ABSTRACT
Pseudomonas aeruginosa is one of the most common biofilm-forming pathogens responsible for lung infections of individuals with cystic fibrosis (CF). P. aeruginosa becomes tolerant to antimicrobials in the biofilm state and is difficult to treat. Production of extracellular polymeric substances (EPS), such as alginate and extracellular DNA (eDNA), can allow adherence to abiotic and biotic surfaces, antimicrobial evasion, and resilience to environmental pressures. Alginate-producing mucoid variants of P. aeruginosa are frequently isolated from CF airway samples and are associated with worsening patient outcomes. While eDNA is a major structural component of nonmucoid P. aeruginosa biofilms, the potential role of eDNA in mucoid biofilms is unclear. Here, we investigate how eDNA contributes to clinical mucoid biofilm physiology and integrity. We predicted that eDNA plays a structural and mechanical role in mucoid biofilms. To test this, we quantified biofilm eDNA in mucoid biofilms and used microscopy and rheology to visualize eDNA and detect changes in biofilm structure and mechanics upon DNaseI treatment. We showed that biofilm eDNA abundance is diverse across clinical mucoid strains and observed a temporal increase in foci of eDNA within intact mucoid biofilms. Increased cell dispersal and reduced biomass were also observed following DNaseI treatment of mucoid biofilms. Degradation of eDNA also impacted the mechanical integrity of mucoid biofilms by increasing the stiffness and decreasing the cohesion of the biofilm. These findings advance our understanding of clinical mucoid P. aeruginosa biofilms and facilitate the development of new approaches to target biofilms by exploiting the functions of EPS components.
IMPORTANCE
Understanding the role of eDNA in mucoid Pseudomonas aeruginosa biofilms will lead to therapeutic strategies that combat the biophysical and structural function of EPS for the eradication of bacteria in mucoid biofilms during chronic infections. This knowledge can be used to further identify unknown matrix component interactions within pathogenic biofilm-forming clinical isolates.
KEYWORDS: alginate, cystic fibrosis, DNaseI, extracellular polymeric substances
INTRODUCTION
Biofilms, which are aggregates of microbial communities encased by an extracellular polymeric substance (EPS), are highly resilient (1 – 4). The EPS provides microorganisms within the biofilm with properties including enhanced attachment to abiotic (i.e., surgical implants) and biotic surfaces (i.e., epithelial cells and mucosal layers) and tolerance to host antimicrobial defenses and antibiotics (2, 5). Therefore, biofilm formation is considered a virulence factor and is a leading cause of chronic infection (5 – 7). Pseudomonas aeruginosa is a significant biofilm-forming pathogen, and it is one of the foremost Gram-negative bacteria responsible for nosocomial infections (8). This opportunistic pathogen is also the leading cause of mortality in immunocompromised individuals, such as those with cystic fibrosis (CF) (7, 9 – 11). In the CF lung, due to an abnormally thick mucus layer, microorganisms are not properly cleared. This consequentially causes a build-up of microorganisms, and toxins and virulence factors produced by these organisms (12). In chronic conditions, the CF lung environment promotes the emergence of patho-adapted P. aeruginosa variants (2, 13). Mucoid variants of P. aeruginosa are associated with chronic infection and worsening patient outcomes (14 – 16). Mucoid variants predominate and become the main P. aeruginosa isolates from sputum samples (9, 17). Mucoid P. aeruginosa variants arise due to the acquisition of mutations in muc genes (most common being mucA22), resulting in overproduction of the exopolysaccharide, alginate (18, 19).
Although there are many studies regarding the role of alginate in mucoid biofilms, the function of other P. aeruginosa biofilm EPS components in mucoid biofilms is limited, largely due to the abundance of alginate within the EPS. Previous work has shown how alginate interaction with calcium impacts the structure and cell organization of mucoid biofilms (20, 21). In addition, another EPS exopolysaccharide produced by P. aeruginosa, Psl, enhances mucoid biofilm attachment and host immune evasion (22). Extracellular DNA (eDNA) is an important EPS component of nonmucoid P. aeruginosa biofilms and is considered a conserved EPS component across microbial biofilms (23). Nonmucoid P. aeruginosa biofilms have abundant eDNA that promotes cell surface attachment and maintains 3D architecture and biophysical properties (23 – 27). In this study, we tested the hypothesis that eDNA will play a significant structural and biophysical role in P. aeruginosa clinical mucoid biofilms. We quantified the eDNA concentration of biofilms formed by several clinical mucoid strains and performed correlative comparisons to alginate production. Fluorescent microscopy and rheology were used to analyze structural and mechanical changes of DNaseI-treated P. aeruginosa mucoid biofilms. We discovered that biofilm eDNA abundance is diverse across clinical mucoid isolates and positively correlates with alginate production. We also showed that eDNA, in the form of punctate foci, temporally increases in clinical mucoid biofilms. Our results show that when eDNA is abundant, DNaseI treatment can disperse cells in clinical mucoid biofilms, as well as decrease biofilm thickness and EPS cohesion, and increase stiffness.
RESULTS AND DISCUSSION
eDNA abundance is diverse across clinical mucoid isolates
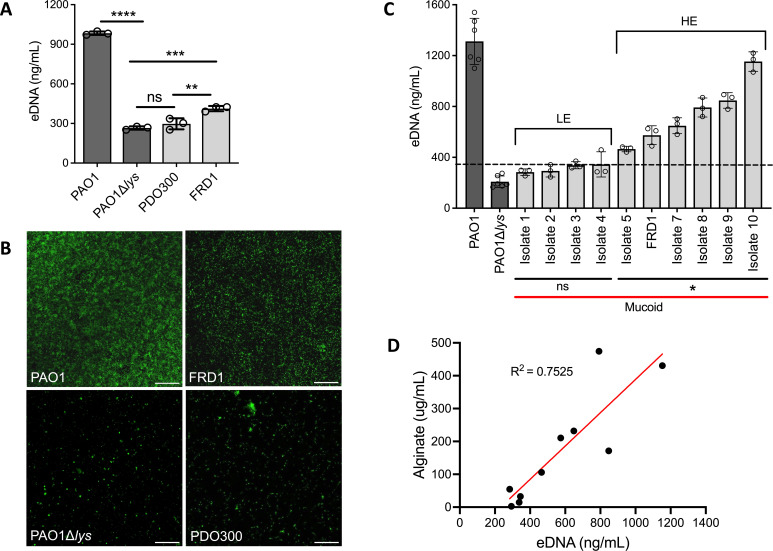
To quantify the abundance of eDNA in mucoid biofilms, we isolated eDNA from colony biofilms of nonmucoid PAO1 (positive eDNA control), PAO1∆lys that has impaired eDNA release (28) (negative eDNA control), and mucoid (∆mucA22) strains PDO300 (lab derived) and FRD1 (clinical isolate) (22, 29). eDNA concentration was quantified by fluorometric analysis. Results show that nonmucoid PAO1 has a high abundance of eDNA, which aligns with previous studies (30). eDNA abundance of the isogenic mucoid PDO300 was lower compared to the parent PAO1 and comparable to the eDNA mutant, PAO1∆lys (Fig. 1A). Although non-isogenic, mucoid clinical isolate FRD1 had higher biofilm eDNA than what was produced by PDO300 and the eDNA mutant (Fig. 1A). To further corroborate these observations, hydrated nonmucoid and mucoid biofilms were visualized by widefield microscopy, and eDNA stained using TOTO-1. Images show that the abundance of eDNA found in colony biofilms reflects the observed level of eDNA in corresponding hydrated biofilms of each strain (Fig. 1B). Since FRD1 is a well-described clinical mucoid isolate (31), it was used as the reference mucoid strain for subsequent experiments.
Fig 1.
eDNA abundance is diverse across clinical mucoid isolates. Fluorometric quantification of eDNA isolated from colony biofilms of nonmucoid (dark gray) PAO1, eDNA mutant PAO1∆lys, and mucoid (light gray) laboratory-derived PDO300 and clinical strain FRD1 (A). Representative images of TOTO-1 eDNA stained 48 h static biofilms of corresponding mucoid and nonmucoid strains; 20x objective, scale bar: 100 µm (B). Quantification of eDNA isolated from colony biofilms of 10 clinical mucoid isolates. The dashed line indicates +3 standard deviations above the eDNA mutant (PAO1∆lys) average (345 ng/mL). The “ns” bar indicates strains with eDNA concentrations comparable to the eDNA mutant, hence, low eDNA (LE). The “*” bar indicates strains with eDNA levels significantly higher than the eDNA mutant, hence, high eDNA (HE) (C). R2 correlation measurement of alginate and eDNA concentrations of the 10 clinical mucoid strains. (D). Each dot represents the average of three technical from three biological replicates of each strain. Analyzed using one-way ANOVA **P < 0.005, ***P < 0.0005, ****P < 0.0001.
As PDO300 and FRD1 mucoid biofilms produced differing eDNA concentrations (Fig. 1A and B), this implies that there may be differing amounts of eDNA produced by biofilms of divergent mucoid strains. Therefore, we measured the eDNA concentration from biofilms of nine other P. aeruginosa mucoid isolates derived from CF sputum samples (Table 1). Quantification of eDNA in these clinical isolates confirmed diversity in the abundance of eDNA across various clinical mucoid strains. Biofilms of six clinical isolates contained high amounts of eDNA, while biofilms of four clinical isolates had similar levels as the eDNA mutant (Fig. 1C). Although the eDNA amount varied, there was no significant difference in the number of cells from the colony biofilms of each clinical strain (Fig. S1). Based on comparison to the eDNA mutant, the clinical mucoid isolates were grouped into those with low eDNA (LE) and high eDNA (HE) abundance (Fig. 1C). Mucoid isolates in the HE group were designated as those that produce eDNA at concentrations reaching 3 standard deviations above the eDNA mutant average concentration, and in contrast, isolates in the LE group produce eDNA at concentrations below this standard (Fig. 1C, dotted line). We also investigated how mucoid reversion (algT/U and algD mutants) could impact eDNA levels in biofilms formed by these isolates. There was no significant difference in the eDNA levels of the mucoid revertant biofilms compared to the mucoid parental clinical isolate, indicating that alginate production or regulation does not appear to be involved in eDNA release (Fig. S2). Collectively, this suggests that eDNA abundance is intrinsic to the parental mucoid strain background of the clinical isolates.
TABLE 1.
Pseudomonas aeruginosa strains and sources
| P. aeruginosa strains | Description | Source or reference |
|---|---|---|
| mPAO1 PAO1Δlys PDO300 FRD1, Isolate 6 FRD1ΔalgD FRD1ΔalgT/U Isolate 1 Isolate 2 Isolate 3 Isolate 4 Isolate 5 Isolate 7 Isolate 8 Isolate 9 Isolate 10 Isolate 11 Isolate 12 |
Nonmucoid, WT, Parent Nonmucoid, eDNA deficient, lys (PA0629) transposon mutant Mucoid, PAO1 mucA22 Clinical mucoid, mucA22 Nonmucoid variant of FRD1 Nonmucoid variant of FRD1 Clinical mucoid Clinical mucoid Clinical mucoid Clinical mucoid Clinical Mucoid Clinical mucoid Clinical mucoid Clinical mucoid Clinical mucoid Clinical mucoid Clinical mucoid |
Manoil transposon (32) Manoil transposon (28, 32, 33) Lab derived (22) CF sputum (29) CF sputum (34) CF sputum (34) CF sputum #3003 (22) CF sputum #2902 (22) CF sputum #2999 (22) CF sputum #2966 (22) CF sputum #2957 (22) CF sputum #2965 (22) CF sputum #2920 (22) CF sputum #3001 (22) CF sputum #2959 (22) CF sputum #2963 (22) CF sputum #2992 (22) |
We then investigated whether there was any correlation between eDNA and alginate concentrations across clinical mucoid isolates. Alginate produced by colony biofilms of the 10 mucoid clinical isolates was quantified, and the concentration was plotted against the corresponding eDNA concentration. Indeed, alginate abundance positively correlated with eDNA levels in these clinical mucoid biofilms (r-squared value of 0.75; Fig. 1D). This indicates that two major EPS components of P. aeruginosa biofilms are coordinately produced. Low EPS-producing P. aeruginosa clinical isolates may utilize other evasive mechanisms for protection, whereas high eDNA and alginate-producing isolates may rely on protection mechanisms attributed to the biofilm EPS. Location, antibiotic exposure, and even co-existing pathogens can influence the diversification of P. aeruginosa strains in CF lung infections (15, 35). In future studies, it will be important to reveal the possible correlation between antimicrobial tolerance, infection duration, and extent of ciliary clearance, with biofilm eDNA levels of clinical isolates.
eDNA abundance increases temporally and is present as punctate foci in early clinical mucoid biofilms
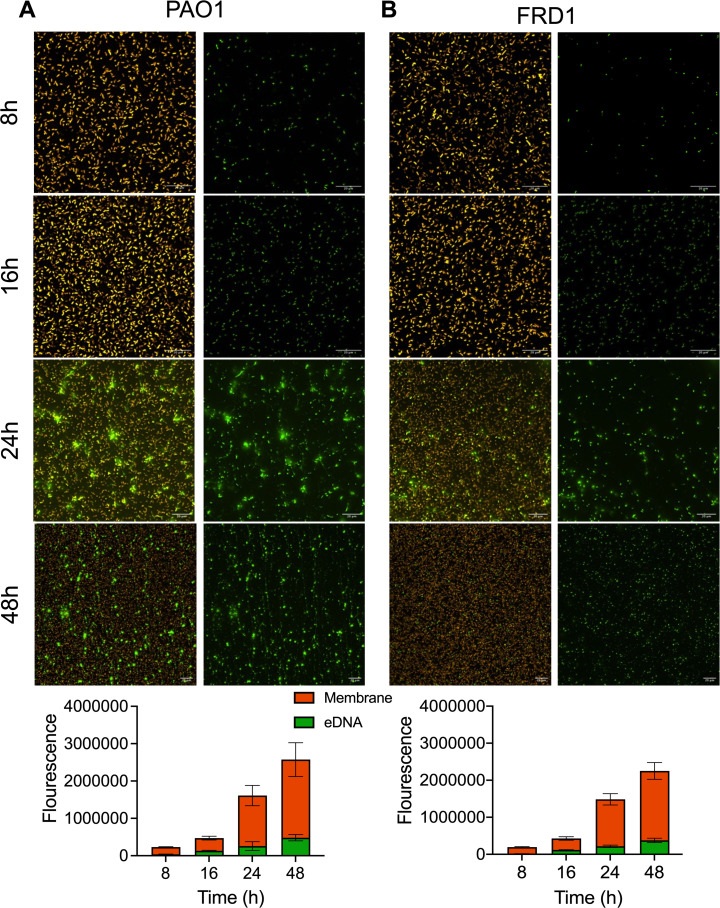
eDNA is important in nonmucoid biofilms at the early stages of biofilm formation (27, 28, 36). However, there are limited studies regarding eDNA abundance during mucoid biofilm formation. To examine the significance of eDNA during early mucoid biofilm formation, static mucoid, and nonmucoid biofilms, grown for 8, 16, 24, and 48 h, were stained with FM4-64 and TOTO-1 to label the cell membrane and eDNA, respectively, and imaged by widefield fluorescent microscopy. As previously reported (27), eDNA appears in nonmucoid PAO1 biofilms during initial attachment and increasingly at later stages (Fig. 2A). eDNA in the mucoid FRD1 biofilms also increased over the course of 48 h (Fig. 2B). Comparable amounts of bacterial cells in nonmucoid and mucoid biofilms were observed at the various timepoints. The abundance of eDNA in the biofilms was elevated after 16 h (monolayer biofilm). Noticeably, at 24 h, punctate eDNA foci began to appear in mucoid and nonmucoid biofilms (Fig. 2A and B). The presence of these eDNA foci is consistent with previous observations visualizing eDNA and its influence on organization patterns within nonmucoid P. aeruginosa biofilms (37). By 48 h, nonmucoid biofilms had both punctate foci and fibrous eDNA forms (Fig. S3A), while the mucoid biofilm still predominately had punctate eDNA foci (Fig. S3B).
Fig 2.
eDNA abundance increases temporally and is present as punctate foci in early clinical mucoid biofilms. Representative images and corresponding image analysis (bottom) using NIS-elements software of static nonmucoid PAO1 (A) and clinical mucoid FRD1 (B) biofilms labeled with FM4-64 (bacterial membrane; orange) and TOTO-1 (eDNA; green). The left panel depicts merged images of FM4-64 and TOTO-1 channels, the right panel depicts the TOTO-1 channel alone; scale bar: 20 µm. Data are depicted as stacked bars of mean ± STDV, N = 3.
There are several mechanisms of eDNA release by P. aeruginosa, including phage lysin, PQS signaling, and cell death (38). In our future work, we will investigate mechanisms of eDNA release to get more insight on eDNA accumulation and staining patterns in mucoid biofilms. Previous reports suggest that the formation of fibrous eDNA strands in P. aeruginosa biofilms is the result of surface motility and migration through eDNA foci (28, 37). In contrast to nonmucoid, mucoid P. aeruginosa are often nonmotile, and alginate production becomes favorable in environments of selective pressure, like in CF lung infections (2, 19, 39). The suppressed motility in mucoid biofilms may explain the lack of eDNA strands in clinical mucoid biofilms. In addition, a prior study showed how eDNA release via cell death from antibiotic treatment enhanced FRD1 biofilm formation and structure (40). Cell death in clinical mucoid biofilms like FRD1 could be a mechanism of eDNA release. Mucoid biofilms may have evolved to utilize this route of eDNA release in an inflammatory host environment, where the bacteria are subjected to antimicrobials from host cells. As a positive feedback loop, increased cell death and the resulting eDNA could augment biofilm integrity and antimicrobial tolerance.
DNaseI disrupts mucoid biofilms and disperses bacterial cells
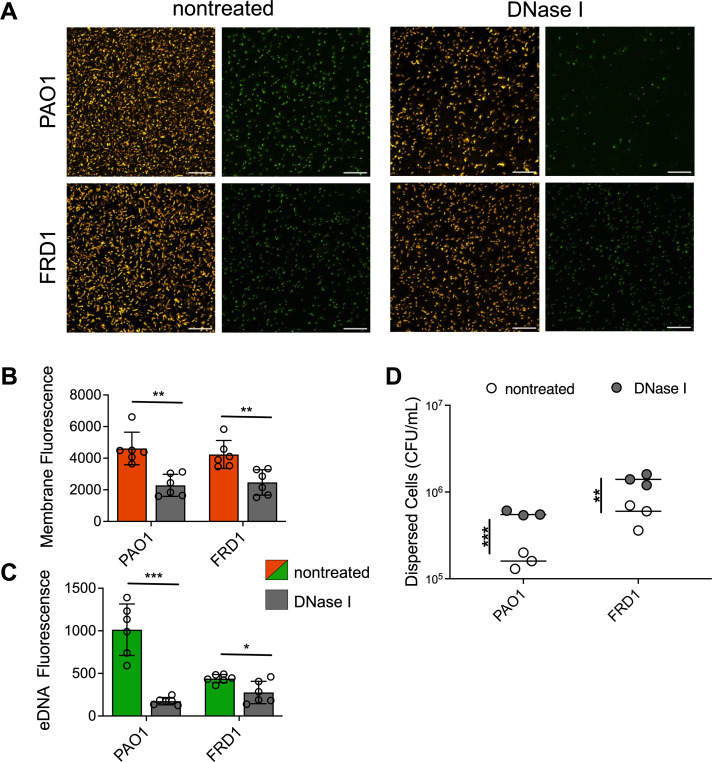
Nonmucoid P. aeruginosa early biofilms, where attachment and microcolonies are first established, are easily disrupted by DNaseI (27, 28). The impact of DNaseI on mature or later-stage biofilms is quite variable (27, 36). Recent studies suggest that this DNaseI resistance is due to the transformation of eDNA to the stable supercoiled Z-DNA form (36, 38). We therefore hypothesized that DNaseI can similarly degrade eDNA in early clinical mucoid biofilms when eDNA is still suspectable to enzymatic degradation. To test this, 16-h static FRD1 mucoid biofilms and PAO1 nonmucoid biofilms were treated with DNaseI. After DNaseI treatment, eDNA level and cell dispersal were assessed using fluorescent image analysis and quantifying cells in the biofilm supernatant. DNaseI treatment decreased bacterial cells in PAO1 and FRD1 biofilms (Fig. 3A and B). Image analysis revealed a significant reduction of bacteria and eDNA staining of DNaseI-treated mucoid and nonmucoid biofilms, compared to nontreated biofilms (Fig. 3B and C). The eDNA signal of PAO1 biofilms was significantly decreased, implying adequate degradation by DNaseI. The eDNA signal of FRD1 biofilms was also reduced after treatment, although to a lesser degree than PAO1 (Fig. 3C). Since total bacterial cells decreased in the biofilms after DNaseI treatment, bacterial cell dispersal was assessed by quantifying the bacteria remaining in the supernatant after DNaseI treatment. Biofilm supernatant of both PAO1 and FRD1 contained more bacterial cells when biofilms were treated with DNaseI, compared to nontreated controls (Fig. 3D).
Fig 3.
DNaseI disrupts mucoid biofilms and disperses bacterial cells. Representative images of bacterial membrane and eDNA-stained nonmucoid PAO1 and clinical mucoid FRD1 biofilms, nontreated (left) or treated (right) with DNaseI (200 µg/mL) after 16-h biofilm formation. The left panel depicts merged images of FM4-64 and TOTO-1 channels, the right panel depicts the TOTO-1 channel alone; the scale bar is 20 µm (A). Quantification of FM4-64 (bacterial membrane; orange) (B) and TOTO-1 (eDNA; green) (C) fluorescence using NIS-elements image analysis software comparing DNaseI treated (gray) counterparts for each strain. CFU quantification of bacterial cells from supernatant of biofilms treated with or without DNaseI (D). N = 6, Analyzed using student t-test *P < 0.05, **P < 0.005, ***P < 0.0005.
As eDNA is degraded, bacterial cells are able to disperse from the early biofilm or detach from the surface. This dispersal was more enhanced in nonmucoid biofilms, compared to clinical mucoid biofilms. This may be due to the diffusion barrier of alginate, the same anti-diffusion mechanisms proposed for antimicrobial agents (41 – 45). This could hinder the effectiveness of DNaseI in mucoid biofilms, in comparison to nonmucoid biofilms which produce minimal amounts of alginate (46). As expected, the reduced impact of DNaseI on mucoid PDO300 biofilms containing low eDNA was observed (Fig. S4). In nonmucoid biofilms, eDNA binds with other EPS components, for example, polysaccharides and DNA-binding proteins that can reinforce and strengthen biofilm structure (36, 47, 48). Degrading eDNA in the biofilm should, therefore, compromise the interactions or bonds with eDNA and other EPS components, and bacterial cells disperse because of the disrupted structural integrity, accounting for our observations here (Fig. 3D).
eDNA in mucoid and nonmucoid biofilms impacts mechanical properties
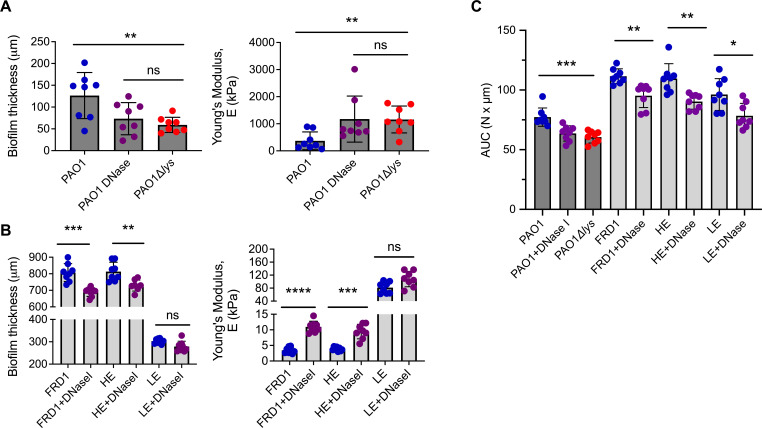
Recent studies suggest that viscoelastic properties of mucoid biofilms augment the persistence of chronic P. aeruginosa infection and challenge lung clearance (49, 50). However, the involvement of bacterial eDNA in influencing the mechanical properties of biofilms formed from clinical mucoid isolates is unknown. To investigate this, uniaxial mechanical indentation (49) was performed on nonmucoid, and clinical mucoid colony biofilms, grown with or without DNaseI treatment. For this analysis, a normal force is applied to the biofilm, and the force required to compress the biofilm is measured. Young’s modulus was used to quantify the stiffness of each biofilm. With this assay, biofilm thickness was also measured. As a control, nonmucoid colony biofilms of PAO1, grown with or without DNaseI, and eDNA mutant (PAO1∆lys) were tested to reveal a benchmark phenotype of eDNA contribution to biofilm mechanics. Results show that eDNA-deficient nonmucoid biofilms (DNaseI treated and eDNA mutant) have an increased Young’s modulus compared to wild-type or nontreated PAO1 biofilms (Fig. 4A). This indicates that eDNA contributes to the stiffness of nonmucoid P. aeruginosa biofilms. Results also show that eDNA-deficient biofilms were thinner than nontreated PAO1 biofilms. In addition, the stiffness and thickness of the DNaseI treated and eDNA mutant biofilms were comparable, also confirming the adequate activity of the DNaseI (Fig. 4A).
Fig 4.
eDNA in mucoid and nonmucoid biofilms impacts mechanical properties. Nonmucoid PAO1 with (purple) or without (blue) DNaseI (200 µg/mL), and eDNA mutant PAO1∆lys (red) biofilms were analyzed by uniaxial mechanical indentation. From these analyses, the biofilm thickness and Young’s modulus were quantified (A). Mucoid biofilms of FRD1, high eDNA (HE; Isolate 9), and low eDNA (LE; Isolate 3) clinical strains with or without DNaseI treatment were also analyzed by uniaxial mechanical indentation where biofilm thickness and Young’s modulus was quantified (B). Squeeze pull-off assay performed on nonmucoid (dark gray bars) PAO1 with or without DNase I, eDNA mutant PAO1∆lys, and mucoid (light gray bars) FRD1, HE, and LE biofilms treated with or without DNase I. Measurements recorded using area under the curve (AUC) (C). Data are depicted as mean ± STDV. N = 8, Analyzed using Student t-test. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
We predict that, like nonmucoid biofilms, if a clinical mucoid strain has abundant eDNA in the biofilm EPS, then removing this eDNA, through enzymatic degradation, should alter the biophysical properties. Therefore, uniaxial mechanical indentation was performed on DNaseI-treated and nontreated clinical HE (Isolate 9), LE (Isolate 3) mucoid biofilms, and the reference FRD1 mucoid biofilm (which is also considered HE) (Fig. 1C). Like nonmucoid biofilms, results show that DNaseI-treated FRD1 and HE biofilms had an increased Young’s modulus and reduced biofilm thickness compared to nontreated controls (Fig. 4B). Although not statistically significant, we also observed a slight increase in Young’s modulus and decreases in the thickness of LE biofilms, compared to nontreated (Fig. 4B). The impact of DNaseI on LE biofilms is reduced likely due to the low abundance of eDNA, suggesting minimal involvement of eDNA in biofilm mechanics compared to HE mucoid biofilms.
We next determined whether eDNA has a role in the cohesion of clinical mucoid biofilms. DNaseI-treated and nontreated nonmucoid (for benchmark comparison) and clinical FRD1, as well as representative HE (Isolate 9) and LE (Isolate 3) mucoid biofilms, were analyzed by a squeeze pull-off assay (49). In this assay, biofilms are compressed, and the force required to raise the probe of the biofilm is measured. The area under the curve (AUC) of the resulting force-displacement curves represents the work required by the probe to raise off the biofilm, indicating the cohesion of the biofilm EPS. Nonmucoid, eDNA-deficient, PAO1 biofilms had low AUC compared to wild type, nontreated PAO1 biofilms, indicating a clear role for eDNA in biofilm EPS cohesion (Fig. 4C). DNaseI-treated FRD1 and HE mucoid biofilms also had significantly lower AUC, than corresponding nontreated biofilms. The effect of DNaseI on LE mucoid biofilms was less pronounced compared to HE biofilms (Fig. 4C). Together, this indicates that eDNA increases the cohesion of biofilm EPS, as eDNA deficiency resulted in reduced cohesiveness of clinical mucoid biofilms.
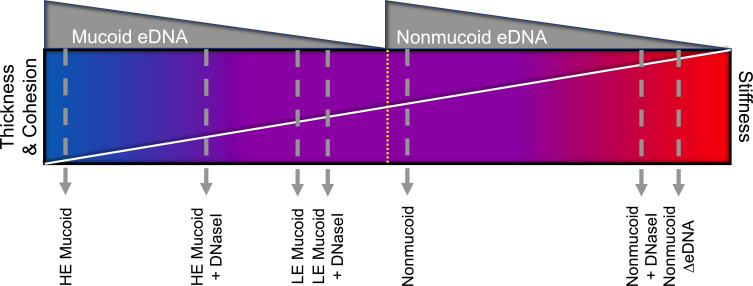
Viscoelastic properties of various EPS components can influence the mechanics of a biofilm and theoretically impede clearance in the CF lung (51 – 56). Moreover, host eDNA increases the viscoelasticity of CF sputum (57, 58). However, it is uncertain if bacterial eDNA plays any role in the ineffective mechanical clearance of the lung in chronic infections. Collectively, we provide evidence that P. aeruginosa mucoid isolates from chronic lung infection are thicker, softer, and more cohesive when eDNA is abundant and intact (Fig. 5). A thick, flexible, and cohesive biofilm would likely further complicate clearance of the biofilms embedded in thick dehydrated mucus. Aerosolized treatment containing saline and human recombinant DNaseI (Pulmozyme) is commonly used as a mucolytic to promote mucus clearance of infected lungs in individuals with CF (59). Here, we show that DNaseI can impact the mechanical properties of clinical mucoid biofilms outside of whole sputum. We presume that in CF lung infections, mucoid strains evolve to produce either low or high eDNA which is influenced by the extent of mechanical stressors in the inflammatory environment. Moreover, previous studies show that most of the eDNA of in vivo P. aeruginosa biofilm infections is derived from the host and can be used as protection for the biofilm against antimicrobials (60). eDNA, exogenous to the bacteria, could impact the mechanical strength of the biofilm and potentially influence the level of eDNA release from the bacteria (61). Bacterial eDNA release may be reduced if the host eDNA is preferentially utilized to enhance biofilm mechanics and protection. Bacterial eDNA may also be increased to compensate for reduced host eDNA in the matrix environment. This could further explain the diversity in eDNA abundance in clinical mucoid biofilms of CF lung infections.
Fig 5.
Summary of the mechanical role of eDNA in clinical mucoid and PAO1 nonmucoid biofilms. This collectively shows the relationship between eDNA abundance (gray) and biofilm thickness, cohesion (upper triangle spectrum) and stiffness (lower triangle spectrum). Stiffness is on the opposite end as it is inversely related to thickness and cohesion. The blue side of the spectrum indicates thick, cohesive, and flexible biofilms, while the red indicates a stiff, thin biofilm with limited cohesion.
EPS produced by P. aeruginosa biofilms strengthens the protection of bacteria continuously faced with selective pressures in the CF lung environment. eDNA is a known EPS produced by nonmucoid P. aeruginosa biofilms. Therefore, the current study evaluated the impact of eDNA in the persistent mucoid biofilms that exacerbate CF lung infections. In this study, we found that biofilm eDNA levels are diverse among CF clinical mucoid strains. We also elucidated the impact of removing biofilm eDNA by enzymatic degradation. DNaseI treatment disrupted clinical mucoid biofilms with abundant eDNA and consequently promoted bacterial cell dispersal. Furthermore, this study revealed a function of eDNA in the mechanical properties of clinical mucoid biofilms, as eDNA in the EPS allows for a softer and more cohesive biofilm. Overall, the function of eDNA is important for the structural and mechanical integrity of clinical mucoid biofilms. Future work will explore if there is a dominant mechanism of eDNA release utilized by mucoid strains isolated from CF lung infections. Future investigation on eDNA levels in relationship to antibiotic and host antimicrobial exposure and infection severity and duration is needed to understand the role of bacterial eDNA in P. aeruginosa fitness during infection.
MATERIALS AND METHODS
P. aeruginosa nonmucoid and mucoid strains
P. aeruginosa strains used are listed in Table 1. PAO1 and PAO1Δlys (eDNA mutant) strains were used for positive and negative eDNA controls, respectively. PDO300 was used as the standard laboratory-derived mucoid strain. FRD1, originally isolated from CF sputum, was used as the reference mucoid strain for experiments. The other 11 clinical mucoid isolates were from CF sputum samples from Nationwide Children’s Hospital (22).
Biofilm formation assays
Luria-Bertani medium, without NaCl (LBNS), was used to culture P. aeruginosa. Pseudomonas isolation agar (PIA) was used throughout experiments for colony biofilms to maintain mucoid phenotype and minimize reversion to nonmucoid.
Static biofilms for microscopy
Mid-log cultures of each strain were grown and normalized to OD600nm0.1 in 10% LBNS. In total, 140 µL of culture was added to each channel of uncoated Ibidi channel slides. Channel slides were placed in a humidified box at 37°C and incubated for 8, 16, 24, and 48 h. Media was replenished after 24 h. Static biofilms were treated with or without DNaseI (200 µg/mL) for 2.5 h.
Colony biofilms for rheology
Mid-log cultures of strains grown in LBNS were diluted to OD600nm0.5 in a final volume of 10 mL. In total, 10 mL of culture of each strain was poured into a sterile petri dish. Nitrocellulose filter membranes (25 mM, 0.45 µm pores from Millipore) were floated on top of the culture in the petri dish for 60 s to seed the filter with bacteria. Filters were placed, seeded side facing up, onto PIA supplemented with or without DNaseI (200 µg/mL), and dried for 2 min prior to incubation. Mucoid biofilms were grown for 24 h at 37°C. Nonmucoid biofilms were grown at 37°C and transferred to new plates every 24 h for 5 days to allow appropriate thickness to be reached for rheology assays.
eDNA isolation and quantification
Sterile inoculation loops were dipped in the mid-log culture of each strain grown in LBNS, normalized to OD600nm0.1. 1 cm streaks were made on PIA and then grown for 24 h at 37°C. Mucoid phenotype maintenance of colony patches was confirmed with brightfield microscopy and reverted strains were discarded. eDNA isolation and quantification was performed using a previously established protocol (28). 24-h biofilms (three biological and three technical replicates) were scraped from PIA and suspended in 1 mL of PBS. In all, 100 µL was drawn for plating CFUs. Suspension was treated with Proteinase K (5 µg/mL) and PNGase F (10 µg/mL) and subjected to vortex pulses to break down biofilm enzymatically and mechanically. Bacterial cells and debris were removed with 13,000 rpm centrifugation, and the supernatant containing eDNA was collected. eDNA was quantified using the fluorometric assay, Qubit.
Alginate isolation and quantification
Mid-log cultures of each strain were grown and normalized to OD600nm0.1 in 10% LBNS. An amount of 100 µL of the diluted culture was pipetted on top of PIA and spread plated evenly with a sterilized metal cell spreader. Plates were dried, inverted, and placed in 37°C for 24-h incubation. Alginate from law-biofilms was isolated and quantified using a previously established protocol (62). Biofilms (three biological and three technical replicates) were resuspended in 1M NaCl and 100 µL was withdrawn for CFUs. Cells were removed via centrifugation (14,000 rpm for 30 min). 2% cetyl pyridinium was added to the supernatant to precipitate alginate in the samples. The precipitated alginate was then pelleted using centrifugation, and then precipitated again in cold isopropanol. After further centrifugation, the purified alginate pellet was resuspended in 1M NaCl overnight at 4°C. Alginate was quantified using a 96-well carbazole assay. Alginate samples and borate-sulfuric acid were heated at 100°C for 15 min. After cooling, 0.1% carbazole in ethanol was added to the wells, and then heated at 100°C for 10 min. Absorbance was detected with a plate reader at 550 nm. Alginate concentration was determined using seaweed alginate to produce a standard curve with r-squared >0.9.
Fluorescent microscopy
Static biofilms were washed with PBS and stained with TOTO-1 (1 µM) and FM4-64 (5 μg mL−1) to label the eDNA and bacterial membrane, respectively. Biofilms were imaged using a Nikon Eclipse Ti2 widefield microscope fitted with 20× objective or 60× oil objective and ORCA-Fusion digital camera. Images of three biological biofilm replicates with two technical images were analyzed. Bacterial cell and eDNA abundance of hydrated biofilms were quantified and analyzed using NIS-elements AR software based on fluorescence threshold and pixel count. Image J was used to process image type conversion, channel split, and scale bar (63).
Rheological assays
TA Instruments Discovery Hybrid Rheometer-2 (HR-2) was used for all rheology experiments.
Uniaxial indentation assay
Indentation was performed using an 8 mM sand-blasted probe with the approach rate set at 1 μm/s. Initial contact between the probe and biofilm was determined at the point where the force began to continuously increase. This point was also determined to be the highest point of the biofilm, therefore the thickness of the biofilm. Eight colony biofilms were analyzed per treatment group of each strain.
Young’s modulus (E) was used to quantify biofilm stiffness, calculated using the forced-displacement relationship equated below (64).
where the slope is from the force-displacement curve (N/m). This slope was measured from the region of the curve where displacement corresponded to 0%–40% through the biofilm (R 2 > 0.9). r is the radius of the probe used (4 mM), and v is the constant defined as Poisson’s ratio of a biofilm (v = 0.5) (65).
Squeeze pull-off assay
This assay was performed using a 25 mM sand-blasted probe. First, the probe was lowered onto the biofilm at an approach rate of 1 μm/s until 0.5N was reached (squeeze). Then, the probe was raised off the biofilm at a rate of 1 μm/s until a height of 850 μm was reached (pull-off). The AUC of the pull-off force-displacement curve was determined in the TRIOS software. One AUC measurement was collected per biofilm. Eight colony biofilms were analyzed per treatment group of each strain.
Statistical analysis
Analysis was performed using GraphPad Prism software. Mean ± STDV represents data bars. Comparisons were made using two-tailed Student’s t-test or one-way ANOVA with P value < 0.05 of normal distribution. Simple linear regression and correlation analysis were used to obtain the line of fit and R2 value from the alginate-eDNA concentration comparison.
ACKNOWLEDGMENTS
We would like to thank Yiwei Liu for valuable feedback on the manuscript and image analysis tutorial. We would also like to thank Pranav Rana and Preston Hill for aiding in microscopy and alginate assays, respectively. We further acknowledge The Ohio State University Comprehensive Cancer Center and the National Institutes of Health under grant number P30 CA016058, which support Campus Microscopy and Imaging Facility (CMIF) Shared Resource.
This study was supported by the National Institute of Allergy and Infectious Diseases (R01AI134895) to DJW and MRP, and the American Heart Association Career Development Award (19CDA34630005) to ESG. This work was supported in part by the Cure CF Columbus Translational Core (C3TC). C3TC is supported by the Division of Pediatric Pulmonary Medicine, the Biopathology Center Core, and the Data Collaboration Team at Nationwide Children’s Hospital (Research Development Program, Grant MCCOY19RO).
Contributor Information
Daniel J. Wozniak, Email: daniel.wozniak@osumc.edu.
George O'Toole, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00238-23.
Supplemental material.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Donlan RM. 2000. Role of biofilms in antimicrobial resistance. ASAIO J 46:S47–52. doi: 10.1097/00002480-200011000-00037 [DOI] [PubMed] [Google Scholar]
- 2. Malhotra S, Limoli DH, English AE, Parsek MR, Wozniak DJ, Bomberger JM, Goldberg J, Vasil M. 2018. Mixed communities of mucoid and nonmucoid Pseudomonas aeruginosa exhibit enhanced resistance to host antimicrobials . mBio 9:mBio doi: 10.1128/mBio.00275-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mah TF, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/s0966-842x(00)01913-2 [DOI] [PubMed] [Google Scholar]
- 4. Ciofu O, Tolker-Nielsen T. 2019. Tolerance and resistance of Pseudomonas Aeruginosa Biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol 10:913. doi: 10.3389/fmicb.2019.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjarnsholt T. 2013. The role of bacterial biofilms in chronic infections. APMIS 121:1–58. doi: 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- 6. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 7. Waters V, Ratjen F. 2006. Multidrug-resistant organisms in cystic fibrosis: management and infection-control issues. Expert Rev Anti Infect Ther 4:807–819. doi: 10.1586/14787210.4.5.807 [DOI] [PubMed] [Google Scholar]
- 8. Obritsch MD, Fish DN, MacLaren R, Jung R. 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 25:1353–1364. doi: 10.1592/phco.2005.25.10.1353 [DOI] [PubMed] [Google Scholar]
- 9. Maurice NM, Bedi B, Sadikot RT. 2018. Pseudomonas Aeruginosa biofilms: host response andclinical implications in lung infections. Am J Respir Cell Mol Biol 58:428–439. doi: 10.1165/rcmb.2017-0321TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther 21:595–599. doi: 10.1016/j.pupt.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King J, Murphy R, Davies JC. 2022. Pseudomonas aeruginosa in the cystic fibrosis lung. Adv Exp Med Biol 1386:347–369. doi: 10.1007/978-3-031-08491-1_13 [DOI] [PubMed] [Google Scholar]
- 12. Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirisits MJ, Prost L, Starkey M, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa Biofilms. Appl Environ Microbiol 71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Govan JRW. 1990. Characteristics of mucoid Pseudomonas aeruginosa in vitro and in vivo. Pseudomonas infection and alginates:50–75. doi: 10.1007/978-94-009-1836-8 [DOI] [Google Scholar]
- 15. Malhotra S, Hayes D, Wozniak DJ. 2019. Mucoid Pseudomonas aeruginosa and regional inflammation in the cystic fibrosis lung. J Cyst Fibros 18:796–803. doi: 10.1016/j.jcf.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farrell PM, Collins J, Broderick LS, Rock MJ, Li Z, Kosorok MR, Laxova A, Gershan WM, Brody AS. 2009. Association between mucoid Pseudomonas infection and bronchiectasis in children with cystic fibrosis. Radiology 252:534–543. doi: 10.1148/radiol.2522081882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JIA, Jensen P, Johnsen AH, Givskov M, Ohman DE, Søren M, Høiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology (Reading) 145 (Pt 6):1349–1357. doi: 10.1099/13500872-145-6-1349 [DOI] [PubMed] [Google Scholar]
- 18. Govan JR, Martin DW, Deretic VP. 1992. Mucoid Pseudomonas aeruginosa and cystic fibrosis: the role of mutations in muc loci. FEMS Microbiol Lett 100:323–329. doi: 10.1111/j.1574-6968.1992.tb14059.x [DOI] [PubMed] [Google Scholar]
- 19. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol 300:557–562. doi: 10.1016/j.ijmm.2010.08.008 [DOI] [PubMed] [Google Scholar]
- 20. Jacobs HM, O’Neal L, Lopatto E, Wozniak DJ, Bjarnsholt T, Parsek MR. 2022. Mucoid Pseudomonas aeruginosa can produce calcium-gelled biofilms independent of the matrix components Psl and CdrA. J Bacteriol 204:e0056821. doi: 10.1128/jb.00568-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sarkisova S, Patrauchan MA, Berglund D, Nivens DE, Franklin MJ. 2005. Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol 187:4327–4337. doi: 10.1128/JB.187.13.4327-4337.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones CJ, Wozniak DJ. 2017. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio 8:e00864-17. doi: 10.1128/mBio.00864-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. doi: 10.5301/ijao.5000051 [DOI] [PubMed] [Google Scholar]
- 24. Mulcahy H, Charron-Mazenod L, Lewenza S, Gilmore MS. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639 [DOI] [PubMed] [Google Scholar]
- 26. Das T, Sehar S, Manefield M. 2013. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep 5:778–786. doi: 10.1111/1758-2229.12085 [DOI] [PubMed] [Google Scholar]
- 27. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 28. Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohman DE, Chakrabarty AM. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun 33:142–148. doi: 10.1128/iai.33.1.142-148.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- 31. Wang D, Hildebrand F, Ye L, Wei Q, Ma LZ. 2015. Genome sequence of mucoid Pseudomonas aeruginosa strain FRD1. Genome Announc 3:e00376-15. doi: 10.1128/genomeA.00376-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x [DOI] [PubMed] [Google Scholar]
- 34. Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol 176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi: 10.1016/j.chom.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. York A. 2022. From B to Z in the matrix. Nat Rev Microbiol 20:2. doi: 10.1038/s41579-021-00662-0 [DOI] [PubMed] [Google Scholar]
- 37. Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. 2013. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A 110:11541–11546. doi: 10.1073/pnas.1218898110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarkar S. 2020. Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA. Appl Microbiol Biotechnol 104:6549–6564. doi: 10.1007/s00253-020-10687-9 [DOI] [PubMed] [Google Scholar]
- 39. DeVries CA, Ohman DE. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tahrioui A, Duchesne R, Bouffartigues E, Rodrigues S, Maillot O, Tortuel D, Hardouin J, Taupin L, Groleau M-C, Dufour A, Déziel E, Brenner-Weiss G, Feuilloley M, Orange N, Lesouhaitier O, Cornelis P, Chevalier S. 2019. Extracellular DNA release, Quorum sensing, and Prrf1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin-enhanced biofilm formation. NPJ Biofilms Microbiomes 5:15. doi: 10.1038/s41522-019-0088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boyd A, Chakrabarty AM. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol 15:162–168. doi: 10.1007/BF01569821 [DOI] [PubMed] [Google Scholar]
- 42. Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gordon CA, Hodges NA, Marriott C. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J Antimicrob Chemother 22:667–674. doi: 10.1093/jac/22.5.667 [DOI] [PubMed] [Google Scholar]
- 44. Lee KY, Mooney DJ. 2012. Alginate: properties and biomedical applications. Prog Polym Sci 37:106–126. doi: 10.1016/j.progpolymsci.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nichols WW, Dorrington SM, Slack MP, Walmsley HL. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother 32:518–523. doi: 10.1128/AAC.32.4.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 100:7907–7912. doi: 10.1073/pnas.1231792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S, Liu X, Liu H, Zhang L, Guo Y, Yu S, Wozniak DJ, Ma LZ. 2015. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep 7:330–340. doi: 10.1111/1758-2229.12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gloag ES, German GK, Stoodley P, Wozniak DJ. 2018. Viscoelastic properties of Pseudomonas aeruginosa variant biofilms. Sci Rep 8:9691. doi: 10.1038/s41598-018-28009-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alcaraz-Serrano V, Fernández-Barat L, Scioscia G, Llorens-Llacuna J, Gimeno-Santos E, Herrero-Cortina B, Vàzquez N, Puig de la Bellacasa J, Gabarrús A, Amaro-Rodriguez R, Menéndez R, Torres A. 2019. Mucoid Pseudomonas aeruginosa alters sputum viscoelasticity in patients with non-cystic fibrosis bronchiectasis. Respir Med 154:40–46. doi: 10.1016/j.rmed.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 51. Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Doring G. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis 192:410–419. doi: 10.1086/431516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marriott C. 1990. Mucus and mucociliary clearance in the respiratory tract. Adv Drug Deliv Rev 5:19–35. doi: 10.1016/0169-409X(90)90005-D [DOI] [Google Scholar]
- 53. King M. 1987. The role of mucus viscoelasticity in cough clearance. Biorheology 24:589–597. doi: 10.3233/bir-1987-24611 [DOI] [PubMed] [Google Scholar]
- 54. Pestrak MJ, Chaney SB, Eggleston HC, Dellos-Nolan S, Dixit S, Mathew-Steiner SS, Roy S, Parsek MR, Sen CK, Wozniak DJ. 2018. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog 14:e1006842. doi: 10.1371/journal.ppat.1006842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCaslin CA, Petrusca DN, Poirier C, Serban KA, Anderson GG, Petrache I. 2015. Impact of alginate-producing Pseudomonas aeruginosa on alveolar macrophage apoptotic cell clearance. J Cyst Fibros 14:70–77. doi: 10.1016/j.jcf.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Laube BL, Sharpless G, Benson J, Carson KA, Mogayzel PJ. 2014. Mucus removal is impaired in children with cystic fibrosis who have been infected by Pseudomonas aeruginosa. J Pediatr 164:839–845. doi: 10.1016/j.jpeds.2013.11.031 [DOI] [PubMed] [Google Scholar]
- 57. Dasgupta B, King M. 1996. Reduction in viscoelasticity in cystic fibrosis sputum in vitro using combined treatment with nacystelyn and rhDNase. Pediatr Pulmonol 22:161–166. doi: [DOI] [PubMed] [Google Scholar]
- 58. King M, Dasgupta B, Tomkiewicz RP, Brown NE. 1997. Rheology of cystic fibrosis sputum after in vitro treatment with hypertonic saline alone and in combination with recombinant human deoxyribonuclease I. Am J Respir Crit Care Med 156:173–177. doi: 10.1164/ajrccm.156.1.9512074 [DOI] [PubMed] [Google Scholar]
- 59. Yang C, Montgomery M. 2018. Dornase alfa for cystic fibrosis. Cochrane Database Syst Rev 9:CD001127. doi: 10.1002/14651858.CD001127.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Alhede M, Alhede M, Qvortrup K, Kragh KN, Jensen PØ, Stewart PS, Bjarnsholt T. 2020. The origin of extracellular DNA in bacterial biofilm infections in vivo. Pathog Dis 78:ftaa018. doi: 10.1093/femspd/ftaa018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rouillard KR, Kissner WJ, Markovetz MR, Hill DB. 2022. Effects of mucin and DNA concentrations in airway mucus on Pseudomonas aeruginosa biofilm recalcitrance. mSphere 7:e0029122. doi: 10.1128/msphere.00291-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cesaretti M. 2003. A 96-well assay for uronic acid carbazole reaction. Carbohydrate Polymers 54:59–61. doi: 10.1016/S0144-8617(03)00144-9 [DOI] [Google Scholar]
- 63. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to imageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Timoshenko S, Goodier J.. Theory of Elasticity, Third ed. McGraw Hill Higher Education. [Google Scholar]
- 65. Rmaile A, Carugo D, Capretto L, Zhang X, Wharton JA, Thurner PJ, Aspiras M, Ward M, Stoodley P. 2013. Microbial tribology and disruption of dental plaque bacterial biofilms. Wear 306:276–284. doi: 10.1016/j.wear.2013.02.010 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material.