Abstract
Alopecia areata (AA) is an autoimmune form of non-scarring hair loss that occurs on a spectrum from patchy loss of hair on the scalp, to complete hair loss. Histology features can vary, but increased abundance of telogen hair and miniaturized hair follicles are classic hallmarks [Clin Cosmet Investig Dermatol. 2015;8:397–403]. Additionally, lymphocytic infiltration of the hair bulb is a commonly observed histology feature of AA which underscores how the disease is an autoimmune-mediated one that results from immune-mediated attack of the hair follicle. In a healthy individual, the hair follicle is one of the body’s immune-privileged sites, but the breakdown of this immune privilege is thought to be an important driver in AA disease development. Diagnosis of AA is usually based on phenotypic manifestations in conjunction with biopsies which can help conclude whether the hair loss is autoimmune based. However, varied manifestation of disease both clinically and histologically makes diagnosis criteria more ambiguous and early identification of disease harder to achieve. A better understanding of genes that are associated with increased AA risk may help elucidate potential gene targets for future therapeutics.
Keywords: General dermatology, Genodermatoses, Alopecia, Hair loss, Immunodeficiency, Immune mediated disease, Inborn errors of immunity
Introduction
Alopecia areata (AA) is an autoimmune disease with a lifetime risk of 1.7–2% globally [1].AA phenotypically manifests as non-scarring hair loss that occurs on a spectrum of severity, with some patients experiencing limited patchy loss in hair-bearing areas and some losing all of the hair on their head and body. The hair follicle is an immune privileged site which limits local immune and inflammatory responses, but in AA, this privilege is lost. The pathogenesis is largely the result of an interferon gamma driven immune response excellent efficacy. We know other mechanisms may play roles in subsets of patients (especially those who do not respond to interferon gamma blockade). Here, we review available data on genetic risk variants identified through single gene studies and genome-wide association studies (GWASs). We also highlight the Mendelian diseases associated with specific gene variants in which AA can be seen. We aim to provide a background of genetic factors related to AA that may lead to improved understanding of disease subsets and perhaps lead to alternate therapeutics.
Genetic Underpinnings of Disease Development
Inheritance of genetic diseases can be categorized as monogenic, chromosomal, or polygenic/multifactorial. There are 5,000–8,000 monogenic diseases that follow simple Mendelian inheritance where pathogenesis is linked to a single gene [2]. Even for monogenic diseases, gene expression can be influenced by the internal and external environment (epigenetics) [3]. Most diseases are polygenic, where pathogenesis is influenced by multiple genes. While AA is thought to be primarily polygenic like other autoimmune diseases, there are examples of single genes leading to the disease. Single gene studies highlight the monogenic variants and GWASs analyze hundreds of thousands of genetic variants (single nucleotide variants, SNPs) across many genomes to find those statistically associated with a specific trait or disease. Genome-wide analysis of copy number variants (CNVs) has shown additional contributions to AA.
Single Gene Studies
Most single genes associated with AA are immune related and involved in CD4+ regulatory T cell (Treg) function, CD8+ T cell effector function and antigen presentation, and natural killer (NK) cell defense. These genes include forkhead box P3 (FOXP3), inducible T cell co-stimulator ligand (ICOSLG), MHC class I polypeptide-related sequence A (MICA), macrophage migration inhibitory factor (MIF), human leukocyte antigens (HLA) subtypes, interleukin-7 receptor subunit alpha (IL7RA), interleukin 1 receptor antagonist (IL1RN), autoimmune regulator (AIRE), keratin 82 (KRT82), suppressor of cytokine signaling 1 (SOCS1), nuclear factor kappa B subunit 1 (NFKB), and recombinase activating genes (RAG). The majority of these genes have been studied in direct relationship to AA through either serum or lesional skin analysis (see Table 1). We highlight how immune dysregulation is intimately associated with autoimmune disease. Table 1 provides an overview of these genes, their function, and related comorbidities.
Table 1.
Genes identified through single-gene studies
| Gene | Function | Method of evaluation | Implication in other autoimmune conditions |
|---|---|---|---|
| FOXP3 | Transcription factor responsible for regulating T-reg cells | Human lesional AA skin analysis (Conteduca et al. [4], 2014; Ben Hmid et al. [5], 2015) | Thyroiditis, hypothyroidism, autoimmune hemolytic anemia, recurrent infections, and membranous nephropathy |
| ICOSLG | Modifies T-reg cell function | Human serum analysis (Conteduca et al. [4], 2014) | Vitiligo, inflammatory bowel disease, celiac disease, and autoimmune thyroid disease |
| MICA | NK and CD8+ cell activation | Human lesional AA skin and serum analysis (Ito et al. [6], 2008; Mingorance et al. [7], 2020) | Type 1 diabetes mellitus, ankylosing spondylitis, and Behçet's disease |
| MIF | Inhibits NK cells in immune-privileged areas | Human lesional AA skin and serum analysis (Salem et al. [8], 2016; Eldesouky et al. [9], 2020; Younan et al. [10], 2015) | Systemic juvenile rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, uveitis, psoriasis, Wegener’s granulomatosis, and systemic sclerosis |
| IL7RA | Transduces signals from immune cells | Mouse lesional AA skin analysis (Dai et al. [11], 2021 ) | Diabetes, Sjögren’s syndrome, rheumatoid arthritis, and multiple sclerosis |
| IL1RN | Prevents IL-1 signaling | Human serum AA analysis (Tarlow et al. [12], 1994) | Systemic lupus erythematosus, ulcerative colitis, and Crohn’s disease |
| AIRE | Eliminates self-reactive T cells | Human serum AA analysis (Tazi-Ahnini et al. [13], 2002) | Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy |
| KRT82 | Hair-specific type II keratin exclusively expressed during anagen phase of hair follicle cycle | Human serum AA analysis (Erjavec et al. [14], 2022) | Only found in alopecia areata |
| SOCS1 | Suppresses type I interferon function/signaling by inhibition of the JAK-STAT pathway | Clinical series | Thyroiditis, celiac disease, psoriasis, spondyloarthritis, hepatitis, systemic lupus erythematosus |
| NFKB | Transcription factor involved in inflammatory immune response, cell growth, and development | Clinical series | Inflammatory bowel disease, rheumatoid arthritis, psoriasis, and multiple sclerosis |
| RAG | Involved in V(D)J recombination to generate mature T and B cells | Clinical series | Autoimmune hemolytic anemia and neutropenia, vitiligo, vasculitis, autoimmune neuropathy, and myopathy |
Forkhead Box P3
FOXP3 encodes for a protein responsible for suppressing Tregs to ensure self-tolerance. AA has been associated with fewer subpopulations of CD39+ Treg [2]. Specific variants of FOXP3 (rs2294020-3675) which lead to lower expression and fewer FOXP3+ Tregs have been found in increased frequencies in AA patients compared to controls [4, 5]. Lower expression of FOXP3 has also been found in other autoimmune disease [2]. [1] Immunodysregulation, polyendocrinopathy, enteropathy, X-linked syndrome is a rare disorder of the immune system caused by pathogenic variants in the FOXP3 gene resulting in defective development of CD4+CD25+ Tregs (OMIM number 304790). In addition to autoimmune thyroid disease and hemolytic anemia, recurrent infections, and membranous nephropathy, some patients develop AA [15].
Inducible T-Cell Costimulator Ligand
ICOSLG and its ligand are critical for activation, proliferation, differentiation, and cytokine production of T cells and for antibody secretion from B cells during secondary immune responses. Polymorphisms in ICOSLG gene (rs378299) have been identified in higher frequency in AA especially in combination with HLADQB1*03 [4, 15]. Immunodeficiency, common variable, 1 CVID1 (OMIM 607594) is an autosomal recessive disorder (AR) with pathogenic gene variants in ICOS. CVID is the most common form of primary immunodeficiency and primary antibody deficiency. Severe AA as well as vitiligo has been reported in patients with CVID1 [16].
MHC Class I Polypeptide-Related Sequence A
MICA is an antigen that is induced by oxidative stress, typically on cells from epithelial origin. When expressed, MICA stimulates the NKG2D receptor on NK and CD8+ T cells, activating them against the stressed cell [6, 17]. In one small study, there was strongly increased MICA expression found in lesioned AA skin [2]. Specific alleles including MICA*009 suggest a protective role [7]. A significant positive correlation between MICA*005.1 and patchy AA and MICA*006 and AA has been reported [18]. GWASs have evaluated the single nucleotide polymorphisms of MICA association with AA (discussed below). Autoimmune Polyendocrine Syndrome Type 2 (OMIM 269200) is an autosomal dominant disorder [19]. This syndrome often presents with AA, as well as autoimmune Addison’s disease, thyroid disease, and diabetes mellitus. Autoimmune polyendocrine syndrome type 2 related Addison’s disease has been associated with the MICA 5.1 and MICB-CA-25 alleles and combination of the MICA 5.1 allele and DRB1*03-DQA1*0501-DQB1*0201 haplotypes may increase risk for Addison disease [20]. MICA variants are also reported in type 1 diabetes, ankylosing spondylitis, and Behçet’s disease [21–24].
Macrophage Migration Inhibitory Factor
MIF is an immunoregulatory cytokine which inhibits NK cells in immune-privileged areas. There have been conflicting studies about whether it is elevated or decreased in serum and tissue of patients with AA [6]. Several studies have found upregulated MIF levels (correlating with severity of disease) in AA serum and lesional skin [8–10]. A lower frequency of the C allele or rs755622 SNP has been found among AA patients in one study but another larger study disputes this [25]. Certain MIF173*C alleles may be a risk factor for early onset AA [26]. MIF has also been implicated in systemic juvenile rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, and uveitis [27, 28].
HLA Subtypes
HLAs are protein markers responsible for presenting antigens to T cells and are classified as either subclass I or subclass I. Subclass molecules I are located on all nucleated cells and are encoded by HLA-A, HLA-B, and HLA-C alleles. Subclass II molecules are located on antigen-presenting cells and are encoded by HLA-DP, HLA-DQ, or HLA-DR alleles. HLA subclass II types, HLA-DQB1*0502 allele, HLA-DQB1*0301 allele, and in combination with HLA-DQB1*0301-HLA-DQB1*0303 have been associated with AA [29]. Combined HLA-DQB1*03 alleles are associated with increased disease severity as were DRB1*16 alleles. HLA-DQB1*0603, DRB1*03, DRB1*13, and DRB1*15 were decreased amongst AA patients [30]. GWASs HLA alleles with AA will also be discussed below. HLA haplotypes have been associated with celiac disease, ankylosing spondylitis, psoriatic arthritis, multiple sclerosis, type 1 diabetes mellitus, system lupus erythematosus [30].
Interleukin-7-Receptor Subunit Alpha
IL7RA encodes a receptor on the cell surface of lymphocytes responsible for transducing signals from immune cells. It signals through JAK-STAT signaling and is key for VDJ recombination seen in T and B cells [31–33]. IL-7 has been shown to play a direct role in mouse models of AA by upregulating cytotoxic T cells. Blocking IL7RA in this model led to reduced cytotoxic T cells (not Tregs) in lesional skin [11]. IL7RA has also been linked type 1 diabetes, Sjögren’s syndrome, rheumatoid arthritis, and multiple sclerosis [11].
Interleukin-1 Receptor Antagonist
IL1RN encodes the interleukin-1 receptor antagonist (IL-1RA; IL-1RA dampens excessive inflammatory response) [34]. A polymorphism in IL1RN, known as allele 2, has been associated with increased severity of AA [12]. Allele 2 for IL1RN is associated with other inflammatory autoimmune diseases including systemic lupus erythematosus, ulcerative colitis, and Crohn’s [35].
Autoimmune Regulator
AIRE encodes the autoimmune regulator protein which typically functions to eliminate self-reactive T cells as they mature in the thymus. It is associated with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. The three main features associated with this disease include recurrent fungal infection by candida, adrenal gland insufficiency, and hypoparathyroidism. AA is a common comorbidity condition in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy with 30% of patients developing it [13]. However, not all variants in AIRE resulted in the same risk or severity of AA. One study identified 20 variants in a cohort of 202 AA patients and 175 matched controls and found variants at position G961C were associated with a >3 risk factor for alopecia universalis and early disease onset [13].
Keratin 82
Keratin 82 is a keratin that is specific to the hair shaft. Keratins are proteins that make up the fibrous structure of hair, nails, and outer layer of skin. These proteins can be divided into two classes: type I and type II, of which KRT82 is classified as. Type II keratins are larger and have been previously associated with other hair-related genetic disorders including ectodermal dysplasia, pseudofolliculitis barbae, and Monilethrix, unlike Type I keratins [14]. KRT82 is exclusively expressed during the anagen (active growth) phase of the hair follicle life cycle, which is extremely relevant in the context of AA since the autoimmune attack that is evident in active disease flares is also exclusive to the anagen phase [14]. Eight hundred and forty-nine patients with AA and 15,640 controls were analyzed with whole-exome sequencing In 6% of patients with AA, KRT82 was identified as an AA risk gene due to damaging variants with notable significance (p = 2.18E-7) [14]. A negative correlation between KRT82 expression and CD8+ infiltrates was also identified.
Suppressor of Cytokine Signaling 1
SOCS1 encodes the suppressor of cytokine signaling 1 protein and is a member of the STAT-induced JAK inhibitors. SOCS1 works to prevent constitutive activation of the JAK-STAT pathway [36]. Clinical phenotype analysis of newly described early onset autoimmunity with SOCS1 deficiency shows AA is one autoimmune manifestation and unpublished data (L.C.S.) has identified multiple family members with a SOCS1 pathogenic variant and AA/universalis as the primary phenotypic manifestation.
Nuclear Factor Kappa B
NFKB is a family of transcription factors that are implicated in inflammatory signaling cascade, immune regulation, and cell proliferation and differentiation [37, 38]. As such, NFKB is responsible for the expression of various pro-inflammatory cytokines, chemokines, and adhesion molecules. B-cell deficiency due to NFKB is associated with AA. Additionally, NFKB signaling has further been implicated asthma, inflammatory bowel disease, rheumatoid arthritis, psoriasis [39].
Recombinase Activating Genes
Recombinase activating genes (RAG), such as RAG1 and RAG2, are responsible for V(D)J recombination, which determines T- and B-cell antigen receptor diversity [40]. Mutations or deficiencies in RAG1 or RAG2 have serious immune effects, including leading to the development of severe combined immunodeficiency disorders (SCID) and Omenn syndrome (OMIM:603554). SCID can often present with lymphocytopenia or absent immunoglobulins, T-cell proliferation and function defects, and thymic dysfunction. Clinically AA can be part of both SCID and Omenn syndromes.
Alopecia Genome Wide-Association Studies
Two GWASs have identified risk variants in genetic loci in AA. The first (2010) included 1,054 North American patients and 3,278 controls [41]. Results yielded 139 SNPs spread across eight loci which included cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), interleukin-2 receptor alpha (IL2RA), zinc finger protein Eos (IKZF4), UL16 binding protein 3/6 (ULBP3/ULBP6), interleukin-2/21 (IL-2/IL-21), HLA subtypessyntaxin 17 (STX17), and peroxiredoxin 5 (PRDX5) [41]. A follow-up study (2012) included 1,702 Central European patients and 1,723 controls confirmed 5 and tested 12 additional ones [42]. [3] The identified susceptibility loci were subdivided into those pertaining to the immune system, those pertaining to the hair follicle, and “other.” The eight genes to be discussed next are the ones found to have the strongest association with AA.
Analyses of copy number variants (CNVs), the number of copies of a given gene an individual possesses, have also been included in GWAS [43]. A study including 16,984 AA patients and controls showed that fourteen genes had varied expression due to CNVs [43].
Immune System Related Genes
It is not a surprise that most of the susceptible loci identified in the first AA GWAS are genes that play a role in leukocyte activity.
Cytotoxic T-Lymphocyte-Associated Protein 4
CTLA-4 encodes cytotoxic T-lymphocyte-associated protein 4 which is highly expressed in Tregs, a subpopulation of T cells that suppress the immune response and reduce inflammation and allow tissue to remain self-tolerant [44]. In the GWAS, CTLA4 was observed with the highest correlation with AA [41]. This is not surprising to those who care for with monogenic disease from CLTA4 deficiency which is marked by autoimmune disease (type-1 diabetes, rheumatoid arthritis, multiple sclerosis, and systemic lupus erythematosus and AA), respiratory infections, and intestinal diseases [45].
Interleukin-2 Receptor Subunit Alpha
IL2RA is a protein receptor present on T cells and B cells. Tregs constitutively express IL2RA, and therefore IL2RA plays a key role in T cell regulation and preventing autoimmunity by immune tolerance. The 10p15.1 region of this gene was found to have the strongest association with AA [41]. Similar to CTLA-4, this gene is also implicated in autoimmune disease, including type-1 diabetes, multiple sclerosis, and Guillain-Barré syndrome [41].
Zinc Finger Protein Eos
IKZF4 is the gene that encodes the zinc finger protein Eos, which is a transcription factor expressed in Tregs. Eos Knock out mice exhibited decreased Treg function and develops autoimmune disease [46]. The 12q13 region on the IKZF4 gene has been the most strongly associated with AA [41]. The follow-up GWAS also confirmed this association [42].
UL16 Binding Protein 3/6
The family of ULBP genes encodes six distinct proteins and is the ligands which bind to NKG2D receptors of NK cells. Ligand binding to NKG2D triggers cytotoxic mediated effects including cytokine secretion and targeted cell death through cytotoxic granule release. It had been previously found that the two classes of NKG2D ligands (MICA/B and ULBP1-6) both have varying degree of polymorphisms which is associated with a higher risk of autoimmunity and ULBP6 was found to be the most polymorphic [47]. In both GWASs, ULBP6 gene was found to have an extremely strong association with AA [41, 42]. ULBP3 was also found to have a strong association with AA [41, 42]. Among all the loci studied in the European GWAS, the strongest association was in the ULBP genes [42].
Interleukin-2/21
IL2/21 encodes protein interleukin-2/21, which binds to IL-2 receptors on lymphocytes. IL-2 is involved in the body’s ability to recognize self from non-self. Therefore, it is not a surprise that both GWASs found this gene to be another SNP target that has an association with AA [41, 42]. It is also found in GWASs of type 1 diabetes, rheumatoid arthritis, Crohn disease [41].
HLA Subtypes
HLA encodes HLA proteins present on leukocytes and recognized by T-cells to help distinguish between self from non-self. The North American GWAS identified correlation between HLA-DRA, HLA-DQA1, HLA-DQA2, HLA-DQB2, and AA [41]. The European GWAS did not further confirm the HLA findings due to initial GWAS producing convincing and definitive results.
Hair Related Genes
Syntaxin 17
STX17 has high correlation with AA in the GWASs. It is expressed within the hair follicle itself, and it had been previously found to cause graying of hair/mane in horses [48]. AA appears to preferentially attack pigmented hair, and one study found that it preferentially attacks dark hair [49]. Whole genome sequencing in 849 AA patients and 15,640 controls identified KRT82 as an associated gene, with 51 of the AA patients who were heterozygous exhibiting damaging variants for this gene [14]. KRT82 encodes a keratin protein that is exclusively expressed in the hair shaft during anagen. In AA, hair follicles experience attack in this phase of hair growth, and patients are found to have decreased KRT82 expression [14].
Other
Peroxiredoxin 5
PRDX5 encodes an antioxidant enzyme which was found to have a high correlation with AA in GWASs. Overexpression of PRDX5 reduces cell death from toxic peroxide and under expression renders cells susceptible to oxidative damage [50]. Lipid peroxidation occurs when toxic oxidants degrade lipids such as those present in cell membranes. Lipid peroxides release a break-down product known as malondialdehyde (MDA), and patients with AA have been shown to have higher serum levels of MDA than controls and levels increase with disease length and severity [51].
Conclusion
Single gene and GWASs show that largely immune genes have been correlated with AA like what has been observed in other autoimmune diseases (Fig. 1). Many of these genes have also been implicated in innate errors of immunity where AA is often part of the phenotype. This suggests we may learn more about pathogenesis of AA and potential new targets for therapeutics from these monogenic diseases (Tables 1, 2). Additional genes identified in hair specific genes and oxidative stress-related genes may provide alternative targets for patients who do not respond to immune mediated therapies. These types of targets may also be considered in the future for combination therapy or maintenance therapy.
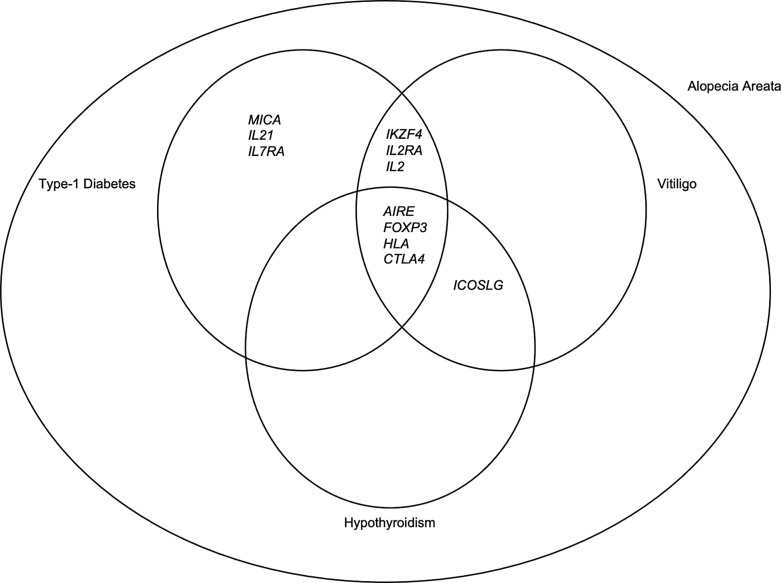
Fig. 1.
This diagram depicts an overview of the overlap of genes implicated in the pathogenesis of alopecia areata, type-1 diabetes, vitiligo, and hypothyroidism.
Table 2.
| Gene | Function | Implication in other autoimmune conditions |
|---|---|---|
| CTLA4 | T-reg activation | Type-1 diabetes, rheumatoid arthritis, multiple sclerosis, systemic lupus erythematosus |
| IL-2RA | Immune tolerance/T-cell proliferation/differentiation | Type-1 diabetes, multiple sclerosis, Guillain-Barré syndrome |
| Eos (IKZF4) | T-reg regulation | Type-1 diabetes, systemic lupus erythematosus |
| ULBP6/ULBP3 | NK cell activation | Only found in alopecia areata |
| IL2/IL21 | Self-tolerance | Type-1 diabetes, rheumatoid arthritis, Crohn's disease, psoriasis |
| HLA | Antigen presentation | Type-1 diabetes, rheumatoid arthritis, celiac disease, multiple sclerosis |
| STX17 | Hair pigmentation | Only found in alopecia areata |
| PRDX5 | Antioxidant defense | Multiple sclerosis |
Conflict of Interest Statement
B.P. and H.E. have no conflicts of interest to declare. L.C.S. has in the past received Honorarium and grant from Pfizer (relationship has ended); Medical Advisory Board for the National Alopecia Areata Foundation (volunteer position).
Funding Sources
L.C.S. and H.E. salary and time were supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. B.P. received a mentorship grant from the American Hair Research Society and this work is part of the mentorship collaboration between B.P. and L.C.S.
Author Contributions
B.P., H.E., and L.C.S. made substantial contributions to concept of the work, all provided final approval of version to be published, and drafted the work. L.C.S. revised it critically for important intellectual content and agreed to be accountable for all aspects of the work in ensuring that questions related to accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding Statement
L.C.S. and H.E. salary and time were supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. B.P. received a mentorship grant from the American Hair Research Society and this work is part of the mentorship collaboration between B.P. and L.C.S.
References
- 1. Villasante Fricke AC, Miteva M. Epidemiology and burden of alopecia areata: a systematic review. Clin Cosmet Investig Dermatol. 2015;8:397–403. 10.2147/CCID.S53985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hamed FN, Astrand A, Bertolini M, Rossi A, Maleki-Dizaji A, Messenger AG, et al. Alopecia areata patients show deficiency of FOXP3+CD39+ T regulatory cells and clonotypic restriction of Treg TCRβ-chain, which highlights the immunopathological aspect of the disease. PLoS One. 2019;14(7):e0210308. 10.1371/journal.pone.0210308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tukker AM, Royal CD, Bowman AB, McAllister KA. The impact of environmental factors on monogenic mendelian diseases. Toxicol Sci. 2021;181(1):3–12. 10.1093/toxsci/kfab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conteduca G, Rossi A, Megiorni F, Parodi A, Ferrera F, Tardito S, et al. Single nucleotide polymorphisms in the promoter regions of Foxp3 and ICOSLG genes are associated with Alopecia areata. Clin Exp Med. 2014;14(1):91–7. 10.1007/s10238-012-0224-3. [DOI] [PubMed] [Google Scholar]
- 5. Ben Hmid A, Belhadj Hmida N, Abdeladhim M, Ben Osman A, Louzir H, Mokni M, et al. FOXP3 transcription is enhanced in lesional and perilesional skin of patients with focal Alopecia areata. Int J Dermatol. 2015;54(8):e319–21. 10.1111/ijd.12857. [DOI] [PubMed] [Google Scholar]
- 6. Ito T, Ito N, Saatoff M, Hashizume H, Fukamizu H, Nickoloff BJ, et al. Maintenance of hair follicle immune privilege is linked to prevention of NK cell attack. J Invest Dermatol. 2008;128(5):1196–206. 10.1038/sj.jid.5701183. [DOI] [PubMed] [Google Scholar]
- 7. Mingorance Gamez CG, Martinez Chamorro A, Moreno Casares AM, Tercedor Sanchez J, Arias-Santiago S, Garcia-Lora E, et al. Joint study of the associations of HLA-B and the transmembrane short tandem repeat polymorphism of MICA protein with alopecia areata shows independent associations of both with the disease. Clin Exp Dermatol. 2020;45(6):699–704. 10.1111/ced.14208. [DOI] [PubMed] [Google Scholar]
- 8. Salem SA, Asaad MK, Elsayed SB, Sehsah HM. Evaluation of macrophage Migration Inhibitory Factor (MIF) levels in serum and lesional skin of patients with alopecia areata. Int J Dermatol. 2016;55(12):1357–61. 10.1111/ijd.13344. [DOI] [PubMed] [Google Scholar]
- 9. Eldesouky F, Ibrahim ASM, Sharaf SM. Macrophage migration inhibitory factor in alopecia areata and vitiligo: a case-controlled serological study. J Clin Aesthet Dermatol. 2020;13(10):24–7. [PMC free article] [PubMed] [Google Scholar]
- 10. Younan DN, Agamia N, Elshafei A, Ebeid N. Serum level of macrophage Migration Inhibitory Factor (MIF) in Egyptians with alopecia areata and its relation to the clinical severity of the disease. J Clin Lab Anal. 2015;29(1):74–9. 10.1002/jcla.21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dai Z, Wang EHC, Petukhova L, Chang Y, Lee EY, Christiano AM. Blockade of IL-7 signaling suppresses inflammatory responses and reverses alopecia areata in C3H/HeJ mice. Sci Adv. 2021;7(14):eabd1866. 10.1126/sciadv.abd1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarlow JK, Clay FE, Cork MJ, Blakemore AI, McDonagh AJ, Messenger AG, et al. Severity of alopecia areata is associated with a polymorphism in the interleukin-1 receptor antagonist gene. J Invest Dermatol. 1994;103(3):387–90. 10.1111/1523-1747.ep12395398. [DOI] [PubMed] [Google Scholar]
- 13. Tazi-Ahnini R, Cork MJ, Gawkrodger DJ, Birch MP, Wengraf D, McDonagh AJ, et al. Role of the Autoimmune Regulator (AIRE) gene in alopecia areata: strong association of a potentially functional AIRE polymorphism with alopecia universalis. Tissue Antigens. 2002;60(6):489–95. 10.1034/j.1399-0039.2002.600604.x. [DOI] [PubMed] [Google Scholar]
- 14. Erjavec SO, Gelfman S, Abdelaziz AR, Lee EY, Monga I, Alkelai A, et al. Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82. Nat Commun. 2022;13(1):800. 10.1038/s41467-022-28343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nieves DS, Phipps RP, Pollock SJ, Ochs HD, Zhu Q, Scott GA, et al. Dermatologic and immunologic findings in the immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Arch Dermatol. 2004;140(4):466–72. 10.1001/archderm.140.4.466. [DOI] [PubMed] [Google Scholar]
- 16. Agarwal S, Cunningham-Rundles C. Autoimmunity in common variable immunodeficiency. Curr Allergy Asthma Rep. 2009;9(5):347–52. 10.1007/s11882-009-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajabi F, Drake LA, Senna MM, Rezaei N. Alopecia areata: a review of disease pathogenesis. Br J Dermatol. 2018;179(5):1033–48. 10.1111/bjd.16808. [DOI] [PubMed] [Google Scholar]
- 18. Barahmani N, de Andrade M, Slusser JP, Zhang Q, Duvic M. Major histocompatibility complex class I chain-related gene A polymorphisms and extended haplotypes are associated with familial alopecia areata. J Invest Dermatol. 2006;126(1):74–8. 10.1038/sj.jid.5700009. [DOI] [PubMed] [Google Scholar]
- 19. Betterle C, Dal Pra C, Mantero F, Zanchetta R. Autoimmune adrenal insufficiency and autoimmune polyendocrine syndromes: autoantibodies, autoantigens, and their applicability in diagnosis and disease prediction. Endocr Rev. 2002;23(3):327–64. 10.1210/edrv.23.3.0466. [DOI] [PubMed] [Google Scholar]
- 20. Gambelunghe G, Falorni A, Ghaderi M, Laureti S, Tortoioli C, Santeusanio F, et al. Microsatellite polymorphism of the MHC class I chain-related (MIC-A and MIC-B) genes marks the risk for autoimmune Addison's disease. J Clin Endocrinol Metab. 1999;84(10):3701–7. 10.1210/jcem.84.10.6069. [DOI] [PubMed] [Google Scholar]
- 21. Kawabata Y, Ikegami H, Kawaguchi Y, Fujisawa T, Hotta M, Ueda H, et al. Age-related association of MHC class I chain-related gene A (MICA) with type 1 (insulin-dependent) diabetes mellitus. Hum Immunol. 2000;61(6):624–9. 10.1016/s0198-8859(00)00118-x. [DOI] [PubMed] [Google Scholar]
- 22. Gambelunghe G, Ghaderi M, Cosentino A, Falorni A, Brunetti P, Falorni A, et al. Association of MHC Class I chain-related A (MIC-A) gene polymorphism with Type I diabetes. Diabetologia. 2000;43(4):507–14. 10.1007/s001250051336. [DOI] [PubMed] [Google Scholar]
- 23. Zhou X, Wang J, Zou H, Ward MM, Weisman MH, Espitia MG, et al. MICA, a gene contributing strong susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2014;73(8):1552–7. 10.1136/annrheumdis-2013-203352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizuki N, Ota M, Katsuyama Y, Yabuki K, Ando H, Goto K, et al. Association analysis between the MIC-A and HLA-B alleles in Japanese patients with Behcet’s disease. Arthritis Rheum. 1999;42(9):1961–6. . [DOI] [PubMed] [Google Scholar]
- 25. Redler S, Albert F, Brockschmidt FF, Herold C, Hanneken S, Eigelshoven S, et al. Investigation of selected cytokine genes suggests that IL2RA and the TNF/LTA locus are risk factors for severe alopecia areata. Br J Dermatol. 2012;167(6):1360–5. 10.1111/bjd.12004. [DOI] [PubMed] [Google Scholar]
- 26. Shimizu T, Hizawa N, Honda A, Zhao Y, Abe R, Watanabe H, et al. Promoter region polymorphism of macrophage migration inhibitory factor is strong risk factor for young onset of extensive alopecia areata. Genes Immun. 2005;6(4):285–9. 10.1038/sj.gene.6364191. [DOI] [PubMed] [Google Scholar]
- 27. Donn RP, Shelley E, Ollier WE, Thomson W; British Paediatric Rheumatology Study Group . A novel 5'-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2001;44(8):1782–5. . [DOI] [PubMed] [Google Scholar]
- 28. Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Welsh EA, Clark HH, Epstein SZ, Reveille JD, Duvic M. Human leukocyte antigen-DQB1*03 alleles are associated with alopecia areata. J Invest Dermatol. 1994;103(6):758–63. 10.1111/1523-1747.ep12412584. [DOI] [PubMed] [Google Scholar]
- 30. Moravvej H, Tabatabaei-Panah PS, Abgoon R, Khaksar L, Sokhandan M, Tarshaei S, et al. Genetic variant association of PTPN22, CTLA4, IL2RA, as well as HLA frequencies in susceptibility to alopecia areata. Immunol Invest. 2018;47(7):666–79. 10.1080/08820139.2018.1480032. [DOI] [PubMed] [Google Scholar]
- 31. Lundtoft CSJ, Jacobsen M. IL7RA genetic variants differentially affect IL-7Rα expression and alternative splicing: a role in autoimmune and infectious diseases? Genes Immun Feb. 2020;21(2):83–90. [DOI] [PubMed] [Google Scholar]
- 32. Alt FW, Oltz EM, Young F, Gorman J, Taccioli G, Chen J. VDJ recombination. Immunol Today. 1992;13(8):306–14. 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 33. Muegge K, Vila MP, Durum SK. Interleukin-7: a cofactor for V(D)J rearrangement of the T cell receptor beta gene. Science. 1993;261(5117):93–5. 10.1126/science.7686307. [DOI] [PubMed] [Google Scholar]
- 34. Mertens M, Singh JA. Anakinra for rheumatoid arthritis. Cochrane Database Syst Rev. 2009;1:CD005121. 10.1002/14651858.CD005121.pub3. [DOI] [PubMed] [Google Scholar]
- 35. Witkin SS, Gerber S, Ledger WJ. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis. 2002;34(2):204–9. 10.1086/338261. [DOI] [PubMed] [Google Scholar]
- 36. Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. 2018;9(1):1558. 10.1038/s41467-018-04013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alharbi KS, Fuloria NK, Fuloria S, Rahman SB, Al-Malki WH, Javed Shaikh MA, et al. Nuclear factor-kappa B and its role in inflammatory lung disease. Chem Biol Interact. 2021;345:109568. 10.1016/j.cbi.2021.109568. [DOI] [PubMed] [Google Scholar]
- 38. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1(6):a001651. 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336(15):1066–71. 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 40. Chinn IK, Shearer WT. Severe combined immunodeficiency disorders. Immunol Allergy Clin North Am. 2015;35(4):671–94. 10.1016/j.iac.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 41. Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–7. 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jagielska D, Redler S, Brockschmidt FF, Herold C, Pasternack SM, Garcia Bartels N, et al. Follow-up study of the first genome-wide association scan in alopecia areata: IL13 and KIAA0350 as susceptibility loci supported with genome-wide significance. J Invest Dermatol. 2012;132(9):2192–7. 10.1038/jid.2012.129. [DOI] [PubMed] [Google Scholar]
- 43. Petukhova L, Patel AV, Rigo RK, Bian L, Verbitsky M, Sanna-Cherchi S, et al. Integrative analysis of rare copy number variants and gene expression data in alopecia areata implicates an aetiological role for autophagy. Exp Dermatol. 2020;29(3):243–53. 10.1111/exd.13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182(2):459–65. 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitsuiki N, Schwab C, Grimbacher B. What did we learn from CTLA-4 insufficiency on the human immune system? Immunol Rev. 2019;287(1):33–49. 10.1111/imr.12721. [DOI] [PubMed] [Google Scholar]
- 46. Gokhale AS, Gangaplara A, Lopez-Occasio M, Thornton AM, Shevach EM. Selective deletion of Eos (Ikzf4) in T-regulatory cells leads to loss of suppressive function and development of systemic autoimmunity. J Autoimmun. 2019;105:102300. 10.1016/j.jaut.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zuo J, Willcox BE, Moss P. ULBPs: regulators of human lymphocyte stress recognition. Oncotarget. 2017;8(63):106157–8. 10.18632/oncotarget.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosengren Pielberg G, Golovko A, Sundstrom E, Curik I, Lennartsson J, Seltenhammer MH, et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nat Genet. 2008;40(8):1004–9. 10.1038/ng.185. [DOI] [PubMed] [Google Scholar]
- 49. Yousaf A, Lee J, Fang W, Kolodney MS. Notice of Retraction. Yousaf A et al. Association Between Alopecia Areata and natural hair color among white individuals. JAMA dermatol. Published online March 10, 2021. JAMA Dermatol. 2021;157(8):1007–8. 10.1001/jamadermatol.2021.1832. [DOI] [PubMed] [Google Scholar]
- 50. Zhou Y, Kok KH, Chun AC, Wong CM, Wu HW, Lin MC, et al. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem Biophys Res Commun. 2000;268(3):921–7. 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]
- 51. Prie BE, Voiculescu VM, Ionescu-Bozdog OB, Petrutescu B, Iosif L, Gaman LE, et al. Oxidative stress and alopecia areata. J Med Life. 2015;8 Spec Issue(Spec Issue):43–6. [PMC free article] [PubMed] [Google Scholar]