Abstract
Coronary artery calcification (CAC), a measure of subclinical atherosclerosis, predicts future symptomatic coronary artery disease (CAD). Identifying genetic risk factors for CAC may point to new therapeutic avenues for prevention. Currently, there are only 4 known risk loci for CAC identified from genome-wide association studies (GWAS) in the general population. Here, we conducted the largest multi-ancestry GWAS meta-analysis of CAC to date, which comprised 26,909 individuals of European ancestry and 8,867 individuals of African ancestry. We identified 11 independent risk loci, of which 8 are novel for CAC and 5 have not been reported for CAD. These novel CAC loci are related to bone mineralization, phosphate catabolism, and hormone metabolic pathways. Several novel loci harbor candidate causal genes supported by multiple lines of functional evidence and are regulators of smooth muscle cell-mediated calcification ex vivo and in vitro. Together, these findings help refine the genetic architecture of CAC, and extend our understanding of the biological and potential druggable pathways underlying CAC.
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in developed and developing countries1,2. Atherosclerosis is the primary etiology of CAD, involving chronic lesion progression and luminal narrowing of arteries3. Subclinical coronary atherosclerosis is associated with an increased risk of developing future clinical CAD in males and females and across populations, which is independent of traditional risk factors4–6. Subclinical coronary atherosclerosis can be detected noninvasively as coronary artery calcification (CAC) by cardiac computed tomography. Detectable CAC has a sensitivity of 97% and a specificity of 72.4% for detection of at least 50% stenosis after adjusting for verification basis7. Current clinical guidelines recommend assessment of CAC as an option to clarify atherosclerotic cardiovascular disease (CVD) risk and to improve management decisions for those at borderline or intermediate atherosclerotic CVD risk8.
The degree of CAC varies widely, with microcalcification or spotty, fragmented calcification associated with unstable plaque, whereas advanced sheet-like calcification is associated with stable plaque9,10. Several studies have demonstrated a key role for smooth muscle cells (SMCs) in vascular calcification as they transition from a contractile to osteochondrogenic phenotype and release matrix vesicles and apoptotic bodies in the necrotic core11,12. Importantly, increased CAC is also associated with increased risk of other age-related diseases such as stroke, dementia, cancer, chronic kidney disease, chronic obstructive pulmonary disease, and hip fractures in the general population13–15.
Based on family data, the estimated heritability for CAC is 30–40%16,17. Prior genome-wide association studies (GWAS) from general population cohorts have identified non-coding single nucleotide polymorphisms (SNPs) at 9p21 (CDKN2B-AS1) and 6p24 (PHACTR1) as well as a protein-coding variant in APOB associated with a greater extent of CAC in individuals of European ancestry18–20. Another protein-coding variant in APOE was associated with CAC in individuals of both European and African ancestries18. These four loci identified for CAC are also associated with a greater risk for CAD21.
We carried out the largest CAC GWAS meta-analysis to date by analyzing 1000 Genomes Phase 3-imputed genotype data from 35,776 individuals of European and African ancestries through a collaboration between the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium22 and other collaborating cohorts. We then performed a series of in silico functional genomic analyses to (1) gain mechanistic and biological insights into how the identified genetic loci impact CAC quantity, (2) prioritize the most clinically relevant CAC loci, and (3) identify potential druggable targets for CAC. Finally, our ex vivo and in vitro experimental studies support our main genetic findings and provide motivation for future mechanistic and translational studies.
RESULTS
Multi-ancestry CAC genome-wide association meta-analysis
We performed a GWAS meta-analysis of CAC quantity expressed in Agatston scores23 from 35,776 individuals of European and African ancestries across cohorts in the CHARGE consortium and collaborating cohorts (Fig. 1 and Supplementary Table 1). We identified 16 lead significant SNPs (Table 1) through linkage disequilibrium (LD) based clumping (at r2 < 0.1 using the 1000G Phase 3 reference) resulting in 11 independent genomic risk loci (Supplementary Figs. 1–3, Supplementary Table 2 and Methods). Among these 11 loci, 8 are novel for CAC at genome-wide significance threshold (Table 1); associations at PHACTR1 (6p24.1), CDKN2B-AS1/CDKN2B (9p21.3), and APOE (19q13.32) replicated known findings in multi-cohort GWAS18,19. We annotated the lead SNPs in the 11 loci and identified two missense lead SNPs in IGFBP3 and APOE, while the remaining SNPs were annotated as non-coding (Table 1). The specific APOB association reported earlier18 was not replicated here, most likely because the Old Order Amish were not included in this study and they have the highest frequency of a rare coding variant (R3500G) associated with CAC24.
Figure 1 |. Study summary.
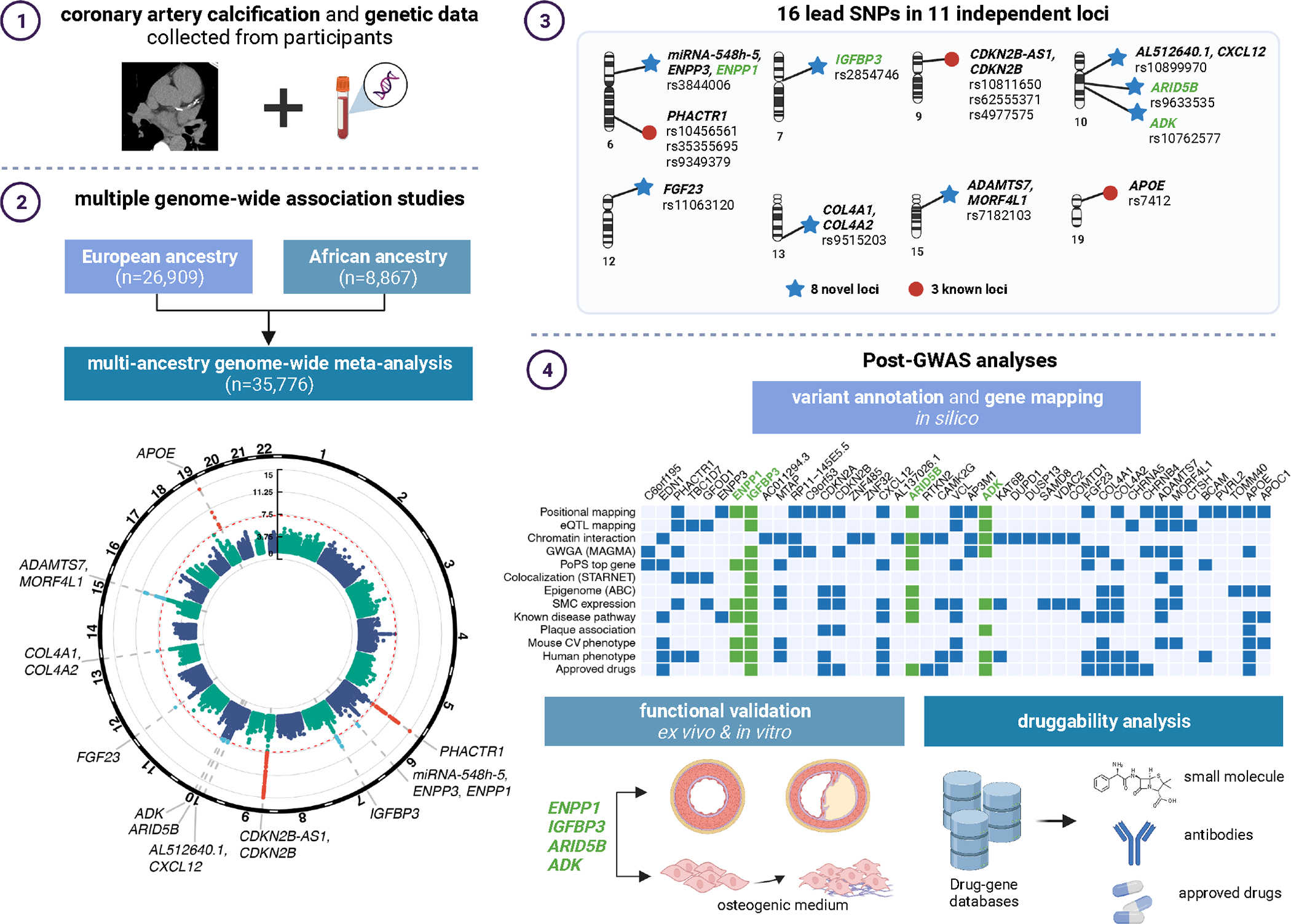
Schematic of study design and meta-analysis for CAC in European and African ancestry participants (1 and 2), main results identifying 16 lead SNPs in 11 loci (3), and post-GWAS analyses involving variant annotation and gene mapping in silico, functional validation in vitro, and druggability analysis (4). Figure created in BioRender.
Table 1 |.
Novel and known independent lead SNPs associated with coronary artery calcification
| rsID | Chr | Pos (hg19) | Effect/ Other allele |
EAF | Effect | SE | P meta | I 2 | P het | Nearest gene(s) | Annotation |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Novel loci for coronary artery calcification quantity | |||||||||||
|
| |||||||||||
| rs3844006 | 6 | 132,095,002 | T/C | 0.221 | −0.114 | 0.020 | 7.56E-09 | 0.5 | 0.453 | miR-548h-5 (dist: 18,309 bp), ENPP3 (dist.: 26,449 bp), ENPP1 (dist.: 34,154 bp) | intergenic |
| rs2854746 | 7 | 45,960,645 | C/G | 0.414 | 0.110 | 0.018 | 5.33E-10 | 0 | 0.760 | IGFBP3 | missense |
| rs10899970 | 10 | 44,515,716 | A/G | 0.474 | 0.095 | 0.017 | 2.91E-08 | 0 | 0.940 | AL512640.1 (dist.:14,271 bp), CXCL12 (dist: 366,225 bp) | intergenic |
| rs9633535 | 10 | 63,836,088 | T/C | 0.371 | 0.098 | 0.018 | 2.57E-08 | 4 | 0.407 | ARID5B | intronic |
| rs10762577 | 10 | 75,917,431 | A/G | 0.258 | −0.107 | 0.019 | 4.09E-08 | 0 | 0.683 | ADK | intronic |
| rs11063120 | 12 | 4,486,618 | A/G | 0.303 | −0.133 | 0.022 | 2.78E-09 | 56 | 0.001 | FGF23 | intronic |
| rs9515203 | 13 | 111,049,623 | T/C | 0.732 | 0.123 | 0.022 | 1.43E-08 | 0 | 0.744 | COL4A1 (dist: 90,119 bp), COL4A2 (dist: -91,464 bp) | intronic |
| rs7182103 | 15 | 79,123,946 | T/G | 0.575 | 0.112 | 0.017 | 1.62E-11 | 4.1 | 0.405 | ADAMTS7 (dist.: -20,140 bp), MORF4L1 (dist.: 21,117 bp) | intronic |
|
| |||||||||||
| Known loci for coronary artery calcification quantity | |||||||||||
|
| |||||||||||
| rs10456561 | 6 | 12,887,465 | A/G | 0.036 | 0.375 | 0.069 | 4.76E-08 | 14.7 | 0.292 | PHACTR1 | intronic |
| rs35355695 | 6 | 12,891,103 | T/G | 0.256 | −0.115 | 0.020 | 4.87E-09 | 5.2 | 0.390 | PHACTR1 | intronic |
| rs9349379 | 6 | 12,903,957 | A/G | 0.623 | −0.218 | 0.020 | 6.11E-29 | 18 | 0.234 | PHACTR1 | intronic |
| rs10811650 | 9 | 22,067,593 | A/G | 0.619 | −0.195 | 0.018 | 2.15E-27 | 53.2 | 0.003 | CDKN2B-AS1 | ncRNA intronic |
| rs72652478 | 9 | 22,102,043 | C/G | 0.957 | −0.479 | 0.081 | 3.82E-09 | 0 | 0.983 | CDKN2B-AS1 | ncRNA intronic |
| rs62555371 | 9 | 22,107,238 | A/T | 0.866 | 0.270 | 0.032 | 5.49E-17 | 20.9 | 0.190 | CDKN2B-AS1 | ncRNA intronic |
| rs4977575 | 9 | 22,124,744 | C/G | 0.455 | −0.264 | 0.018 | 7.49E-47 | 45 | 0.012 | CDKN2B-AS1 (dist.: -3,651 bp), CDKN2B (dist: 115,440 bp) | intergenic |
| rs7412 | 19 | 45,412,079 | T/C | 0.090 | −0.313 | 0.039 | 4.42E-16 | 0 | 0.657 | APOE | missense |
Top lead SNP in genomic risk loci associated with coronary artery calcification quantity at a significance level of P < 5 × 10−8 for the combined-ancestry meta-analysis (up to 35,776 individuals from 22 studies). SNP effect sizes (Beta) and two-sided P−values (PMETA) were derived from weighted Z−scores in fixed effects model and central association P−values determined from chi-square test statistics. rsID, rsID of the lead SNP. Lead SNP chromosome (Chr) and position (Pos) are provided in hg19/b37 and hg38. Effect/Other allele indicates the effect and other (non-effect) allele. EAF, effect allele frequency. Effect, effect size. SE, standard error of the effect. Pmeta, P−value of association of the lead SNP with CAC after multi-ancestry meta-analysis. I2, heterogeneity statistic indicating the variation between CAC quantity across the studies, expressed as a percent. Phet, P−value of the heterogeneity test; P > 0.05 is indicative of study homogeneity for a given SNP. Nearest gene(s), nearest gene upstream or downstream and nearest protein-coding gene(s) to lead SNP, with distance to canonical TSS for intergenic SNPs or intronic with equidistant protein-coding genes. Annotation, functional annotation of lead SNP.
Reported is either the gene that overlaps with the SNP or the nearest gene(s) up- and downstream of the sentinel variant (separated by a comma).
SNP rs4977575 and rs10811650 reside 57,151 bp apart (r2 = 0.116, D′ = 0.643).
SNP rs4977575 and rs62555371 reside 17,506 bp apart (r2 = 0.076, D′ = 0.983).
SNP rs9349379 and rs35355695 reside 12,854 bp apart (r2 = 0.088, D′ = 0.988).
SNP rs9349379 and rs10456561 reside 16,492 bp apart (r2 = 0.027, D′ = 0.981).
We also performed a sex-stratified GWAS and SNP-sex interaction tests (Methods) for the 11 lead SNPs using a subset of the cohorts with available data (Supplementary Table 3). Despite the lower sample sizes, we found genome-wide significant associations with CAC at PHACTR1 for both males and females and at CDKN2B-AS1/CDKN2B for males. We found two significant SNP-sex interaction signals (P < 4.53 × 10−3) at the ARID5B and CDKN2B-AS1/CDKN2B loci, with a stronger allelic effect in males compared to females despite similar allele frequencies.
Conditional and credible set analysis for CAC loci
We performed conditional analyses on the summary statistics from the European ancestry cohorts25, which identified three additional conditionally independent significant SNPs, not identified through LD-clumping, at CDKN2B-AS1/CDKN2B and CXCL12 loci (Supplementary Table 4 and Methods). We then performed credible set analyses to refine the association signals26. As expected, the 95% credible set reduced the number of candidate causal variants at most loci, notably including only a single candidate variant at PHACTR1, FGF23, and APOE loci (Supplementary Table 5). By leveraging ancestry-stratified analyses, the African ancestry meta-analyzed results reduced the credible set size for 8 of 11 loci, particularly for loci with broad association signals (e.g., CXCL12) (Supplementary Table 5 and Supplementary Fig. 3).
CAC loci to gene annotation
We identified 38 candidate genes using FUMA27 through a combination of positional gene mapping, expression quantitative trait loci (eQTL), and chromatin interaction mapping (see Methods and Supplementary Table 6). We identified another two candidate genes (C9orf53 and C6orf195) through a genome-wide gene association analysis (MAGMA28; Supplementary Table 7 and Supplementary Figs. 4 and 5). We also identified three candidate genes (ENPP1, ENPP3, and CXCL12 in Table 1) by annotating the nearest protein-coding genes that were not mapped through other methods (Methods). These 43 candidate causal CAC genes identified using locus-specific methods (Supplementary Fig. 5) were further annotated through a Polygenic Priority Score (PoPS)29 analysis (Supplementary Table 8). This provided support for several genes that were non-significant using MAGMA, emphasizing the need for orthogonal gene prioritization methods.
Prioritization of candidate causal CAC genes
To prioritize candidate causal CAC genes, we performed Summary-based Mendelian Randomization (SMR)30 and colocalization31. The SMR-HEIDI test determines whether the effect size on the GWAS trait is mediated by gene expression using eQTLs30. By integrating the European-ancestry CAC meta-analysis summary statistics and cardiometabolic tissue cis-eQTLs from STARNET32,33, we identified 11 and 18 gene expression-trait associations using eQTLs in atherosclerotic aortic root (AOR) and subclinical/non-atherosclerotic internal mammary artery (MAM) tissues, respectively (Fig. 2 and Supplementary Table 9). This supports that the effect of the CAC variants are likely mediated by gene expression differences. To provide additional functional fine-mapping evidence, we performed colocalization using coloc31, which revealed colocalization of CAC variants with cis-eQTLs in 22, 25, and 7 genes, in AOR, MAM, and liver (LIV), respectively (Fig. 2 and Supplementary Table 10). We observed the strongest evidence of colocalization at known CAD loci PHACTR1 and ADAMTS7 in AOR, consistent with recent fine-mapping studies34. As expected, we observed substantial overlap between prioritized genes associated with CAC and CAD (PP4 > 0.80 for both traits). However, we also identified a subset of genes with strong evidence of colocalization with CAC (PP4 > 0.80) but not CAD (PP4 < 0.50), such as IGFBP3. Notably, IGFBP3 was identified as a target gene using SMR in both AOR and MAM, suggesting a causal role in both early and advanced atherosclerosis.
Figure 2 |. Prioritization of CAC causal genes using STARNET eQTLs.
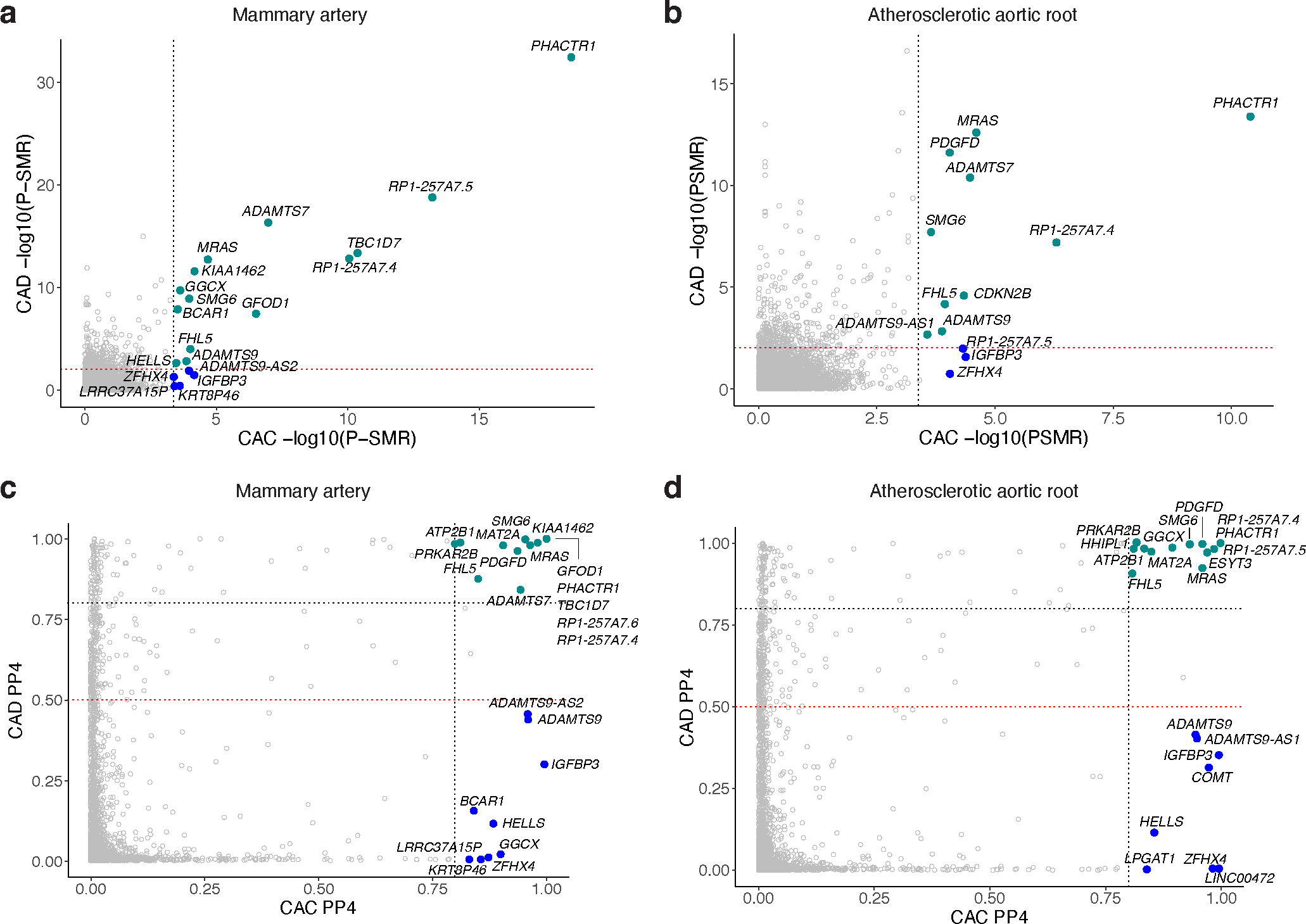
a,b, Summary-based Mendelian Randomization (SMR) to identify causal CAC genes using subclinical atherosclerotic mammary artery (MAM, a) and atherosclerotic aortic root (AOR, b) tissue cis-eQTLs in STARNET. Dashed black and red lines indicate SMR P-value significance thresholds for tested CAC and CAD candidate genes, respectively. Teal green dots represent significant genes for both CAC and CAD, while blue dots represent those only significant for CAC. SMR P-value determined by approximate chi-square test statistic for mediating effect of gene expression on CAC or CAD. c,d, Coloc-based colocalization analysis of CAC and CAD candidate genes using STARNET MAM (c) and AOR (d) cis-eQTLs. Dashed black lines indicate high colocalization (PP4 > 0.8) and dashed red lines indicate low colocalization (PP4 < 0.5) thresholds, respectively. Teal green dots represent high colocalized genes (PP4 > 0.8) for both CAC and CAD, whereas blue dots represent those highly colocalized for CAC (PP4 > 0.8) but not CAD (PP4 < 0.5).
To further resolve the regulatory mechanisms of GWAS variants35,36, we performed epigenomic fine-mapping of the combined European- and African-ancestry summary statistics using activity-by-contact (ABC)37 and enhancer-gene linking38 methods. Using a suggestive threshold for CAC-associated variants (P < 1 × 10−5), we identified 42 and 54 variants (among 1,526 candidate variants) overlapping enhancer-promoter contacts for predicted target genes in human coronary artery smooth muscle cells (HCASMCs) and coronary artery, respectively (Supplementary Table 11 and Supplementary Fig. 6). Notably, this provided additional support for novel CAC variants regulating IGFBP3 (rs2854746 and rs924140), ENPP1 (rs3844006), and ARID5B, and a known variant in the 9p21 locus (rs1537373) regulating CDKN2B-AS1/CDKN2B (Supplementary Table 11a,b). We confirmed these findings using our recent single-nucleus chromatin accessibility dataset in healthy and diseased coronary arteries39, which identified credible CAC variants at IGFBP3, ARID5B, and ENPP1 loci overlapping cell type-specific peak-to-gene links (Fig. 3 and Supplementary Fig. 6). Notably, CAC-associated variants were most enriched in SMC accessible chromatin regions compared to other coronary artery cell types (Supplementary Table 12). These results demonstrate that several identified CAC GWAS signals map to relevant genes, particularly in SMCs in the vascular wall, and implicate candidate regulatory mechanisms for the CAC associations.
Figure 3 |. Single-nucleus coronary epigenomic annotation of ARID5B and IGFBP3 CAC loci.
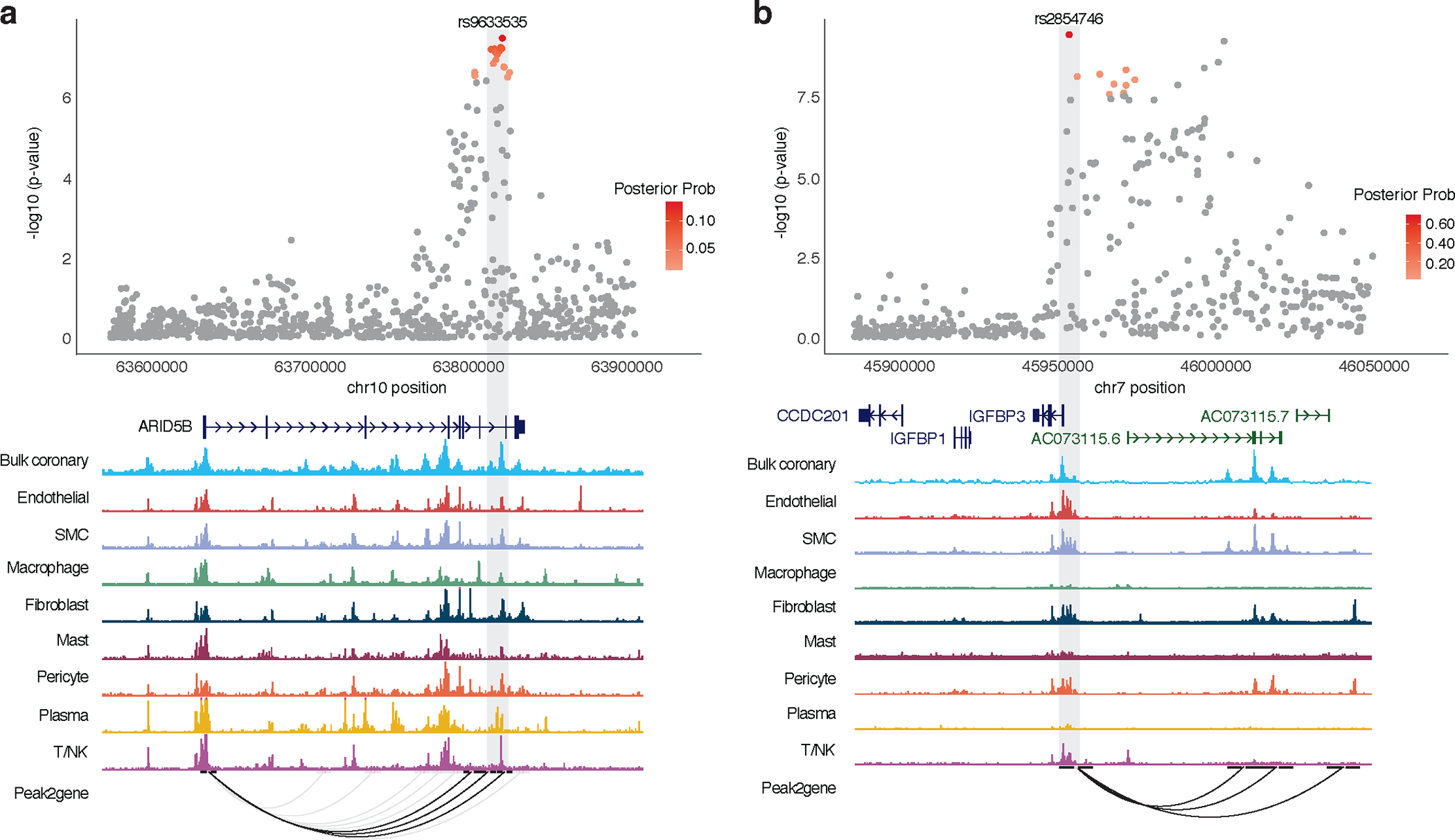
a,b, Top, ARID5B (a) and IGFBP3 (b) locus association plots showing CAC meta-analysis results in European and African American ancestry individuals with credible set SNPs color coded by posterior probability (red). Meta-analysis P-values determined from weighted Z-scores in fixed effects model and central association P-values determined from chi-square test statistic. Bottom, overlapping chromatin accessibility profiles in coronary artery cell types determined by bulk and single-nucleus ATAC-seq. Peak2gene links highlight predicted enhancer promoter interactions across all cell types with those overlapping CAC SNPs shown in black. Light grey box highlights lead SNP at each locus. SMC, smooth muscle cells; T/NK: T-cells or natural killer cells.
Mapping target pathways, cell types, and plaque phenotypes
Using gene-set pathway enrichment analysis for the candidate CAC genes, we identified significant enrichment of bone mineralization regulation, vitamin D receptor, and riboflavin metabolism pathways (Supplementary Table 13). We also identified enrichment of phosphate catabolism/homeostasis (ENPP1/ENPP3 and FGF23) and hormone secretion (FGF23) pathways, suggesting that there could be unique genetic risk factors disrupting essential hormonal metabolic pathways that have been linked to mineralization and/or plaque stability40.
We next investigated the overall expression profiles for the candidate CAC genes in bulk GTEx tissues (Supplementary Fig. 7). Many of the candidate genes (e.g., COL4A1/2, IGFBP3, ENPP1, ADAMTS7) were expressed together in artery tissues relative to other tissues, suggesting shared cell-type expression profiles. Thus, we explored the cellular distribution in single-cell gene expression data from atherosclerotic coronary artery41–43 and carotid plaques44. Among the identified CAC genes, ARID5B, COL4A2, and CXCL12 were specifically expressed in SMCs and/or pericytes and IGFBP3 was expressed in mesenchymal-like endothelial cells as well as fibroblast-like SMCs (Supplementary Fig. 8)45. ENPP1 and ENPP3 were expressed in small proportions of SMCs and mast cells, respectively.
To gain insight in the pathobiology of the 16 lead CAC SNPs from FUMA, we assessed the association with advanced plaque morphology. We examined seven plaque morphological characteristics measured in atherosclerotic carotid plaques in the Athero-Express Biobank Study46. APOE was associated with increased intraplaque fat content and vessel density, and decreased collagen content, all known features of increased plaque vulnerability (Supplementary Table 14a). Individual variant analyses at the remaining loci were most significant for plaque calcification (ADK, PHACTR1), smooth muscle cell (ADK, CDKN2B-AS1) and macrophage content (CXCL12), collagen deposition (IGFBP3), and intraplaque neovessel density (PHACTR1) (Supplementary Table 14b). This suggests that there may be overlap in the pathological hallmarks between the CAC- and CAD-associated genes in advanced plaques.
Heritability, genetic correlations, and Mendelian randomization
We applied linkage disequilibrium score regression (LDSC)47 to estimate the heritability of CAC in European ancestry study participants and observed that the genome-wide set of variants account for 16% (SE = 2.5%) of the variance in CAC. This estimate represents almost half of the heritability estimated from phenotypic correlations among relatives16,17. We expect the CAC heritability to be equivalent for individuals of African ancestry, based on a recent multi-ancestry GWAS meta-analysis for CAD48.
We estimated the genetic correlation between CAC and clinical CVD, subclinical atherosclerosis, selected CVD risk factors, and family history of CVD in individuals of European ancestry. There were significant genetic correlations between CAC and carotid plaque and abdominal aortic calcification as well as several clinical outcomes—including CAD and myocardial infarction—and risk factors such as high cholesterol, use of cholesterol lowering medication, hypertension, body mass index (BMI), waist circumference, and whole-body fat mass. There were also correlations with family history of CVD and age at parental death (Fig. 4 and Supplementary Table 15).
Figure 4 |. Genetic correlations for CAC and Mendelian randomization for cardiovascular disease risk factors.
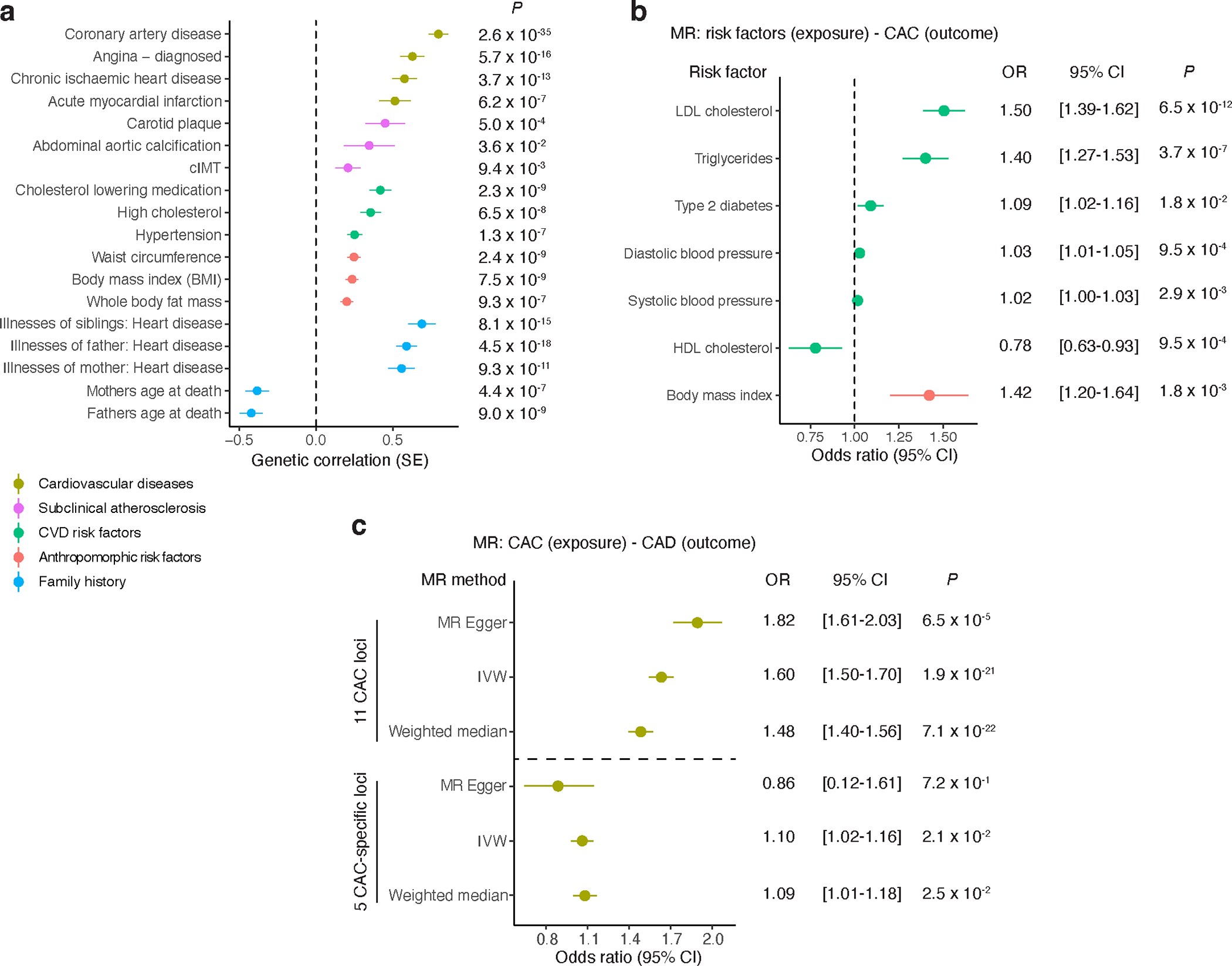
a, Cross-trait LD-score regression-based genetic correlation of CAC quantity with cardiovascular disease risk factors, anthropomorphic risk factors, family history, subclinical and clinical cardiovascular disease using European ancestry CAC and UK Biobank trait associations. Vertical dashed line set at genetic covariance = 0. Values represent the genetic correlation estimates using the slope from the regression of the product of Z-scores from two GWAS studies on the LD score and error bars represent the standard error estimates of the LD score. P-values (two-tailed) are computed from chi-square test statistics. b, Mendelian randomization (MR) results showing causal effects of cardiovascular disease and anthropomorphic risk factors on CAC quantity using the inverse variance-weighted method (IVW). Values represent the mean odds ratio and error bars reflect the 95% confidence intervals. P-values (two-tailed t test) < 7.14 × 10−3 (0.05/7) are considered statistically significant. Vertical dashed line set at odds ratio = 1.0. c, MR results showing causal effects of CAC quantity on coronary artery disease (CAD) using either 16 independent lead SNPs at the 11 CAC loci or 5 lead SNPs from the 5 CAC-specific loci (separated by horizontal dashed line). Values represent the mean odds ratio and error bars reflect the 95% confidence intervals. P-values (two-tailed t test) < 0.05 are considered statistically significant. Different MR methods used are shown, including MR-Egger, IVW, and Weighted median. Sample sizes for a-c are provided in Supplementary Tables 15 and 16. LDL, low-density lipoprotein; HDL, high-density lipoprotein; cIMT: carotid intima-media thickness.
We then performed a Mendelian randomization (MR) analysis to assess the potential causality of CVD risk factors with CAC. Low-density lipoprotein (LDL) cholesterol, triglycerides, systolic and diastolic blood pressure, BMI, and type 2 diabetes were causally associated with an increase in CAC, while an increase in high-density lipoprotein (HDL) cholesterol was causally associated with a decrease in CAC (Fig. 4 and Supplementary Table 16a). Moreover, CAC was causally associated with clinical CAD when the 16 independently significant lead SNPs from FUMA in 11 different loci for CAC were considered (Supplementary Table 16b). However, as expected, the association was diminished when restricted to the five CAC-specific loci (Supplementary Table 16c), suggesting that the effects on clinical CAD from all loci is likely driven by independent SNPs with large effects on CAD (e.g., 9p21). We performed weighted median estimator (WME) and MR Egger regression analyses as sensitivity analyses to rule out potential bias caused by horizontal pleiotropy49. While the associations showed robust effect estimates in similar directions, we cannot entirely rule out potential pleiotropic effects through shared cardiometabolic risk factors.
Functional characterization of CAC genes
Given the strong association between CAC and CAD, we further examined whether each of the candidate CAC genes are also associated at genome-wide significance with clinical CAD based on large-scale GWAS data from the CARDIoGRAMplusC4D consortium, UK Biobank, and Million Veteran Program (MVP)21,50. While a few of the candidate genes were associated with CAD as expected, among our eight novel CAC loci (Table 1), several (ENPP1/ENPP3, IGFBP3, ARID5B, ADK, and FGF23) have not yet been reported to be associated with clinical CAD (Supplementary Table 17). We observed similar results in our PheWAS analysis considering only the eight novel CAC loci (Supplementary Table 18).
We next evaluated the protein localization of the novel CAC genes in subclinical and advanced atherosclerotic coronary artery sections ex vivo. ENPP1, IGFBP3, and ADK proteins were expressed in the medial vessel layer of both subclinical and advanced atherosclerotic human coronary arteries, as shown by immunofluorescence staining (Fig. 5). FGF23 was not detected in the coronary artery specimens, consistent with recent reports that its associations with CVD may be mediated by endocrine and paracrine effects51. ENPP1, IGFBP3, ARID5B, and ADK proteins were all highly expressed in the neointima of diseased vessels, with ARID5B more restricted to the intimal layer (Fig. 5). This provides evidence that some of these CAC-associated proteins are present at the early stages of atherosclerosis and may directly participate in intimal calcification.
Figure 5 |. Immunofluorescence staining showing localization of ENPP1, IGFBP3, ARID5B and ADK in control and atherosclerotic human coronary arteries.
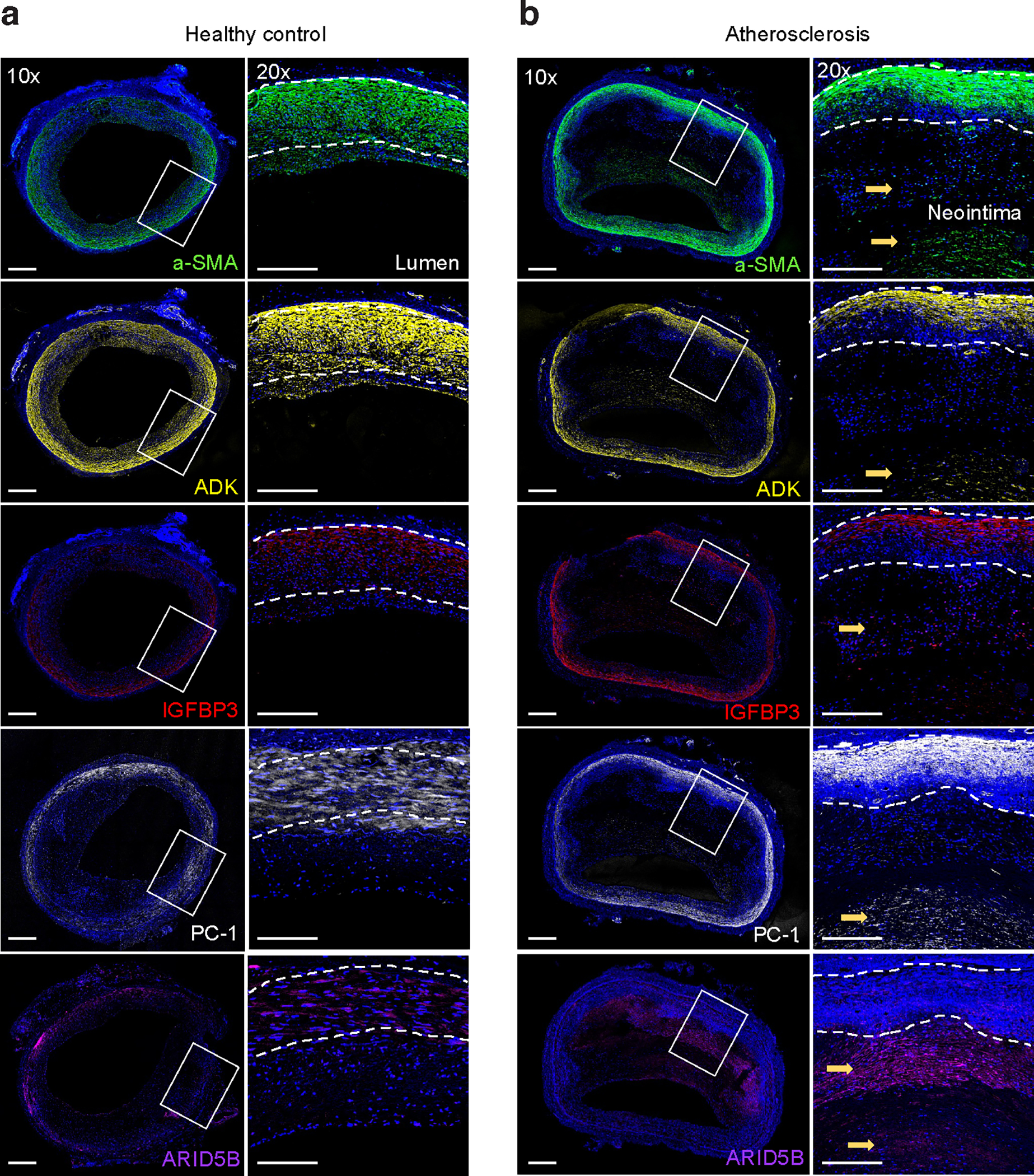
a,b, Transverse sections of healthy control (a) and atherosclerotic (b) human coronary arteries were stained for alpha-smooth muscle actin (α-SMA) (green), DAPI nuclei marker (blue), ENPP1/PC-1 (white), IGFBP3 (red), ARID5B (purple), and ADK (yellow). High levels of ENPP1/PC-1, IGFBP3, ARID5B, and ADK were observed in the neointimal layer of atherosclerotic diseased coronary arteries. Whole artery images were captured at 10× magnification and regions of interest were captured at 20×. Images are representative of n = 4 independent donors per group.
To examine the functional role of these four genes (ENPP1, IGFBP3, ARID5B, and ADK) in vitro, human coronary artery smooth muscle cells (HCASMCs) were treated with siRNAs directed against these genes in the presence and absence of osteogenic media. FGF23 was not evaluated given its undetectable expression in HCASMCs. Compared to HCASMCs incubated in control media, cells grown in osteogenic media for 48 h exhibited a two-fold increase in mRNA levels of RUNX2, a known master regulator of osteogenic phenotype switch in SMCs (Fig. 6a). Treatment of cells with siARID5B and siADK prevented the osteogenic media-induced increase in RUNX2; treatment with siRNA directed against IGFBP3 also attenuated this increase (Fig. 6a, Supplementary Fig. 9). At the protein level, increased RUNX2 levels were observed with cells grown in osteogenic media compared to control media, while treatment with siRNA directed against IGFBP3, ARID5B, or ADK attenuated this increase in RUNX2 (Fig. 6b). Similarly, treatment with siIGFBP3 and siADK prevented the osteogenic media-induced decrease in CNN1 mRNA expression, thus promoting the contractile SMC phenotype (Fig. 6c). Treatment with siIGFBP3, siARID5B, and siADK resulted in inhibition of calcification, as assessed by Alizarin Red staining compared to siCTRL-treated cells (Fig. 6d). Of note, knockdown of ENPP1 had no effect on RUNX2, CNN1, or calcification levels. These results implicate IGFBP3, ARID5B, and ADK in the development of an osteogenic phenotype, which is characterized by an increase in cell proliferation and migration. Treatment with siIGFBP3 resulted in decreased cell proliferation, as measured by MTT assay (Fig. 6e). Treatment with siIGFBP3 and siADK led to an expected decrease in cell migration, while siARID5B treatment was associated with greater migration (Fig. 6f,g). Taken together, these results indicate that IGFBP3, ARID5B, and ADK play a key role in regulating VSMC phenotype and promoting VSMC calcification, which is consistent with the direction of the effect alleles from the CAC GWAS and eQTL signals for these genes.
Figure 6 |. Functional assays of ENPP1, IGFBP3, ARID5B, and ADK in coronary artery calcification and vascular smooth muscle cell phenotype.
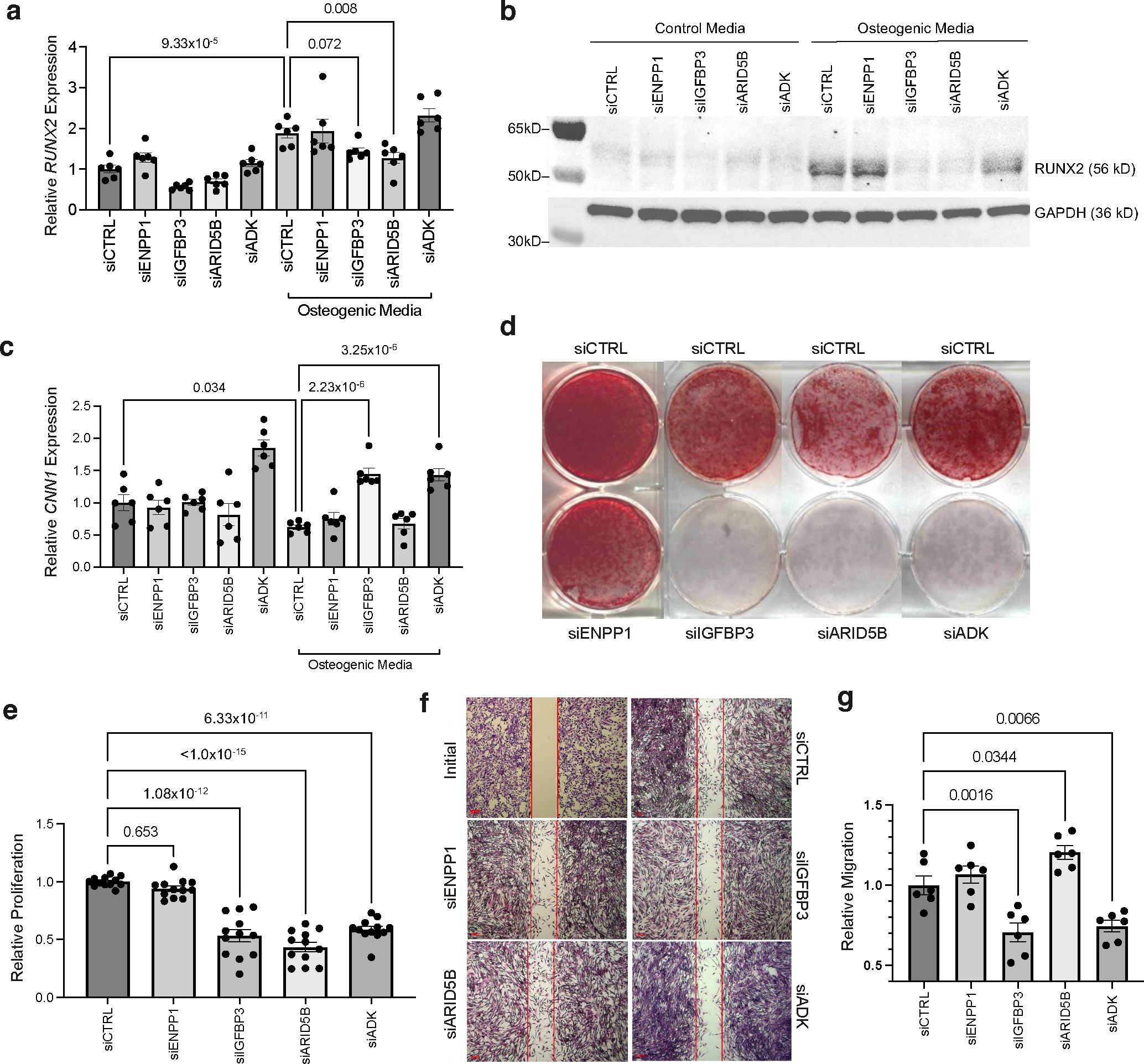
a, Effects of siRNA-mediated knockdown of ENPP1, IGFBP3, ARID5B, and ADK on osteogenic marker RUNX2 gene expression in human coronary artery smooth muscle cells (HCASMCs) cultured in control or osteogenic media (OM). n = 6 biological replicates per group. b, Western blot analysis of RUNX2 protein levels in HCASMCs cultured in basal control or OM and transfected with siCTRL, siENPP1, siIGFBP3, siARID5B, or siADK. GAPDH was used as a loading control. c, Effects of siRNA knockdown on CNN1 mRNA expression. n = 6 biological replicates per group. d, Calcification assessed by Alizarin Red staining in HCASMCs cultured in OM and transfected with siCTRL, siENPP1, siIGFBP3, siARID5B, or siADK. Representative images from three independent experiments. e, MTT assay-based proliferation in HCASMCs transfected with siCTRL, siENPP1, siIGFBP3, siARID5B or siADK. n = 12 biological replicates per treatment and control group; each individual sample read three times; value reported represents the mean of three technical replicates. f,g, Scratch wound-based migration assay in HCASMCs transfected with siCTRL, siENPP1, siIGFBP3, siARID5B or siADK, as quantified in g. An initial image of the assay is provided to demonstrate creation of a cell-free gap, prior to incubation with OM. n = 6 biological replicates per treatment and control group. Scale bars, 200 μm. All statistical comparisons shown were made using a two-tailed one-way ANOVA with Sidak’s test for multiple comparisons. Values in a-g represent mean ± standard error of the mean. Full-length blots are provided as Source Data.
Druggability analysis
To investigate the potential clinical utility of CAC candidate genes at the 11 independent risk loci, we performed an integrative druggability analysis (Supplementary Table 19a,b). Three loci were identified as targets of clinically actionable compounds (CDKN2B, ARID5B and FGF23), which have potential for repurposing approved drugs and informing clinical trials for CAC. Notably, ENPP1/ENPP3, IGFBP3, ARID5B, ADK, and FGF23 represent targets of the druggable genome, which should be considered for preclinical studies of CAC. By querying the drug-gene interaction database (DGIdb) and DrugBank database, we found approved compounds under investigation targeting IGFBP3, ADK, and FGF23 (Fig. 7). For example, IGFBP3 is a target of the biosynthetic hormone mecasermin and non-steroidal anti-inflammatory drugs (NSAIDs). We identified many targets of nutrient supplementation late-stage clinical trials for nutrient deficiencies associated with chronic metabolic diseases. For example, Burosumab (monoclonal antibody against FGF23) is being evaluated for hypophosphatemic rickets and chronic pain (NCT03581591) and X-linked hypophosphatemia (NCT04146935). Also, ARID5B was identified as a target of ascorbic acid (Vitamin C). These findings offer preclinical, repurposing, and prevention opportunities to modulate CAC via these targets, to slow CAD progression or even to promote plaque stability in advanced stages52.
Figure 7 |. Schematic of CAC candidate genes and approved or investigational drugs.
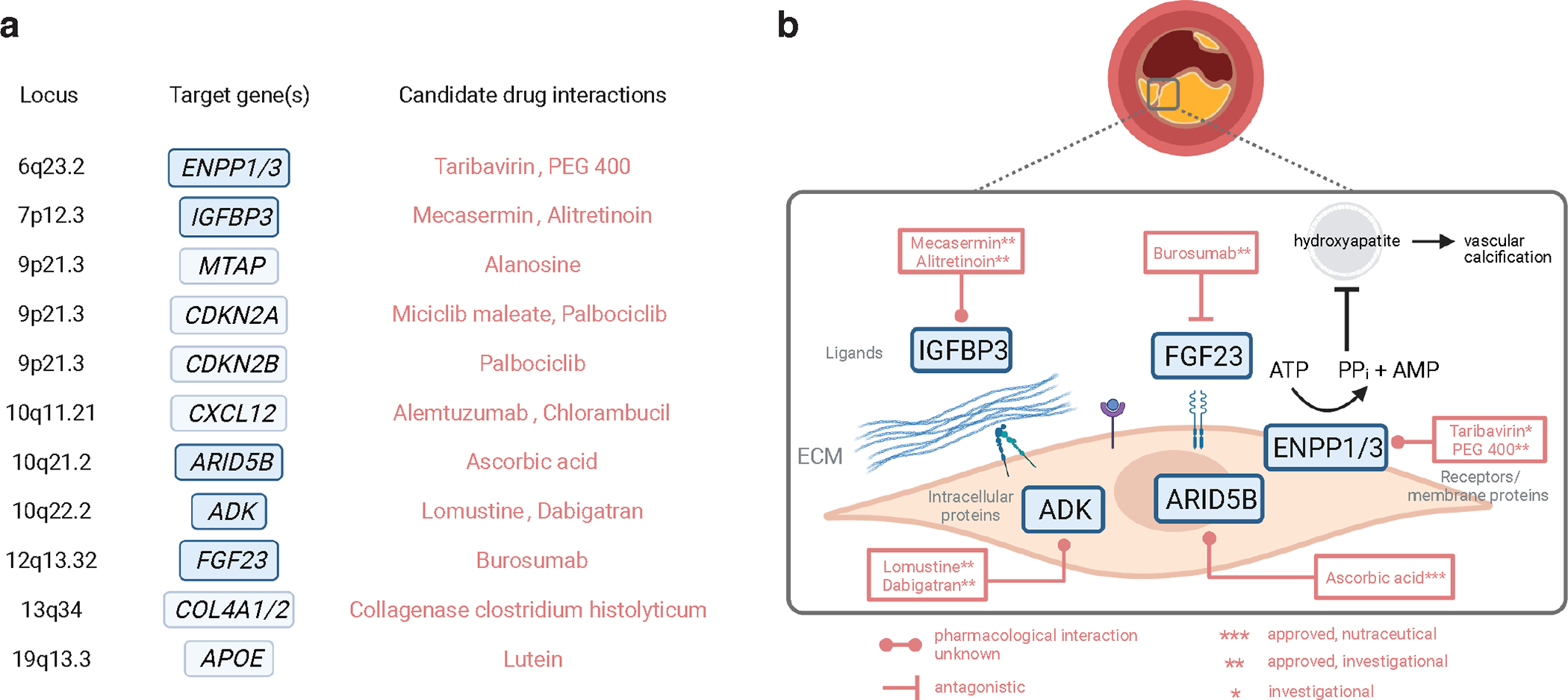
a, Summary of gene-drug interactions for CAC candidate genes using DGIdb and DrugBank databases. Novel CAC-specific genes are shown in blue and other genes are shown in light blue corresponding to each genomic locus. Generic names for the top interacting drugs/compounds are shown for each target in red. b, Schematic showing the cellular location of the protein targets for novel CAC genes. Names of interacting approved drugs or compounds under investigation are shown along with predicted pharmacological interaction if known. ECM, extracellular matrix; ATP, adenosine triphosphate; AMP, adenosine monophosphate; PPi, inorganic pyrophosphate; PEG, polyethylene glycol. Figure created in BioRender.
DISCUSSION
We identified 16 independent genome-wide significant variant associations for CAC at 11 distinct genomic loci in FUMA, including 8 loci not previously reported to be associated with CAC, by leveraging genome-wide genotypes from 35,776 participants (Table 1 and Fig. 1). Through integrating functional data with GWAS results as well as gene-based analyses, we identified a total of 43 candidate genes for CAC (Supplementary Table 20). Top pathways identified involve bone mineralization regulation, vitamin and phosphate metabolism, and hormone secretion. Notably, 5 of our 8 novel CAC loci have not been previously reported to be associated with clinical CAD. We provide ex vivo and/or in vitro functional validation for four of these genes (ENPP1, IGFBP3, ARID5B, ADK), supporting their causal role in CAC. Several of our CAC-associated genes are targets of drug or supplement interactions, revealing opportunities to study how these compounds may promote or inhibit CAC at different stages of atherosclerosis.
The CAC locus on 6q23.2 resides 26 kb and 34 kb upstream of paralog genes ENPP3 and ENPP1 (Ectonucleotide Pyrophosphatase/Phosphodiesterase 3 and 1), respectively. ENPP1 is one of the causal genes (along with ABCC6) of the spectrum of rare Mendelian arterial calcification diseases known as generalized arterial calcification of infancy (GACI) and pseudoxanthoma elasticum (PXE)53,54. ENPP1 and ENPP3 have both been associated with phosphate levels in multiple studies55, while ENPP1 was associated with venous thromboembolism56, and ENPP3 with type 2 diabetes57 and vitamin D levels55. ENPP1 and ENPP3 are involved in bone mineralization and riboflavin metabolism pathways, suggesting common variants in this locus impact CAC through phosphate catabolism/homeostasis. While we did not observe direct functional effects of perturbing ENPP1 expression in HCASMCs, it is expressed in the intima of atherosclerotic coronary arteries, suggesting enzymatic perturbation may be more fruitful. The druggability of these genes presents an opportunity to translate the genetic findings to treatments for CAC in the general population, particularly in the setting of type 2 diabetes.
The associated variant on 7p12.3 within IGFBP3, rs2854746, was supported by several lines of evidence, including positional and eQTL mapping, gene-based analyses, and single-cell epigenomics. IGFBP3 was previously associated with blood pressure traits58,59, cellular fibronectin and NT-proBNP60, anthropometric measures61, appendicular lean mass62, and bone mineral density63. Our fine-mapping analyses provide strong evidence for IGFBP3 to be implicated in vitamin D receptor metabolism. Consistently, our functional studies demonstrate direct effects on SMC calcification, proliferation, and migration. In addition, IGFBP3 is a target of the approved biosynthetic compound mecasermin. Given that IGFBP3 protein levels have been previously associated with the presence and extent of coronary atherosclerosis in a small study64, there are both prognostic and therapeutic opportunities to explore further.
ARID5B (AT-Rich Interaction Domain 5B) is known to transcriptionally activate or repress metabolic gene expression65 involved in adipogenesis and lipid metabolism66 and mesenchymal cell differentiation during SMC phenotypic modulation67. ARID5B is expressed in ACTA2+ SMCs and in endothelial cells marked by endothelial-to-mesenchymal transition45, and epigenome-based fine-mapping identified ARID5B as a target gene in coronary and aortic tissue. Variants in ARID5B are associated with coronary atherosclerosis68 and type 2 diabetes69 in smaller studies of a Japanese population, but also with measures of height and heel bone mineral density63. ARID5B has been shown to promote chondrogenesis70 and consistently our perturbation studies demonstrated potent effects on osteogenic marker expression, calcification, and proliferation in SMCs. Our druggability analyses identified ARID5B as a clinically actionable target of phosphate lowering vitamin C supplementation to evaluate variant angina and atheroma regression in a clinical trial (NCT03228238). Interestingly, ARID5B was the only novel CAC locus not previously associated with CAD, harboring a significant gene-sex interaction, with a stronger effect in males. Future studies could explore whether this difference is related to distinct atherosclerotic plaque profiles in males compared to females.
Extensive positional and functional mapping analyses consistently point to ADK (Adenosine Kinase) as the causal gene for the CAC locus on 10q22.2. ADK is a protein-coding gene involved in regulating extracellular adenosine and intracellular adenine nucleotide levels. Adenosine has widespread effects on the cardiovascular, nervous, respiratory, and immune systems71. Endothelial intracellular adenosine and its key regulator, ADK, play important roles in endothelial inflammation and vascular inflammatory diseases72. Loss of endothelial Adk in mice reduces atherosclerosis and leads to protection against ischemia/reperfusion injury in the cerebral cortex72. Adk knockout in myeloid cells and the resulting augmented intracellular adenosine levels protects ApoE−/− mice against atherosclerosis73. Further, Adk was upregulated along with Enpp1 in the thoracic aorta of the Abcc6 knockout mouse model of PXE, providing a functional link to arterial calcification74. This aligns with our in vitro functional studies demonstrating strong effects on SMC calcification and related osteogenic SMC phenotypes. ADK-modulating strategies could act as anti-inflammatory agents for treating atherosclerotic diseases75.
FGF23 (12p13.32) is a well-studied protein-coding gene linked to vascular calcification in chronic kidney disease patients on hemodialysis76. It has been suggested that this may represent a paracrine-mediated homeostatic mechanism to maintain phosphorus levels77. While FGF23 may not be causally related to CAC78, consistent with the absent expression in coronary artery cells/tissues, follow-up pre-clinical studies could investigate the effects of monoclonal antibody burosumab on calcification secondary to renal outcomes.
As the hallmark of atherosclerosis, CAC is strongly associated with future CAD events. Consistent with the shared etiology between CAC and CAD, several of our novel CAC genes were previously reported in association with CAD. Our colocalization analyses also indicated substantial overlap between prioritized genes in CAC and CAD. This is expected given that CAC is an integral component of both early intimal thickening and advanced atherosclerotic plaque, likely representing a ‘healing’ phase12,79. Our MR analyses using 16 significant lead SNPs for CAC as instrumental variables provided evidence for a causal association between CAC and CAD events. However, several of our novel CAC-associated genes, including IGFBP3, FGF23, ENPP1/ENPP3, ARID5B, and ADK, have so far not been reported to be associated with CAD. In line with this, these genes showed strong evidence for colocalization with CAC but not with CAD. Clinical studies have shown that increased CAC is associated with future acute coronary syndrome; however, pathological evidence has demonstrated that CAC represents a marker of the extent of disease80. It is suggested that stable, slowly progressive large plaques, leading to a negative remodeling of the vessel, do not readily correlate with onset of symptoms and clinical CAD events81. In contrast, unstable plaques at high risk of producing symptomatic rupture carry highly inflamed fibrous caps and are not necessarily more stenotic82. Notably, some of our CAC candidate genes were implicated in pathways underlying bone mineralization regulation and associated with plaque stability features in advanced carotid plaque tissues. We also identified genes that may be involved in early lesions, based on expression in mesenchymal or fibroblast-like SMCs that may precede osteogenic transitions.
CAC reflects the vessel’s accumulation of lifetime exposure to risk factors. While observational studies suggest a role for hypertension, higher BMI, and type 2 diabetes in development and progression of CAC83, evidence regarding the association of an unfavorable lipid profile with coronary calcification, in particular progression of CAC, remains unclear84,85. Moreover, important questions remain about causality of these risk factors. We found evidence of significant genetic correlations between CAC quantity and high cholesterol, cholesterol lowering medication use, hypertension, BMI, waist circumference, and whole-body fat mass. Our MR analyses further provided evidence for a causal association between modifiable CVD risk factors, including LDL and HDL cholesterol, triglycerides, type 2 diabetes, and BMI, with CAC quantity. These findings emphasize the value of optimal risk factor control for reducing atherosclerosis burden and further extend our knowledge of the pathways underlying coronary calcification.
Our GWAS meta-analysis requires replication and includes only individuals of European and African ancestry. Future multi-ancestry meta-analyses will benefit from larger sample sizes, diverse ancestral populations, and use of larger, more diverse reference imputation panels such as TOPMed to provide replication of our significant novel loci as well as discovery of new CAC loci. Future studies should also include the X chromosome since a very recent study identified new variants on the X chromosome associated with CAD48. Future stratified analyses based on ancestry, sex, and risk factors (e.g., smoking) may reveal further insights into interindividual differences in CAC risk. Smoking is particularly important to consider given the interaction between a variant upstream of the ADAMST7 gene and smoking on clinical CAD in a candidate gene study86. A small GWAS of gene-by-smoking interaction also demonstrated the potential to identify novel genes for CAC87.
There are many strengths to our study, which employed a series of complementary statistical, functional fine-mapping, and ex vivo and in vitro experimental validation studies. We utilized several unique atherosclerotic tissue biobanks, which were derived from patients during surgical procedures and provide a more relevant context to the effector genes, plaque phenotypes and cell types. Finally, we employed ex vivo and in vitro functional validation assays and druggability analyses to help inform translational strategies and identify cell-specific mechanisms for these candidate targets. Notably, IGFBP3, ARID5B, and ADK were shown to promote calcification in HCASMCs, consistent with changes in gene expression, related SMC phenotypes, and protein expression in atherosclerotic coronary arteries.
In summary, we discovered eight novel loci associated with CAC, thus doubling the total known CAC loci to date. Extensive post-GWAS fine-mapping and annotation provided evidence for cell- and disease-specific expression of IGFBP3, FGF23, ENPP1/ENPP3, ADK, and ARID5B in coronary and carotid arterial plaque tissue. Importantly, many of these genes encode proteins identified as predicted targets of approved drugs or investigational compounds. While we provide evidence supporting the causal role for some of these CAC-associated genes, additional functional analyses may support other candidate genes. Future studies should elucidate the molecular mechanisms of these genes in the cells of the arterial wall, evaluate their function in preclinical animal models of atherosclerosis, and focus on creating a saturated map of common and rare variants influencing CAC through the inclusion of diverse ancestries.
METHODS
Ethics statement.
All human research was approved by the relevant institutional review boards for each study and conducted according to the Declaration of Helsinki. All participants provided written informed consent (see Supplementary Table 22 for details).
Study populations and CAC assessment.
The GWAS for CAC included 16 different cohorts. These cohorts contributed 26,909 participants of exclusively European ancestry and 8,867 participants of African ancestry. The descriptive characteristics of the participants are shown in Supplementary Table 1. All cohorts followed standardized protocols for the ascertainment of CAC quantity and statistical analyses. CAC was evaluated using computed tomography as explained in Supplementary Table 1. We used data from the baseline examination or the first examination in which CAC was assessed.
Genotyping, imputation, and study-level quality control.
Association analyses were performed using standardized protocols (Supplementary Note). Within each study, linear regression was used to model CAC quantity (i.e., log(CAC+1)) with an additive genetic model (SNP dosage) adjusted for age, sex, and up to 10 principal components. Extensive quality control (QC) was applied to the data. Genotyping arrays and QC pre-imputation are shown in Supplementary Table 1. Each study conducted genome-wide imputation using a Phase 1 integrated (March 2012 release) reference panel from the 1000G Consortium using IMPUTE or MaCH/minimac and used Human Reference Genome Build 37. There was little evidence for population stratification in any of the studies (Supplementary Table 21). Sample QC was performed with exclusions based on call rates, extreme heterozygosity, sex discordance, cryptic relatedness, and outlying ancestry. SNP QC excluded variants based on call rates across samples and extreme deviation from Hardy–Weinberg equilibrium (Supplementary Table 21). Non-autosomal SNPs were excluded from imputation and association analysis. We used the EasyQC R package (v23.8) to perform QC for each study before the meta-analysis and excluded markers absent in the 1000G reference panel: no- A/C/G/T/D/I markers; duplicate markers with low call rate; monomorphic SNPs and those with missing values in alleles, allele frequency, and/or beta estimates; SNPs with large effect estimates or standard error (SE) ≥ 10; and SNPs with allele frequency difference > 0.3 compared to 1000G global reference panel.
Meta-analysis.
A joint meta-analysis of all available CAC GWAS was performed using fixed-effects meta-analysis in METAL, using SNP P-values weighted by sample size. Summary statistics from each study were combined using an inverse variance weighted meta-analysis. Additional filters were applied during meta-analyses based on imputation quality (MACH r2 < 0.3 and IMPUTE info < 0.4), a minor allele frequency (MAF) < 0.01, and SNPs that were not present in at least four studies or in both ancestry groups. Moreover, the variants with heterogeneity I2 ≥ 70% in the meta-analysis were excluded, leaving 8,586,047 variants. The genome-wide significance threshold was considered at P < 5.0 × 10−8.
We further conducted trans-ancestry meta-analyses using MR-MEGA (v0.2)88 to account for potential heterogeneity at our lead SNPs. MR-MEGA uses multidimensional scaling of allele frequencies across all the cohorts to derive principal axes of genetic variation that can be used for ancestry adjustment. Using one principal component, which captured all of the population structure in the dataset, we estimated the heterogeneity of ancestry-associated (P-value_ancestry_het) and residuals (P-value_residual_het) for each lead SNP.
Gene-sex interaction analysis.
We performed a sex-stratified meta-analysis at our 11 genome-wide significant CAC loci using the lead SNPs for a subset of cohorts (~75% total sample; 10 European ancestry cohorts with 9,058 males and 10,132 females and 3 African ancestry cohorts with 1,816 males and 1,873 females) with reported sex-stratified GWAS results, following a similar approach used for the sex combined analyses with all cohorts (same additive model and fixed-effects model in METAL). We tested the difference of the results between the two strata using EasyStrata89,90. The SNP-sex interaction was assessed by extracting a two-tailed P-value from the Z-score using the following equation:
Beta1 and Beta2 are the estimated CAC effect sizes for the SNP in males and females separately, and se(Beta1) and se(Beta2) are the standard errors for the Beta estimated from males and females. The interaction test of the two groups was performed within the combined ancestry populations and the results are shown in Supplementary Table 3. Sex-specific GWAS and SNP-sex interaction analyses were determined to be significant at P < 4.54 × 10−3 (Bonferroni correction for 11 tests).
Genomic risk loci definition.
We used FUMA version v1.3.6a to obtain the genomic risk loci and functional information for the relevant SNPs in these loci based on 1000G phase 3 (version 5 based on ALL populations). FUMA combines several external data sources to provide comprehensive annotation information. First, independent significant SNPs, at P < 5 × 10−8 and a linkage disequilibrium (LD) at r2 < 0.6 were identified through LD-based clumping in FUMA. The independent genomic risk loci were defined by identifying all SNPs in LD (r2 ≥ 0.6) with, and in a region of 250 kilobases (kb) around one of the independent significant SNPs. We further defined the lead SNPs in each genomic risk locus as a subset of the independent significant SNPs that were in approximate LD with each other at r2 < 0.1 through LD-based clumping. In naming the nearest genes for the independent loci, we mapped the nearest gene (protein coding or non-coding) to the lead SNPs based on physical location to the transcription start sites (TSS). We also assigned the nearest protein-coding genes at these loci using canonical TSS from GENCODE (v30) in ANNOVAR functional annotator (version 2019–10-24)91.
Genome-wide gene-based analysis.
We performed gene-based analyses to summarize SNP associations at the gene level to map these gene sets to biological pathways. We used MAGMA v1.0828 through FUMA to perform gene-based analyses using the summary statistics of the combined meta-analysis in a window of 50 kb around 19,177 protein-coding genes (mapped to GRCh37/hg19 based on Ensembl 92). We used default settings to calculate empirical P-values derived from 1,000 permutations and set a nominal P-value conservatively at 0.05/19,177 = 2.61 × 10-6. MAGMA leverages the per-variant test statistics by applying a multiple regression model to derive an empirical P-value for association of individual genes considering the LD structure that exists among variants and potential multi-marker effects28.
To further prioritize candidate genes, we also ran Polygenic Priority Score (PoPS v2.0)29 using gene expression information from relevant human tissues. The above MAGMA gene annotation and scores for European-ancestry-only GWAS summary statistics were generated with FUMA and features included from GTEx v8 coronary artery, aorta and tibial artery tissues and no controls were used; otherwise, default settings were used as described in https://github.com/FinucaneLab/pops (accessed March 15, 2023).
eQTL based fine-mapping.
In order to identify causal genes at CAC loci, we used two eQTL-based statistical fine-mapping methods, summary level Mendelian randomization (SMR)30 and coloc (v5.1.0)31. We integrated the European ancestry CAC GWAS summary statistics with eQTL summary data from the STARNET cohort (European ancestry) of 7 cardiometabolic tissues: atherosclerotic aortic root (AOR), whole blood (Blood), liver (LIV), mammary artery (MAM), subcutaneous fat (SF), visceral fat (VF), and skeletal muscle (SKLM), derived from ~600 individuals32, with a focus on AOR, MAM, and LIV tissues. We first used SMR to test whether top CAC GWAS variants influence the phenotype through perturbation of gene expression in these atherosclerosis relevant tissues. We considered only genes with at least one cis-eQTL P < 5 × 10−5 for colocalization. To account for a model of linkage, where two distinct signals drive the association with gene expression and CAC, we used the 1000G EUR LD reference panel and the post-hoc heterogeneity in dependent instruments (HEIDI) test 1 to exclude loci with evidence of linkage or heterogeneity in the genetic instruments. We performed the SMR/HEIDI test on 5,233 and 5,293 eGenes (PeQTL < 5 × 10−5) in AOR and MAM tissues, respectively, to identify genes that passed a q-value92 threshold < 0.10 (AOR: PSMR < 3 × 10−4, MAM: PSMR < 4 × 10−4) and HEIDI test (PHEIDI > 0.01).
We also performed a colocalization analysis using the R based package, coloc31. This Bayesian statistical approach calculates the posterior probability that the CAC GWAS and eQTL data from different STARNET tissues share a common signal under the one causal variant assumption. Following a filtering step to include only eQTLs with P < 0.05, we tested for colocalization between overlapping variants in the CAC GWAS and STARNET expression data. We considered PP4 > 0.8 as strong evidence of colocalization. We considered strongly colocalized loci for CAC as those having PP4 > 0.8 for CAC but PP4 < 0.5 for CAD, and strongly shared colocalized loci as those having PP4 > 0.8 for both traits.
snATAC-seq analysis.
Atherosclerotic coronary artery segments were obtained from explanted hearts from 41 patients undergoing heart transplantation or donor hearts procured for research purposes at Stanford University. All samples were collected under Institutional Review Board (IRB) approval and written informed consent. Frozen tissues were transferred to the University of Virginia through a material transfer agreement and IRB approved protocols and stored at −80 °C until day-of-processing. For snATAC-seq analysis of human coronary artery samples, nuclei were isolated from frozen tissues, purified over an Opti-Prep sucrose gradient as described39 and subjected to 10X Genomics based library preparation and sequencing on a NovaSeq 6000 (paired end, 2 × 50 bp) to achieve ~50,000 unique fragments per cell. Initial pre-processing was performed using the 10X Genomics pipeline (Cell Ranger ATAC v1.2.0). All ATAC-seq reads were mapped to the human genome reference hg38 build using the default parameters. Approximately 28,000 cells were included in the clustering analysis after filtering for high-quality cells with TSS enrichment >7.0 and >10,000 fragments using the ArchR package (v.1.0.1) package93. ArchR was also used for downstream analyses including dimensionality reduction, clustering, calculation of imputed gene scores, and Peak2gene links as described39. snATAC tracks were visualized on the UCSC browser and compared with existing bulk coronary artery ATAC-seq tracks.
We used the LDSC package (https://github.com/bulik/ldsc) to perform LDSC on our published snATAC-seq peaks and the European ancestry CAC summary statistics. The munge_sumstats.py script was used to convert the summary statistics to a compatible format for LDSC. For each coronary artery cell type, we lifted over bed file peak coordinates from hg38 to hg19. The hg19 bed files were then used to make annotation files for each cell type. We performed LDSC according to the cell-type-specific analysis tutorial (https://github.com/bulik/ldsc/wiki/Cell-type-specific-analyses).
Gene-set pathway enrichment analysis.
Gene set pathway enrichment analyses were first performed in MAGMA28 using both European ancestry and African ancestry CAC summary statistics (both unfiltered and subset to P < 1 × 10−5). Gene-based P-values were computed based on the total sample size, and a gene annotation window of 2 kb upstream and 1 kb downstream of candidate genes. Mapped genes were used for MAGMA gene-set enrichment analyses against the 10,678 gene sets (curated, GO terms and MsigDB v 6.2). GTEx v8 tissues, including blood vessel tissues (coronary, aorta, and tibial artery) were used for the MAGMA gene-property analysis for tissue specificity. We also imported CAC strongly colocalized and CAC/CAD shared colocalized gene sets into enrichR (2020 update) to determine enriched pathways using a combination of databases including BioPlanet, BioCarta, KEGG, WikiPathways, Reactome, and PANTHER94.
Carotid plaque analyses.
Atherosclerotic plaques were obtained from patients undergoing a carotid endarterectomy (CEA) procedure and included in the Athero-Express Biobank Study (AE, approved and registered under number TME/C-01.18), an ongoing biobank study in Utrecht, The Netherlands95. The study design of the AE has been described before95; however, in brief: during surgery both blood and plaques are obtained, stored at −80 °C and plaque material is routinely used for standardized (immuno)histochemical analysis95,96. The number of macrophages (CD68), smooth muscle cells (alpha-SMA), intraplaque vessel density (CD34), intraplaque hemorrhage, intraplaque fat, calcification, collagen, and plaque vulnerability97 were scored as previously described98. Genotype data was used to perform regression analyses of histological plaque vulnerability and morphology characteristics, adjusted for age, sex, and principal components. Detailed methods are in the Supplementary Note, and scripts are provided here: https://github.com/CirculatoryHealth/CHARGE_1000G_CAC.
Heritability estimation of CAC.
We used LD-score regression47 to estimate the proportion of variance in CAC that could be explained by the aggregated effect of the SNPs in those of European ancestry. The method assumes that an estimated SNP effect includes effects of all SNPs in LD with that SNP. On average, a SNP that tags many other SNPs will have a higher probability of tagging a causal variant than a SNP that tags few other SNPs. Thus, SNPs with a higher LD-score have, on average, stronger effect sizes than SNPs with lower LD-scores. By regressing the effect size obtained from the GWAS against the LD-score for each SNP, the slope of the regression line will provide an estimate of heritability based on the analyzed SNPs. We included 1,167,424 SNPs with available betas. After merging with the European reference panel, 1,164,129 SNPs remained. SNP heritability was estimated using European LD scores from 1000G phase 3 data for the HapMap3 SNPs, downloaded from https://data.broadinstitute.org/alkesgroup/.
Genetic correlations.
We used cross-trait LD-score regression to estimate the genetic covariation between traits using GWAS summary statistics99. The genetic covariance is estimated using the slope from the regression of the product of z-scores from two GWAS studies on the LD-score. The estimate obtained from this method represents the genetic correlation between the two traits based on all polygenic effects captured by SNPs. Standard LD-scores were used as provided by Bulik-Sullivan et al. based on the 1000 Genomes reference set100, restricted to European ancestry populations. Genetic correlations and specific sources of the GWAS studies are provided in Supplementary Table 15.
Mendelian randomization analyses.
Two-sample Mendelian randomization (MR) using summary-level data was applied to investigate potential causality between CVD risk factors and a higher CAC quantity101, and a higher CAC quantity with CAD. We selected SNPs associated with each CVD risk factor at the genome-wide level of significance (P < 5 × 10−8) as our exposure. SNPs were identified from publicly available genome-wide association studies and only studies with European ancestry populations were considered. We excluded SNPs that had LD with other variants, were absent from the LD reference panel or were palindromic with intermediate allele frequencies. A total of 75 independent genetic instruments for LDL cholesterol, 83 for HDL cholesterol102, 54 for triglycerides102, 375 for systolic blood pressure103, 378 for diastolic blood pressure103, 74 for body mass index104 and 108 for type 2 diabetes were included. As an outcome, the European ancestry summary statistics of our CAC GWAS were used. Significance was defined as P < 0.05/7 = 7.14 × 10−3 after adjusting for multiple tests. For the association of CAC with CAD, we selected the lead 16 independent SNPs in 11 different loci associated with CAC quantity in the current European GWAS as the exposure and summary statistics of CAD as our outcome50. If a requested SNP was not present in the CAD GWAS or being palindromic with intermediate allele frequencies, a proxy SNP that was in LD from the European reference panel with the requested SNP would be used instead. This was done using the LDproxy Tool from LDLink105. Alternatively, we selected the 5 CAC lead SNPs at loci that are not associated with CAD to repeat the MR analysis. Inverse variance-weighted (IVW) analyses were used in which combined effects of the individual genetic instruments on the outcome, here being CAC quantity, result in a weighted mean estimate of a genetically determined increase in exposure on the outcome106. Moreover, we performed weighted median estimator (WME) and MR Egger regression analyses as sensitivity analyses to rule out potential bias caused by directional pleiotropy107. The analyses and data visualizations were performed using MRCIEU/TwoSampleMR (v0.5.6)108 and ggplot2 R packages.
Immunofluorescence analysis in coronary arteries.
Freshly isolated coronary arteries were obtained from consented heart transplant recipients or heart donors as described39,109. Briefly, hearts were arrested in cardioplegic solution, transferred on ice, and left anterior descending (LAD), left circumflex artery and right coronary arteries were dissected from the epicardium, with surrounding adipose and myocardial tissue carefully removed. Coronary artery segments were grossly scored for presence of lesions, embedded in OCT, snap frozen in liquid nitrogen, and stored at −80 °C until analysis. OCT blocks were cryosectioned at 8 μm. Frozen sections were washed, fixed in 4% formaldehyde, permeabilized with triton X-100 at 0.05%, blocked with donkey serum and incubated overnight with primary antibodies as follows: mouse anti-ENPP1 (SC-166649, Santa Cruz Biotechnology, Dallas, TX), rabbit anti-IGFBP3 (#10189–2-AP, Proteintech, Rosemont, IL), rabbit anti-ARID5B (#HPA015037, Atlas Antibodies, Stockholm, SWE), or mouse anti-ADK (SC-514588, Santa Cruz Biotechnology) at 1:100 dilution and mouse anti-α-SMA (SC-53142, Santa Cruz Biotechnology) at 1:100 dilution. Slides were washed with PBS-Tween and incubated with appropriate secondary antibodies. More detailed methods are included in the Supplementary Note.
Calcification assay.
Calcification was induced in siRNA-transfected cells (20 nM) after 24 h, using osteogenic conditions as described above. Media was changed every 48–72 h and cells were retreated with siRNA 5 days after initial transfection, again at 20 nM final concentration. Cells were grown in osteogenic conditions for a total of 18 days, after which they were fixed with 4% paraformaldehyde and stained with Alizarin Red to detect calcification. For Alizarin Red staining, cells were incubated with 1% Alizarin Red solution (pH 4.1–4.3) for 10 minutes, washed multiple times with distilled water, and then images were captured.
Proliferation assay.
HCASMCs were plated in a 96-well format and treated with either siCTRL, siENPP1, siIGFBP3, siARID5B, or siADK at 20 nM final concentration for 24 h. Media was then changed, and cells were incubated for an additional 48 h, at which time MTT assay (#30–1010K, ATCC, Manassas, VA) was performed according to manufacturer instructions. Briefly, MTT reagent was added to each well, and the plate was incubated at 37 °C in the dark for 2 h, at which point a purple precipitate was visible inside the cells via light microscopy. Detergent reagent was then added to each well, and the plate was incubated at room temperature in the dark for an additional 2 h before measurement of absorbance at 570 nm and 650 nm.
Migration assay.
HCASMCs were plated in a 6-well format and treated with either siCTRL, siENPP1, siIGFBP3, siARID5B, or siADK at 20 nM final concentration. After 24 h, media was changed and cells were allowed to incubate for an additional 24 h. Cells were then trypsinized, counted, and seeded in silicone insert with defined cell-free gap (#80209, Ibidi, Gräfelfing, DE) at a density of 4 × 105 cells/mL. Cells were then incubated overnight, dividers were removed, osteogenic media was added, and cells were observed until sufficient migration had taken place in the control group. After 6 h, all wells were fixed with 4% PFA, stained with Giemsa, and then washed with deionized water. Images were obtained using the Leica DMI 4000B inverted microscopy station and digitized with the Leica Application Suite X software. ImageJ was used to calculate the percentage of the cell-free gap occupied by cells.
Druggability analysis.
We used several databases to explore the potentially druggability of CAC candidate gene targets prioritized using the various statistical and functional fine-mapping methods. Candidate genes were first annotated for predicted function using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We then queried gene targets in the druggable genome using the most recent druggable genome list established from the NIH Illuminating the Druggable Genome Project (https://github.com/druggablegenome/IDGTargets) that are also available via the Pharos web platform. Druggability target categories and predicted drug-gene interactions were queried using an API script for the latest Drug-Gene Interaction database (DGIdb v4.0) to retrieve the top interacting drugs from 43 databases. We also queried protein targets for available active ligands in ChEMBL. We performed a more comprehensive druggability analysis for the identified approved drugs by querying the DrugBank, ChEMBL and ClinicalTrials.gov databases to provide drug annotations, and information on late-stage clinical trials, and disease indications.
Statistical analysis.
Details on the statistical tests performed are listed in the respective Methods sections or figure legends. For the functional experiments in cultured smooth muscle cells, statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, La Jolla, CA). Comparisons of more than two groups were performed using a two-tailed one-way ANOVA with Sidak’s post-hoc correction for multiple comparisons. Data are reported as mean ± standard error of mean (SEM), unless otherwise specified. A P-value ≤ 0.05 (adjusted for multiple comparisons) was considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by grants from: the National Institutes of Health (R01HL148239 and R01HL164577 to C.L.M.; R01HL142809 and R01HL159514 to R.M.; F31HL156463 to D.W.; R01HL125863 to J.L.M.B.; R01 HL146860 to P.S.d.V., K01HL164687 to C.L.L.C., N.R.H., P.A.P., and L.F.B.; R01 HL163972 to N.F.; P30 DK063491 to J.I.R.; R01 DK114183 to T.L.A.; European Union funded H2020 TO_AITION (grant 848146 to S.W.v.d.L.); Netherlands CardioVascular Research Initiative of the Netherlands Heart Foundation (CVON 2011/B019 and CVON 2017–20 to S.W.v.d.L.: Generating the best evidence-based pharmaceutical targets for atherosclerosis [GENIUS I&II]), the ERA-CVD program ‘druggable-MI-targets’ (01KL1802 to S.W.v.d.L.), and the Leducq Foundation (‘PlaqOmics’ 18CVD02 to C.L.M., J.L.M.B. and S.W.v.d.L.). The CHARGE consortium is supported by NHLBI grant R01HL105756. A full list of the funding support for each study is provided in the Supplementary Note. M.d.W. is supported by the Netherlands Heart Foundation (CVON 2011/B019, CVON 2017–20); the Netherlands Heart Foundation and Spark-Holding BV (2019B016); Leducq Foundation (LEAN 16CVD01); Amsterdam UMC; ZonMW (Open competition 09120011910025). A complete list of acknowledged funding support for individual studies is provided in Supplementary Table 23.
Footnotes
Competing Interests
S.W.v.d.L. has received Roche funding for unrelated work. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. R.M. receives research funding from Angea Biotherapeutics and Amgen and serves as a consultant for Myokardia/BMS, Renovacor, Epizon Pharma, and Third Pole, all unrelated to the current project. C.L.M. has received funding from AstraZeneca on an unrelated project. J.C.K. is the recipient of an Agilent Thought Leader Award (January 2022), which includes funding for research that is unrelated to the current manuscript. All other co-authors confirm that they have no conflicts of interest related to the presented work.
Code availability
General post-GWAS analysis scripts: https://github.com/CirculatoryHealth/CHARGE_1000G_CAC
Post-GWAS fine-mapping scripts: https://github.com/MillerLab-CPHG/Fine_mapping/
Coronary artery snATAC: https://github.com/MillerLab-CPHG/Coronary_scATAC
Data availability
The GWAS meta-analysis summary statistics are available on the EMBL-EBI GWAS catalog (accession numbers: GCST90278455 and GCST90278456) and the Downloads page of the Cardiovascular Disease Knowledge Portal (CVDKP), and will be integrated into CVDKP. Locus zoom plots are available at https://my.locuszoom.org/gwas/125033/ and FUMA output results are available through the FUMA website.
STARNET eQTL data are available in the Database for Genotypes and Phenotypes (dbGaP) via accession: phs001203.v1.p1, as well as web browser: http://starnet.mssm.edu. Coronary artery snATAC data are available in the Gene Expression Omnibus (GEO) database via accession: GSE175621. Athero-Express scRNAseq data are available at https://doi.org/10.34894/TYHGEF. snATAC and scRNAseq processed datasets are also available on the PlaqView web portal. Athero-Express GWAS, and phenotype data are available at https://doi.org/10.34894/4IKE3T.
Genetic variants for imputation were obtained from 1000 Genomes Phase 3 (v5). Variant annotations were obtained from Ensembl (v92). PheWAS data were obtained from the GWAS Atlas. Gene annotations were obtained from GENCODE (v30). eQTL data were also obtained from Genotype Tissue Expression (GTEx v8). Epigenomics data were obtained from Roadmap Epigenomics (release 9) and ENCODE (v4). Pathway annotations were obtained from MsigDB (v6.2) and Kyoto Encyclopedia of Genes and Genomes (KEGG2). Druggability annotations were obtained from: DGIDb (v4.0), IDGTargets, Pharos, ChEMBL, DrugBank, and ClinicalTrials.gov.
REFERENCES
- 1.Timmis A et al. European society of cardiology: Cardiovascular disease statistics 2017. Eur. Heart J. 39, 508–579 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Tsao CW et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation 145, e153–e639 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Libby P, Ridker PM & Hansson GK Progress and challenges in translating the biology of atherosclerosis. Nature 473, 317–325 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Baber U et al. Prevalence, impact, and predictive value of detecting subclinical coronary and carotid atherosclerosis in asymptomatic adults: the BioImage study. J. Am. Coll. Cardiol. 65, 1065–1074 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Polonsky TS et al. Coronary artery calcium score and risk classification for coronary heart disease prediction. JAMA 303, 1610–1616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kavousi M et al. Evaluation of Newer Risk Markers for Coronary Heart Disease Risk Classification. Annals of Internal Medicine 156, 438–444 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Bielak LF, Rumberger JA, Sheedy PF 2nd, Schwartz RS. & Peyser PA. Probabilistic model for prediction of angiographically defined obstructive coronary artery disease using electron beam computed tomography calcium score strata. Circulation 102, 380–385 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Grundy SM et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 139, e1082–e1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H-Y et al. The Relationship Between Coronary Calcification and the Natural History of Coronary Artery Disease. JACC Cardiovasc. Imaging 14, 233–242 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Jinnouchi H et al. Calcium deposition within coronary atherosclerotic lesion: Implications for plaque stability. Atherosclerosis 306, 85–95 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Durham AL, Speer MY, Scatena M, Giachelli CM & Shanahan CM Role of smooth muscle cells in vascular calcification: implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 114, 590–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakahara T et al. Coronary Artery Calcification: From Mechanism to Molecular Imaging. JACC Cardiovasc. Imaging 10, 582–593 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Fujiyoshi A et al. Coronary Artery Calcium and Risk of Dementia in MESA (Multi-Ethnic Study of Atherosclerosis). Circ. Cardiovasc. Imaging 10, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handy CE et al. The Association of Coronary Artery Calcium With Noncardiovascular Disease: The Multi-Ethnic Study of Atherosclerosis. JACC Cardiovasc. Imaging 9, 568–576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann DM et al. Coronary artery calcification is an independent stroke predictor in the general population. Stroke 44, 1008–1013 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Peyser PA et al. Heritability of coronary artery calcium quantity measured by electron beam computed tomography in asymptomatic adults. Circulation 106, 304–308 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Wojczynski MK et al. Genetics of coronary artery calcification among African Americans, a meta-analysis. BMC Med. Genet. 14, 75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan P et al. Multiethnic Exome-Wide Association Study of Subclinical Atherosclerosis. Circ. Cardiovasc. Genet. 9, 511–520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donnell CJ et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 124, 2855–2864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Setten J et al. Serum Lipid Levels, Body Mass Index, and Their Role in Coronary Artery Calcification. Circ. Cardiovasc. Genet. 8, 327–333 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Nelson CP et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 49, 1385–1391 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Psaty BM et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2, 73–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agatston AS et al. Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 15, 827–832 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Shen H et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 170, 1850–1855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Taskesen E, van Bochoven A & Posthuma D Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks EM et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. Nat. Genet. 55, 1267–1276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franzén O et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 353, 827–830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franceschini N et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat. Commun. 9, 5141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao K et al. Integrative Prioritization of Causal Genes for Coronary Artery Disease. Circ. Genom. Precis. Med. 15, e003365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cano-Gamez E & Trynka G From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 11, 424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B et al. Genetic Regulatory Mechanisms of Smooth Muscle Cells Map to Coronary Artery Disease Risk Loci. Am. J. Hum. Genet. 103, 377–388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fulco CP et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Sarkar A, Kheradpour P, Ernst J & Kellis M Evidence of reduced recombination rate in human regulatory domains. Genome Biol. 18, 193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner AW et al. Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nat. Genet. 54, 804–816 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen J et al. Regulation of Vascular Calcification by Growth Hormone-Releasing Hormone and Its Agonists. Circ. Res. 122, 1395–1408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan H et al. Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation 142, 2060–2075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirka RC et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alsaigh T, Evans D, Frankel D & Torkamani A Decoding the transcriptome of calcified atherosclerotic plaque at single-cell resolution. Commun Biol 5, 1084 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slenders L et al. Intersecting single-cell transcriptomics and genome-wide association studies identifies crucial cell populations and candidate genes for atherosclerosis. Eur. Heart J. Open 2, oeab043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depuydt MAC et al. Microanatomy of the Human Atherosclerotic Plaque by Single-Cell Transcriptomics. Circ. Res. 127, 1437–1455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Laan SW et al. Genetic Susceptibility Loci for Cardiovascular Disease and Their Impact on Atherosclerotic Plaques. Circ Genom Precis Med 11, e002115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tcheandjieu C et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat. Med. 28, 1679–1692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemani G, Bowden J & Davey Smith G Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 27, R195–R208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Harst P & Verweij N Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 122, 433–443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donovan K et al. Fibroblast Growth Factor-23 and Risk of Cardiovascular Diseases: A Mendelian Randomization Study. Clin. J. Am. Soc. Nephrol. 18, 17–27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall KT et al. Catechol-O-Methyltransferase and Cardiovascular Disease: MESA. J. Am. Heart Assoc. 8, e014986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nitschke Y et al. Generalized arterial calcification of infancy and pseudoxanthoma elasticum can be caused by mutations in either ENPP1 or ABCC6. Am. J. Hum. Genet. 90, 25–39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ralph D et al. ENPP1 variants in patients with GACI and PXE expand the clinical and genetic heterogeneity of heritable disorders of ectopic calcification. PLoS Genet. 18, e1010192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinnott-Armstrong N et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrera-Rivero M et al. Single- and Multimarker Genome-Wide Scans Evidence Novel Genetic Risk Modifiers for Venous Thromboembolism. Thromb. Haemost. 121, 1169–1180 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Spracklen CN et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 582, 240–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato N et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 47, 1282–1293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giri A et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat. Genet. 51, 51–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thériault S et al. Identification of Circulating Proteins Associated With Blood Pressure Using Mendelian Randomization. Circ Genom Precis Med 13, e002605 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Kichaev G et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 104, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez Cordero AI et al. Genome-wide Associations Reveal Human-Mouse Genetic Convergence and Modifiers of Myogenesis, CPNE1 and STC2. Am. J. Hum. Genet. 105, 1222–1236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SK Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS One 13, e0200785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuler-Lüttmann S et al. Insulin-Like Growth Factor–Binding Protein-3 Is Associated With the Presence and Extent of Coronary Arteriosclerosis. Arterioscler. Thromb. Vasc. Biol. 20, e10–e15 (2000). [PubMed] [Google Scholar]
- 65.Claussnitzer M et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 373, 895–907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitson RH Jr, Li S-L., Zhang G., Larson GP. & Itakura K. Mice with Fabp4-Cre ablation of Arid5b are resistant to diet-induced obesity and hepatic steatosis. Mol. Cell. Endocrinol. 528, 111246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe M et al. Regulation of smooth muscle cell differentiation by AT-rich interaction domain transcription factors Mrf2alpha and Mrf2beta. Circ. Res. 91, 382–389 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Wang G et al. Genetic variations of Mrf-2/ARID5B confer risk of coronary atherosclerosis in the Japanese population. Int. Heart J. 49, 313–327 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Wang G et al. Associations of variations in the MRF2/ARID5B gene with susceptibility to type 2 diabetes in the Japanese population. J. Hum. Genet. 57, 727–733 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Hata K et al. Arid5b facilitates chondrogenesis by recruiting the histone demethylase Phf2 to Sox9-regulated genes. Nat. Commun. 4, 2850 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Paganelli F, Gaudry M, Ruf J & Guieu R Recent advances in the role of the adenosinergic system in coronary artery disease. Cardiovasc. Res. 117, 1284–1294 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Xu Y et al. Regulation of endothelial intracellular adenosine via adenosine kinase epigenetically modulates vascular inflammation. Nat. Commun. 8, 943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M et al. Ablation of Myeloid ADK (Adenosine Kinase) Epigenetically Suppresses Atherosclerosis in ApoE−/− (Apolipoprotein E Deficient) Mice. Arterioscler. Thromb. Vasc. Biol. 38, 2780–2792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kauffenstein G et al. Alteration of Extracellular Nucleotide Metabolism in Pseudoxanthoma Elasticum. J. Invest. Dermatol. 138, 1862–1870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boison D Adenosine kinase: exploitation for therapeutic gain. Pharmacol. Rev. 65, 906–943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozkok A et al. FGF-23 associated with the progression of coronary artery calcification in hemodialysis patients. BMC Nephrol. 14, 241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murali SK et al. FGF23 Regulates Bone Mineralization in a 1,25(OH)2 D3 and Klotho-Independent Manner. J. Bone Miner. Res. 31, 129–142 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Donovan K et al. Fibroblast growth factor-23 and risk of cardiovascular diseases: A Mendelian randomisation study. Clin. J. Am. Soc. Nephrol. 18, 17–27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexopoulos N & Raggi P Calcification in atherosclerosis. Nat. Rev. Cardiol. 6, 681–688 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Arbab-Zadeh A & Fuster V The myth of the ‘vulnerable plaque’: transitioning from a focus on individual lesions to atherosclerotic disease burden for coronary artery disease risk assessment. J. Am. Coll. Cardiol. 65, 846–855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schoenhagen P et al. Extent and direction of arterial remodeling in stable versus unstable coronary syndromes : an intravascular ultrasound study. Circulation 101, 598–603 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Mauriello A et al. Coronary calcification identifies the vulnerable patient rather than the vulnerable Plaque. Atherosclerosis 229, 124–129 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Nicoll R, Zhao Y, Ibrahimi P, Olivecrona G & Henein M Diabetes and Hypertension Consistently Predict the Presence and Extent of Coronary Artery Calcification in Symptomatic Patients: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 17, 1481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Toorn JE et al. Arterial calcification at multiple sites: sex-specific cardiovascular risk profiles and mortality risk-the Rotterdam Study. BMC Med. 18, 263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kronmal RA et al. Risk factors for the progression of coronary artery calcification in asymptomatic subjects: results from the Multi-Ethnic Study of Atherosclerosis (MESA). Circulation 115, 2722–2730 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Saleheen D et al. Loss of Cardioprotective Effects at the ADAMTS7 Locus as a Result of Gene-Smoking Interactions. Circulation 135, 2336–2353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polfus LM et al. Genome-wide association study of gene by smoking interactions in coronary artery calcification. PLoS One 8, e74642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 88.Mägi R et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 26, 3639–3650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winkler TW et al. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics 31, 259–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Randall JC et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 9, e1003500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Granja JM et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Verhoeven BAN et al. Athero-express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur. J. Epidemiol. 19, 1127–1133 (2004). [DOI] [PubMed] [Google Scholar]
- 96.van Lammeren GW et al. Atherosclerotic plaque vulnerability as an explanation for the increased risk of stroke in elderly undergoing carotid artery stenting. Stroke 42, 2550–2555 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Verhoeven B et al. Carotid atherosclerotic plaques in patients with transient ischemic attacks and stroke have unstable characteristics compared with plaques in asymptomatic and amaurosis fugax patients. J. Vasc. Surg. 42, 1075–1081 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Hellings WE et al. Intraobserver and interobserver variability and spatial differences in histologic examination of carotid endarterectomy specimens. J. Vasc. Surg. 46, 1147–1154 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Zheng J et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lawlor DA Commentary: Two-sample Mendelian randomization: opportunities and challenges. International journal of epidemiology 45, 908–915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Willer CJ et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Evangelou E et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yengo L et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Machiela MJ & Chanock SJ LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bowden J & Holmes MV Meta-analysis and Mendelian randomization: A review. Res Synth Methods 10, 486–496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burgess S & Thompson SG Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hemani G et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hartiala JA et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 42, 919–933 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS meta-analysis summary statistics are available on the EMBL-EBI GWAS catalog (accession numbers: GCST90278455 and GCST90278456) and the Downloads page of the Cardiovascular Disease Knowledge Portal (CVDKP), and will be integrated into CVDKP. Locus zoom plots are available at https://my.locuszoom.org/gwas/125033/ and FUMA output results are available through the FUMA website.
STARNET eQTL data are available in the Database for Genotypes and Phenotypes (dbGaP) via accession: phs001203.v1.p1, as well as web browser: http://starnet.mssm.edu. Coronary artery snATAC data are available in the Gene Expression Omnibus (GEO) database via accession: GSE175621. Athero-Express scRNAseq data are available at https://doi.org/10.34894/TYHGEF. snATAC and scRNAseq processed datasets are also available on the PlaqView web portal. Athero-Express GWAS, and phenotype data are available at https://doi.org/10.34894/4IKE3T.
Genetic variants for imputation were obtained from 1000 Genomes Phase 3 (v5). Variant annotations were obtained from Ensembl (v92). PheWAS data were obtained from the GWAS Atlas. Gene annotations were obtained from GENCODE (v30). eQTL data were also obtained from Genotype Tissue Expression (GTEx v8). Epigenomics data were obtained from Roadmap Epigenomics (release 9) and ENCODE (v4). Pathway annotations were obtained from MsigDB (v6.2) and Kyoto Encyclopedia of Genes and Genomes (KEGG2). Druggability annotations were obtained from: DGIDb (v4.0), IDGTargets, Pharos, ChEMBL, DrugBank, and ClinicalTrials.gov.


