Summary:
Beetle daisies evolved novel floral spots that mimic female bee flies to entice mate-seeking males for pollination. This study shows that these deceptive spots emerged through stepwise co-option of multiple genetic elements, shedding light on the origin of complex phenotypic novelties.
Dispatch:
Many flowers display stunning adaptations to their specialized pollinators. Among the most captivating examples are sexually deceptive flowers that mimic the appearance, texture, and/or sexual pheromones of a female insect, attracting males and ultimately tricking them into mating to achieve pollination. While such sexually deceptive pollination is relatively common in orchids1, the South African Beetle daisy (Gorteria diffusa) is the only known plant outside of the orchid family to employ this strategy2. Unlike the orchids, which rely primarily on olfactory cues for the attraction of pollinators3, G. diffusa deception is based primarily on visual and tactile cues.
G. diffusa exhibits a range of floral trait combinations, called morphotypes, that are found in largely non-overlapping zones across its geographic range2. Some morphotypes display patterns that mimic resting female bee flies (Megapalpus capensis) on one to four ray florets4 (Figure 1). It is a convincing mimic—these spots are raised to give a three-dimensional appearance, and the greenish-black pigmentation is intermixed with small UV reflective spots that gives the appearance of sunlight glaring off a bee fly exoskeleton4. The remaining morphotypes each have some (but not all) elements of this pattern, and with reduced complexity comes a corresponding reduction in the number of visitations by mate-seeking males2. Curiously, pollinator surveys reveal that all morphotypes in this continuum are pollinated by the same bee fly species, indicating that a pollinator shift was not the driving factor for floral diversification5. However, the evolution of deceptive features by some morphotypes was not in vain—plants that put on a convincing ruse are rewarded with increased visitation by mate-seeking male bee flies4,6 and increased pollen export2.
Figure 1.
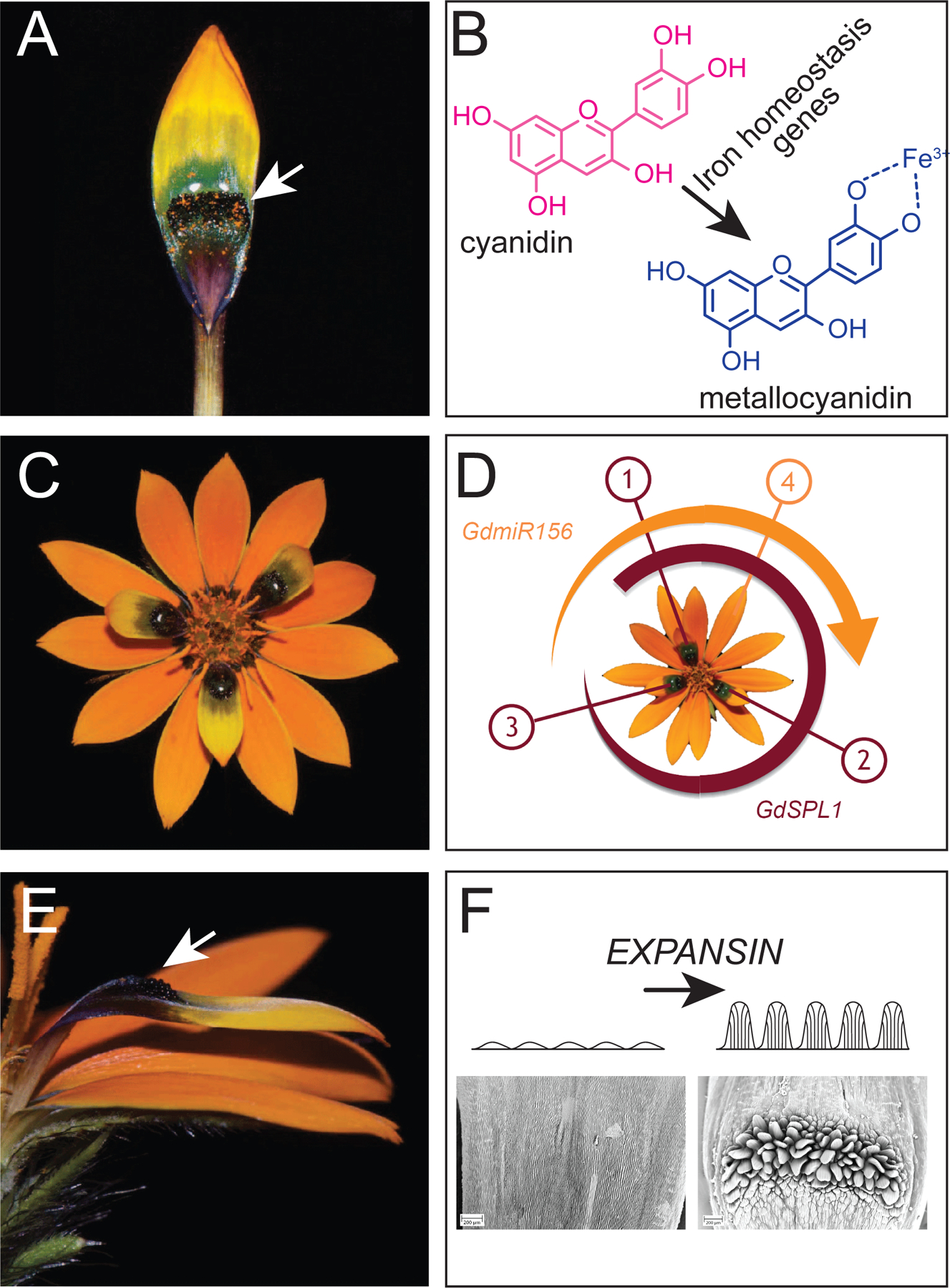
(A) Photograph of pigmented Gorteria diffusa Springbok ray floret with white arrow showing the greenish-black coloration and (B) the chemical structures of cyanidin and metallocyanidin associated with the shift in coloration from reddish to greenish-black. Chemical structures were generated using PubChem Sketcher v2.4; (C) photograph depicting the spatial arrangement of pigmented florets in the Springbok morphotype and (D) model showing how the miR156-SPL1 module regulates their spatial arrangement; (E) close-up of the textured section of the Springbok ray floret and (F) CryoSEM images taken on the center of the petal spot of the Naries morphotype (lower left) and Springbok morphotype (lower right). Naries has typical flat epidermal cells, while Sprinkbok has elongated cells that form multicellular papillae, as shown in the illustrations above the images. Panel A, C, and E images by Dr. Samuel Brockington, Associate Professor & Curator, Department of Plant Sciences, University of Cambridge.
A new study in this issue of Current Biology by Kellenberger et al.7 breaks down the highly deceptive phenotype of G. diffusa into three primary components—the greenish-black pigmentation that mimics the color of the bee fly exoskeleton; the spatial arrangement of spotted florets that gives the appearance of individual female visitors; and the texture of the spots to provide dimensionality (Figure 1)—and aims to determine the genetic origin of each one. Often, novel structures or patterns are the result of one gene or genetic network taking on a new function elsewhere within the organism, in a phenomenon known as “gene co-option”8,9. In the deceptive morphotypes of G. diffusa, these three components are found to have originated through separate co-option events of completely unrelated genes and/or gene networks.
The ability to break a complex novel phenotype into its composite parts and determine their individual origins is a unique advantage of this study system. Most phenotypic novelties we study today have an evolutionary origin in the distant past (e.g., turtle shell, beetle horn, angiosperm flower), making it infeasible to elucidate the order and causal factors of the initial phenotypic transitions. The G. diffusa radiation is relatively young on an evolutionary timescale (0.6–2.2 MYA), and as such, intermediate morphotypes with various combinations of those composite parts are still available2. The existence of these intermediates enabled the ancestral reconstruction of each component10, and thus allowed Kellenberger et al.7 to present a logical hypothesis regarding the order in which these co-option events occurred.
The first step was to achieve the greenish-black color that mimics the exoskeleton of a female bee fly (Figure 1A), and in the genus Gorteria, this is accomplished entirely through pigmentary color. Pigments called anthocyanins are common in floral patterns and typically impart pink, red, or blue hues11. The authors show that genes associated with iron homeostasis (i.e., OBP3-RESPONSIVE GENEs ORG1 and ORG2) and iron transport (i.e., OLIGOPEPTIDE TRANSPORTER 3, NRAMP3/4 and VACUOLAR IRON TRANSPORTER1) are more strongly expressed in spotted ray florets compared to unspotted ones. Concordantly, iron accumulation was highest at the base of spotted florets and was specifically localized to the vacuoles of papillae and the surrounding adaxial epidermal cells. Iron forms complexes with an anthocyanin called cyanidin, causing the pigment to shift from its typical pink color to a bluish hue (Figure 1B). This blue pigment is overlaid with the uniform orange carotenoid-based color of the floret, resulting in a greenish-black hue.
This greenish-black coloration is observed as a contiguous nectar guide in several morphotypes and closely related Gorteria species, indicating that this co-option is ancestral to the G. diffusa complex10. The authors logically hypothesize that the next step in the generation of the sexually deceptive morphotypes was to limit the greenish-black color to just a few spatially non-adjacent florets, thus breaking the continuous ring and giving the appearance that a few female bee flies are resting on the capitulum (Figure 1C). Ray florets of the beetle daisy mature in a golden angle phyllotactic pattern, such that the oldest floret is closest to the center of the capitulum with new florets emerging every ~137.5º apart (Fig 1D)12. Analysis of differentially expressed genes in spotted versus unspotted florets implicates the co-option of a well-known developmental regulatory module, the miR156-SPL module, as the key to this careful timing of spot deposition. Members of the SPL transcription factor family are involved in an antagonistic relationship with a microRNA known as miR156, and together they are generally associated with age-dependent developmental transition from the juvenile to reproductive phase in plants13. GdSPL1 is expressed specifically in spotted florets, whereas GdmiR156 is accumulated at high levels in unspotted florets. Thus, it seems likely that this developmental phase transitioning module has been co-opted to regulate the duration of spotted floret initiation during capitulum development, although the precise molecular mechanisms through which GdSPL1 enables spot formation on the first few florets remain unknown.
The production of greenish-black color and the subsequent arrangement into discrete spots appears to have been sufficient for attracting more mate-seeking males2. However, true sexual deception only occurs in the morphotypes that display a raised spot2,6. This three-dimensional elaboration is due to elongated epidermal cells that come together to form tongue-like papillae in a horseshoe-shape around the distal edge of the spot (Figure 1F)12. Kellenberger et al.7 identify a member of the EXPANSIN gene family as a strong candidate, being highly differentially expressed between deceptive and non-deceptive morphotypes. This gene, GdEXPA7, shares a homolog with Arabidopsis that associated with the regulation of root hair elongation14,15. The authors confirmed that GdEXPA7 does have a root-hair specific cis-regulatory element in its promoter and that it is expressed in both roots and developing spotted florets, thus supporting a root-based origin. Interestingly, root hair development is normally controlled by developmental signals or environmental cues; however, none of the genes normally associated with those signals are upregulated in G. diffusa florets. Therefore, the authors conclude that GdEXPA7 has been co-opted for the papillae gene network and is no longer responding signals associated with root hair development. Thus, at least three different genetic co-option events—that of the iron homeostasis network, the miR156-SPL developmental regulatory module, and GdEXPA7—are required for the emergence of sexual deception in G. diffusa.
These results have laid a solid foundation for subsequent studies to address many interesting questions on the mechanisms behind the co-option of these genes/modules. For example, how many mutations were required for these independent co-option events, and what is the nature of those mutations? Did these genetic changes result from de novo mutations in the deceptive morphotypes, or rather by the fixation of standing genetic variation within the ancestral populations? Can we recreate the deceptive spots in the non-deceptive morphotypes, either through introgression by serial backcrossing, or through direct transgenic manipulations? Further, is there a regulatory hierarchy between these three modules, or is the composite phenotype the result of phenotypic integration? Lastly, what determines the spatial patterning of the spots within an individual floret, and could this within-floret patterning be due to additional co-option events? Truly, G. diffusa is a seductive emerging model system for the study of composite phenotypic novelty.
And just in case anyone is still left worrying about the hapless bee fly, fear not. Male bee flies do learn to recognize the patterns associated with sexually deceptive morphotypes and will avoid them for at least a short time after the encounter16.
References:
- 1.Schiestl FP (2005). On the success of a swindle: pollination by deception in orchids. Naturwissenschaften 92, 255–264. [DOI] [PubMed] [Google Scholar]
- 2.Ellis AG, and Johnson SD (2010). Floral mimicry enhances pollen export: the evolution of pollination by sexual deceit outside of the Orchidaceae. The American Naturalist 176, E143–E151. [DOI] [PubMed] [Google Scholar]
- 3.Bohman B, Flematti GR, Barrow RA, Pichersky E, and Peakall R (2016). Pollination by sexual deception—it takes chemistry to work. Current Opinion in Plant Biology 32, 37–46. [DOI] [PubMed] [Google Scholar]
- 4.Johnson SD, and Midgley JJ (1997). Fly pollination of Gorteria diffusa (Asteraceae), and possible mimetic function for dark spots on the capitulum. American Journal of Botany 84, 429–436. [PubMed] [Google Scholar]
- 5.Ellis AG, and Johnson SD (2009). The evolution of floral variation without pollinator shifts in Gorteria diffusa (Asteraceae). American Journal of Botany 96, 793–801. [DOI] [PubMed] [Google Scholar]
- 6.De Jager ML, and Ellis AG (2012). Gender-specific pollinator preference for floral traits. Functional Ecology 26, 1197–1204. [Google Scholar]
- 7.Kellenberger RT, Ponraj U, Delahaie B, Fattorini R, Balk J, Lopez-Gomollon S, Muller KH, Ellis AG, and Glover BJ (2023). Multiple gene co-options underlie the rapid evolution of sexually deceptive flowers in Gorteria diffusa. Current Biology [DOI] [PubMed]
- 8.True JR, and Carroll SB (2002). Gene co-option in physiological and morphological evolution. Annual Review of Cell and Developmental Biology 18, 53–80. [DOI] [PubMed] [Google Scholar]
- 9.McQueen E, and Rebeiz M (2020). On the specificity of gene regulatory networks: How does network co-option affect subsequent evolution? . Current Topics in Developmental Biology 139, 375–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delahaie B, Mellers G, Kellenberger RT, Fernandez-Mazuecos M, Fattorini R, Brockington SF, Ellis AG, and Glover BJ (2022). The phylogenetic history of the Gorteria diffusa radiation sheds light on the origins of plant sexual deception. bioRxiv 10.1101/2022.12.22.521170. [DOI]
- 11.Davies KM, Albert NW, and Schwinn KE (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Functional Plant Biology 39, 619–638. [DOI] [PubMed] [Google Scholar]
- 12.Thomas MM, Rudall PJ, Ellis AG, Savolainen V, and Glover BJ (2009). Development of a complex floral trait: the pollinator‐attracting petal spots of the beetle daisy, Gorteria diffusa (Asteraceae). American Journal of Botany 96, 2184–2196. [DOI] [PubMed] [Google Scholar]
- 13.Xu M, Hu T, Park M-Y, Earley KW, Wu G, Yang L, and Poethig RS (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLOS Genetics 12, e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H-T, and Cosgrove DJ (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. The Plant Cell 14, 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C, Choi H-S, and Cho H-T (2011). Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Molecules and Cells, 393–397. [DOI] [PMC free article] [PubMed]
- 16.De Jager ML, and Ellis AG (2014). Costs of deception and learned resistance in deceptive interactions. Proceedings of the Royal Society B 281, 20132861. [DOI] [PMC free article] [PubMed] [Google Scholar]


