ABSTRACT
Streptococcus pneumoniae is a commensal bacterium and invasive pathogen that causes millions of deaths worldwide. The pneumococcal vaccine offers limited protection, and the rise of antimicrobial resistance will make treatment increasingly challenging, emphasizing the need for new antipneumococcal strategies. One possibility is to target antioxidant defenses to render S. pneumoniae more susceptible to oxidants produced by the immune system. Human peroxidase enzymes will convert bacterial-derived hydrogen peroxide to hypothiocyanous acid (HOSCN) at sites of colonization and infection. Here, we used saturation transposon mutagenesis and deep sequencing to identify genes that enable S. pneumoniae to tolerate HOSCN. We identified 37 genes associated with S. pneumoniae HOSCN tolerance, including genes involved in metabolism, membrane transport, DNA repair, and oxidant detoxification. Single-gene deletion mutants of the identified antioxidant defense genes sodA, spxB, trxA, and ahpD were generated and their ability to survive HOSCN was assessed. With the exception of ΔahpD, all deletion mutants showed significantly greater sensitivity to HOSCN, validating the result of the genome-wide screen. The activity of hypothiocyanous acid reductase or glutathione reductase, known to be important for S. pneumoniae tolerance of HOSCN, was increased in three of the mutants, highlighting the compensatory potential of antioxidant systems. Double deletion of the gene encoding glutathione reductase and sodA sensitized the bacteria significantly more than single deletion. The HOSCN defense systems identified in this study may be viable targets for novel therapeutics against this deadly pathogen.
IMPORTANCE
Streptococcus pneumoniae is a human pathogen that causes pneumonia, bacteremia, and meningitis. Vaccination provides protection only against a quarter of the known S. pneumoniae serotypes, and the bacterium is rapidly becoming resistant to antibiotics. As such, new treatments are required. One strategy is to sensitize the bacteria to killing by the immune system. In this study, we performed a genome-wide screen to identify genes that help this bacterium resist oxidative stress exerted by the host at sites of colonization and infection. By identifying a number of critical pneumococcal defense mechanisms, our work provides novel targets for antimicrobial therapy.
KEYWORDS: Tn-seq, oxidative stress, lactoperoxidase, pneumonia, myeloperoxidase
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a bacterium that asymptomatically colonizes the mucosal surfaces of the upper respiratory tract as part of the commensal flora (1). While nasopharyngeal S. pneumoniae colonization is normally asymptomatic, invasion to other areas of the body including the lungs can lead to serious pathological infections, most notably pneumonia. During S. pneumoniae infection in the lungs, the human peroxidase enzymes, lactoperoxidase (LPO) and myeloperoxidase, will convert thiocyanate (SCN−), present at concentrations of up to 650 µM (2, 3), and pneumococcus-derived hydrogen peroxide (H2O2) (4), to hypothiocyanous acid (HOSCN) (5, 6). As such, the tolerance of S. pneumoniae to HOSCN may provide this bacterium with a survival advantage over co-localized bacteria. Indeed, we have recently reported the relatively high tolerance of S. pneumoniae to HOSCN-mediated killing (7). We hypothesize that targeting HOSCN defenses in these bacteria may have therapeutic value.
HOSCN targets redox-sensitive proteins by oxidizing critical cysteine residues (8). An evaluation of antioxidant mechanisms in S. pneumoniae known to protect other organisms from HOSCN identified the low molecular weight thiol glutathione (9) and a previously uncharacterized HOSCN reductase enzyme (10) as critical components of the pneumococcal HOSCN defense. In the present study, we used transposon mutagenesis in combination with high-throughput transposon sequencing (Tn-seq) as an unbiased screen to identify other genes that help S. pneumoniae withstand HOSCN.
Tn-seq involves the insertion of a single mini-transposon randomly within the genome of each bacterium to create a library of different single-site mutants, which collectively represent transposons inserted into all genes (or intergenic regions) that are non-essential under optimal growth conditions (11, 12). Any mutation in essential genes will make the bacterium non-viable, so it will not be able to be investigated. Tn-seq in S. pneumoniae utilizes the natural genetic transformation of these bacteria to generate a saturated transposon insertion library (11). Tn-seq has previously investigated genes involved in S. pneumoniae survival in human saliva (13), survival upon desiccation (14), mammalian host transmission (15), and antibiotic stress (16). Additionally, small non-coding RNAs (sRNAs) important in gene regulation have been investigated with Tn-seq (17), emphasizing the wide applicability of this method to identify gene responses in S. pneumoniae.
Here, we report the use of Tn-seq to investigate the genes involved in S. pneumoniae oxidant tolerance. This investigation was performed using two transposon mutant libraries in the D39 strain of S. pneumoniae and led to the identification of 37 genes that are involved in S. pneumoniae HOSCN tolerance. These genes may be useful drug targets for sensitizing this bacterium to oxidative stress at sites of colonization and infection.
RESULTS AND DISCUSSION
Generation and screening of S. pneumoniae transposon libraries
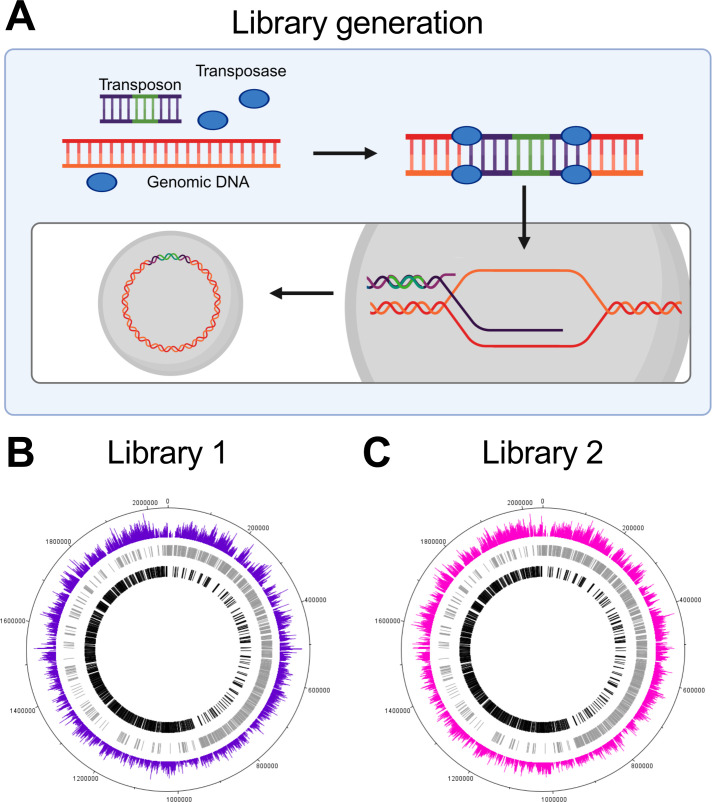
To investigate genes involved in S. pneumoniae HOSCN tolerance, a transposon library (Library 1) was generated and exposed to HOSCN alongside a second previously published library (Library 2) (14). The libraries, which served as biological replicates, were generated by combining S. pneumoniae genomic DNA with transposon DNA and a transposase enzyme, which led to the incorporation of transposon DNA into bacterial genomic DNA (Fig. 1A). This DNA was integrated into the DNA of competent S. pneumoniae cells via homologous recombination (Fig. 1A). Technical replicates of each library were then simultaneously exposed to 800 µM of HOSCN for 60 min; conditions that do not affect the viability of wild-type (WT) bacteria but result in a significant decrease in the viability of a glutathione-deficient mutant strain (9). After HOSCN treatment, bacteria were grown for three to four generations before genomic DNA was extracted, processed, and sequenced. The transposon junctions from the control and treatment DNA libraries were deep sequenced on the Illumina platform and mapped to the S. pneumoniae D39 genome to locate transposon insertion sites. A visual representation of transposon insertion coverage in Library 1 and 2 across the S. pneumoniae D39 genome indicates that uniform genome coverage was achieved in both libraries (Fig. 1B and C). The average number of unique insertions was ~42,000 and ~73,000 for Library 1 and 2, respectively. As the S. pneumoniae D39 genome consists of 2,046 kbp, this corresponds to a unique insertion (transposon insertion) on average every 49 bp in Library 1 and 28 bp in Library 2.
Fig 1.
Transposon library generation and genome coverage. (A) S. pneumoniae genomic DNA (orange) was combined with transposon DNA (purple, with an antibiotic resistance gene in green) and the transposase enzyme (blue oval). Transposon DNA is inserted at TA nucleotide sites within the genomic DNA by transposase enzyme. Double crossover homologous recombination integrates the mutated DNA into the wild-type bacterial genome. This is done in parallel to generate a library of mutants with every non-essential gene mutated across the genome. This diagram was created with Biorender.com. (B and C) There was uniform transposon coverage in the genome of (B) Library 1 and (C) Library 2, with a single transposon insertion represented every 49 and 28 bp for Libraries 1 and 2, respectively. Genes on the forward and reverse strands are shown in gray and black, respectively. Colored bars designate the location and frequency of transposon insertions.
Identification of genes involved in S. pneumoniae HOSCN tolerance
To identify genes important for HOSCN tolerance, the transposon insertion sites in HOSCN-treated samples were compared with untreated controls using differential expression analysis software BioTraDIS and edgeR (18, 19). To designate genes as significantly different between control and treated conditions, cutoffs of an adjusted P-value of <0.05 and a log2[fold change (FC)] of ≤−1 or ≥1 were imposed. There were 20 and 30 genes that were designated as significant in Library 1 and Library 2, respectively. Of these, 13 genes were significant in both libraries, while the other 24 were only significant in one of the two libraries (Table S1). Where possible, gene names that were not annotated in GenBank have been added from the literature.
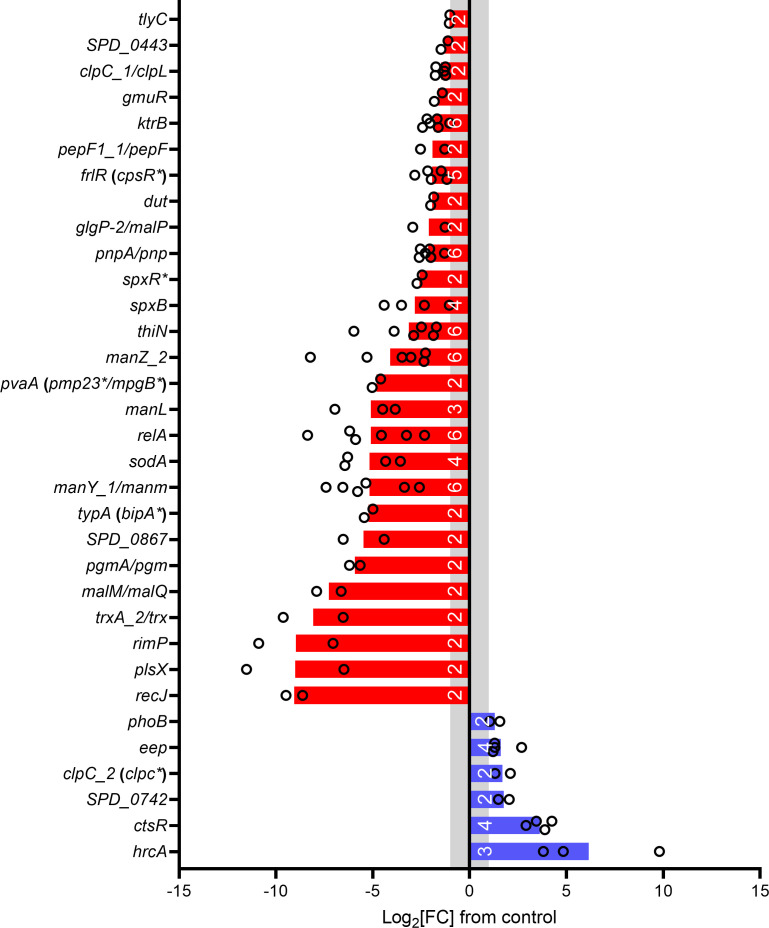
The log2[FC] of significant genes in either or both libraries was averaged to identify whether genes were involved in the sensitivity or tolerance of S. pneumoniae to HOSCN. Twenty-seven genes were identified that help S. pneumoniae tolerate HOSCN, i.e., were underrepresented in the HOSCN-treated samples compared to controls; and six genes that make S. pneumoniae more sensitive to HOSCN, i.e., overrepresented in the HOSCN-treated samples (Fig. 2; Table 1). Four additional genes were significantly different between control and treated samples, but the direction of change was variable, so more investigation is needed to determine their contribution to the effects of HOSCN (Table 1). The gene data output from each replicate in each library can be examined in “Library 1 data.xlsx” and “Library 2 data.xlsx” worksheets in the supplementary information.
Fig 2.
Genes that are involved in S. pneumoniae HOSCN tolerance. Genes that make S. pneumoniae tolerant (red) or sensitive (blue) to HOSCN are plotted. Gene names with a slash had different gene names depending on the reference genome annotation (GenBank: LS483374.1/CP000410.2). Genes without a name have the locus tag from GenBank from the annotation CP000410.2. Gene names identified from the literature are designated by *. White numbers on the bars designate the total number of replicates the gene was significant in (out of a total of six). The log2[FC] values are the average of the replicates. A gray band denotes the log2[FC] significance cutoff of ≤ −1 or ≥1.
TABLE 1.
Genes involved in S. pneumoniae HOSCN stress and their putative function c
| Locus tag | Gene | Putative gene function | No. of replicates significant in | Mean | SD |
|---|---|---|---|---|---|
| Log2[FC] a | |||||
| Genes that allow S. pneumoniae to tolerate HOSCN | |||||
| SPD_0532 | recJ | Single-stranded DNA-specific exonuclease | 2 | −9.05 | 0.61 |
| SPD_0043 | plsX | Fatty acid/phospholipid synthesis protein | 2 | −9 | 3.55 |
| SPD_0478 | rimP | SP14.3 protein | 2 | −8.97 | 2.71 |
| SPD_1567 | trx | Thioredoxin | 2 | −8.08 | 2.19 |
| SPD_1933 | malQ | 4-Alpha-glucanotransferase (amylomaltase) | 2 | −7.27 | 0.89 |
| SPD_1326 | pgmA | Phosphoglucomutase | 2 | −5.92 | 0.39 |
| SPD_0867 | SPD_0867 | O-methyltransferase | 2 | −5.47 | 1.49 |
| SPD_0593 | bipA b | GTP-binding protein TypA/elongation factor Tu family protein | 2 | −5.22 | 0.32 |
| SPD_0263 | manM | PTS d system mannose-specific transporter subunit IIC | 6 | −5.17 | 1.85 |
| SPD_0667 | sodA | Superoxide dismutase | 4 | −5.16 | 1.43 |
| SPD_1458 | relA | GTP pyrophosphokinase, stringent response protein | 6 | −5.1 | 2.19 |
| SPD_0264 | manL | PTS system mannose-specific transporter subunit IIAB | 3 | −5.1 | 1.65 |
| SPD_0912 | mpgB b | Pneumococcal vaccine antigen A | 2 | −4.81 | 0.31 |
| SPD_0262 | manZ_2 | PTS system mannose/fructose/sorbose family transporter subunit IID | 6 | −4.11 | 2.3 |
| SPD_1779 | thiN | Thiamine pyrophosphokinase | 6 | −3.14 | 1.6 |
| SPD_0636 | spxB | Pyruvate oxidase | 4 | −2.83 | 1.46 |
| SPD_0969 | spxR b | CBS domain-containing transcription factor | 2 | −2.58 | 0.19 |
| SPD_0512 | pnp | Polyribonucleotide nucleotidyltransferase | 6 | −2.13 | 0.48 |
| SPD_1932 | malP | Maltodextrin phosphorylase | 2 | −2.10 | 1.18 |
| SPD_0027 | dut | Deoxyuridine 5'-triphosphate nucleotidohydrolase | 2 | −1.93 | 0.11 |
| SPD_0064 | cpsR b | GntR family transcriptional regulator | 5 | −1.92 | 0.65 |
| SPD_0866 | pepF | Oligoendopeptidase F | 2 | −1.91 | 0.88 |
| SPD_0076 | ktrB | Trk transporter membrane-spanning protein - K+ transport | 6 | −1.83 | 0.5 |
| SPD_1829 | gmuR | GntR family transcriptional regulator | 2 | −1.62 | 0.29 |
| SPD_0308 | clpL | Group II intron maturase or ATP-dependent Clp protease, ATP-binding subunit | 2 | −1.46 | 0.27 |
| SPD_0443 | SPD_0443 | Sodium-dependent phosphate transporter/Na/Pi-cotransporter II-related protein | 2 | −1.3 | 0.25 |
| SPD_1761 | tlyC | CBS domain membrane protein | 2 | −1.03 | 0.02 |
| Genes that make S. pneumoniae sensitive to HOSCN | |||||
| SPD_0458 | hrcA | Heat-inducible transcription repressor | 3 | 6.15 | 3.20 |
| SPD_2023 | ctsR | CtsR family transcriptional regulator | 4 | 3.66 | 0.50 |
| SPD_0742 | SPD_0742 | Sugar ABC transporter, membrane-spanning permease | 2 | 1.77 | 0.40 |
| SPD_2022 | clpC b | ATP-dependent Clp protease, ATP-binding subunit | 2 | 1.71 | 0.56 |
| SPD_0245 | eep | Metalloprotease or zinc metalloprotease | 4 | 1.62 | 0.71 |
| SPD_2020 | phoB | DNA-binding response regulator | 2 | 1.3 | 0.37 |
| Genes involved in S. pneumoniae HOSCN response that were variable in the screen | |||||
| Log2[FC] values | |||||
| SPD_0150 | tcyA (gshT b ) | ABC transporter substrate-binding lipoprotein | 4 | −1.16, −1.26, 2.41, 2.47 | |
| SPD_0373 | ahpD b | Alkylhydroperoxidase D | 2 | −3.47, 1.92 | |
| SPD_0256 | yafQ | RelE/StbE family addiction module toxin/conserved hypothetical protein TIGR00053 | 2 | −5.31, 6.10 | |
| SPD_1290 | yxeN | Aspartate/glutamate ABC transporter permease | 2 | −1.31, 2.63 | |
Mean log2[FC] was calculated from the average fold-change across both libraries.
Gene name identified from the literature not by GenBank.
Locus tags are from reference genome annotation for S. pneumoniae D39 (GenBank: CP000410.2). Gene names are from GenBank: CP000410.2 or LS483374.1.
PTS, phosphotransferase system.
Genes identified here to confer tolerance or susceptibility to HOSCN were classified into the main categories of metabolism, genetic information processing, environmental information processing and cellular processes, and BRITE hierarchies using the KEGG Orthology database (KEGG Orthology.xlsx, supplementary information). Some genes could not be assigned by the KEGG database, while others were identified as being a part of multiple pathways. The number of pathways identified indicates that diverse molecular functions are affected in S. pneumoniae by HOSCN exposure. Indeed, multiple bacterial processes are known to be altered by HOSCN, including substrate transport, glycolysis, the pentose phosphate pathway, respiration, and pH adaption [reviewed in reference (20)]. Similarly, recent gene expression studies examining the response of Pseudomonas aeruginosa to HOSCN have shown that HOSCN affects cellular systems involved in metabolism, macromolecule repair and detoxification, export of toxic compounds, and virulence (21, 22). In light of these complex cellular impacts of HOSCN, it is to be expected that the bacterial HOSCN defense is multifactorial, as illustrated by the wide-ranging functions of HOSCN tolerance genes identified here.
A number of genes found to play a role in HOSCN tolerance in the present study were also identified in similar screening experiments that looked at S. pneumoniae desiccation tolerance (14), survival in human saliva (13), transmission in a ferret model (15) and survival in vitro and in vivo (mice) models (23) (Table S2). The spxB gene was identified to be important in all studies (Table S2). The largest overlap between the data from this screen and other Tn-seq studies was with genes identified from exposure to human saliva (13), with eight genes in common. Saliva is rich in HOSCN due to the high concentrations of the precursor SCN− (24). Genes involved in HOSCN tolerance will aid survival in this environment and are potential targets for limiting S. pneumoniae colonization.
Validation of the transposon mutant screen
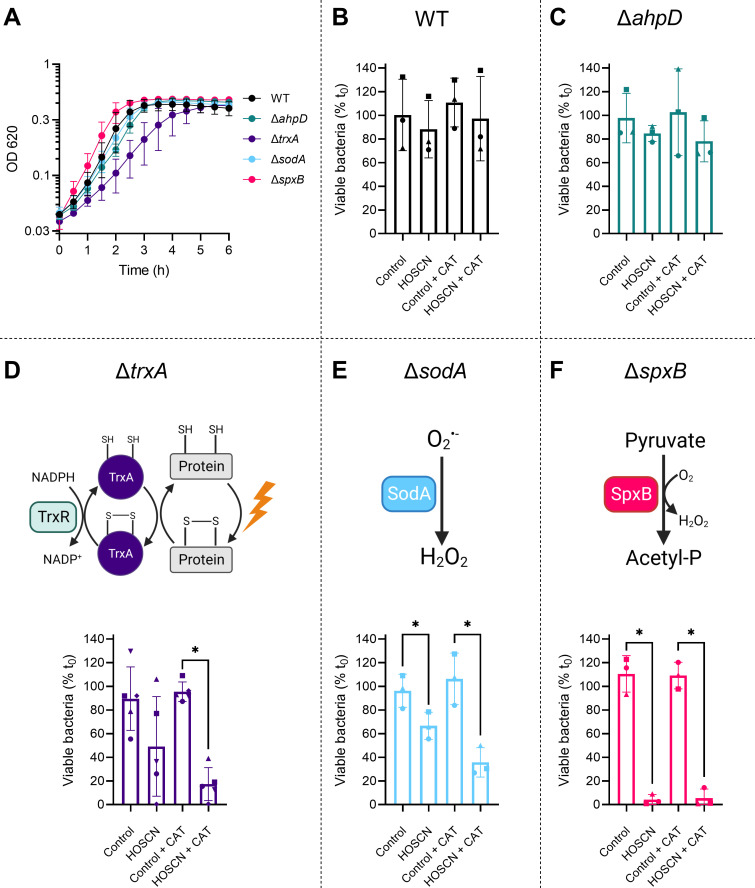
To validate the screen, we focused on genes known to have a function in antioxidant defense and generated single-gene deletion mutants by replacing the gene of interest with the antibiotic resistance cassette for spectinomycin. These genes were trxA (thioredoxin), sodA (superoxide dismutase), spxB (pyruvate oxidase), and ahpD (putative alkylhydroperoxidase D). Once generated, the growth characteristics and HOSCN sensitivity of the mutants were compared to WT (Fig. 3).
Fig 3.
Validation of Tn-seq using single-gene deletion mutants. (A) S. pneumoniae WT and mutant strains were grown in a 96-well plate in brain heart infusion media for 6 hours at 37°C in the presence of 5% CO2. Absorbance measurements (OD620) were taken every 30 min, following which the media-alone sample values were subtracted. Data are presented as the mean ± SD from at least three independent experiments, the y-axis is log10 scale. (B–F) S. pneumoniae strains (B) WT, (C) ΔahpD, (D) ΔtrxA, (E) ΔsodA, and (F) ΔspxB (OD620 0.02) were incubated in the presence or absence of 800 µM HOSCN ± catalase (CAT, 20 µg/mL) for 90 min in Hank’s balanced salt solution buffer, pH 6.8. Viable bacteria were determined by counting colony forming units and expressed relative to those before incubation (t 0). Data are presented as the mean ± SD from at least three independent experiments, with symbols designating matching replicate data. A significant difference to the respective control was determined by paired t-tests and is indicated by * for P < 0.05. (D–F) Schematic representation of the function of proteins TrxA, SodA, and SpxB, in reducing oxidized thiols in proteins, detoxifying superoxide, and metabolizing pyruvate, respectively. The schematic representations were created with Biorender.com.
While one mutant had a longer lag phase when grown in liquid culture, all mutants reached a similar absorbance to the WT strain at stationary phase, suggesting that these genes are not essential for growth in rich media (Fig. 3A). To quantify the impact of HOSCN on bacterial viability, mutants were exposed to 800 µM of HOSCN for 90 min and plated to determine colony-forming units (Fig. 3B through F). S. pneumoniae generate H2O2 during metabolism that can also oxidize thiol proteins, but this will not accumulate in vivo due to the presence of host peroxidases. Catalase was added to mimic the in vivo environment and limit any combined effects of HOSCN and H2O2. The viability of WT bacteria was not affected by the presence of HOSCN or catalase (Fig. 3B), consistent with our previous findings (7, 9).
The gene ahpD had an inconsistent HOSCN response in the screen (log2[FC] of 1.92 and −3.47 from Library 2 replicates) so was mutated to determine whether it has a protective or detrimental role in HOSCN stress. AhpD enzymes convert peroxides to alcohol and water, and while S. pneumoniae AhpD has unique structural characteristics compared to AhpD proteins from other species, this enzyme displays low reactivity with H2O2 (25). Bacterial survival has previously shown to be increased in a S. pneumoniae ΔahpD mutant when treated with H2O2, which was thought to be due to the upregulation of compensatory antioxidant systems (26). In contrast, we did not detect any significant change in bacterial viability of our ΔahpD mutant in the presence of HOSCN, indicating that AhpD is not involved in HOSCN tolerance (Fig. 3C).
The gene trxA had a log2[FC] of −8.08, the largest change of any of the antioxidant genes and the fourth largest of all genes. TrxA is an oxidoreductase in the thioredoxin/thioredoxin reductase (Trx/TrxR) system. Trx uses an active site dithiol group to reduce disulfides in proteins, with the resultant oxidized Trx being reduced by TrxR using NADPH as the electron donor (Fig. 3D). When trxA was deleted, bacterial growth was slowed, and viability in the presence of HOSCN and catalase was significantly decreased (Fig. 3A and D). HOSCN is a thiol-specific oxidant (27), and as such it is not surprising that a system that reduces thiol proteins is important for tolerance. The Trx/TrxR system has previously been identified as part of the survival strategy of S. pneumoniae in vivo, with decreased bacterial load and mice mortality observed when mice were administered a potent TrxR inhibitor (28). Together, these results suggest that TrxA may be important for S. pneumoniae to tolerate HOSCN stress at sites of infection.
The gene sodA had a log2[FC] of −5.16 in the screen. SodA is a manganese-dependent enzyme that dismutates superoxide to hydrogen peroxide and oxygen (Fig. 3E). As such, it plays an important role in protecting organisms from superoxide generated during aerobic metabolism (29). In agreement with the Tn-seq data, the deletion of sodA sensitized S. pneumoniae to HOSCN (Fig. 3E). Previous studies have reported that S. pneumoniae ΔsodA mutants are hypersensitive to paraquat, which stimulates intracellular superoxide generation (30, 31). Furthermore, an S. pneumoniae ΔsodA mutant was less virulent in a mouse model of S. pneumoniae infection (31). Our present study highlights that this enzyme is also important in protecting from HOSCN exposure. HOSCN may increase the generation of superoxide inside bacteria, or damage caused by superoxide during normal bacterial metabolism may be exacerbated by the addition of HOSCN. Further work is required to investigate the relevance of superoxide to HOSCN toxicity.
The gene-encoding pyruvate oxidase, spxB, had a log2[FC] of −2.83 in the screen. SpxB plays a central role in S. pneumoniae metabolism, where it generates H2O2 and acetyl-phosphate from pyruvate and oxygen (32) (Fig. 3F). Acetyl-phosphate is used for several cellular processes through its conversion to acetate and acetyl-CoA (32). SpxB is responsible for the majority of the H2O2 produced by S. pneumoniae (32 – 35). Loss of SpxB also leads to a decrease in ATP by ~85%, which is thought to account for an increased sensitivity of this mutant to H2O2 (34). Here, we show that SpxB is also essential for S. pneumoniae HOSCN tolerance, with a significant decrease in viability in the presence of HOSCN upon deletion of spxB (Fig. 3F). The importance of SpxB in S. pneumoniae virulence has been shown in multiple studies using rabbit, ferret, mouse, and rat models, with SpxB involved in transmission, colonization, pneumonia, and bacteremia (15, 32, 33, 36, 37). In addition, SpxB alters capsule production in a strain-dependent manner, with a greater or similar level of capsule production in strain D39 but significantly less in TIGR4 (35, 38). Regulation of spxB is proposed to be by the protein SpxR sensing the metabolic and energy state of S. pneumoniae (33). The gene spxR was also identified as important for HOSCN tolerance in this screen (log2[FC] of −2.58), further emphasizing the importance of SpxB in S. pneumoniae HOSCN tolerance.
Hypothiocyanous acid reductase and glutathione reductase are increased in S. pneumoniae mutants
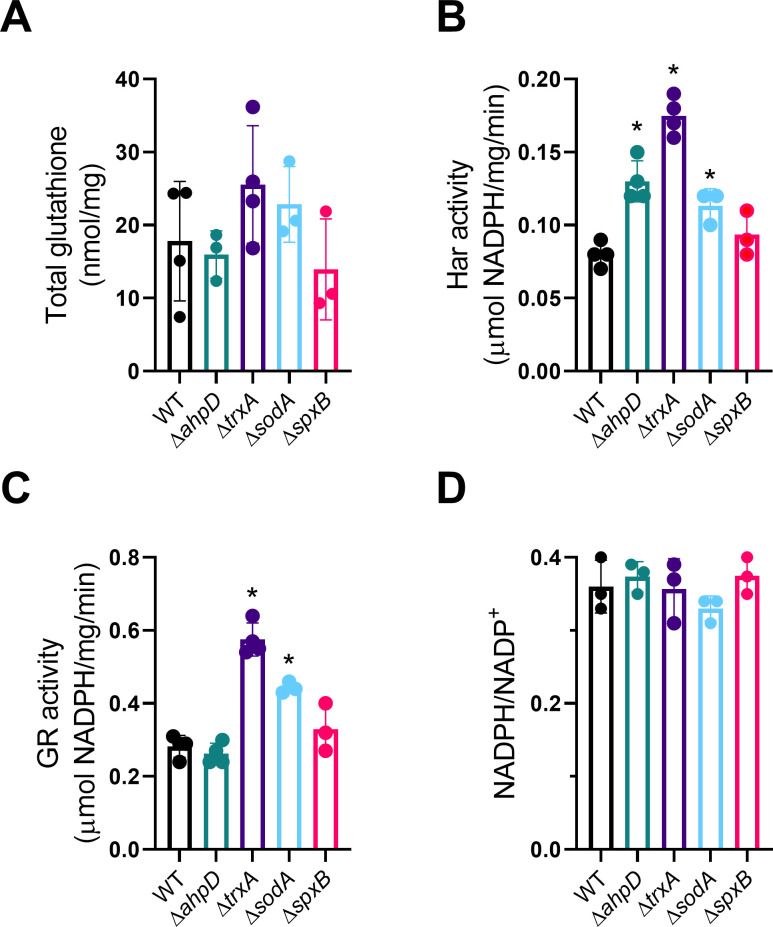
Mutation of antioxidant genes may result in the compensatory upregulation of other protective pathways, driven by increased levels of oxidative stress. For example, H2O2 activates the expression of thiol peroxidase tpxD in S. pneumoniae under H2O2 stress via its activator CodY (39). In our study, the combination of HOSCN and catalase treatment decreased bacterial viability more than HOSCN alone in all mutant strains, except for ΔspxB where HOSCN was already maximally effective, consistent with bacterial H2O2 increasing bacterial defenses. We, therefore, investigated whether known HOSCN defenses are upregulated in the mutant strains. Glutathione import is an integral part of S. pneumoniae HOSCN tolerance (9, 10). To determine whether the mutants imported more glutathione, we measured total glutathione in bacterial lysates (Fig. 4A); however, we did not detect a significant difference in the total glutathione content in these strains.
Fig 4.
Glutathione content, HOSCN reductase (Har) and glutathione reductase (GR) activity, and NADPH levels in WT and mutant strains. (A) Total glutathione (B), Har activity (C), GR activity, and (D) NADPH/NADP+ ratios were measured in bacterial lysates as described in the Materials and Methods. Data are presented as the mean ± SD from at least three independent experiments. A significant difference when compared with lysates from WT was determined by one-way ANOVA with Dunnett’s multiple comparisons test and is indicated by * for P < 0.05.
Next, we assessed the activities of the NADPH-dependent enzymes hypothiocyanous acid reductase (Har) and glutathione reductase (GR), both known to contribute to S. pneumoniae HOSCN defense (9, 10). Har activity was significantly higher in the ΔahpD, ΔtrxA, and ΔsodA mutants (Fig. 4B), while GR activity was greater in the ΔtrxA and ΔsodA mutants (Fig. 4C). The compensatory upregulation of Har activity may be masking a possible protective role of AhpD itself. To determine whether the observed increased reductase activity was due to the presence of higher levels of endogenous NADPH, its concentration was measured in bacterial lysates. No difference in NADPH levels (data not shown) or NADPH/NADP+ ratios (Fig. 4D) was detected in any of the mutants compared to WT, suggesting that the observed increase in reductase activity was due to increased enzyme expression. There was no increase in Har or GR activity in the ΔspxB mutant, which had the greatest decrease in viability (Fig. 4B and C). One explanation for this observation might be that H2O2, which is generated to a considerably lower degree in the spxB mutant, is required for the upregulation of HOSCN protective genes. An impaired compensatory response may also explain why the ΔspxB bacteria were most sensitive to HOSCN.
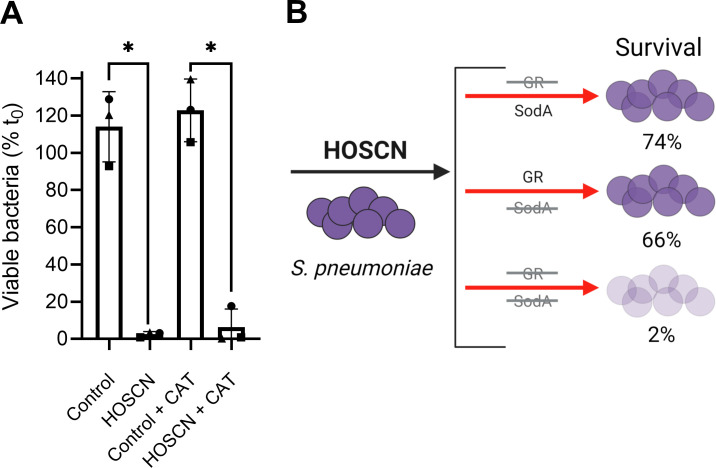
Bacterial compensatory mechanisms may be overcome by targeting more than one gene at once. Since the ΔtrxA and ΔsodA mutants displayed significantly increased glutathione reductase activity, we investigated whether double deletion mutants involving the glutathione reductase gene, gor, would be further sensitized to HOSCN. We were unable to generate a double mutant with gor and trxA, suggesting that while either the Trx/TrxR or glutathione/GR system can be impaired, it is vital to have one of the systems. In support, functional crosstalk between these systems is known to take place under stress conditions (40, 41). However, we could generate a Δgor-ΔsodA mutant, which as hypothesized was significantly impacted by HOSCN (Fig. 5A). Bacterial killing was much more pronounced in the double mutant (2%) than in the single ΔsodA mutant (Fig. 3E) or the Δgor mutant (9), which had 66% and 74% viable bacteria after 90 min, respectively (Fig. 5B).
Fig 5.
Viability of the double mutant strain Δgor-ΔsodA upon HOSCN treatment. (A) S. pneumoniae strain Δgor-ΔsodA (OD620 0.02) was grown and treated with 800 µM HOSCN for 90 min, and viability was measured via plate counts as described in Fig. 3. Data are presented as the mean ± SD from three independent experiments, with symbols designating matching replicate data. A significant difference to the respective control was determined by paired t-tests and is indicated by * for P < 0.05. (B) Summary diagram showing the impact of Δgor (9), ΔsodA (Fig. 3E), and Δgor-ΔsodA mutations on S. pneumoniae HOSCN viability. The diagram was created in Biorender.com.
Collectively, these results show that S. pneumoniae possess multiple defense mechanisms that underscore the ability to tolerate HOSCN. Novel antimicrobial strategies may need to target more than one protein to work effectively.
Additional HOSCN genes of interest identified in the screen
Our validation experiments were focused on a selected number of antioxidant defense genes; however, many other genes were identified in our screen that warrant future investigation. The genes manY_1/manM, relA, manZ_2, thiN, pnp/pnpA, and ktrB would be particularly interesting as they were significant hits in all replicates.
The genes manZ_2 (manN) and manY_1/manM, along with the gene manL which was significant in three replicates, are encoded on the same operon, called manLMN (42, 43). The ManLMN proteins form a transporter in the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system (PTS) superfamily. ManLMN is the major glucose transporter in S. pneumoniae and can also transport other sugars including mannose, fructose, galactose, N-acetylglucosamine, and glucosamine (43, 44). The importance of the ManLMN proteins in HOSCN tolerance suggests that the bacteria need to efficiently uptake glucose to protect themselves from oxidative stress. Glucose is needed for numerous cellular processes, including the generation of NADPH, which is a reducing equivalent required for Har, TrxR and GR activity. The ManLMN proteins might also play additional roles in S. pneumoniae, with PTS reported to have regulatory functions in addition to transport and phosphorylation [reviewed in reference (45)]. Of interest, PTSs are ubiquitous in bacteria but are not present in eukaryotes, and as such they have been suggested as targets for antimicrobial drugs (46 – 49). As deletion of any of the three genes in the manLMN operon led to significantly reduced bacterial survival upon HOSCN treatment, the manLMN operon PTS system might be an appropriate drug target in S. pneumoniae. However, as S. pneumoniae have multiple sugar transporters (43) that can take up other sugars when in a more complex environment, manLMN mutants would warrant additional testing in a non-glucose-limiting environment to determine their applicability as drug targets.
RelA is a stringent response protein that is the main source of the alarmones guanosine-pentaphosphate and -tetraphosphate ((p)ppGpp) in S. pneumoniae (50). (P)ppGpp plays important roles in bacteria, including coordinating oxidative stress responses (51). Induction of the stringent response in a ΔrelA strain of S. pneumoniae D39 using the antibiotic mupirocin resulted in a decrease in the relative transcript expression of genes including sodA, tpxD, and spxB (50). This highlights the interplay between stress genes in S. pneumoniae. RelA has also been shown to be essential for S. pneumoniae survival in mouse pneumonia models both by direct mutations (50) and site-directed mutagenesis screening (52).
The gene thiN encodes a thiamine pyrophosphokinase, which is responsible for transferring a phosphate group to vitamin B1, forming thiamine pyrophosphate (TPP), an essential coenzyme for metabolic functions (53). Enzymes dependent on TPP as a co-factor include pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, transketolase, and acetohydroxyacid synthase, which are essential in many metabolic pathways in bacteria (53). Dependence on TPP for pyruvate oxidase activity has been reported in Streptococcus sanguis and some other bacteria (54, 55). Additionally, a single missense mutation in the spxB TPP-binding site in S. pneumoniae reduced H2O2 production to the level of a full ΔspxB mutant and led to H2O2 hypersensitivity, demonstrating the importance of TPP for bacterial oxidant tolerance (33).
The gene ktrB encodes a potassium transporter membrane-spanning protein, called TrkH, that is required for potassium uptake in S. pneumoniae (56). Due to the importance of potassium for cell growth, this effect may have been a combined effect, where the extra stress imposed by the oxidant was too great for the bacteria to overcome. Similarly, the exact role of the gene pnpA in S. pneumoniae HOSCN tolerance is difficult to discern. The gene pnpA encodes a polyribonucleotide nucleotidyltransferase, and the KEGG annotation of this gene assigns a role in RNA degradation. The fact that disruption of pnpA and ktrB robustly decreased survival of HOSCN stress in all replicates points to their important role in HOSCN tolerance, but to fully understand the response observed by deletion of these genes, specific deletion mutants would need to be generated for further investigation.
In addition to investigating genes important for increasing HOSCN tolerance in S. pneumoniae, genes that were identified to prevent optimal HOSCN tolerance should be studied to gain a better understanding of the S. pneumoniae HOSCN response. The two largest Log2[FC] genes in this category were hrcA and ctsR, which encode heat shock protein repressors. Heat shock proteins are induced by various stresses such as temperature, pH, or oxidative stress to protect the proteome (57). HrcA acts as a repressor on the dnaK and groESL operons, while CtsR acts as a repressor on the groESL operon and clpP/C/B/E/L genes (58 – 61). Mutation of the repressor protein genes hrcA and ctsR in this screen would have increased the transcription of heat shock proteins, thereby enhancing tolerance to HOSCN. The genes clpL and clpC were also identified in this screen, further emphasizing the importance of a functional heat shock response to repair the protein damage caused by HOSCN stress.
Limitations
While this screen allowed us to identify many genes involved in HOSCN tolerance, there are caveats to the use of mutant libraries that mean some genes involved in HOSCN tolerance will be missed. First, only non-essential genes can be investigated in mutant libraries as any mutation in essential genes will render the bacterium non-viable and so will not be present in the library. This limitation may be overcome by using inducible CRISPR interference libraries. Additionally, polar effects made during transposon mutagenesis may disrupt the function of downstream genes that will be masked.
Our screening approach was designed to identify genes that promote bacterial survival, i.e., the number of viable mutants dropped during the period of HOSCN exposure. Therefore, mutations that are important for allowing bacterial growth in the presence of HOSCN will be more difficult to detect in our experimental model as the period of bacterial growth to amplify survivors occurred post-exposure to HOSCN. Modification of experimental conditions, such as a long period in the presence of lactoperoxidase, could reveal previously unidentified genes essential for bacterial growth.
The reductases gor and har have previously been shown to be important in S. pneumoniae HOSCN tolerance (9, 10) but were not identified in our screen. Enzymes responsible for scavenging oxidants might not be detected by screening mutant libraries if mutants are protected by neighboring bacteria. This is unlikely to be an issue for HOSCN, however, because extracellular HOSCN is only removed slowly by bacterial cultures (7). The more likely limitation is that compensatory effects mask the impact of single mutants. Har mutation alone only had a limited impact on the ability of S. pneumoniae to grow in the presence of HOSCN, but growth was completely blocked with a combined gor/har mutation (10). Transposon mutant libraries generated from a single-gene deletion mutant, thereby creating a double-mutant genetic interaction screen, might lead to the identification of additional genes of interest that were masked in our libraries.
Conclusion
Even with the use of vaccines and antibiotics, S. pneumoniae is still a major pathogen. The results presented here provide insight into the mechanisms by which S. pneumoniae tolerates HOSCN produced by the immune system and highlight the interplay between key antioxidant proteins under oxidative stress. We identified 37 genes involved in HOSCN tolerance and validated 4 genes involved in antioxidant defense. The antioxidant defense systems identified in this study could be investigated as novel therapeutic targets to aid the immune system in clearing this deadly pathogen.
MATERIALS AND METHODS
Materials
LPO from bovine milk [ε412 = 112,000 M−1 cm−1 (62)], Hank’s balanced salt solution (HBSS), and phosphate buffered saline (PBS) for cell culture, sodium thiocyanate, EC-oxyrase, calcium chloride dihydrate, spectinomycin dihydrochloride pentahydrate, N-ethylmaleimide (NEM) and Amicon ultra 0.5 centrifugal filter units (MWCO 10 k Da) were purchased from Sigma-Aldrich (Merck). Competence stimulating peptide-1 was purchased from AnaSpec. Brain heart infusion (BHI) media were from Oxoid. Todd Hewitt media supplemented with 1.5% (wt/vol) yeast extract (THY) was made from Bacto Todd Hewitt Broth and yeast extract (Becton Dickinson). Columbia sheep blood and tryptic soy blood agar plates were either purchased from Fort Richard Laboratories or made from defibrinated sheep’s blood (Fort Richard Laboratories) and Difco Columbia Blood Agar Base (BD). Bovine serum albumin was from New England Biolabs or Thermo Fisher Scientific. Hydrogen peroxide (30% vol/vol) [H2O2, ε240 = 43.6 M−1 cm−1 (63)] was from LabServ. 2-Nitro-5-thiobenzoate (TNB) was prepared from 5,5'-dithiobis-(2-nitrobenzoic acid) (Sigma-Aldrich) through alkaline hydrolysis as described (64). HOSCN was generated from lactoperoxidase, thiocyanate, and H2O2, and quantified at A412 using TNB as described in reference (9), kept on ice, and used within 30 min of quantification.
Bacterial strains and culture conditions
Bacterial strains were maintained on Columbia or tryptic soy broth sheep blood agar plates. Mutant strains were grown on Columbia sheep blood agar plates with 150 µg/mL spectinomycin. For experiments, bacteria were statically grown overnight in BHI media at 37°C with 5% CO2, diluted in fresh media, and grown to mid-log phase before experiments.
Transposon mutant library generation
The S. pneumoniae transposon library (Library 1) was constructed as described previously (11, 12, 14). Briefly, the mini-transposon magellan6, containing a gene to confer spectinomycin resistance, was inserted into S. pneumoniae strain D39 genomic DNA by the enzyme transposase MarC9. DNA was ligated together, and DNA containing the transposon was transformed into competent S. pneumoniae cells. Transposon-containing mutants were selected for Columbia sheep blood agar plates containing 150 µg/mL spectinomycin. Mutants were scraped off the plates using a sterile cell spreader, pooled together in THY media, diluted to OD600 0.05 and grown to OD600 0.3. The culture was then concentrated into 5 mL THY media with 12% glycerol and frozen at −80°C in 500 µL aliquots.
Tn-seq HOSCN tolerance screen
Library 1 was used alongside a second previously generated library (Library 2) (14) to investigate genes involved in S. pneumoniae HOSCN tolerance. An aliquot of each transposon mutant library (Library 1 and Library 2) was defrosted and added to THY media containing 200 µg/mL spectinomycin. Cultures were grown until mid-log phase, then washed twice in PBS, and suspended in HBSS, pH 6.8. Duplicate tubes of each library were then set up, and bacteria (5 × 107 CFU/mL) were exposed to 800 µM HOSCN or buffer for 60 min at 37°C with 5% CO2. After incubation, bacteria were centrifuged at 12,000 g, and each cell pellet was frozen on dry ice and stored at −80°C for DNA isolation.
DNA sequencing
Genomic DNA from the HOSCN tolerance screen was isolated using the DNeasy Blood and Tissue Kit (Qiagen), and samples were prepared for sequencing using the HTML-PCR protocol (12, 14). Briefly, genomic DNA was sheared via sonication until DNA fragments were between 100 and 600 bp. DNA ends were then blunted using the Quick Blunting Kit (NEB), and genomic DNA was purified and concentrated using the HighPrep PCR Clean-Up System magnetic beads (MagBio). Poly-C tails were then added to the 3′ ends of the DNA using terminal deoxynucleotide transferase (Promega), and DNA was re-purified and concentrated using the magnetic beads. Nested PCR reactions were then carried out to amplify transposon junctions and add unique index barcode sequences to the DNA for each sample [Table S3; (12, 14)]. Once each sample was barcoded by its unique index sequence, samples were pooled for each library for simultaneous sequencing. Pooled libraries consisting of all 12 samples per library were sequenced with Illumina HiSeq single-end 50 cycle sequencing by the Otago Genomics Facility, University of Otago, New Zealand.
Sequencing analysis
Sequence analysis was conducted using a modified method from references (65, 66) and the BioTraDIS pipeline (19). Quality control analysis on fastq files was run on all raw sequencing files using FastQC (67). Cutadapt was used to remove poly-C tail adapters from the samples (68). For this, the adapter (-a) was defined as “C{10}N{10}” [10 Cs followed by 10 of any base (CCCCCCCCCCNNNNNNNNNN)]. Once the poly-C tails were removed, all reads were filtered so that any reads shorter than 15 nucleotides were removed (-m 15).
The BioTraDIS pipeline (19) (bioconda version 1.4.5) was used to map reads to the S. pneumoniae D39 genome (GenBank: LS483374.1). The bacteria_tradis script was used with the following code: “bacteria_tradis --smalt --smalt_k 10 --smalt_s 1 --smalt_y 0.96 --smalt_r -1 -v -n 5 -f filelist.txt -r GCF_900475305.1_43721_E02_genomic.fna.gz.” This code designates that the SMALT read mapper will be used to map the reads to the reference genome (--smalt) (69). The minimum length of an exact match between the read and genome to attempt to align the read needed to be 10 (this designates k-mer size) (smalt_K 10), and every k-mer word was indexed along the reference [step size for k-mers of 1 (smalt_s 1)]. Only reads with at least 96% sequence identity to the genome were mapped (smalt_y 0.96), reads that mapped equally well to more than one location were discarded so as to not influence downstream differential analyses (smalt_r -1), and the number of reads is specified by (-n 5).
The tradis_gene_insert_sites script, which assigns the insertion information (.plot file) to the annotation file (.embl file) was used for post-processing analysis in R. First, reads that map to the first 10% from the 3′ end of each feature were removed from the final output as many genes can tolerate insertions at the 3′ end (70, 71). To compare read frequencies between treated and control samples in each replicate, the tradis_comparision.R script which uses edgeR was used (18). The script for comparisons was “tradis_comparison.R --controls <.txt> --conditions <.txt> -o day1.txt -p day1.pdf -f -t 15,” with day1.txt and day1.pdf being changed to the files corresponding to each replicate and library. In this script, read filtering was allowed (-f), and a minimum of 15 reads was needed to analyze the gene (-t 15). The output for this script was in log2 fold change in the treated samples compared to the untreated controls. It also provided adjusted P-values to account for false discovery rates using Benjamini and Hochberg correction. Genes with an adjusted P-value of <0.05 and a log2[FC] of ≤−1 or ≥1 were identified as significant. Genes that met these criteria in at least two of the three replicates within each library were reproducibly significant and were used for further analyses. Functional annotation of genes was performed using the KEGG Ontology database. Visualization of the number of reads and gene coverage was performed in Artemis 18.1.0 (72).
Generation of S. pneumoniae gene deletion mutants
S. pneumoniae mutants were generated by transformation of DNA constructs (synthesized by Genscript Biotech, USA). These genes mutated were trxA (NCTC7466_01600/SPD_1567), sodA (NCTC7466_00694/SPD_0667), spxB (NCTC7466_00663/SPD_0636), and ahpD (NCTC7466_00383/SPD_0373). Constructs were designed to carry a spectinomycin resistance (SpecR) cassette sandwiched between adjacent arms 5′ and 3′ of the target gene sequence to be knocked out (Method S1). Relevant primers (Table S4) were used to PCR the homologous arms with their SpecR insert. PCR products were purified, quantified, and used for transformation of S. pneumoniae as previously described (10). Briefly, DNA (0.1–1 µg) was transformed into S. pneumoniae D39, and mutants were selected for on spectinomycin-containing blood agar plates. Transformants were cultured and their genomic DNA extracted, and PCR amplified using primers external to the construct sequences. NotI restriction endonuclease cleavage of DNA intrinsic to the inserted constructs was used to identify successful transformation and recombination, and gene knockouts were confirmed by Sanger sequencing. The primers used for these PCR and sequencing reactions are detailed in Table S4.
Bacterial growth characterization
S. pneumoniae WT and mutant strains were grown overnight, diluted in fresh media, and then grown to mid-log phase. Bacteria were washed twice in PBS, suspended in BHI media, diluted to OD620 0.1, and grown for 6 h at 37°C with 5% CO2 in BHI media in a 96-well plate. OD620 measurements were taken every 30 min using a BioTek Synergy Neo2 Hybrid Multi-Mode Microplate Reader (Winooski, VT). A BHI media sample was used as a control and subtracted from all other values.
Bacterial survival after exposure to HOSCN
Survival of S. pneumoniae strains in the presence or absence of 800 µM HOSCN was performed as described before (9), with modifications. S. pneumoniae (OD620 0.02) were incubated for 90 min in the presence or absence of 800 µM HOSCN ± catalase (20 µg/mL) in HBSS, pH 6.8, at 37°C with 5% CO2. Catalase controls were included to determine whether H2O2 generated by the bacterial strains was impacting viability. After 90 min, bacteria were diluted in PBS, plated and counted the following day. Bacterial viability was determined as the percentage of colonies at 90 min compared to the 0-time point.
HOSCN and glutathione reductase activity in bacterial lysates
S. pneumoniae WT and mutants were grown to an OD620 of 0.4–0.7, pelleted by centrifugation at 10,000 g for 5 min at 4°C, washed once with PBS, and then resuspended in 2.5 mL of 100 mM phosphate (pH 7.4), 1 mM EDTA. S. pneumoniae was lysed by pulse sonication on ice for 10 min. Bacterial debris was removed by centrifugation at 10,000 g for 10 min at 4°C. Activity of HOSCN and glutathione reductase (Har and GR, respectively) in clarified lysates was determined as described before (10). Briefly, the consumption of NADPH (200 µM) after the addition of HOSCN (100 µM) or GSSG (1 mM) was measured by monitoring the loss of absorbance at 340 nm. The reductase activity was defined as the rate of NADPH consumption and was calculated using ɛ340 = 6,220 M−1cm−1 for NAD(P)H (73), then expressed relative to the protein concentration in bacterial lysates as determined by Bradford assay (74).
Quantification of NADPH/NADP+ and total glutathione
S. pneumoniae lysates were prepared as described for reductase assays above. NAPDH and NADP+ levels in lysates were measured using a NAD/NADH-Glo Assay Kit (Promega; catalog no. G9081), according to the manufacturer’s instructions. For this, lysates were diluted 1:1 with bicarbonate base buffer (100 mM sodium bicarbonate, pH 10–11, 10 mM nicotinamide, 0.05% Triton X-100) and then split into two for heat treatment (60°C) in acidic and basic conditions to destroy NADPH and NADP+, respectively. A NADPH standard curve was prepared using a buffer containing the same final proportions of lysis buffer/bicarbonate base buffer/acid/base as samples.
To measure total glutathione, DTT (1 mM) was added to bacterial lysates and incubated at 60°C for 15 min, then N-ethylmaleimide (NEM) (20 mM) was added and incubated at 20–22°C for 15 min. An equal volume of acetonitrile was added to precipitate protein which was removed by centrifugation. The supernatant was diluted 1:5 with 0.25% formic acid in water and spiked with isotopically labeled heavy standards (13C2,15N1-GSH-NEM and 13C4,15N2-GSSG). Ten microliters of sample along with standards of GSH-NEM were injected for multiple-reaction-monitoring-based LC-MS analysis using an Infinity 1290 LC system (Agilent, Santa Clara, CA) coupled to a 6500 QTrap mass spectrometer (Sciex, Framingham, MA) as described previously (75).
Statistical analyses
Graphs were plotted, and the statistical analyses stated in the figure legends were performed using GraphPad Prism (Version 8.2.1). A P-value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This study was supported by the Canterbury Medical Research Foundation (Project Grant #05/20), a University of Otago Research Grant (#3579), a Sir Charles Hercus Health Research Fellowship from the Health Research Council of New Zealand to N.D., University of Otago Doctoral Scholarships (H.L.S. and L.M.S.) and a Professor Sandy Smith Memorial Scholarship from the Dunedin Basic Medical Sciences Trust to H.L.S. Development of Tn-seq methodologies by P.C.F. and L.M.S. were supported by the School of Biomedical Sciences Bequest Fund from the University of Otago and Te Pūtea Rangahau a Marsden, Marsden Fund from the Royal Society Te Apārangi, New Zealand.
All authors edited the manuscript and approved its final version.
The authors have no conflict of interest to declare.
Contributor Information
Heather L. Shearer, Email: heather.shearer@otago.ac.nz.
Nina Dickerhof, Email: nina.dickerhof@otago.ac.nz.
Tina M. Henkin, Ohio State University, Columbus, Ohio, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00208-23.
Spreadsheets with gene data from Library 1 replicates.
Spreadsheets with gene data from Library 2 replicates.
Four supplemental tables and one supplemental method.
KEGG gene orthology table with pathway and gene links.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Bogaert D, De Groot R, Hermans PWM. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- 2. Wijkstrom-Frei C, El-Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, Salathe M, Conner GE. 2003. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol 29:206–212. doi: 10.1165/rcmb.2002-0152OC [DOI] [PubMed] [Google Scholar]
- 3. Thomson E, Brennan S, Senthilmohan R, Gangell CL, Chapman AL, Sly PD, Kettle AJ, Australian Respiratory Early Surveillance Team for Cystic Fibrosis (AREST CF), Balding E, Berry LJ, Carlin JB, Carzino R, de Klerk N, Douglas T, Foo C, Garratt LW, Hall GL, Harrison J, Kicic A, Laing IA, Logie KM, Massie J, Mott LS, Murray C, Parsons F, Pillarisetti N, Poreddy SR, Ranganathan SC, Robertson CF, Robins-Browne R, Robinson PJ, Skoric B, Stick SM, Sutanto EN, Williamson E. 2010. Identifying peroxidases and their oxidants in the early pathology of cystic fibrosis. Free Radic Biol Med 49:1354–1360. doi: 10.1016/j.freeradbiomed.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 4. Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aune TM, Thomas EL. 1977. Accumulation of hypothiocyanite ion during peroxidase‐catalyzed oxidation of thiocyanate ion. Eur J Biochem 80:209–214. doi: 10.1111/j.1432-1033.1977.tb11873.x [DOI] [PubMed] [Google Scholar]
- 6. Thomas EL, Fishman M. 1986. Oxidation of chloride and thiocyanate by isolated leukocytes. J Biol Chem 261:9694–9702. [PubMed] [Google Scholar]
- 7. Shearer HL, Kaldor CD, Hua H, Kettle AJ, Parker HA, Hampton MB. 2022. Resistance of Streptococcus pneumoniae to hypothiocyanous acid generated by host peroxidases. Infect Immun 90:e0053021. doi: 10.1128/IAI.00530-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pattison DI, Davies MJ, Hawkins CL. 2012. Reactions and reactivity of myeloperoxidase-derived oxidants: differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic Res 46:975–995. doi: 10.3109/10715762.2012.667566 [DOI] [PubMed] [Google Scholar]
- 9. Shearer HL, Paton JC, Hampton MB, Dickerhof N. 2022. Glutathione utilization protects Streptococcus pneumoniae against lactoperoxidase-derived hypothiocyanous acid. Free Radical Biology and Medicine 179:24–33. doi: 10.1016/j.freeradbiomed.2021.12.261 [DOI] [PubMed] [Google Scholar]
- 10. Shearer HL, Pace PE, Paton JC, Hampton MB, Dickerhof N. 2022. A newly identified flavoprotein disulfide reductase Har protects Streptococcus pneumoniae against hypothiocyanous acid. J Biol Chem 298:102359. doi: 10.1016/j.jbc.2022.102359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Opijnen T, Lazinski DW, Camilli A. 2014. Genome‐wide fitness and genetic interactions determined by Tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Curr Protoc Mol Biol 106:7. doi: 10.1002/0471142727.mb0716s106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verhagen LM, de Jonge MI, Burghout P, Schraa K, Spagnuolo L, Mennens S, Eleveld MJ, van der Gaast-de Jongh CE, Zomer A, Hermans PWM, Bootsma HJ. 2014. Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS One 9:e89541. doi: 10.1371/journal.pone.0089541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matthews AJ, Rowe HM, Rosch JW, Camilli A, Freitag NE. 2021. A Tn-seq screen of Streptococcus pneumoniae uncovers DNA repair as the major pathway for desiccation tolerance and transmission. Infect Immun 89:00713–00720. doi: 10.1128/IAI.00713-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rowe HM, Karlsson E, Echlin H, Chang T-C, Wang L, van Opijnen T, Pounds SB, Schultz-Cherry S, Rosch JW. 2019. Bacterial factors required for transmission of Streptococcus pneumoniae in mammalian hosts. Cell Host Microbe 25:884–891. doi: 10.1016/j.chom.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Opijnen T, Dedrick S, Bento J. 2016. Strain dependent genetic networks for antibiotic-sensitivity in a bacterial pathogen with a large pan-genome. PLoS Pathog. 12:e1005869. doi: 10.1371/journal.ppat.1005869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mann B, van Opijnen T, Wang J, Obert C, Wang Y-D, Carter R, McGoldrick DJ, Ridout G, Camilli A, Tuomanen EI, Rosch JW, Cossart P. 2012. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog 8:e1002788. doi: 10.1371/journal.ppat.1002788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J. 2016. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32:1109–1111. doi: 10.1093/bioinformatics/btw022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shearer HL, Hampton MB, Dickerhof N. 2021. Bactericidal activity of the oxidants derived from mammalian heme peroxidasestericidal, p 171. In Mammalian heme peroxidases: diverse roles in health and disease. doi: 10.1201/9781003212287 [DOI] [Google Scholar]
- 21. Farrant KV, Spiga L, Davies JC, Williams HD. 2020. Response of Pseudomonas aeruginosa to the innate immune system-derived oxidants hypochlorous acid and hypothiocyanous acid. J Bacteriol 203:e00300-20. doi: 10.1128/JB.00300-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groitl B, Dahl JU, Schroeder JW, Jakob U. 2017. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol 106:335–350. doi: 10.1111/mmi.13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Opijnen T, Camilli A. 2012. A fine scale phenotype–genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pruitt KM. 1987. The salivary peroxidase system: thermodynamic, kinetic and antibacterial properties. J Oral Pathol Med 16:417–420. doi: 10.1111/j.1600-0714.1987.tb02078.x [DOI] [PubMed] [Google Scholar]
- 25. Meng Y, Sheen CR, Magon NJ, Hampton MB, Dobson RCJ. 2020. Structure-function analyses of alkylhydroperoxidase D from Streptococcus pneumoniae reveal an unusual three-cysteine active site architecture. J Biol Chem 295:2984–2999. doi: 10.1074/jbc.RA119.012226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paterson GK, Blue CE, Mitchell TJ. 2006. An operon in Streptococcus pneumoniae containing a putative alkylhydroperoxidase D Homologue contributes to virulence and the response to oxidative stress. Microb Pathog 40:152–160. doi: 10.1016/j.micpath.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 27. Winterbourn CC, Kettle AJ, Hampton MB. 2016. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85:765–792. doi: 10.1146/annurev-biochem-060815-014442 [DOI] [PubMed] [Google Scholar]
- 28. Aguinagalde L, Díez-Martínez R, Yuste J, Royo I, Gil C, Lasa Í, Martín-Fontecha M, Marín-Ramos NI, Ardanuy C, Liñares J, García P, García E, Sánchez-Puelles JM. 2015. Auranofin efficacy against MDR Streptococcus pneumoniae and Staphylococcus aureus infections. J Antimicrob Chemother 70:2608–2617. doi: 10.1093/jac/dkv163 [DOI] [PubMed] [Google Scholar]
- 29. Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- 30. Eijkelkamp BA, Morey JR, Ween MP, Ong CY, McEwan AG, Paton JC, McDevitt CA. 2014. Extracellular zinc competitively inhibits manganese uptake and compromises oxidative stress management in Streptococcus pneumoniae. PLoS One 9:e89427. doi: 10.1371/journal.pone.0089427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun 68:2819–2826. doi: 10.1128/IAI.68.5.2819-2826.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x [DOI] [PubMed] [Google Scholar]
- 33. Ramos-Montañez S, Tsui H-CT, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham L-T, Winkler ME. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS‐HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol 67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x [DOI] [PubMed] [Google Scholar]
- 34. Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Echlin H, Frank MW, Iverson A, Chang T-C, Johnson MDL, Rock CO, Rosch JW. 2016. Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLoS Pathog 12:e1005951. doi: 10.1371/journal.ppat.1005951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis 190:1661–1669. doi: 10.1086/424596 [DOI] [PubMed] [Google Scholar]
- 37. Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. 2007. Spxb is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J Bacteriol 189:6532–6539. doi: 10.1128/JB.00813-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carvalho SM, Farshchi Andisi V, Gradstedt H, Neef J, Kuipers OP, Neves AR, Bijlsma JJE, Chi J-TA. 2013. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PLoS ONE 8:e68277. doi: 10.1371/journal.pone.0068277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hajaj B, Yesilkaya H, Shafeeq S, Zhi X, Benisty R, Tchalah S, Kuipers OP, Porat N. 2017. CodY regulates thiol peroxidase expression as part of the pneumococcal defense mechanism against H2O2 stress. Front. Cell. Infect. Microbiol 7. doi: 10.3389/fcimb.2017.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnér ESJ. 2022. Chapter 10 - Thioredoxin and glutathione reductases, p 197–218. In Alvarez B, Comini MA, Salinas G, Trujillo M (ed), Redox chemistry and biology of thiols. Academic Press. doi: 10.1016/B978-0-323-90219-9.00009-1 [DOI] [Google Scholar]
- 41. Tuggle CK, Fuchs JA. 1985. Glutathione reductase is not required for maintenance of reduced glutathione in Escherichia coli K-12. J Bacteriol 162:448–450. doi: 10.1128/jb.162.1.448-450.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paixão L, Caldas J, Kloosterman TG, Kuipers OP, Vinga S, Neves AR. 2015. Transcriptional and metabolic effects of glucose on Streptococcus pneumoniae sugar metabolism. Front Microbiol 6:1041. doi: 10.3389/fmicb.2015.01041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. doi: 10.1371/journal.pone.0033320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fleming E, Camilli A. 2016. Manlmn is a glucose transporter and central metabolic regulator in Streptococcus pneumoniae. Mol Microbiol 102:467–487. doi: 10.1111/mmi.13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deutscher J, Aké FMD, Derkaoui M, Zébré AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kok M, Bron G, Erni B, Mukhija S. 2003. Effect of enzyme I of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology (Reading) 149:2645–2652. doi: 10.1099/mic.0.26406-0 [DOI] [PubMed] [Google Scholar]
- 47. Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA 104:2384–2389. doi: 10.1073/pnas.0608775104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Magoch M, Nogly P, Grudnik P, Ma P, Boczkus B, Neves AR, Archer M, Dubin G. 2020. Crystal structure of mannose specific IIA subunit of phosphotransferase system from Streptococcus pneumoniae Molecules 25:4633. doi: 10.3390/molecules25204633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang K-J, Lin S-H, Lin M-R, Ku H, Szkaradek N, Marona H, Hsu A, Shiuan D. 2013. Xanthone derivatives could be potential antibiotics: virtual screening for the inhibitors of enzyme I of bacterial phosphoenolpyruvate-dependent phosphotransferase system. J Antibiot 66:453–458. doi: 10.1038/ja.2013.30 [DOI] [PubMed] [Google Scholar]
- 50. Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. 2009. Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol 72:590–611. doi: 10.1111/j.1365-2958.2009.06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p) ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hava DL, Camilli A. 2002. Large‐scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.t01-1-03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du Q, Wang H, Xie J. 2011. Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets Int J Biol Sci 7:41–52. doi: 10.7150/ijbs.7.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carlsson J, Kujala U. 1984. Pyruvate oxidase activity dependent on thiamine pyrophosphate, flavin adenine dinucleotide and orthophosphate in Streptococcus sanguis . FEMS Microbiol Lett 25:53–66. doi: 10.1111/j.1574-6968.1984.tb01374.x [DOI] [Google Scholar]
- 55. Chen L, Ge X, Dou Y, Wang X, Patel JR, Xu P. 2011. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology 157:13–20. doi: 10.1099/mic.0.039669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. 2014. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol 196:614–623. doi: 10.1128/JB.01041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kwon H-Y, Kim S-W, Choi M-H, Ogunniyi AD, Paton JC, Park S-H, Pyo S-N, Rhee D-K. 2003. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae . Infect Immun 71:3757–3765. doi: 10.1128/IAI.71.7.3757-3765.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim S-N, Bae Y-G, Rhee D-K. 2008. Dual regulation of dnaK and groE operons by HrcA and Ca++ in Streptococcus pneumoniae. Arch Pharm Res 31:462–467. doi: 10.1007/s12272-001-1179-4 [DOI] [PubMed] [Google Scholar]
- 59. Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram‐positive bacteria. Mol Microbiol 63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x [DOI] [PubMed] [Google Scholar]
- 60. Chastanet A, Prudhomme M, Claverys J-P, Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J Bacteriol 183:7295–7307. doi: 10.1128/JB.183.24.7295-7307.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Derre I, Rapoport G, Msadek T. 1999. CtsR, a novel regulator of stress and heat shock response, controls Clp and molecular chaperone gene expression in Gram‐positive bacteria. Mol Microbiol 31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x [DOI] [PubMed] [Google Scholar]
- 62. Paul KG, Ohlsson PI.. 1985. p 15-29. In P K.M., T J.O. (ed), The lactoperoxidase system, chemistry and biological significance. Marcel Dekker, New York. [Google Scholar]
- 63. BEERS RF Jr, SIZER IW. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem 195:133–140. [PubMed] [Google Scholar]
- 64. Nagy P, Jameson GNL, Winterbourn CC. 2009. Kinetics and mechanisms of the reaction of hypothiocyanous acid with 5-thio-2-nitrobenzoic acid and reduced glutathione. Chem Res Toxicol 22:1833–1840. doi: 10.1021/tx900249d [DOI] [PubMed] [Google Scholar]
- 65. Smith LM, Jackson SA, Gardner PP, Fineran PC. 2021. SorTN-Seq: a high-throughput functional genomics approach to discovering regulators of bacterial gene expression. Nat Protoc 16:4382–4418. doi: 10.1038/s41596-021-00582-6 [DOI] [PubMed] [Google Scholar]
- 66. Smith LM, Jackson SA, Malone LM, Ussher JE, Gardner PP, Fineran PC. 2021. The Rcs stress response inversely controls surface and CRISPR–Cas adaptive immunity to discriminate plasmids and phages. Nat Microbiol 6:162–172. doi: 10.1038/s41564-020-00822-7 [DOI] [PubMed] [Google Scholar]
- 67. Andrews S. 2010. A quality control tool for high throughput sequence data, On babraham bioinformatics. Babraham Institute. [Google Scholar]
- 68. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 69. Ponstingl H, Ning Z. 2010. SMALT-a new mapper for DNA sequencing reads. F1000 Posters 1.
- 70. Le Breton Y, Belew AT, Valdes KM, Islam E, Curry P, Tettelin H, Shirtliff ME, El-Sayed NM, McIver KS. 2015. Essential genes in the core genome of the human pathogen Streptococcus pyogenes. Sci Rep 5:9838. doi: 10.1038/srep09838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang ZR, Bullifent HL, Moore K, Paszkiewicz K, Saint RJ, Southern SJ, Champion OL, Senior NJ, Sarkar-Tyson M, Oyston PCF, Atkins TP, Titball RW. 2017. A noise trimming and positional significance of transposon insertion system to identify essential genes in Yersinia pestis. Sci Rep 7:41923. doi: 10.1038/srep41923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. doi: 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. HORECKER BL, KORNBERG A. 1948. The extinction coefficients of the reduced band of pyridine nucleotides. J Biol Chem 175:385–390. [PubMed] [Google Scholar]
- 74. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 75. Dickerhof N, Isles V, Pattemore P, Hampton MB, Kettle AJ. 2019. Exposure of Pseudomonas aeruginosa to bactericidal hypochlorous acid during neutrophil phagocytosis is compromised in cystic fibrosis. J Biol Chem 294:13502–13514. doi: 10.1074/jbc.RA119.009934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spreadsheets with gene data from Library 1 replicates.
Spreadsheets with gene data from Library 2 replicates.
Four supplemental tables and one supplemental method.
KEGG gene orthology table with pathway and gene links.