Abstract
Small round cell sarcoma is a group of undifferentiated malignancies arising in the bone and soft tissue, notable for Ewing sarcoma. Recently, a new World Health Organization classification has been introduced, including an additional subset of these sarcomas, named CIC-rearranged sarcoma. Within this group, CIC-FOXO4 translocation is an exceedingly rare fusion that has been reported only 4 times in the literature. Herein, we report in-depth the pathological, clinical, and molecular features of a CIC-FOXO4 translocation-driven tumor in a 46-year-old woman.
Keywords: Small round blue cell sarcoma, CIC-FOXO4 translocation, Transcriptome
Introduction
Small round cell sarcoma (SRCS) is a group of undifferentiated malignancies arising in bone and soft tissue. Ewing sarcoma (ES) is the most well-known malignancy in this group. ES harbors the prototypical translocation involving the ESWR1 gene with genes from the ETS family [1]. Until recently, SRCS without the ESWR1-ETS translocation was classified as a subgroup of the ES family. However, accumulating data from pathological, molecular, and clinical studies suggest that SRCS is a heterogenic group that includes entities other than ES. In the 5th edition of the WHO classification of soft tissue and bone tumors, the SRCS group was renamed as undifferentiated round cell sarcomas, with the incorporation of new subgroups, including the newly recognized entity of “CIC-rearranged sarcoma” [2].
The capicua (CIC) gene is a transcriptional repressor that regulates different receptor tyrosine kinase pathways such as the epidermal growth factor pathway [3]. CIC is evolutionary conserved and has two highly conserved domains, the high-mobility group (HMG) box and the DNA-binding domain C1. CIC activity is regulated by phosphorylation which prevents its nuclear import and weakens its ability to bind DNA. The most common fusion partner of CIC in SRCS is DUX4. In a study covering 115 cases of CIC-rearranged sarcomas, CIC-DUX4 was present in more than 50% of the tumors [4]. There are additional rare partners such as NUTM1, NUTM2A, SYK, and FOXO4 [5]. Several studies identified CIC-rearranged sarcoma as tumors with an aggressive clinical course, but these studies are significantly limited by the extreme rarity of these lesions [4, 6]. CIC-rearranged sarcomas differ from classic ES in their transcriptional landscape. Two independent studies were able to compare between CIC-rearranged tumors and ES, showing the specific upregulation of VGF, DLK1, CRH, Zic1 in CIC-rearranged tumors compared to ES [7, 8]. On the other hand, both studies demonstrated upregulation of the PEA3 transcription factor family, a previously described player in ES tumor progression and a potential fusion partner for ESWR1 [9]. These studies involving murine models are of high importance, shedding new light on molecular mechanisms and promoting understanding, which is critical for the development of targeted therapies in this uncommon entity. However, it is noteworthy that most tumors that were investigated are CIC-DUX4. Whether other translocations involving CIC act differently is still an open question.
The CIC-FOXO4 translocation is a rare occurrence, and its similarity to CIC-DUX4 tumors remains unknown due to the rarity of both tumors. This distinction is of great importance as it has implications for follow-up and treatment strategies. Currently, only four reports have been published describing this uncommon translocation. The first case was described by Yustein et al. [10]. The described patient was a 10-year-old boy presenting with a large intra-abdominal mass, bearing a t(X;19)(Q13;Q13.3) translocation. The tumor involved the liver, colon, and right hemidiaphragm. The patient underwent surgery and a complete gross excision. Imaging studies did not reveal distant metastatic disease. Histologically, the tumor was composed of small round blue cells embedded in fibrotic tissue. It is noteworthy that the translocation was found using fluorescent in situ hybridization. Sequencing of the translocation was not performed. Follow-up time and whether adjuvant therapy was administered were not reported. Sugita et al. [11] reported in 2014 a case of a 63-year-old man with an intramuscular mass in his right posterior neck, also with a t(X;19)(Q13;Q13.3) translocation. Histologically, the tumor was composed of sheets of undifferentiated small round blue cells with abundant desmoplastic fibrous stroma. The cells had irregular round nuclei, with coarse chromatin and conspicuous nucleoli. Specifically, CIC-FOXO4 translocation was identified using RNA sequencing which was further validated by Sanger sequencing of tumor DNA. Similar to the previous case, no metastatic disease was seen and therefore the patient underwent surgery followed by chemo-radiation. At a 6-month follow-up, the patient had no evidence of disease. The third case published by Solomon et al. [12] described a 13-year-old boy presenting with a scalp mass with a similar translocation. Histologically, the tumor was composed of sheets of mitotically active small round blue cells with areas of necrosis with no well-defined matrix. The cells had irregular round nuclei, with coarse chromatin and conspicuous nucleoli. The translocation was found using genomic analysis (the exact method was not reported) [12]. The patient was treated with resection followed by adjuvant chemotherapy. Eleven months after diagnosis, the disease relapsed with lung metastases. Due to his poor performance status at that point, the patient received supportive care alone. In a recent study, CIC-FOXO4 was identified in a 27-year-old male with a thigh mass [13]. Again, the diagnosis was performed using genomic sequencing without further details. This patient underwent surgical removal with involved margins followed by radiation. Then, due to disease progression (without additional details disclosed), he received three lines of chemotherapy – docetaxel with cyclophosphamide, irinotecan plus temozolomide and vincristine, doxorubicin and cyclophosphamide alternating with ifosfamide plus etoposide (VDE\IE). The progression-free survival of all chemotherapy regimens was 8.9 months combined. Yet, the full follow-up after the VDE\IE regimen was not reported. Altogether, all 4 cases were of male gender and with a limited follow-up duration.
In the present report, we describe the detailed clinical outcomes of a female patient with an aggressive tumor harboring CIC-FOXO4 translocation, with metastasis. The patient was treated with several systemic treatment lines and was followed up from diagnosis until death. In addition, we have characterized the tumor phenotypically and molecularly.
Case Presentation
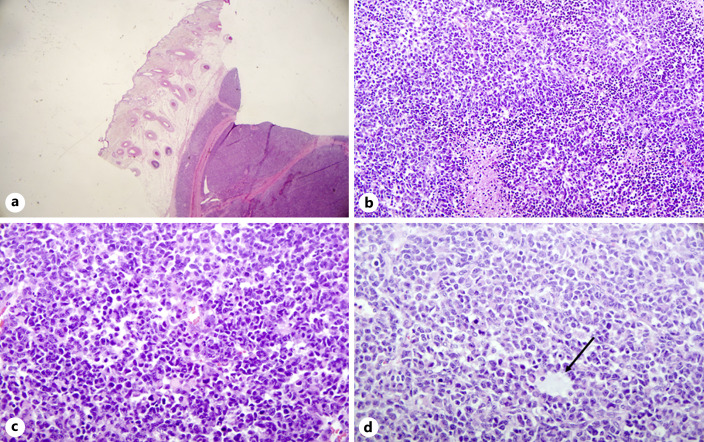
A 46-year-old female, previously healthy, presented with an oval, rapidly growing scalp lesion, with no systemic symptoms. A full-body CT scan showed a 5.5*5-cm heterogeneous hyper-vascular soft tissue lesion in the scalp without clear bone involvement. Due to the rapid growth of the lesion, she underwent a wide excision. Upon pathology examination, a 7-cm mass penetrating the bone with deep margin involvement and clear circumferential margins was seen. Microscopic examination revealed a multinodular, highly cellular neoplasm (Fig. 1a). The neoplastic cells had predominantly oval nuclei with molding and cell-cell wrapping (Fig. 1b). Rhabdoid-like cells were also seen. The neoplasm was highly vascular with scattered branching vessels and occasional staghorn-like appearance (Fig. 1c). Scattered ill-formed rosettes were also seen. There was high mitotic activity and foci of necrosis (Fig. 1d); the Ki67 proliferation index was 40%. Lymphovascular invasion was present. The immunohistochemical profile was not diagnostic; tumor cells were positive for vimentin, MAP2, BCL2, CD99 and patchy positive for panCK, TLE1, Bcl1, and FLI1. Other stains performed – CK20, CK7, EMA, CD57, BCL1, SSTR2A, STAT6, WT1, NKX2.2, BCOR, C-KIT, SATB2, TTF1, GATA3, MART1, HMB45, S-100, SOX10, CD34, CD20, CD3, CD56, INSM1, ALDH1, LIN28A, desmin, smooth muscle alpha actin, synaptophysin, chromogranin, GFAP, PAX8, ER, HER2, p53 – were all negative. BRG1 and INI-1 nuclear expression was retained.
Fig. 1.
Histologic features of CIC-FOXO4 sarcoma. Hematoxylin and eosin stain. a Multinodular subcutaneous mass. b Oval nuclei with molding, cell-cell wrapping and rhabdoid-like cells. c Highly vascular tumor with scattered branching vessels and occasional staghorn-like appearance. d Foci of necrosis and ill formed rosettes (arrow).
Two weeks post-surgery, there was a rapid local recurrence of the tumor. The patient underwent local electron radiotherapy with concomitant cisplatin treatment, followed by a wide re-excision with clear margins and partial resection of the underlying bone. Tumor tissue was submitted for next-generation sequencing (FoundationOne Heme© assay), which revealed a CIC-FOXO4 fusion. The case was discussed at a multidisciplinary tumor board which recommended adjuvant systemic treatment with vincristine, doxorubicin, and ifosfamide. Of note, the treatment was stopped after the second cycle due to infection of the surgical wound and progression to septic shock.
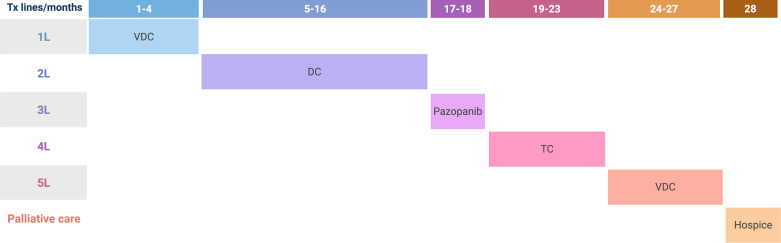
Three months later, the patient was diagnosed with three lung lesions. The patient received stereotactic radiation to the lesions and initiated systemic therapy with vincristine, doxorubicin, and cyclophosphamide (VDC). Under this treatment, the patient was stable for approximately 7 cycles (i.e., 4 months). Then, due to disease progression, treatment was changed to dacarbazine and irinotecan and SBRT to a lesion in the pelvis. During this course of treatment, the disease remained stable, and there was observed regression of the pelvic lesion. Due to repeated low blood counts, dacarbazine and irinotecan treatment was changed after a year to oral pazopanib with a daily dose of 800 mg. After 2 months of pazopanib, the disease progressed, and the treatment was switched again to a combination of topotecan and cyclophosphamide (TC). In addition, the patient received a palliative radiotherapy course to an obstructive mass in the right bronchus. On the TC regimen, the patient was clinically and radiographically stable for 5 months. Then, new multiple brain metastases were identified, a finding which warranted whole-brain radiation therapy and a change in systemic therapy to VDC. After four cycles of VDC, the patient developed a convulsive disorder secondary to the progressing brain lesions. Due to her poor performance status at that point, it was decided in common agreement with the patient and her family to switch to palliative care. The patient was admitted to an in-patient hospice and passed away a month later. Overall, the patient lived 3 years since she was diagnosed. A schematic presentation of the different treatment lines is summarized in Figure 2.
Fig. 2.
Treatment lines and timeline. A schematic representation of treatment lines. Time is represented horizontally in months. Treatment lines are shown vertically. Every line of therapy was changed due to disease progression under the previous line. L, line; VDC, vincristine, doxorubicin, and cyclophosphamide; DC, dacarbazine and irinotecan; TC, topotecan and cyclophosphamide.
Molecular Findings
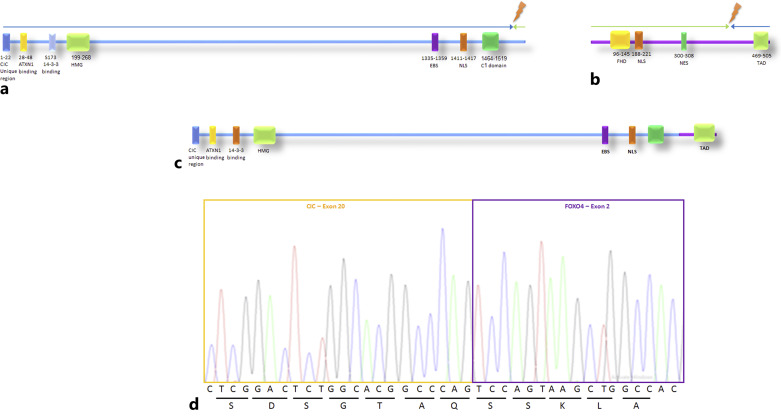
As mentioned, FoundationOne Heme© identified a CIC-FOXO4 translocation (Fig. 3a). The breakpoint is located on exon 20 of the CIC gene (NM_015125) and exons 2–3 of the FOXO4 gene (NM_005938). The chimeric gene can be identified as 5′ – CIC (ex. 1–20 UTR NM_015125 – FOXO4 (ex. 2-3 NM_005938) (Fig. 3b). Thus, the predicted protein will consist of CIC almost by its entirety at the N terminus, followed by the C terminus of FOXO4 in the protein’s C terminus. Regarding CIC, the position of the translocation does not interrupt any known functional regions of CIC such as the HMG box [3]. By contrast, the translocation does not include FOXO4 in its entirety. For instance, the predicted protein will not include the FH domain – a domain that is essential for DNA recognition.
Fig. 3.
Establishing CIC-FOXO4 fusion and its suggested protein. a Schematic representation of capicua (CIC) protein. The different domains are shown in rectangles in different colors (text below rectangles, numbers representing the position of the amino acids). The blue arrow represents the part that is included in the suggested protein. The green arrow represents the part that is lost due to the CIC-FOXO4 fusion. The lighting represents the breakpoint by which the translocation occurs. b Schematic presentation of the FOXO4 protein. The different domains are shown in rectangles in different colors (text below rectangles, numbers representing the position of the amino acids). The blue arrow represents the part that is included in the suggested protein. The green arrow represents the part that is lost due to the CIC-FOXO4 fusion. The lighting represents the breakpoint by which the translocation occurs. c Schematic presentation of the suggested fused protein. The blue line represents the part that is derived from the CIC protein and the purple line represents the part that is derived from FOXO4. d Sanger sequencing confirming CIC-FOXO4 fusion. The letters below the peaks represent the different nucleotides. The letters below the horizontal line represent the different amino acids: s – serine; d – aspartate; g – glycine; t – threonine; a – alanine; q – glutamine; k – lysine; l – leucine. ATXN1, Ataxin 1; HMG, high-mobility group; EBS, ERK-binding site; NLS, nuclear localization sequence; FHD, forkhead winged-helix DNA-binding domain; NES, nuclear export sequence; TAD, transactivation domain.
Next, we verified the results of the molecular testing using RT-PCR, followed by Sanger sequencing using the primer CIC 5′ – TGACCCCACCTCACCCAG – 3′ and FOXO 4 5′ – TTGGCTTGCAGCTTCAGTAG – 3′. We were able to show an in-frame fusion between exon 20 of CIC and exon 2 of FOXO4 (Fig. 2c).
Frozen tumor pieces were submitted for RNA extraction using a High Pure FFPET RNA Isolation Kit (Roche Diagnostics, Indianapolis, IN, USA). To gain further insight into the biology and gene expression pattern, we performed RNA sequencing. We ranked the genes according to their expression levels and performed a single-sample Gene Set Enrichment Assay (ssGSEA). The gene sets that were used were the hallmark gene sets taken from Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/). The five pathways with the highest expression are shown in Table 1 and mostly involve proliferation-related pathways. Conversely, the five pathways with the lowest expression are shown in Table 2, related to genes that are downregulated by KRAS, UV damage-related DNA repair, and immune response.
Table 1.
List of the five most positively enriched pathways and their process category
| Position | Hallmark | Process category |
|---|---|---|
| 1 | Myc targets V1 | Proliferation |
| 2 | Unfolded protein response | Pathway |
| 3 | Myc targets V2 | Proliferation |
| 4 | E2F targets | Proliferation |
| 5 | G2M checkpoint | Proliferation |
Higher positions represent pathways with higher expression. The process category was taken from the hallmark gene set found in https://www.gsea-msigdb.org/gsea/msigdb/.
Table 2.
List of the five most negatively enriched pathways and their process category
| Position | Hallmark | Process category |
|---|---|---|
| 1 | Kras signaling down | Signaling |
| 2 | UV response | DNA damage |
| 3 | Inflammatory response | Immune |
| 4 | Interferon alpha response | Immune |
| 5 | Interferon gamma response | Immune |
Higher positions represent pathways with less expression. The process category was taken from the hallmark gene set found in https://www.gsea-msigdb.org/gsea/msigdb/.
Discussion
Herein, we report a case of a young adult female with a CIC-FOXO4 translocation-driven tumor. Molecular analysis of the tumor was the key diagnostic test. Using this case, we wish to highlight and raise awareness about the novel CIC-rearranged subgroup of SRCS. A high level of suspicion is paramount, especially in highly aggressive malignancies. In a recent case series from Australia, summarizing the experience of multiple hospitals in a 5-year period (2014–2019), the 5-year overall survival of patients with CIC-rearranged sarcoma was only 34% in patients who presented with localized disease [13]. This study was in line with older studies, where the 5-year overall survival ranged from 17% to 43% [4, 6]. Additionally, CIC-rearranged tumors were shown to have a very high relapse rate despite adjuvant chemotherapy (e.g., 55% in the Australian series, at a median of 10.5 months) and have short-lived responses to various multi-agent chemotherapy regimens [13]. As mentioned earlier, most CIC-rearranged tumors primarily involve CIC-DUX4. Hence, caution should be exercised when extrapolating findings from CIC-DUX4 to CIC-FOXO4 as the differences between these tumor types are not fully understood. However, the aggressive nature of the tumor presented in this case underscores the importance of pursuing aggressive treatment strategies, like those employed for CIC-DUX4. In the current case, adjuvant chemotherapy was interrupted early due to severe infection. Given the information presented in the aforementioned studies and the notable aggressive nature of the tumor presented here, which shares similarities with CIC-DUX4, it is possible that greater attention should have been given to the continuation of adjuvant chemotherapy, despite the presence of grade 3–4 toxicity. This approach represents the only potential intervention capable of influencing the likelihood of a fatal recurrence of the disease. In line with this aggressive clinical phenotype, the transcriptional landscape of the tumor described here showed that like in other CIC-rearranged sarcomas, proliferation-related pathways’ expression was enriched. Interestingly, a recent report discovered that Wee1 inhibition in CIC-DUX4 tumors led to increased apoptosis and reduced tumor growth in both in vitro and in vivo models [14]. Wee1 is a key cell cycle kinase that regulates CDK1 and CDK2, both essential for tumor cells’ mitotic progression. Furthermore, CCNE1, a known CDK2 interactor, was reported to be upregulated in clinical samples of CIC-DUX4 tumors [7]. In our case, CCNE1, CCND1, Wee1, CDK1, and CDK2 were also highly upregulated (higher than the 85th percentile – data not shown).
It is noteworthy that in the previously described cases, the translocation involved similar regions of both proteins. In Sugita’s report [11], the breakpoint was in exon 19 of CIC, whereas in Solomon’s report [12] and in our report, the breakpoint is in exon 20. This observation might suggest that the functional domains of the CIC protein are essential for tumorigenesis. On the other hand, the breakpoint on FOXO4 is exon 2, which leads to a loss of most of the functional domains within FOXO4. This finding implies that FOXO4 may be a less essential factor of tumorigenesis. Interestingly, in additional CIC-rearranged sarcomas which underwent molecular characterization, CIC rearrangement occurs on exon 19 or 20 [5]. Since CIC acts as a transcriptional repressor [3] and the fused protein in CIC-rearranged sarcoma functions as a transcription activator [5], it is plausible that the C terminus of the CIC protein plays a crucial role in transcriptional suppression. Further studies could provide insights into the specific roles of different domains in the CIC protein and their significance in the exact mechanism of tumorigenesis in these malignancies.
An additional important question to consider is whether different CIC rearrangements have varying outcomes. On one hand, it could be assumed that since all tumors involve the CIC gene, the biology, molecular landscape, and clinical course might be similar across these tumors. However, these rearrangements also involve another gene, which could potentially impact the molecular and clinical outcomes. Additionally, it is well-established that ESWR1 fusions not only lead to varying clinical outcomes but also give rise to different types of tumors, such as ES, clear cell sarcoma of soft tissue, desmoplastic small round cell tumor, and several others [15]. Further studies utilizing preclinical models and analyzing patient outcomes are needed to gain a deeper understanding of this question.
In conclusion, to the best of our knowledge, this is the first CIC-FOXO4-rearranged sarcoma female patient with a detailed follow-up for the entire disease course. Future studies will help delineate the similarities and differences between the different CIC rearrangements, paving the way to a better treatment of these aggressive malignancies. The CARE checklist has been completed by the authors for this case report, attached as online supplementary material (for all online suppl. material, see https://doi.org/10.1159/000533519 for details).
Acknowledgments
We are deeply grateful to I.D. and her family for their support of this report. May her memory be a blessing.
We thank Dr. Sharona Elgavish, Dr. Yuval Nevo, Dr. Hadar Benyamini, and Shmuel Ruppo from the I-CORE Bioinformatics Unit of the Hebrew University of Jerusalem for ssGSEA data analysis.
Statement of Ethics
Written informed consent was obtained from the patient for the publication of the details of their medical case and any accompanying images prior to their passing away.
The Ethics Committee of Hadassah Medical Center approved this study (protocol 346-12).
Conflict of Interest Statement
The authors declare that they have no conflicts of interest with the contents of this report.
Funding Sources
This research received no specific funding from any funding agency in the public, commercial, or not-for-profit sectors funding sections.
Author Contributions
Concept and design: Drs. Aryeh Babkoff and Albert Grinshpun. Data acquisition, analysis, and interpretation: Drs. Aryeh Babkoff, Yael Berner-Wygoda, Judith Diment, Anatoli Kustanovich, Aviad Zick, Daniela Katz, and Albert Grinshpun. Writing: Drs. Aryeh Babkoff, Albert Grinshpun, Aviad Zick, and Judith Diment. Review and approval of the manuscript: Drs. Aryeh Babkoff, Yael Berner-Wygoda, Judith Diment, Anatoli Kustanovich, Aviad Zick, Daniela Katz, and Albert Grinshpun.
Funding Statement
This research received no specific funding from any funding agency in the public, commercial, or not-for-profit sectors funding sections.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author.
Supplementary Material
References
- 1. Sbaraglia M, Righi A, Gambarotti M, Dei Tos AP. Ewing sarcoma and Ewing-like tumors. Virchows Arch. 2020;476(1):109–19. 10.1007/s00428-019-02720-8. [DOI] [PubMed] [Google Scholar]
- 2. Sbaraglia M, Bellan E, Dei Tos AP. The 2020 WHO classification of soft tissue tumours: news and perspectives. Pathologica. 2021;113(2):70–84. 10.32074/1591-951X-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee Y. Regulation and function of capicua in mammals. Exp Mol Med. 2020;52(4):531–7. 10.1038/s12276-020-0411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antonescu CR, Owosho AA, Zhang L, Chen S, Deniz K, Huryn JM, et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a clinicopathologic and molecular study of 115 cases. Am J Surg Pathol. 2017;41(7):941–9. 10.1097/PAS.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Linos K, Dermawan JK, Bale T, Rosenblum MK, Singer S, Tap W, et al. Expanding the molecular diversity of CIC-rearranged sarcomas with novel and very rare partners. Mod Pathol. 2023 May;36(5):100103. 10.1016/j.modpat.2023.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida A, Goto K, Kodaira M, Kobayashi E, Kawamoto H, Mori T, et al. CIC-rearranged sarcomas: a study of 20 cases and comparisons with Ewing sarcomas. Am J Surg Pathol. 2016;40(3):313–23. 10.1097/PAS.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 7. Specht K, Sung YS, Zhang L, Richter GHS, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 2014;53(7):622–33. 10.1002/gcc.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshimoto T, Tanaka M, Homme M, Yamazaki Y, Takazawa Y, Antonescu CR, et al. CIC-DUX4 induces small round cell sarcomas distinct from ewing sarcoma. Cancer Res. 2017;77(11):2927–37. 10.1158/0008-5472.CAN-16-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qi T, Qu Q, Li G, Wang J, Zhu H, Yang Z, et al. Function and regulation of the PEA3 subfamily of ETS transcription factors in cancer; 2020. Available from: www.ajcr.us/. [PMC free article] [PubMed] [Google Scholar]
- 10. Yustein JT, Rednam S, Bertuch AA, Goss JA, Brandt ML, Eldin K, et al. Abdominal undifferentiated small round cell tumor with unique translocation (X;19)(q13;q13.3). Pediatr Blood Cancer. 2010;54(7):1041–4. 10.1002/pbc.22437. [DOI] [PubMed] [Google Scholar]
- 11. Sugita S, Arai Y, Tonooka A, Hama N, Totoki Y, Fujii T, et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol. 2014;38(11):1571–6. 10.1097/PAS.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 12. Solomon DA, Brohl AS, Khan J, Miettinen M. Clinicopathologic features of a second patient with ewing-like sarcoma harboring CIC-FOXO4 gene fusion. Am J Surg Pathol. 2014;38(12):1724–5. 10.1097/PAS.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connolly EA, Bhadri VA, Wake J, Ingley KM, Lewin J, Bae S, et al. Systemic treatments and outcomes in CIC-rearranged Sarcoma: a national multi-centre clinicopathological series and literature review. Cancer Med. 2022 Apr;11(8):1805–16. 10.1002/cam4.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ponce RKM, Thomas NJ, Bui NQ, Kondo T, Okimoto RA. WEE1 kinase is a therapeutic vulnerability in CIC-DUX4 undifferentiated sarcoma. JCI Insight. 2022;7(6):e152293. 10.1172/jci.insight.152293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romeo S, Dei Tos AP. Soft tissue tumors associated with EWSR1 translocation. Virchows Arch. 2010 Feb;456(2):219–34. 10.1007/s00428-009-0854-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author.