malaria risk in Rwanda is categorised into four ecologic zones. Therefore, study findings may not be generalizable to all Rwandan sites. That notwithstanding, our findings provide the most recent accurate surveillance data for key CQ and SP resistance- mediating polymorphisms at two sites of variable malaria transmission intensities.
Conclusions
Overall, sustained high levels of SP resistance and a slow recovery of CQ susceptible parasites were found in our study conducted after 7 and 14 years after complete SP and CQ drug withdrawal, respectively. Most likely, the high prevalence of SP resistant parasites is due to the continued selection by use of Pfdhfr/Pfdhps inhibitors like TS (among HIV infected individuals) and IPTp-SP (for malaria prevention among pregnant women). Interestingly, the prevalence for the two high-level SP resistance imparting pfdhfr 164L and pfdhps 581G mutants were observed to decline with the pfdhfr 164L mutant noted to be restricted to the Eastern Rwanda site Continued surveillance of P. falciparum polymorphisms and characterization of the determinants of anti-malarial drug sensitivity epidemiology is recommended for guiding future rational drug policy-making and mitigation of future risk of anti-malaria drug resistance development.
Highlights.
Antimalarial drugs chloroquine and sulphodoxine – pyrimethamine resistance was a setback to malaria control.
Studies reveal that withdrawal of there can lead to recovery of sensitivity of parasites over time.
In Rwanda chloroquine susceptibility recovery appears slow but sulphodoxine – pyrimethamine resistance remains high after years of withdrawal.
Elucidating the determinants of these drug-related genomic adaptations over time and exposure to varying drug pressures in Rwanda is needed
Acknowledgments.
We thank study participants at both Ruhuha and Mubuga health centres who provided samples for analysis and the health facility leadership, study laboratory personnel for supporting study conduct.
Financial support.
This work was financially supported by a Fogarty International Center, National Institutes of Health, Training grant #TW007375. Supplementary support was received from The Netherlands Organisation for Scientific Research (NWO-WOTRO) under Grant# AMC A1050243 to the Academic Medical Centre – The Netherlands.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors have declared that they have no competing interests.
References
- Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB, Vestergaard LS, Ishengoma D, et al. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009; 80:523–527. [PubMed] [Google Scholar]
- Allen EN, Little F, Camba T, Cassam Y, Raman J, Boulle A, at al. Efficacy of sulphadoxine-pyrimethamine with or without artesunate for the treatment of uncomplicated Plasmodium falciparum malaria in southern Mozambique: a randomized controlled trial. Malar J. 2009; 8:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in P. falciparum malaria. N Engl J Med. 2015; 371:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliraine FN, Rosenthal PJ. Prolonged Selection of pfmdr1 Polymorphisms After Treatment of Falciparum Malaria With Artemether-Lumefantrine in Uganda. J Infect Dis. (2011) 204 (7): 1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraka V, Ishengoma DS, Fransis F, Minja DT, Madebe RA, Ngatunga D, Van Geertruyden JP. High-level Plasmodium falciparum sulfadoxine-pyrimethamine resistance with the concomitant occurrence of septuple haplotype in Tanzania. Malaria J. 14:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Rempis E, Schnack A, Decker S, Rubaihayo J, Tumwesigye NM, et al. Lack of effect of intermittent preventive treatment for malaria in pregnancy and intense drug resistance in western Uganda. Malar J. 2015; 14:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde AA, Fofana B, Sagara I, Sidibe B, Toure S, Dembele D, et al. Efficacy, safety, and selection of molecular markers of drug resistance by two ACTs in Mali. Am J Trop Med Hyg. 2008; 78: 455–461. [PubMed] [Google Scholar]
- Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether–lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006; 50:1893–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009; 361: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enosse S, Magnussen P, Abacassamo F, Gómez-Olivé X, Rønn AM, Thompson R, et al. Rapid increase of Plasmodium falciparum dhfr/dhps resistant haplotypes, after the adoption of sulphadoxine-pyrimethamine as first line treatment in 2002, in southern Mozambique. Malar J. 2008; 7:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen J, Nsimba SE, Minzi OM, Sanga AJ, Petzold M, Gustafsson LL, et al. Adoption of the new antimalarial drug policy in Tanzania–a cross-sectional study in the community. Trop Med Int Health. 2005; 10:1038–1046. [DOI] [PubMed] [Google Scholar]
- Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 2000;6:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Lehners N, Mayengue PI, Gabor J, Dal-Bianco M, Kombila DU et al. A thirteen-year analysis of Plasmodium falciparum populations reveals high conservation of the mutant pfcrt haplotype despite the withdrawal of chloroquine from national treatment guidelines in Gabon. Malaria J, 2011; 10: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch AE, Venkatesan M, Laufer MK. Patterns of chloroquine use and resistance in sub-Saharan Africa: a systematic review of household survey and molecular data. Malar J. 2011; 10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahutu JB, Steininger C, Shyirambere C, Zeile I, Cwinya-Ay N, Danquah I, et al. Prevalence and risk factors of malaria among children in southern highland Rwanda. Malar J. 2011; 10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasasira AF, Kamya MR, Ochong EO, Vora N, Achan J, Charlebois E, et al. Effect of trimethoprim-sulphamethoxazole on the risk of malaria in HIV-infected Ugandan children living in an area of widespread antifolate resistance. Malar J. 2010; 9:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009; 4(2): e4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailemeskel E, Kassa M, Taddesse G, Mohammed H, Woyessa A, Tasew G, et al. Prevalence of sulfadoxine-pyrimethamine resistance-associated mutations in dhfr and dhps genes of Plasmodium falciparum three years after SP withdrawal in Bahir Dar, Northwest Ethiopia. Acta Trop. 2013; 128:636–641. [DOI] [PubMed] [Google Scholar]
- Harinasuta T, Suntharasamai P, Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965; 2:657–660. [DOI] [PubMed] [Google Scholar]
- Harrington WE, Mutabingwa TK, Muehlenbachs A, Sorensen B, Bolla MC, Fried M, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci U S A. 2009;106: 9027–9032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MD, Bates SJ, Blackstone EA, Monks SM, Mutabingwa TK, Sibley CH. Highly Pyrimethamine-resistant alleles of dihydrofolate reductase in isolates of Plasmodium falciparum from Tanzania. Trans R Soc Trop Med Hyg. 2002; 96(6):674–676. [DOI] [PubMed] [Google Scholar]
- Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, et al. Amodiaquine and artemether–lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007; 51:991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriemenam NC, Shah M, Gatei W, van Eijk AM, Ayisi J, Kariuki S et al. Temporal trends of sulphadoxine-pyrimethamine (SP) drug-resistance molecular markers in Plasmodium falciparum parasites from pregnant women in western Kenya. Malar J. 2012; 11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer JK, Milhous WK, Cortese JF, Kublin JG, Plowe CV. Plasmodium falciparum cross-resistance between trimethoprim and pyrimethamine. The Lancet. 2001; 358:1066–1067. [DOI] [PubMed] [Google Scholar]
- Kain KC, Lanar DE. Determination of genetic variation within P. falciparum by using enzymatically amplified DNA from filter paper disks impregnated with whole blood. J Clin Microbiol. 1991; 29:1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamugisha E, Bujila I, Lahdo M, Pello-Esso S, Minde M, et al. Large differences in prevalence of Pfcrt and Pfmdr1 mutations between Mwanza, Tanzania and Iganga, Uganda-a reflection of differences in policies regarding withdrawal of chloroquine? Acta Trop. 2012; 121:148–151. [DOI] [PubMed] [Google Scholar]
- Karema C, Aregawi MW, Rukundo A, Kabayiza A, Mulindahabi M, Fall IS, et al. Trends in malaria cases, hospital admissions and deaths following scale-up of anti-malarial interventions, 2000–2010, Rwanda. Malar J. 2012; 11:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karema C, Imwong M, Fanello CI, Stepniewska K, Uwimana A, Nakeesathit S, et al. Molecular Correlates of High-Level Antifolate Resistance in Rwandan Children with Plasmodium falciparum Malaria. Antimicrob Agents Chemother. 2010; 54: 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I, Ronn AM, Alifrangis M, Gabar HA, Satti GM, Bygbjerg IC. Dihydrofolate reductase and dihydropteroate synthase genotypes associated with in vitro resistance of Plasmodium falciparum to pyrimethamine, trimethoprim, sulfadoxine, and sulfamethoxazole. Am J Trop Med Hyg. 2003; 68:586–589. [DOI] [PubMed] [Google Scholar]
- Koenderink JB, Kavishe RA, Rijpma SR, Russel FG. The ABCs of multidrug resistance in malaria. Trends Parasitol. 2010; 26:440–446. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003; 187:1870–1875. [DOI] [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006; 355:1959–1966. [DOI] [PubMed] [Google Scholar]
- LeClair NP, Conrad MD, Baliraine FN, Nsanzabana C, Nsobya SL, Rosenthal PJ. Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of plasmodium falciparum. J Clin Microbiol. 2013; 51:2564–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo E, de Sousa B, Rosa S, Figueiredo P, Lobo L, Pateira S, et al. Prevalence of pfmdr1 alleles associated with artemether-lumefantrine tolerance/resistance in Maputo before and after the implementation of artemisinin-based combination therapy. Malar J. 2014; 13:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C, Pearce R, Pota H, Cox J, Abeku TA, Rwakimari J, et al. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J Infect Dis. 2008; 197:1598–1604. [DOI] [PubMed] [Google Scholar]
- Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, et al. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J. 2013; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matondo SI, Temba GS, Kavishe AA, Kauki JS, Kalinga A, van Zwetselaar M, et al. High levels of sulphadoxine-pyrimethamine resistance Pfdhfr-Pfdhps quintuple mutations: a cross sectional survey of six regions in Tanzania. Malar J. 2014; 13: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, et al. Temporal Changes in Prevalence of Molecular Markers Mediating Antimalarial Drug Resistance in a High Malaria Transmission Setting in Uganda. Am J Trop Med Hyg. 2014; 91(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minja D, Schmiegelow C, Mmbando B, Boström S, Oesterholt M, Magistrado P,et al. Plasmodium falciparum Mutant Haplotype Infection during Pregnancy Associated with Reduced Birthweight, Tanzania. Emerg Infect Dis. 2013. Sep; 19(9): 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed A, Ndaro A, Kalinga A, Manjurano A, Mosha JF, Mosha DF, et al. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J. 2013; 12:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009; 8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nankoberanyi S, Mbogo GW, LeClair NP, Conrad MD, Tumwebaze P, Tukwasibwe S, et al. Validation of the ligase detection reaction fluorescent microsphere assay for the detection of Plasmodium falciparum resistance mediating polymorphisms in Uganda. Malar J. 2014; 13:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye M, Faye B, Tine R, Ndiaye JL, Lo A, Abiola A et al. Assessment of the Molecular Marker of Plasmodium falciparum Chloroquine Resistance (Pfcrt) in Senegal after Several Years of Chloroquine Withdrawal. Am J Trop Med Hyg. 2012; 87(4):640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob Agents Chemother. 2010; 54:1200–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila-Mounda A, Mberu EK, Sibley CH, Plowe CV, Winstanley PA, Watkins WM: Kenyan Plasmodium falciparum field isolates: correlation between pyrimethamine and chlorcycloguanil activity in vitro and point mutations in the dihydrofolate reductase domain. Antimicrob Agents Chemother 1998, 42:164–169, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okombo J, Kamau AW, Marsh K, Sutherland CJ, Ochola-Oyier LI. Temporal trends in prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. Int J Parasitol Drugs Drug Resist. 2014; 4 (3):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord R, Alexander N, Dunyo S, Hallett R, Jawara M, Targett G, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J Infect Dis. 2007; 196:1613–1619. [DOI] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, Bâ el-H, Mombo-Ngoma G, Malisa AL, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS. Med. 2009; 6(4):e1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PMI/MOH-Rwanda. President’s Malaria Initiative Rwanda Malaria Operational Plan FY 2015. http://www.pmi.gov/docs/default-source/default-document-library/malaria-operational-plans/fy-15/fy-2015-rwanda-malaria-operational-plan.pdf?sfvrsn=3.
- Raman J, Sharp B, Kleinschmidt I, Roper C, Streat E, Kelly V, et al. Differential effect of regional drug pressure on dihydrofolate reductase and dihydropteroate synthetase mutations in southern Mozambique. Am J Trop Med Hyg. 2008; 78:256–261. [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PJ. The interplay between drug resistance and fitness in malaria parasites. Mol Microbiol. 2013; 89:1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Omosun Y, Lal A, Lal A, Odero C, Gatei W, Otieno K, et al. Assessment of molecular markers for anti-malarial drug resistance after the introduction and scale-up of malaria control interventions in western Kenya. Malar J. 2015; 14:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009. Mar 1;199(5):750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether–lumefantrine (Coartem). J Infect Dis. 2005; 191:1014–1017. [DOI] [PubMed] [Google Scholar]
- Spalding MD, Eyase FL, Akala HM, Bedno SA, Prigge ST, Coldren RL, et al. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu, Kenya. Malar J. 2010; 9:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, et al. Complex polymorphisms in a ~330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 1997;91:593–603. [DOI] [PubMed] [Google Scholar]
- Tessema SK, Kassa M, Kebede A, Mohammed H, Leta GT, Woyessa A et al. Declining trend of Plasmodium falciparum dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant alleles after the withdrawal of Sulfadoxine-Pyrimethamine in North Western Ethiopia. PLoS ONE. 2015; 10(10): e0126943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Menting JGT, Wilson C, Cowman AF: Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA 1997, 94:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, et al. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis. 2010; 201:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ. Antimalarial drug resistance and combination chemotherapy. Philos Trans R Soc Lond B Biol Sci. 1999; 354:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, World Malaria Report 2015. http://apps.who.int/iris/bitstream/10665/200018/1/9789241565158_eng.pdf.
- Young MD, Moore DV. Chloroquine resistance in Plasmodium falciparum. Am J Trop Med Hyg. 1961; 10:317–320. [DOI] [PubMed] [Google Scholar]
- Zeile I, Gahutu JB, Shyirambere C, Steininger C, Musemakweri A, Sebahungu F, et al. Molecular markers of Plasmodium falciparum drug resistance in southern highland Rwanda. Acta Tropica. 2012; 121:50–54. [DOI] [PubMed] [Google Scholar]


