Abstract
Odd ploidy-level cytotypes in sexually reproducing species are considered a dead end due to absent or reduced fertility. If sterility is only partial, however, their contribution to the population gene pool can be augmented by longevity and clonal growth. To test this, we investigated the cytotype origin and spatial pattern, and pollen viability in three relict shrub species of the genus Daphne (Thymelaeaceae Juss.) in central Europe. Daphne cneorum subsp. cneorum is a widespread European species that has a broad ecological amplitude, whereas D. cneorum subsp. arbusculoides and D. arbuscula are narrow endemics of the western Pannonian Plain and the Western Carpathians, respectively. Our study confirmed that all three taxa are diploid. However, of more than a thousand analysed individuals of D. cneorum subsp. cneorum, five in four different populations were triploid. Our data indicate that these triploids most likely originate from recurrent autopolyploidization events caused by the fusion of reduced and unreduced gametes. High pollen viability was observed in all three taxa and in both diploid and triploid cytotypes, ranging from 65 to 100 %. Our study highlights the significant role of odd ploidy-level cytotypes in interploidy gene flow, calling for more research into their reproduction, genetic variability, and overall fitness. Interestingly, while the endemic D. arbuscula differs from D. cneorum based on genetic and genome size data, D. cneorum subsp. arbusculoides was indistinguishable from D. cneorum subsp. cneorum. However, our study reveals that the subspecies differ in the number of flowers per inflorescence. This is the first comprehensive cytogeographic study of this intriguing genus at a regional scale, and in spite of its karyological stability, it contributes to our understanding of genomic evolution in plant species with a wide ecological amplitude.
Keywords: Carpathians, Daphne, endemics, genome size stasis, ITS, Pannonian Basin, pollen fertility, polyploidy, relicts, triploids
The presented study sheds light on the emergence of odd-ploidy cytotypes in diploid species, specifically in long-lived relic shrubs from the genus Daphne (Thymeleaceae). Rare odd-ploidy cytotypes are generally considered maladaptive in sexual diploid species due to reduced fitness and fertility. However, our research revealed the independent and recurring evolution of triploid cytotypes, most likely arising via the fusion of reduced and unreduced gametes in exclusively diploid systems and exhibiting significant pollen fertility. Their even partial pollen fertility, especially, in the context of their longevity, can contribute to a more diverse gene pool in their parental diploid populations or even to the spontaneous emergence of new cytotypes. Additionally, we demonstrated that although Daphne cneorum exhibits a large ecological amplitude, spanning various bedrock substrates and altitudinal ranges, its diploid genomes remain highly stable with no ecologically driven genome size expansions or reductions, as previously observed in many species with large ecological amplitudes.
Introduction
Quaternary climate change, coupled with human influence in the Anthropocene, has had a crucial effect on the evolution of biomes and the present-day distribution patterns of all organisms (Hewitt 1996, 2000; Lewis and Maslin 2015). Extensive environmental disturbances have induced species range shifts and habitat fragmentation, posing a substantial threat to the survival of plant populations and natural communities. Numerous plant species could only withstand such significant environmental perturbations in isolated, albeit ecologically stable habitat islands in areas that are inaccessible to humans and livestock (Larson et al. 2000; Lavergne et al. 2005; Huang et al. 2015; Tang et al. 2018). The complex evolutionary histories of these relict populations may augment their evolutionary dynamics, resulting in genomic rearrangements, genome size diversification through the accumulation of transposable elements and whole-genome duplications (Soltis et al. 2003; Macas et al. 2015; Schubert and Vu 2016; Van de Peer et al. 2017). Genome size variation is often evidenced in widespread and ecologically tolerant species, with structural genome rearrangements driven by environmental heterogeneity (Šmarda and Bureš 2006; Slovák et al. 2009; Nunvářová Kabátová et al. 2019). Polyploid cytotypes are also common in relict species (e.g. Siljak-Yakovlev et al. 2008; Besnard and Baali-Cherif 2009; García-Verdugo et al. 2013). Morphological, anatomical and physiological changes associated with polyploidization may lead to improved fitness and a shift in ecological requirements in novel cytotypes, enhancing their ability to survive and adapt to environmental changes (Levin 2002; Otto 2007). Odd ploidy cytotypes may emerge either through the fusion of reduced and unreduced gametes within diploid species or through evolution as a result of heteroploid hybridization (Ramsey and Schemske 1998; Husband et al. 2013). In certain heteroploid systems, gene flow between different cytotypes can contribute to the establishment of newly formed polyploids (Burton and Husband 2001; Čertner et al. 2017; Kolář et al. 2017). Occasional triploids are typically considered maladaptive in sexual diploid species due to their decreased fitness and fertility (Ramsey and Schemske 1998; Joly and Bruneau 2004). Nevertheless, their persistence, frequency and potential evolutionary significance in exclusively diploid systems may be remarkably greater in long-lived species such as shrubs, trees and clonal species, and may even exhibit better adaptation to more stressful environments than their diploid progenitors (Mock et al. 2012). Consequently, a longer lifespan and at least partial fertility in triploids can contribute to an enriched gene pool in their parental diploid populations (Lefort et al. 2000; Blakesley et al. 2002; Dzialuk et al. 2007; Besnard and Baali-Cherif 2009; Mock et al. 2012).
We investigate the karyological variability of three relict taxa of the genus Daphne (Thymelaeaceae Juss.) assumed to have evolved in the Quaternary or even the Tertiary epoch (Browicz and Gostyńska-Jakuszewska 1967; Muller 1997; Halda 2001; Melnyk and Baransky 2020), namely D. arbuscula, D. cneorum subsp. cneorum and D. cneorum subsp. arbusculoides. All three taxa are evergreen, long-lived small shrubs with strongly fragrant, pinkish, insect-pollinated flowers and fruits (drupes) dispersed by animals. They are reported to be allogamous, lacking noticeable long-distance dispersal systems, and to have reduced seed production and germination, accompanied by a high level of clonal spread (e.g. Erdelská and Turis 1995; Šedivá and Žlebčík 2010; Gajdošová 2020; Di Sacco et al. 2021). The phylogenetic relationships and even clear taxonomic identities of D. arbuscula and D. cneorum subsp. arbusculoides have never been clarified using genetic data. Daphne arbuscula is a narrow endemic to the Western Carpathians (Muránska Planina Mts.), occurring in mostly small, isolated patches at mid-altitude on relict limestone cliffs (Fig. 1A–C). Despite its limited distribution, D. arbuscula is not stenotopic; distinct groups of micro-localities show different plant associations and microclimatic conditions in regard to temperature and humidity (Valachovič and Jarolímek 1994; Erdelská and Turis 1995; Uhlířová and Bernátová 2003, 2004; Kochjarová et al. 2004). In contrast, D. cneorum is widespread and can be found across southern, central, and eastern Europe. Its island-type distribution has likely been influenced by habitat fragmentation associated with anthropogenic factors (Browicz and Gostyńska-Jakuszewska 1967; Muller 1997; Orsenigo et al. 2019; Melnyk and Baransky 2020). The nominate subspecies occurs in a variety of relict habitats, from the lowlands (≥120 m a.s.l.) to the subalpine zone (≤1796 m a.s.l., seeSupporting Information—Table S1), also occurring in various bedrock types. It prefers relictual grasslands and oak or pine forests on limestone bedrock, but it can also be found in siliceous grasslands, winded sands, and occasionally also on serpentine or gypsum bedrock localities (Fig. 1D–J; Webb and Ferguson 1968; Halda 1976; Orsenigo et al. 2019). In contrast, D. cneorum subsp. arbusculoides is narrowly distributed in the south-westernmost part of the Pannonian Basin, with micro-localities in SW Hungary, NE Slovenia, and SE Austria (Fig. 1K and L), occurring in grassland-forest habitats on siliceous bedrock. Morphologically, D. cneorum subsp. arbusculoides differs from the nominate subspecies by having more erect branches and leaves with revolute margins (Tuzson 1911; Soó 1971; Halda 2001).
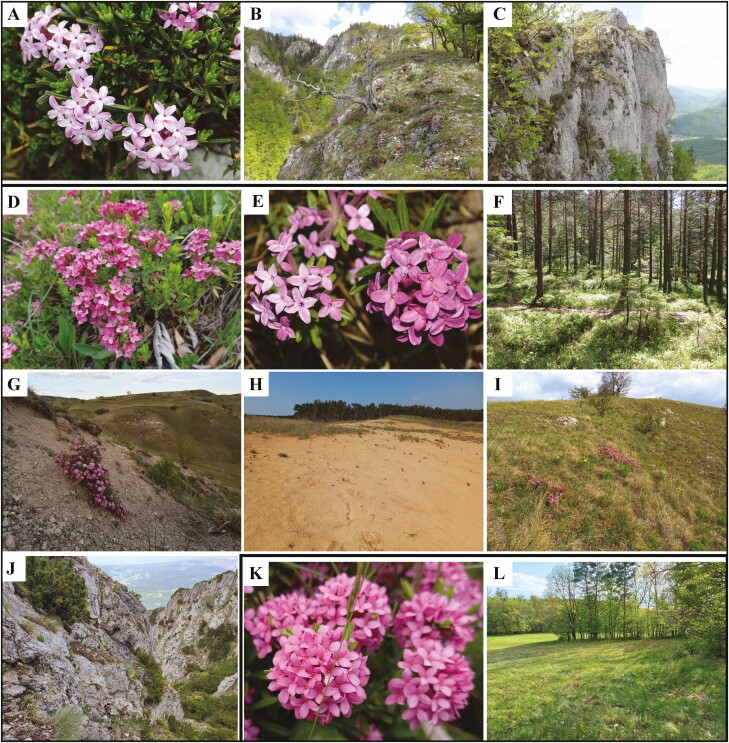
Figure 1.
Studied taxa and their habitats: (A) Daphne arbuscula; (B and C) calcareous cliffs in Muránska Planina Mts., Western Carpathians (SK-DA 7 and SK-DA 8); (D) D. cneorum subsp. cneorum—diploid cytotype; Sokol, Veľká Fatra Mts., Western Carpathians (SK-DC 6); (E) D. cneorum subsp. cneorum—triploid cytotype; Záhorská nížina Lowland, Pannonian Basin (SK-DC 1); (F) calcareous relict pine forest, Karlschutt, Hochschwab Mts., Eastern Alps Mts. (AT-DC 11); (G) steppic grasslands on gypsum, Sfăraș-Jebucu, Transylvanian Plateau, Romania (RO-DC 19); (H) siliceous sands, Záhorská nížina Lowland, Pannonian Basin (SK-DC 1); (I) steppic calcareous grasslands, Gánt, Transdanubian Mts., Pannonian Basin (HU-DC 17); (J) subalpine calcareous rocks, Turnu peak, Piatra Craiului Mts., Southern Carpathians (RO-DC 25); (K) D. cneorum subsp. arbusculoides (HU-DCA 2); (L) grassland and forest margins on the siliceous substrate, Őrség region, Pannonian Basin (HU-DCA 2). Photos A–D, F, H, I, K, L credits to Z. Gajdošová; E credit to J. Kučera, G credit to B.-I. Hurdu; J credit to A. Indreica.
The vast majority of members of the genus Daphne, including D. arbuscula and D. cneorum subsp. cneorum, are diploids with 2n = 2x = 18, with only a few published instances of polyploidy, including triploids (Goldblatt and Johnson 1979; Rice et al. 2015; seeSupporting Information—Table S2). No karyological data for D. cneorum subsp. arbusculoides have been published to date. During our pilot investigation on the karyological diversity of D. cneorum subsp. cneorum from the Slovak populations, we identified two individuals with relative genome sizes (RGSs) that deviated from the diploid stage, indicating a triploid level. However, it is unclear whether these individuals are true triploids resulting from the fusion of unreduced and reduced gametes or heteroploid hybrids between diploid and possibly tetraploid individuals (Soltis and Soltis 2000; Kolář et al. 2017). Although the reported karyological variability of D. cneorum subsp. cneorum and D. arbuscula is limited, their distribution patterns and ecological setup might indicate a niche shift driven by genome size and ploidy-level diversification (Slovák et al. 2009; Nunvářová Kabátová et al. 2019). This provides an ideal opportunity to investigate ploidy level and genome size variability of three Daphne taxa in relation to spatial patterns in the Carpathians, Eastern Alps, and the Pannonian Plain. Furthermore, because triploid individuals may not be completely sterile and thus contribute to population ploidy-level diversification through gene flow, we also investigate the pollen viability of all studied taxa and cytotypes to determine their evolutionary potential with respect to their contribution to parental gene pools. Notably, our study is the first to provide insight into the genetic variability and divergence of D. arbuscula and D. cneorum.
Material and Methods
Plant material and study sites
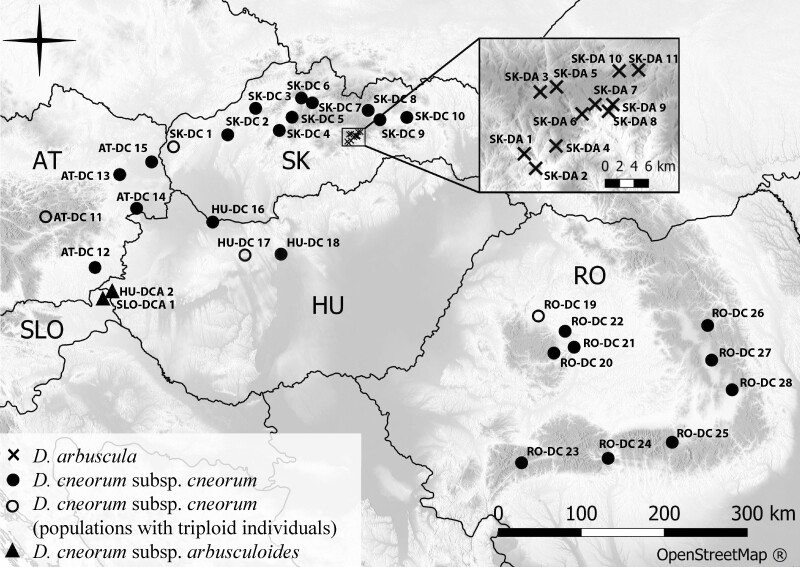
We collected material from the entire distribution range of both endemic taxa, namely D. arbuscula from the Western Carpathians (11 populations) and D. cneorum subsp. arbusculoides from the SW Pannonian Basin (two populations) (Fig. 2; seeSupporting Information—Table S1). Daphne cneorum subsp. cneorum, a pan-European species distributed from Spain in the west to Ukraine in the east (Halda 2001), was sampled only in the central and eastern parts of its distribution range, including the Carpathians, the Pannonian Basin, and, to a lesser extent, the NE Alps. However, the 28 sampled populations cover the entire habitat and ecological range reported for this taxon (Fig. 2; seeSupporting Information—Table S1; Webb and Ferguson 1968; Halda 1976; Orsenigo et al. 2019). For the purpose of our analyses, we used both exsiccated and fresh material (flowers or flower buds, young leaves, and root samples). The sampling effort aimed to cover the entire genetic diversity across each population while avoiding clonality effects (maximizing distance between individuals), reducing any negative impact on the population, and mitigating any damage to individual shrubs by collecting only the number of floral buds necessary for analyses per individual.
Figure 2.
Map of sampled populations from the analysed Daphne taxa: DA—D. arbuscula; DC—D. cneorum subsp. cneorum; DCA—D. cneorum subsp. arbusculoides. Country codes: AT—Austria, HU—Hungary, RO—Romania, SK—Slovakia, SLO—Slovenia. Taxa and cytotypes are indicated by different symbols. Population codes follow Supporting Information—Table S1.
Ploidy level, and relative and absolute genome size estimation
To evaluate ploidy level and estimate RGS, we analysed 5–30(−250) individuals per population, depending on population size (89 individuals in D. arbuscula, 1149 diploid and 5 triploid individuals in Daphne cneorum subsp. cneorum, and 177 individuals in D. cneorum subsp. arbusculoides; seeSupporting Information—Table S1). Absolute genome size (AGS) was estimated using at least three individuals per population from selected populations (9 individuals in D. arbuscula, 28 diploid individuals, and 1 triploid individual in Daphne cneorum subsp. cneorum, and 10 individuals in D. cneorum subsp. arbusculoides; seeSupporting Information—Table S1). Both measurements were performed using Partec CyFlow ML (Partec GmbH, Münster, Germany) equipped with a green solid-state laser (Cobolt Samba 532 nm, 150 mW; Cobolt, Stockholm, Sweden) as the excitation source for AGS or a UV lamp or HBO 100 W mercury arc lamp for RGS. Samples were prepared according to the standard protocol using a general-purpose buffer and propidium iodide (PI) for AGS (Loureiro et al. 2007), or the two-step procedure with Otto buffers and 4ʹ,6-diamidino-2-phenylindole (DAPI) for RGS (Otto 1990; Doležel et al. 2007). Bellis perennis was chosen as an internal standard (2C DNA = 3.38 pg; J. Suda unpubl. in: Schönswetter et al. 2007). For RGS, three to five samples were simultaneously evaluated in larger populations. If samples showed traces of RGS variation, we aimed to detect the real genome size difference using simultaneous analyses. At least 5000 nuclei for AGS and 3000 nuclei for RGS were measured, with the coefficients of variation of the G0/G1 peaks for both samples and the internal standard never exceeding 5 %. For AGS, at least three independent measurements were performed on consecutive days for each individual, with the inter-day variation threshold of the three iterations per sample being set at 3 % (Greilhuber and Obermayer 1997). If this variance was greater than 3 %, the outlying value was discarded and the corresponding sample was reanalysed. The AGS (2C value) estimation was computed as the ratio of the sample G0/G1 peak position to the standard G0/G1 peak position multiplied by the 2C DNA content of the standard (pg DNA; Doležel and Bartoš 2005). RGS was calculated as the ratio between the positions of the sample and standard G0/G1 peaks. Flow cytometric histograms (both PI and DAPI) were assessed using FloMax version 2.70 software (Partec Gmbh, Münster, Germany). The GC content was calculated separately for each pair of RGS and AGS measurements using the protocol by Šmarda et al. (2008) and subsequently averaged for each taxon and cytotype.
Chromosome counting
Somatic chromosome numbers (2n) were counted in fresh roots or very young flower bud meristems obtained from wild plants or rooting shoots of D. arbuscula, diploid and triploid cytotypes of D. cneorum subsp. cneorum, and D. cneorum subsp. arbusculoides. Individuals with counted chromosome numbers were simultaneously analysed for AGS and RGS and served as references for estimating ploidy in the remaining cytotypes and taxa. Freshly collected meristems were pre-treated in a 0.002 M water solution of 8-hydroxyquinoline for 3–5 h at low temperature (4 °C), washed in distilled water, fixed in a 3:1 mixture of 96 % ethanol and 98 % acetic acid for 24 h, and stored in 75 % ethanol. Samples were washed for 10 min in distilled water before and after being macerated in a 1:1 mixture of 35 % HCl and 96 % ethanol for 3 min prior to analyses. Microscopic slides and squashes were prepared using the cellophane square technique (Murín 1960). The slides were stained with a 7 % Giemsa stain solution, dried, and observed microscopically in a drop of immersion oil.
All chromosome spreads were analysed under 1000-fold magnification using a light microscope, and micrographs were taken using a ZEISS Axiocam 105 colour (Carl Zeiss, Vienna, Austria) and AxioVision LE64 v. 4.9.1.0 (Carl Zeiss, Microscopy GmbH).
Pollen viability and flower number
Fresh flower buds were dried and stored in silica gel. Pollen viability was analysed using a modified Alexander’s stain, following the standard protocol (Peterson et al. 2010). Viable and non-viable pollen grains were discriminated based on shape, size, and colour. Specifically, magenta–red pollen grains of regular shape were considered viable, while bluish–green, greenish–purple, and shrunken grains were considered non-viable (seeSupporting Information—Fig. S1; Peterson et al. 2010). A light microscope was used to examine pollen grains at 400-fold magnification, and micrographs were taken using a ZEISS Axiocam 105 colour (Carl Zeiss, Vienna, Austria) and AxioVision LE64 v. 4.9.1.0 (Carl Zeiss, Microscopy GmbH). Pollen viability was calculated as the percentage of non-aborted pollen grains from at least 100 analysed grains per flower, individualizing the standard deviation. Where possible, we examined five individuals per population with one flower per individual (18 individuals in D. arbuscula, 52 diploid and 3 triploid individuals in Daphne cneorum subsp. cneorum, and 10 individuals in D. cneorum subsp. arbusculoides; seeSupporting Information—Table S1).
In addition, to provide further evidence about the potential shifts in fitness and fertility of triploids, we tested for ploidy-induced shifts in flower number per inflorescence between cytotypes and taxa. Flowers were counted in one inflorescence per individual; usually from 5 to 15 individuals per selected population, or exceptionally more (40 individuals in D. arbuscula, 158 diploid and 3 triploid individuals in Daphne cneorum subsp. cneorum, and 95 individuals in D. cneorum subsp. arbusculoides; seeSupporting Information—Table S1). Only three out of five triploid individuals were examined for pollen viability and number of flowers, as the other two lacked flower buds and/or complete inflorescences at the time of collection. In triploids, pollen viability was determined using four buds per individual, whereas flower number was evaluated using 1–3 inflorescences per plant.
Molecular Analysis
DNA extraction, molecular markers and PCR amplification
Total genomic DNA was extracted from silica gel-conserved, young, intact leaves using a GeneAll® ExgeneTM Plant SV mini kit (GeneAll Biotechnology, Songpa-gu, Seoul, Korea), following the manufacturer’s protocol, with minor modifications (added PVP and 6 µl of RNase A). Genetic variability was tested using the ITS1-5.8S-ITS2 region (ITS) of nuclear ribosomal DNA (nrDNA; 26 individuals, 3 taxa) and the ndhF-rpl32 region of chloroplast DNA (cpDNA; 23 individuals, 3 taxa). The ITS region was amplified using primers ITS4 and ITS5, and internal primers ITS2 and ITS3, when necessary (White et al. 1990). The ndhF-rpl32 region was amplified using ndhF and rpl32-R primers (Shaw et al. 2007). Polymerase chain reaction (PCR) amplifications were performed in a total volume of 10 µl containing 1 µl of template DNA, 0.25 U DreamTaq DNA polymerase (Thermo Fisher Scientific Inc., Waltham, MA, USA), 1 µl of 10× DreamTaq buffer, 0.2 µl of dNTP Mix (10 mM each), and 0.2 µl of 10 mM each forward and reverse primers. The following PCR conditions were used: 95 °C for 3 min, 30 cycles (95 °C for 30 s, 51 °C for 30 s, 72 °C for 1 min), and 72 °C for 10 min for the ITS amplification; 80 °C for 5 min, 30 cycles (95 °C for 1 min, 52 °C for 1 min, followed by a 5 % ramp to 65 °C, 65 °C for 4 min), and 65 °C for 5 min for the ndhF-rpl32 region. PCR products were purified using a mixture (1:2) of Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific Inc.) or using NucleoSpin® Gel and PCR Clean-up (Macherey-Nagel GmbH & Co. KG, Germany) and sequenced by the ABI 3730xl DNA analyser at Eurofins Genomics Company (Konstanz, Germany).
Sequence processing and alignment
Sequences were edited and aligned using Geneious software R7.1.9 (Biomatters Ltd., Auckland, New Zealand). Both the ITS and ndhF-rpl32 datasets were supplemented with sequences of other related Daphne species and a selection of Thymelaeaceae outgroups available in GenBank. Between- and within-species variability was analysed using Bayesian analyses as conducted in MrBayes v.3.2.7a (Ronquist and Huelsenbeck 2003; Ronquist et al. 2012) and randomized accelerated maximum likelihood (ML, RAxML) algorithm (Stamatakis 2014). Prior to carrying out phylogenetic inferences, the best-fit models of nucleotide substitutions were assessed independently for each partition of the nucleotide data partition in jModelTest v.2.1.10 (Darriba et al. 2012) using the Akaike information criterion (AIC; Akaike 1974). The nrDNA dataset included three partitions and nucleotide substitution models: (i) non-coding ITS1 and ITS2 (HKY + I); (ii) coding 5.8S rDNA (SYM + I); (iii) indels within the ITS dataset. The ndhF-rpl32 plastid region had a single partition and the TVM + G model. Indels present in the sequence alignments were coded as binary characters using FastGap 1.2 software (Borchsenius 2009) according to the simple indel coding approach (Simmons and Ochoterena 2000). Bayesian analyses (BI) were conducted with four Markov chain Monte Carlo (MCMC) and two independent runs for 10–15 million generations, with a sampling frequency of every 100th generation. The first 10 % of trees were discarded as ‘burn-in’. RAxML analyses were run using rapid bootstrapping for 1000 replicates. Analyses were run on the CIPRES Science Gateway (Miller et al. 2010). Phylogenetic trees were visualized, and bootstrap support (BS) and posterior probabilities (PP) values were appended to trees using FigTree (v1.4.4). Bootstrap support was categorized according to the following criteria: strong (>85 %), moderate (70–85 %), weak (50–69 %) and poor (<50 %). Posterior probability values of 0.90 and above were considered significant, and those values below 0.90 were regarded as non-significant.
Data analysis
Inter-specific and ploidy-level variability in RGS, AGS, pollen viability, and the number of flowers among the three Daphne taxa were assessed by generalized linear mixed models (GLMMs; Bolker et al. 2009). Differences in non-integer values of RGS and AGS were tested using GLMMs with a Gaussian distribution and identity link function. A Poisson model with a logarithmic link was used to compare differences in flower counts. Finally, a binomial GLMM with a logit link was employed to evaluate pollen viability, that is, the proportion of viable pollen grains out of the total grains examined. Since our sampling design involved measurements of several plants at each site, we treated the population as a random intercept in the models to deal with a potential autocorrelation within sites. An additional random effect was included in the binomial GLMM to account for multiple pollen samples from the same plants nested within populations. The performance of each model was evaluated using a simulation-based approach to residual diagnostic (Dunn and Smyth 1996); no major deviations from the assumptions behind the models were observed. The statistical significance of the Gaussian GLMMs was evaluated using F-tests with Kenward-Roger adjusted degrees of freedom (Kenward and Roger 1997). The significance of the Poisson and binomial GLMM was assessed using likelihood ratio tests. Significant overall tests were followed by pairwise comparisons based on estimated marginal means with Tukey’s adjustments (Lenth 2016). The analyses were performed in R v. 4.1.2 (R Core Team 2021) using the libraries DHARMa (Hartig 2022), emmeans (Lenth 2022), ggplot2 (Wickham 2016), lme4 (Bates et al. 2015) and lmerTest (Kuznetsova et al. 2017).
Results
Chromosome numbers
We confirmed the diploid level as 2n = 2x = 18 for D. arbuscula and both subspecies of D. cneorum[seeSupporting Information—Fig. S2A–C]. In the presumed polyploid cytotype of D. cneorum subsp. cneorum, chromosome counting analyses uncovered a triploid cytotype with 2n = 3x = 27 [seeSupporting Information—Fig. S2D and Table S1].
Genome size and ploidy level
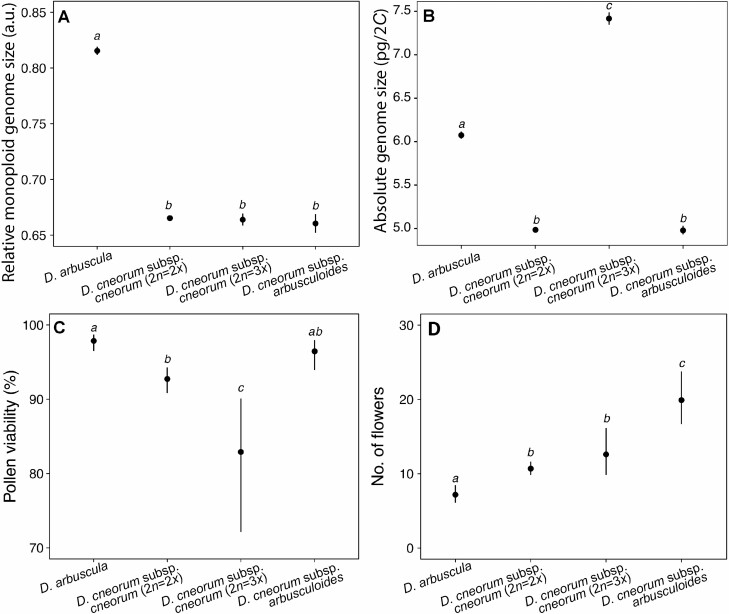
Both RGS and AGS analyses showed that all samples were diploid (Table 1) with the exception of five individuals from four different populations of D. cneorum subsp. cneorum (SK-DC 1, AT-DC 11, HU-DC 17, and RO-DC 19) where both AGS and RGS indicated a triploid ploidy level [seeSupporting Information—Fig. S3A–B and Table S1]. The GLMM revealed significant differences in RGS among investigated taxa (F(3, 53.7) = 1485, P < 0.0001). Daphne arbuscula showed significantly higher RGS than the other taxa whose RGS values were statistically indistinguishable (Fig. 3A; Table 1; seeSupporting Information—Fig. S3B). The mean RGS values of D. cneorum subsp. cneorum DNA-diploids at the monoploid level did not differ statistically from those of DNA-triploids (Fig. 3A; Table 1). Likewise, the monoploid RGS of D. cneorum subsp. arbusculoides fully overlapped with the RGS of the nominate subspecies (Fig. 3A; Table 1). The intraspecific variation in RGS exceeded 8.59 % in total in the diploid cytotype of D. cneorum subsp. cneorum[seeSupporting Information—Fig. S4 and Table S1], whereas the interpopulation RGS variability was no more than 5.34 %. However, higher RGS intrapopulation variability was observed, reaching 7.86 % in the SK-DC 1 population [seeSupporting Information—Table S1]. Nevertheless, we were not able to support these RGS differences by using simultaneous analyses. No within- or between-population variation in RGS was found in D. arbuscula.
Table 1.
Relative genome size (RGS), absolute genome size (AGS), GC content, pollen viability, and flower number in studied Daphne taxa and cytotypes. Mean, standard deviation (SD), and minimum and maximum values are shown. *Two triploid individuals were analysed.
| Taxa/cytotype | RGS euploid (a.u.) ± SD | RGS monoploid (a.u.) ± SD | AGS (pg) ± SD |
GC content (%) ± SD | Pollen viability (%) | No. of flowers |
|---|---|---|---|---|---|---|
| D. arbuscula | 1.63 ± 0.01 (1.61‒1.66) | 0.82 ± 0.01 (0.80‒0.83) | 6.09 ± 0.03 (6.05‒6.14) | 41.42 ± 0.14 (41.22‒41.64) | 96.84 ± 3.6 (89‒100) | 7.2 ± 1.0 (5‒9) |
| D. cneorum subsp. cneorum diploid | 1.33 ± 0.02 (1.28‒1.39) | 0.66 ± 0.01 (0.64‒0.69) | 4.99 ± 0.05 (4.90‒5.11) | 41.48 ± 0.32 (40.54‒42.11) | 90.78 ± 7.9 (65‒99) | 10.9 ± 2.7 (6‒21) |
| D. cneorum subsp. cneorum triploid | 1.98 ± 0.04 (1.91‒2.04) | 0.66 ± 0.01 (0.64‒0.68) | 7.38, 7.47* | 41.26; 41.47* | 79.19 ± 7.2 (66‒91) | 12.3 ± 1.7 (9‒14) |
| D. cneorum subsp. arbusculoides | 1.32 ± 0.01 (1.29‒1.35) | 0.66 ± 0.01 (0.65‒0.68) | 4.98 ± 0.06 (4.90‒5.12) | 41.69 ± 0.24 (41.32‒42.22) | 95.93 ± 1.99 (93‒99) | 23.0 ± 6.5 (9‒45) |
Figure 3.
Overall variability in parametric analyses in Daphne arbuscula, D. cneorum subsp. cneorum diploids (2n = 2x) and triploids (2n = 3x) and D. cneorum subsp. arbusculoides: (A) relative monoploid genome size [a.u.]; (B) absolute genome size [pg]; (C) pollen viability [%]; (D) number of flowers per inflorescences. GLMM-estimated mean values (dots) and their 95 % confidence intervals (error bars) are displayed. The estimates labelled with the same lowercase letters were not significantly different according to Tukey’s pairwise comparison, and vice-versa.
The mean 2C values of AGS (Fig. 3B, Table 1) differed significantly among taxa (F(3, 17.6) = 2210, P < 0.0001). Two analysed triploids of D. cneorum subsp. cneorum showed significantly higher AGS (7.38 and 7.47 pg) than the other taxa followed by D. arbuscula (6.09 ± 0.03 pg), and diploids of D. cneorum (4.99 ± 0.05 pg) and D. cneorum subsp. arbusculoides (4.98 ± 0.06 pg) which had statistically indistinguishable AGS.
The differences in the GC content among the analysed cytotypes and taxa were negligible, with values ranging from 40.54 to 42.22 % (Table 1).
Pollen viability
Pollen viability was relatively high in all analysed taxa and cytotypes and ranged between 79 and 97 % on average, but it significantly differed among taxa (χ2(3) = 26.9, P < 0.0001). The highest pollen viability was found in D. arbuscula (96.84 ± 3.6 %; Fig. 3C; Table 1 and seeSupporting Information—Table S1). No significant differences were found between D. arbuscula and D. cneorum subsp. arbusculoides (z = 1.35, P = 0.53), and between diploid cytotypes of D. cneorum subsp. cneorum and D. cneorum subsp. arbusculoides (90.78 ± 7.9 % and 95.93 ± 1.9 %, respectively; z = 2.4, P = 0.076). The pollen viability of three studied triploid individuals of D. cneorum subsp. cneorum was high (79.19 ± 7.2 %), but still significantly lower than in the diploid cytotype of D. cneorum and D. arbuscula (z = 2.8, P = 0.026).
Number of flowers in the inflorescences
The number of flowers in inflorescences varied significantly among all three taxa (χ2(3) = 30.1, P < 0.0001). The lowest number of flowers was observed in D. arbuscula (7 ± 1.0), while D. cneorum subsp. arbusculoides had a significantly higher number of flowers (23 ± 6.5) than nominate subspecies D. cneorum subsp. cneorum (11 ± 2.7). Triploid individuals of D. cneorum subsp. cneorum did not differ in flower number from diploid D. cneorum subsp. cneorum (12 ± 1.7; z = 1.3, P = 0.54; Fig. 3D; Table 1).
Phylogenetic analyses
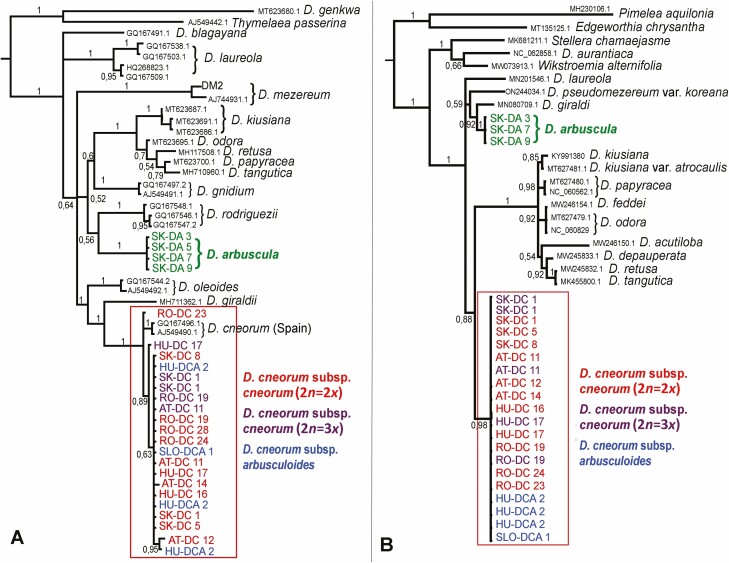
Both BI and ML analyses recovered hierarchically shallow topologies with largely unresolved relationships among the studied taxa in the ITS and plastid phylogenies. We show, however, that D. arbuscula and D. cneorum are genetically clearly distinct entities, based on both ITS and ndhF-rpl32 regions (Fig. 4A and B; seeSupporting Information—Fig. S5A–B). In the ITS phylogeny, D. cneorum appeared clustered with D. giraldii and D. oleoides (BS = 70, PP = 1). Although D. arbuscula forms a strongly supported clade, its relationship with other species remains unresolved (Fig. 4A; seeSupporting Information—Fig. S5A). Phylogenetic analyses based on the ndhF-rpl32 region uncovered an even less resolved topology than ITS; however, D. arbuscula and D. cneorum formed two well-supported clades (BS = 93, PP = 1, and BS = 99, PP = 0.98, respectively; Fig. 4B; seeSupporting Information—Fig. S5B). Both nuclear and plastid data indicate that D. cneorum subsp. arbusculoides is identical to the nominate subspecies (Fig. 4A and B; seeSupporting Information—Fig. S5A–B). The ITS region of the widespread D. cneorum subsp. cneorum showed minute intraspecific variation; distinct ribotypes were discovered in Spanish and Romanian accessions (RO-DC 23; Fig. 4A; seeSupporting Information—Fig. S5A).
Figure 4.
Bayesian inference as recovered by MrBayes: (A) phylogeny based on sequences of the ITS1-5.8S-ITS2 region of nuclear ribosomal DNA; (B) phylogeny based on sequences of the ndhF-rpl32 region of chloroplast DNA. A combination of sequences generated within this study and those retrieved from GenBank were used in both phylogenies. Bayesian posterior probabilities are indicated near the branches. Population codes of samples generated in this study follow Supporting Information—Table S1.
Discussion
Relict Daphne species exhibit extensive karyological stasis
This is the first comprehensive cytogeographic study of this fascinating genus, which has been substantially understudied from a karyological and genetic perspective despite its evolutionary significance due to its relict distribution pattern, and its richness in horticulturally attractive endemic or threatened taxa (Webb and Ferguson 1968; Halda 2001). We uncovered rather limited cytotype variability in all studied taxa; basic diploid ploidy was confirmed in most accessions. However, we found a few triploid individuals from four populations of D. cneorum subsp. cneorum[seeSupporting Information—Fig. S2D and Table S1]. RGS analyses indicate a moderate level of within-species RGS variation in D. cneorum subsp. cneorum (≤8.59 %), possibly caused by the occurrence of rare aneuploid cytotypes originating from the fusion of reduced and aneuploid gametes. Aneuploid formation appears feasible, especially given the existence of triploids and the fact that diploid individuals with the greatest RGS difference were found in populations with mixed cytotypes (particularly SK-DC 1, seeSupporting Information—Fig. S4). However, since we were unable to confirm these minor changes in RGS using simultaneous analyses, we cannot rule out the possibility that at least some of these deviating values were caused by instrumental errors (Greilhuber 1998, 2005) or/and secondary metabolites that caused problems during flow cytometry analyses (cf. Walker et al. 2006).
Our results suggest that although D. cneorum occurs in small, often highly isolated relict populations along large altitudinal gradients and on various substrates with diverse environmental conditions, its genomes appear to be largely stable, with no extensive genomic rearrangements, or accelerated dynamics in transposable element evolution (Lysák et al. 2000; Auckland et al. 2001; Bogunic et al. 2007; Vít et al. 2016). We can only hypothesize about the causes of this genomic stasis in spite of the potential effect of environmental heterogeneity and patchy distribution pattern across its range. One cause could be related to the life strategies of the studied taxa. Even though polyploidization has been found in many woody species, the rate of whole-genome duplication in herbaceous species has been estimated to be about six times higher than in woody species (Zenil-Ferguson et al. 2017). Conversely, long-lived species may have more opportunities for somatic duplication than short-lived taxa (Stebbins 1938; Grant 1981), and the formation of unreduced gametes and their successful fusion with (un)reduced gametes may be more common in species reproducing regularly over multiple years (Otto and Whitton 2000). Similarly, in the case of D. cneorum, the survival and persistence of triploid cytotypes in natural populations are likely to be facilitated by a longer lifespan, typical of woody perennials with clonal growth (Lynch et al. 1998; Nakagawa et al. 1998; Joly and Bruneau 2004; Truong et al. 2015; Morgan et al. 2021). In fact, clonality can contribute to the survival and vegetative reproduction of these triploids, albeit to a limited extent. In contrast, rare cytotypes, including those with an odd ploidy level in annual or short-lived species, have ephemeral persistence and rely on recurrent de novo formation (e.g. Čertner et al. 2017).
Occurrence and origin of triploid cytotypes in D. cneorum
While polyploidy has not played a prominent role in the evolutionary history of the genus Daphne (Bolkhovskikh et al. 1969; Murín 1990; Dawson and Beuzenberg 2013), several instances of autopolyploids and allopolyploids have been reported (Malla et al. 1977; Landolt and Hauser 1981; Urbani 1992). Our study reveals a unique example of a triploid cytotype reoccurring in several populations of D. cneorum in central Europe. Exceptional cases of triploid cytotypes have already been detected in D. mezereum (Pustahija et al. 2013) and D. odora Thunb. (2n = 27, 28, 30; Osawa 1913; Yamaha 1927; Takenaka 1931; Hiraoka 1958; Okura and Kono 1959). Triploid cytotypes may form either as a result of the fusion of reduced and unreduced gametes in purely diploid populations or heteroploid hybridization between predominantly diploid and tetraploid cytotypes (Ramsey and Schemske 1998; Husband et al. 2013). The following observations in our study lend support to the former hypothesis: (i) No tetraploid cytotypes were found in any of the studied populations to suggest heteroploid gene flow and the formation of triploid cytotypes. Although we cannot definitively rule out the existence of an unsampled tetraploid cytotype in D. cneorum, it is not very likely as polyploid cytotypes have never been detected in this species (Goldblatt and Johnson 1979; Rice et al. 2015; seeSupporting Information—Table S2). In addition, if triploids would result from heteroploid hybridization, indicating the presence of a triploid bridge between cytotypes (Kolář et al. 2017), we would anticipate a higher prevalence of triploid cytotypes in given populations (cf. Burton and Husband 2001; Čertner et al. 2017); (ii) The monoploid genome size of diploids and triploids were identical (Fig. 3A), indicating that they most likely originated from the same parental taxon; (iii) The triploids were morphologically similar to their diploid counterparts and even had the same number of flowers per inflorescence (Figs. 1D, E and 3D), ruling out the possibility of hybridization with another related, but morphologically distinct polyploid taxon; (iv) Likewise, the results of our phylogenetic analyses do not indicate the presence of other genomes than that of D. cneorum in detected triploid cytotypes (Fig. 4A and B; seeSupporting Information—Fig. S5A–B). Although none of these observations explicitly reject the allopolyploid origin of triploids in D. cneorum, the combination of factors mentioned above makes this alternative hypothesis highly improbable. Furthermore, the detected triploids likely evolved independently because they appear in different, often geographically distant, populations with diverse environmental conditions, that is, various altitudes and bedrock types (Figs. 1F–I and 2; seeSupporting Information—Table S1).
Pollen viability of triploids in D. cneorum and their frequency and persistence in diploid populations
Triploids in sexually reproducing species have typically been regarded as a dead end due to low fertility caused by irregular chromosomal pairing and unequal meiotic division, resulting in the formation of a range of euploid and aneuploid gametes (Ramsey and Schemske 1998; Husband 2004). Unbalanced chromosomal compositions of triploids and aneuploids typically lead to sterility (Eckert 2001; Husband 2004). Irregularities in meiosis and aneuploid gamete production were demonstrated, for instance, in the Asian congener, D. odora (Osawa 1913; Yamaha 1927; Takenaka 1931; Hiraoka 1958; Okura and Kono 1959). In contrast to D. odora (Osawa 1913), however, the pollen viability in triploids and diploids of D. cneorum is comparable, ranging from 66 to 91 % (Fig. 3C; Table 1). This indicates that triploids in D. cneorum subsp. cneorum could theoretically be considered at least semi-fertile due to their high pollen viability (Schinkel et al. 2017). The pollen fertility of triploids in angiosperms is highly variable and species-specific, ranging from 0 to 97 % (Ramsey and Schemske 1998). Low pollen viability was detected, for instance, in triploid hybrid individuals of Populus (2.78 %; Wang et al. 2017), Betula (8–9 %; Anamthawat-Jónsson et al. 2021), and Tamarix cf. kermanensis (28.5 %; Samadi et al. 2011). In contrast, triploids of Arabidopsis arenosa can produce even 82 % viable pollen grains (Morgan et al. 2021). The pollen viability revealed in both cytotypes and subspecies of D. cneorum and D. arbuscula is markedly higher than that of their congeners, D. gnidium (41 %; Roccotiello et al. 2009), D. genkwa (51 %; Liu et al. 2011) and D. aurantiaca (35–75 %; Liu et al. 2018). Our results are also not in line with the assumption that low pollen viability might be a potential factor responsible for the low success of fruit production in D. arbuscula and D. cneorum (Erdelská and Turis 1995; Gajdošová 2020).
The frequency of D. cneorum triploids in three populations varied between 0.71 and 1.3 %, whereas in population RO-DC 19 this incidence was 10 %. Although bedrock type was not indicative of the occurrence of triploid individuals, most populations harbouring odd cytotypes are characterized by harsh drought conditions (RO-DC 19 on gypsum, SK-DC 1 on siliceous sands, or HU-DC 17 on the calcareous substrate; Fig. 1G–I; seeSupporting Information—Table S1). We may hypothesize that extreme microclimate conditions could act as a major environmental driver in shifting survival strategies and promoting clonality (e.g. Ramsey and Schemske 1998; Mock et al. 2012). In addition, it is known that the formation of unreduced gametes is more frequent under stressful conditions, which in turn can lead to an increase in triploid cytotype formation in woody perennials (e.g. Harlan and deWet 1975; Ramsey and Schemske 1998). More data on both in situ microclimate particularities and the extent of clonality in these populations are required in order to fully test this hypothesis. Conversely, this result is also attributable to the small sample sizes belonging to these populations [seeSupporting Information—Table S1]. Comparatively low frequencies of triploid individuals within diploid populations have also been reported in other woody species, specifically 2.8 % of triploids in Olea europaea (Besnard and Baali-Cherif 2009) and from 0.25 to 0.57 % in various Quercus species (Dzialuk et al. 2007 and citations therein). There are multiple explanations for the rarity of triploids in sexually reproducing diploid plant species. In fact, their absence may be due to the very low incidence of unreduced gametes, with production in native populations rarely exceeding 2 %. It is reported to be higher in asexual species than in sexual species with selfing, mixed mating, or outcrossing pollination (Ramsey 2007; Kreiner et al. 2017). Even after the successful fusion of 2n and n gametes, seeds could have significantly reduced viability and germination abilities (Burton and Husband 2000; Truong et al. 2015; Wang et al. 2016). Furthermore, even after successful germination (e.g. Rounsaville et al. 2011), their fitness and competitiveness in the environment in a given population can be weak (Burton and Husband 2000; Baack 2005; Truong et al. 2015; Morgan et al. 2021), and they may not survive to the stage of a fully developed, reproducible individual (Truong et al. 2015; Wang et al. 2016). Our study calls for further research to determine whether triploids in D. cneorum produce their own seeds, their viability, and whether potential seedlings survive to the reproductive stage.
Notes on evolutionary relationships and taxonomy of studied Daphne taxa
Phylogenetic analyses revealed that D. cneorum and D. arbuscula are well-defined species. Even though the overall relationships between the taxa were only partially resolved, there appears to be little support for the previously predicted close relationship between D. arbuscula and D. cneorum (Tuzson 1911; Halda 1976, 2001). Intriguingly, D. cneorum subsp. arbusculoides, which has been reported as separate endemic subspecies within D. cneorum, cannot be distinguished from nominate subspecies based on ITS and cpDNA sequences or karyological traits like ploidy and genome size. As reported in taxonomic literature, both subspecies should differ not only in terms of ecology and distribution range but also with respect to morphological traits (Tuzson 1911; Webb and Ferguson 1968; Soó 1971; Halda 1976). Our study shows that D. cneorum subsp. arbusculoides accessions differ significantly in flower number from those of D. cneorum subsp. cneorum. Another important diagnostic trait delimiting this taxon should be a more ascending habitus and the shape of the leaf margins, which we did not formally test here. To draw sound taxonomic conclusions, further taxonomic research using a combination of morphological, ecological, and robust molecular markers (NGS-based) analyses is necessary.
Supporting Information
The following additional information is available in the online version of this article –
Table S1. Details of localities, genome size values, chromosome numbers, pollen viability, and number of flowers in inflorescences for sampled populations of Daphne taxa from the present study.
Table S2. A synthesis of published chromosome numbers and genome size values within the genus Daphne.
Figure S1. Stained pollen grains of studied Daphne taxa considered as viable (magenta–red) and non-viable (blue–green).
Figure S2. Microphotographs of chromosome mitotic metaphase of studied Daphne taxa.
Figure S3. Differences in RGS of studied Daphne taxa.
Figure S4. Boxplots of RGSs for populations of studied Daphne taxa.
Figure S5. Maximum likelihood analyses as recovered by RaxML.
Acknowledgements
We thank Viktória Chilová (Veľká Fatra National Park Administration, Martin, Slovakia) for accompanying during field sampling of several populations of D. cneorum from Slovakia; Adrian Indreica (Transylvania University, Brașov, Romania) for sampling one population of D. cneorum from Romania and providing helpful information regarding other populations from the Eastern Carpathians and Transylvania; Marius Ailincăi for assistance during field sampling of several populations in Romania; and Claudia Danau (Retezat Mts. National Park Administration), Liliana Varga, Corina Steiu, and Anca-Maria Cruceat Răduleț for providing valuable information regarding several localities of D. cneorum in the Southern Carpathians.
Evolution & Diversity. Section Editor: Jeremy Beaulieu
Contributor Information
Zuzana Gajdošová, Department of Evolution and Systematics, Institute of Botany, Plant Sciences and Biodiversity Centre, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 23 Bratislava, Slovak Republic.
Marek Svitok, Department of Biology and General Ecology, Faculty of Ecology and Environmental Sciences, Technical University in Zvolen, Ul. T. G. Masaryka 24, SK-960 01 Zvolen, Slovak Republic; Department of Forest Ecology, Czech University of Life Sciences Prague, CZ-16 521 Suchdol, Praha 6, Czech Republic.
Veronika Cetlová, Department of Evolution and Systematics, Institute of Botany, Plant Sciences and Biodiversity Centre, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 23 Bratislava, Slovak Republic.
Lenka Mártonfiová, Botanical Garden of Pavol Jozef Šafárik University in Košice, Mánesova 23, SK-043 52 Košice, Slovak Republic.
Jaromír Kučera, Department of Evolution and Systematics, Institute of Botany, Plant Sciences and Biodiversity Centre, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 23 Bratislava, Slovak Republic.
Vladislav Kolarčik, Institute of Biology and Ecology, Faculty of Science, Pavol Jozef Šafárik University in Košice, Mánesova 23, SK-041 54 Košice, Slovak Republic.
Bogdan-Iuliu Hurdu, Department of Taxonomy and Evolution, Institute of Biological Research, 48 Republicii St., R-400015 Cluj-Napoca, Romania.
Ioana-Minodora Sîrbu, Faculty of Biology, University of Bucharest, Splaiul Independenței 91–95, R-050095, Bucharest, Romania.
Ingrid Turisová, Department of Biology and Ecology, Faculty of Natural Sciences, Matej Bel University in Banská Bystrica, Tajovského 40, SK-974 01 Banská Bystrica, Slovak Republic.
Peter Turis, Department of Biology and Ecology, Faculty of Natural Sciences, Matej Bel University in Banská Bystrica, Tajovského 40, SK-974 01 Banská Bystrica, Slovak Republic.
Marek Slovák, Department of Evolution and Systematics, Institute of Botany, Plant Sciences and Biodiversity Centre, Slovak Academy of Sciences, Dúbravská cesta 9, SK-845 23 Bratislava, Slovak Republic; Department of Botany, Faculty of Science, Charles University, Benátská 2, CZ-128 01 Praha, Czech Republic.
Sources of Funding
This work was supported by the Slovak Research and Development Agency (no. APVV-22-0365), by the Scientific Grant Agency of the Ministry of Education, Science, Research and Sports of the Slovak Republic and the Slovak Academy of Sciences (VEGA no. 2/0098/22) and by Grant programme for Slovak Academy of Science PhD students (no. APP0267). M.SV. was supported by the Operational Programme Integrated Infrastructure (OPII), funded by the European Regional Development Fund (ERDF; ITMS 313011T721).
Contributions by the Authors
Z.G. was involved in sampling and mapping, performed and interpreted flow cytometric analyses, conducted direct chromosome counting, pollen viability testing, DNA extraction and sequencing, and carried out statistical analyses and plotting. M.SL. conceived the study, participated in sampling, and contributed to writing the manuscript. L.M. conducted chromosome number counting analyses. J.K. participated in sampling and mapping and conducted flow cytometric analyses. V.C. participated in DNA sequencing and molecular analyses. I.T. and P.T. participated in sampling and mapping in Slovakia, while B.I.H. and I.M.S. did so in Romania. V.K. participated in flow cytometric analyses (PI). M.SV. performed statistical data analyses and contributed to writing the manuscript. Z.G. wrote the text with input from all authors, and approved the final version of the manuscript.
Conflict of Interest Statement
None declared.
Data Availability
Data are provided as Supporting Information. The datasets generated for this study can be found in online repositories (https://www.ncbi.nlm.nih.gov/genbank/) with GenBank accession numbers OQ269406–OQ269431 for 26 ITS sequences and OQ320711–OQ320733 for 23 cpDNA sequences (ndhF–rpl32 region).
Literature Cited
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19:716–723. [Google Scholar]
- Anamthawat-Jónsson K, Karlsdóttir L, Thórsson ÆT, Jóhannsson M.. 2021. Naturally occurring triploid birch hybrids from woodlands in Iceland are partially fertile. New Forests 52:659–678. [Google Scholar]
- Auckland L, Johnston J, Price H, Bridgwater F.. 2001. Stability of nuclear DNA content among divergent and isolated populations of Fraser fir. Canadian Journal of Botany 79:1375–1378. [Google Scholar]
- Baack EJ. 2005. Ecological factors influencing tetraploid establishment in snow buttercups (Ranunculus adoneus, Ranunculaceae): minority cytotype exclusion and barriers to triploid formation. American Journal of Botany 92:1827–1835. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S.. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67:1–48. [Google Scholar]
- Besnard G, Baali-Cherif D.. 2009. Coexistence of diploids and triploids in a Saharan relict olive: evidence from nuclear microsatellite and flow cytometry analyses. Comptes Rendus Biologies 332:1115–1120. [DOI] [PubMed] [Google Scholar]
- Blakesley D, Allen A, Pellny TK, Roberts AV.. 2002. Natural and induced polyploidy in Acacia dealbata Link. and Acacia mangium Wild. Annals of Botany 90:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunic F, Muratovic E, Ballian D, Siljak-Yakovlev S, Brown S.. 2007. Genome size stability among five subspecies of Pinus nigra Arnold s.l. Environmental and Experimental Botany 59:354–360. [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White JSS.. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends in Ecology & Evolution 24:127–135. [DOI] [PubMed] [Google Scholar]
- Bolkhovskikh ZV, Grif VG, Zakharieva OI, Matveeva TS.. 1969. Chromosome numbers of flowering plants. Leningrad: Nauka, V. L. Komarov Botanical Institute of the Academy of Sciences of the U.S.S.R. [Google Scholar]
- Borchsenius F. 2009. FastGap 1.2. Denmark: Department of Biosciences, Aarhus University. [Google Scholar]
- Browicz K, Gostyńska-Jakuszewska M.. 1967. 133. Daphne cneorum L.—Wawrzynek główkowy. In: Browicz K, Gostyńska-Jakuszewska M, eds. Atlas of distribution of trees and shrubs in Poland. Part 6. Poznań: Państwowe Wydawnictwo Naukowe, 19–21. [Google Scholar]
- Burton TL, Husband BC.. 2000. Fitness differences among diploids, tetraploids, and their triploid progeny in Chamerion angustifolium: mechanisms of inviability and implications for polyploid evolution. Evolution 54:1182–1191. [DOI] [PubMed] [Google Scholar]
- Burton TL, Husband BC.. 2001. Fecundity and offspring ploidy in matings among diploid, triploid and tetraploid Chamerion angustifolium (Onagraceae): consequences for tetraploid establishment. Heredity 87:573–582. [DOI] [PubMed] [Google Scholar]
- Čertner M, Fenclová E, Kúr P, Kolář F, Koutecký P, Krahulcová A, Suda J.. 2017. Evolutionary dynamics of mixed-ploidy populations in an annual herb: dispersal, local persistence and recurrent origins of polyploids. Annals of Botany 120:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MI, Beuzenberg EJ.. 2013. Contributions to a chromosome atlas of the New Zealand flora—36. Miscellaneous families. New Zealand Journal of Botany 38:1–23. [Google Scholar]
- Di Sacco A, Gajdošová Z, Slovák M, Turisová I, Turis P, Kučera J, Müller JV.. 2021. Seed germination behaviour of the narrow endemic Daphne arbuscula (Thymelaeaceae), compared to the more widespread Daphne cneorum. Folia Geobotanica 56:13–25. [Google Scholar]
- Doležel J, Bartoš J.. 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Annals of Botany 95:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J.. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2:2233–2244. [DOI] [PubMed] [Google Scholar]
- Dunn KP, Smyth GK.. 1996. Randomized quantile residuals. Journal of Computational and Graphical Statistics 5:1–10. [Google Scholar]
- Dzialuk A, Chybicki I, Welc M, Sliwinska E, Burczyk J.. 2007. Presence of triploids among oak species. Annals of Botany 99:959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG. 2001. The loss of sex in clonal plants. Evolutionary Ecology 15:501–520. [Google Scholar]
- Erdelská O, Turis P.. 1995. Biology of Daphne arbuscula Čelak. (Thymelaeaceae). Biologia Bratislava 50:333–348. [Google Scholar]
- Gajdošová Z. 2020. Reprodukčné mechanizmy, genetická a karyologická variabilita Daphne arbuscula Čelak. (Thymelaeaceae). Master Thesis, Matej Bel University in Banská Bystrica, Slovakia. [Google Scholar]
- García-Verdugo C, Calleja JA, Vargas P, Silva L, Moreira O, Pulido F.. 2013. Polyploidy and microsatellite variation in the relict tree Prunus lusitanica L.: how effective are refugia in preserving genotypic diversity of clonal taxa? Molecular Ecology 22:1546–1557. [DOI] [PubMed] [Google Scholar]
- Goldblatt P, Johnson DE.. 1979. Index to plant chromosome numbers (IPCN). St. Louis: Missouri Botanical Garden. [Google Scholar]
- Grant VP. 1981. Plant speciation, 2nd edn. New York: Columbia University Press. [Google Scholar]
- Greilhuber J. 1998. Intraspecific variation in genome size: a critical reassessment. Annals of Botany 82:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J. 2005. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany 95:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Obermayer R.. 1997. Genome size and maturity group in Glycine max (soybean). Heredity 78:547–551. [Google Scholar]
- Halda JJ. 1976. Lýkovce. In: Holzbecher J, ed. Skalky, skalničky: Zpravodaj klubu skalničkářů ČOZS Brno. Brno: Klub skalničkářů, 11–68. [Google Scholar]
- Halda JJ. 2001. The genus Daphne. Hronov: SEN Dobré. [Google Scholar]
- Harlan JR, deWet JMJ.. 1975. On Ö. Winge and a Prayer: the origins of polyploidy. Botanical Review 41:361–390. [Google Scholar]
- Hartig F. 2022. DHARMa: Residual Diagnostics for Hierarchical (Multi-level/Mixed) Regression Models. R Package Version 0.4.6. Available fromhttp://florianhartig.github.io/DHARMa/ [accessed by 05 September 2023].
- Hewitt GM. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society 58:247–276. [Google Scholar]
- Hewitt GM. 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. [DOI] [PubMed] [Google Scholar]
- Hiraoka T. 1958. Somatic syndesis in Daphne odora I. The chromosome behaviour in mitosis. Proceedings of the Japan Academy 34:700–705. [Google Scholar]
- Huang Y, Jacques FMB, Su T, Ferguson DK, Tang H, Chen W, Zhou Z.. 2015. Distribution of Cenozoic plant relicts in China explained by drought in dry season. Scientific Reports 5:14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC. 2004. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society 82:537–546. [Google Scholar]
- Husband BC, Baldwin SJ, Suda J.. 2013. The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes. In: Greilhuber J, Doležel J, Wendel J, eds. Plant genome diversity, Vol. 2. Vienna: Springer, 255–276. [Google Scholar]
- Joly S, Bruneau A.. 2004. Evolution of triploidy in Apios americana (Leguminosae) revealed by the genealogical analysis of the histone H3-D gene. Evolution 58:284–295. [PubMed] [Google Scholar]
- Kenward MG, Roger JH.. 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53:983–997. [PubMed] [Google Scholar]
- Kochjarová J, Turis P, Blanár D, Hrivnák R, Kliment J, Vlčko J.. 2004. Vascular plants of the Muránska Planina Mts. In: Kochjarová J, Uhrín M, eds. Biodiversity of the Muránska Planina National Park. Revúca: Reussia, Supplement 1, 91–190. [Google Scholar]
- Kolář F, Čertner M, Suda J, Schönswetter P, Husband BC.. 2017. Mixed-ploidy species: progress and opportunities in polyploid research. Trends in Plant Science 22:1041–1055. [DOI] [PubMed] [Google Scholar]
- Kreiner JM, Kron P, Husband BC.. 2017. Frequency and maintenance of unreduced gametes in natural plant populations: associations with reproductive mode, life history and genome size. The New Phytologist 214:879–889. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB.. 2017. lmerTest Package: tests in linear mixed effects models. Journal of Statistical Software 82:1–26. [Google Scholar]
- Landolt E, Hauser E.. 1981. Daphne reichesteinii sp. nov., eine neue hybridogene Daphne-Art aus dem Gardasee-Gebiet. Bericht über das Geobotanische Institut an der ETH Stiftung Rübel 48:36–47. [Google Scholar]
- Larson DW, Matthes U, Kelly PE.. 2000. Cliff ecology: pattern and process in cliff ecosystems. New York: Cambridge University Press. [Google Scholar]
- Lavergne S, Thuiller W, Molina J, Debussche M.. 2005. Environmental and human factors influencing rare plant local occurrence, extinction and persistence: a 115-year study in the Mediterranean region. Journal of Biogeography 32:799–811. [Google Scholar]
- Lefort F, Douglas GC, Thompson D.. 2000. Microsatellite DNA profiling of phenotypically selected clones of Irish oak (Quercus spp.) and ash (Fraxinus excelsior L.). Silvae Genetica 49:21–28. [Google Scholar]
- Lenth RV. 2016. Least-squares means: the R package lsmeans. Journal of Statistical Software 69:1–33. [Google Scholar]
- Lenth RV. 2022. emmeans: Estimated Marginal Means, Aka Least-squares Means. R Package Version 1.7.2. Avaiable fromhttps://CRAN.R-project.org/package=emmeans[accessed by 05 September 2023].
- Levin DA. 2002. The role of chromosomal change in plant evolution. Oxford series in ecology and evolution. Oxford and New York: Oxford University Press. [Google Scholar]
- Lewis S, Maslin M.. 2015. Defining the Anthropocene. Nature 519:171–180. [DOI] [PubMed] [Google Scholar]
- Liu F, Wang T, Wang Y, Shen Y.. 2011. Morphological observation and analyses of viability and germination rate of Daphne genkwa pollen. Journal of Plant Resources and Environment 20:94–96. [Google Scholar]
- Liu S, Yang A, Zhou H, Yu F.. 2018. Reproductive characteristics of Daphne aurantiaca. Guihaia 38:626–634. [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C.. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100:875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AJJ, Barnes RW, Vaillancourt RE, Cambecèdes J.. 1998. Genetic evidence that Lomatia tasmanica (Proteaceae) is an ancient clone. Australian Journal of Botany 46:25–33. [Google Scholar]
- Lysák MA, Rostková A, Dixon JM, Rossi G, Doležel J.. 2000. Limited genome size variation in Sesleria albicans. Annals of Botany 86:399–403. [Google Scholar]
- Macas J, Novák P, Pellicer J, Čížková J, Koblížková A, Neumann P, Fuková I, Doležel J, Kelly LJ, Leitch IJ.. 2015. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS One 10:e0143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla SB, Bhattarai S, Gorkhali M, Saiju H, Singh MP.. 1977. Reports. In: Löve Á, ed. IOPB Chromosome number reports LVII. Taxon 26:444–446. [Google Scholar]
- Melnyk VI, Baransky AR.. 2020. Genesis and dynamics of the range of Daphne cneorum (Thymelaeaceae) within Ukraine. Ukrainian Botanical Journal 77:349–362. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE). New Orleans, LA: Institute of Electrical and Electronics Engineers, 1–8. [Google Scholar]
- Mock KE, Callahan CM, Islam-Faridi MN, Shaw JD, Rai HS, Sanderson SC, Rowe CA, Ryel RJ, Madritch MD, Gardner RS, et al. 2012. Widespread triploidy in Western North American aspen (Populus tremuloides). PLoS One 7:e48406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EJ, Čertner M, Lučanová M, Deniz U, Kubíková K, Venon A, Kovářík O, Placette CL, Kolář F.. 2021. Disentangling the components of triploid block and its fitness consequences in natural diploid–tetraploid contact zones of Arabidopsis arenosa. New Phytologist 232:1449–1462. [DOI] [PubMed] [Google Scholar]
- Muller S. 1997. The post-glacial history of Pulsatilla vernalis and Daphne cneorum in Bitcherland, inferred from the phytosociological study of their current habitat. Global Ecology and Biogeography Letters 6:129–137. [Google Scholar]
- Murín A. 1960. Substitution of cellophane for glass covers to facilitate preparation of permanent squashes and smears. Stain Technology 35:351–353. [PubMed] [Google Scholar]
- Murín A. 1990. Karyology of an endemic species Daphne arbuscula Čelak. Botanica 37:35–40. [Google Scholar]
- Nakagawa T, Garfì G, Reille M, Verlaque R.. 1998. Pollen morphology of Zelkova sicula (Ulmaceae), a recently discovered relic species of the European Tertiary flora: description, chromosomal relevance, and palaeobotanical significance. Review of Palaeobotany and Palynology 100:27–37. [Google Scholar]
- Nunvářová Kabátová K, Kolář F, Jarolímová V, Krak K, Chrtek J.. 2019. Does geography, evolutionary history or ecology drive ploidy and genome size variation in the Minuartia verna group (Caryophyllaceae) across Europe? Plant Systematics and Evolution 305:1019–1040. [Google Scholar]
- Okura E, Kono M.. 1959. Cytogenetical consideration of Daphne odora Thunb. based on its karyotype. Biological Journal of Okayama University 5:51–56. [Google Scholar]
- Orsenigo S, Adorni M, Alessandrini A, Armiraglio S, Castello M, Forte L, Gennai M, Magrini S, Medagli P, Montagnani C, et al. 2019. Global and regional IUCN Red List assessments: 7. Italian Botanist 7:107–124. [Google Scholar]
- Osawa J. 1913. On the development of the pollen-grain and embryo-sac of Daphne, with special reference to the sterility of Daphne odora. Journal of the College of Agriculture, Imperial University of Tokyo 4:37–264. [Google Scholar]
- Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods in Cell Biology 33:105–110. [DOI] [PubMed] [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 131:452–462. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34:401–437. [DOI] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C.. 2010. A simplified method for differential staining of aborted and non-aborted pollen grains. International Journal of Plant Biology 1:e13. [Google Scholar]
- Pustahija F, Brown SC, Bogunic F, Bašic N, Muratovic E, Ollier S, Hidalgo O, Bourge M, Stevanovic V, Sijak-Yakovev S.. 2013. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant and Soil 373:427–453. [Google Scholar]
- R Core Team .2021. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramsey J. 2007. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae). Heredity 98:143–150. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29:467–501. [Google Scholar]
- Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I.. 2015. The chromosome counts database (CCDB)—a community resource of plant chromosome numbers. New Phytologist 206:19–26. [DOI] [PubMed] [Google Scholar]
- Roccotiello E, Casazza G, Galli L, Cornara L, Moncalvo A, Minuto L.. 2009. The flower biology of Daphne gnidium L. (Thymelaeaceae). Plant Systematics and Evolution 279:41–49. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsaville TJ, Touchell DH, Ranney TG.. 2011. Fertility and reproductive pathways in diploid and triploid Miscanthus sinensis. HortScience 46:1353–1357. [Google Scholar]
- Samadi N, Ghaffari SM, Akhani H.. 2011. Meiotic behaviour, karyotype analyses and pollen viability in species of Tamarix (Tamaricaceae). Willdenowia 43:195–203. [Google Scholar]
- Schinkel CCF, Kirchheimer B, Dullinger S, Geelen D, De Storme N, Hörandl E.. 2017. Pathways to polyploidy: indications of a female triploid bridge in the alpine species Ranunculus kuepferi (Ranunculaceae). Plant Systematics and Evolution 303:1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann C.. 2007. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Molecular Phylogenetics and Evolution 42:92–103. [DOI] [PubMed] [Google Scholar]
- Schubert I, Vu GTH.. 2016. Genome stability and evolution: attempting a holistic view. Trends in Plant Science 21:749–757. [DOI] [PubMed] [Google Scholar]
- Šedivá J, Žlebčík J.. 2010. Vegetative and generative propagation of the endangered species Daphne cneorum L. Acta Pruhoniciana 96:15–18. [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL.. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 94:275–288. [DOI] [PubMed] [Google Scholar]
- Siljak-Yakovlev S, Stevanović V, Tomašević M, Brown SC, Stevanović B.. 2008. Genome size variation and polyploidy in the resurrection plant genus Ramonda: cytogeography of living fossils. Environmental and Experimental Botany 62:101–112. [Google Scholar]
- Simmons MP, Ochoterena H.. 2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49:369–381. [PubMed] [Google Scholar]
- Slovák M, Vít P, Urfus T, Suda J.. 2009. Complex pattern of genome size variation in a polymorphic member of the Asteraceae. Journal of Biogeography 36:372–384. [Google Scholar]
- Šmarda P, Bureš P.. 2006. Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany 98:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. 2008. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany 101:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2000. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences of the United States of America 97:7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ.. 2003. Evolution of genome size in the angiosperms. American Journal of Botany 90:1596–1603. [DOI] [PubMed] [Google Scholar]
- Soó R. 1971. Species et combinations novae Florae Europae praecipue Hungariae X. Acta Botanica Academiae Scientiarum Hungaricae. Budapest 17:115–125. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1938. Cytological characteristics associated with the different growth habits in the dicotyledons. American Journal of Botany 25:189–198. [Google Scholar]
- Takenaka Y. 1931. Further reports of the cytological investigations on the sterile plants. Journal of Chosen Natural History Society 12:25–41. [Google Scholar]
- Tang CQ, Matsui T, Ohashi H, Dong Y-F, Momohara A, Herrando-Moraira S, Qian S, Yang Y, Ohsawa M, Luu HT, et al. 2018. Identifying long-term stable refugia for relict plant species in East Asia. Nature Communication 9:4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong NX, Song Y-S, Kim N-S, Park J-W, Kim J-H, Wakana A.. 2015. Occurrence and survival of autotriploids in natural diploid populations of Lilium lancifolium Thunb. Journal of the Faculty of Agriculture Kyushu University 60:73–80. [Google Scholar]
- Tuzson J. 1911. A Daphne génusz Cneorum subsectíójáról. (De subsectione „Cneorum“ generis Daphnes.). Botanikai Közlemények 10:135–152. [Google Scholar]
- Uhlířová J, Bernátová D.. 2003. Príspevok k flóre a vegetácii skalných stanovíšť Muránskej planiny. Acta Rerum Naturalium Musei Nationalis Slovaci 49:55–67. [Google Scholar]
- Uhlířová J, Bernátová D.. 2004. A new syntaxonomical view on the association Pulsatillo slavicae–Caricetum humilis. Annotationes Zoologicae et Botanicae 227:3–12. [Google Scholar]
- Urbani M. 1992. Ricerche biosistematiche e corologiche sulle Thymelaeaceae in Italia:1. Daphne alpina L. Webbia 46:203–217. [Google Scholar]
- Valachovič M, Jarolímek I.. 1994. Rastlinné spoločenstvá s výskytom Daphne arbuscula Čelak. na Muránskej planine. Bulletin Slovenskej Botanickej Spoločnosti 16:75–82. [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nature Reviews 18:411–424. [DOI] [PubMed] [Google Scholar]
- Vít P, Krak K, Trávníček P, Douda J, Lomonosova MN, Mandák B.. 2016. Genome size stability across Eurasian Chenopodium species (Amaranthaceae). Botanical Journal of the Linnean Society 182:637–649. [Google Scholar]
- Walker J, Moñino I, Correal E.. 2006. Genome size in Bituminaria bituminosa (L.) C.H. Stirton (Fabaceae) populations: separation of ‘true’ differences from environmental effects on DNA determination. Environmental and Experimental Botany 55:258–265. [Google Scholar]
- Wang X, Cheng ZM, Zhi S, Xu F.. 2016. Breeding triploid plants: a review. Czech Journal of Genetics and Plant Breeding 52:41–54. [Google Scholar]
- Wang J, Huo B, Liu W, Li D, Liao L.. 2017. Abnormal meiosis in an intersectional allotriploid of Populus L. and segregation of ploidy levels in 2x × 3x progeny. PLoS One 12:e0181767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DA, Ferguson IK.. 1968. Daphne. In Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA, eds. Flora Europaea Vol. 2, Rosaceae to Umbelliferae. Cambridge: Cambridge University Press, 256–258. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR Protocol: a guide to methods and applications. San Diego, California: Academic Press, Inc., 315–322. [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Yamaha G. 1927. Experimentelle zytologische Beiträge III. Mitteilung. Über die Wirkung einiger Chemikalien auf die Pollenmutterzellen von Daphne odora Thunb. The Botanical Magazine, Tokyo 41:181–211. [Google Scholar]
- Zenil-Ferguson R, Ponciano JM, Burleigh JG.. 2017. Testing the association of phenotypes with polyploidy: an example using herbaceous and woody eudicots. Evolution; International Journal of Organic Evolution 71:1138–1148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are provided as Supporting Information. The datasets generated for this study can be found in online repositories (https://www.ncbi.nlm.nih.gov/genbank/) with GenBank accession numbers OQ269406–OQ269431 for 26 ITS sequences and OQ320711–OQ320733 for 23 cpDNA sequences (ndhF–rpl32 region).