Abstract
Human islet antigen reactive CD4 + memory T cells (IAR T cells) from peripheral blood have been studied extensively for their role in the pathogenesis of autoimmune type 1 diabetes (T1D). However, IAR T cells are rare, and it remains poorly understood how they affect T1D progression in the pancreas. Using single cell RNA-sequencing coupled with a multiplexed activation induced marker (AIM) enrichment assay, we identified paired TCR alpha/beta (TRA/TRB) T cell receptors (TCRs) in IAR T cells from the blood of healthy, at-risk, new onset, and established T1D donors. Using TCR sequences as barcodes, we measured infiltration of IAR T cells from blood into pancreas of organ donors with and without T1D. We detected extensive TCR sharing between IAR T cells from peripheral blood and pancreatic infiltrating T cells (PIT), with perfectly matched or single mismatched TRA junctions and J gene regions, comprising ~ 34% of unique IAR TCRs. PIT-matching IAR T cells had public TRA chains that showed increased use of germline-encoded residues in epitope engagement and a propensity for cross-reactivity. The link with T cells in the pancreas implicates autoreactive IAR T cells with shared TRA junctions and increased levels in blood with the prediabetic and new onset phases of T1D progression.
Introduction
Many studies have investigated the role of islet antigen reactive (IAR) CD4 + and CD8 + T cells in peripheral blood of subjects with Type 1 Diabetes (T1D). Because of their specificity for islet autoantigens, IAR T cells are thought to be involved in autoimmune destruction of the pancreas. IAR T cells have been investigated for their role in disease mechanisms and as therapeutic targets and biomarkers for beta cell destruction 1–7. Levels of IAR T cells may be increased in the pancreas especially during the active phases of islet autoimmunity, which may last months to years before and after clinical diagnosis 2,3. Since pancreatic biopsy is not tenable in humans, most efforts have focused on peripheral blood. IAR CD4 + and CD8 + T cells are found in blood of at-risk and T1D subjects, but also often in healthy control (HC) subjects 8–10. Distinctive phenotypic properties of IAR T cells in T1D subjects suggest their association with disease, yet because IAR T cells are rare in the blood (1:30,000 to 1:160,000 for GAD65-specific IAR T cells) 9, it remains difficult to ascribe a role for them in pathogenesis in the pancreas.
A defining feature of T cells is the presence of T cell receptors (TCRs) on their cell surfaces. T cells proliferate in response to TCR recognition of antigenic peptides, resulting in clonal expansion of a population of cells with identical TCR sequences at both the nucleic acid and protein sequence levels and the same antigen specificity 11. Using paired TCR sequences identified by single cell RNA-sequencing (scRNA-seq), we previously demonstrated expanded clones of IAR T cells in blood of T1D patients 12. More recently, we identified a population of expanded IAR T cells with diverse specificities determined by TCRs with restricted TRA junctions and germline-constrained antigen recognition properties 13. In combination, these previous studies suggest the possibility that expanded IAR T cell TCR sequences in the blood of T1D patients represent the ontogeny of T cell autoimmune responses during disease. While this suggestion is attractive for both fundamental and translational investigations in T1D, evidence that IAR T cells with these characteristics are found in the pancreas is lacking. Studies have characterized islet infiltrating T cells in the pancreas from organ donors with T1D 14–16, but their linkage to IAR T cells in the blood is unclear.
In this study, we have examined IAR T cells in the pancreas by comparing TCR junction sequences between circulating IAR T cells and pancreatic infiltrating T cells (PIT). We show that a significant fraction of IAR TCRs in peripheral blood share matching TRA chains with PIT TCRs, and vice versa. We characterize PIT-matching TCRs and show that they have features of germline-like (innate) mode(s) of epitope engagement that are associated with TCRs displaying multi-specificity. Finally, we show expansion of PIT-matching TRA chains in blood prior to and near the time of T1D diagnosis, strengthening the link between IAR T cells in blood and pancreatic destruction.
Results
Isolation of TCR sequences from IAR and PIT T cells.
Our central hypothesis is that in vivo expansion of IAR T cells seen in peripheral blood 12 reflects autoimmune destruction of the pancreas during T1D. This hypothesis predicts the presence of IAR T cells in the pancreas during disease. To test this prediction, we utilized the extreme sequence diversity of TCRs to enable their use as “barcodes” for clonal populations of T cells recognizing specific antigens 17. We reasoned that because of the sequence diversity of TCRs and the rarity of IAR T cells in blood, it would be unlikely for individual IAR T cell TCR sequences to be present at high levels in PIT cells by chance. Significant overlap between IAR and PIT cell TCR sequences would therefore suggest a biological role for IAR T cells in the pancreas.
We utilized TCRs from IAR T cells isolated from the peripheral blood of two subject cohorts (Table S1 and Materials and Methods). Cohort 1 was from our previous scRNA-seq comparisons of IAR T cells from HC subjects (n = 11), new onset T1D subjects (newT1D) < 100 days from diagnosis ( n = 26), and established T1D (T1D, n = 16 subjects) 12,13. Cohort 2 comprised additional HC (with no islet-directed autoantibodies) (n = 6) and newT1D subjects(n = 11); subjects with single autoantibodies (moderate risk for developing T1D, n = 8); and subjects having multiple autoantibodies (high risk, n = 6). We used Cohort 2 to validate and expand results obtained with Cohort 1.
We isolated IAR T cells from peripheral blood using an overnight activation induced marker (AIM) assay based on upregulation of CD154 and CD69 12,13. For subjects in cohort 1, PBMC were stimulated with a pool of 35 immunodominant peptides from GAD65, IGRP, ZnT8, and preproinsulin restricted to the high-risk HLA class II DRB1*0401, *0301, or DQ8 molecules 12,13. PBMC from subjects in cohort 2 were stimulated in an HLA agnostic approach with overlapping peptide libraries (20 amino acids (AA) in length, 12 AA overlap) from the above islet proteins (Table S1). For both groups, CD154 + islet peptide activated cells were magnetically enriched, isolated as CD154 + CD69 + by single cell flow sorting and subjected to scRNA-seq to identify paired TRA and TRB chains in IAR T cells. In most experiments, we used TCRs from Cohort 1, comprising 4,331 TCR junctions (2,174 TRA and 2,136 TRB) from 2,784 cells and 53 donors (Tables S1 and S2). Where indicated, we combined Cohorts 1 and 2 and subset to donors with HLA-DRB*04 alleles, yielding 2,967 junctions (1,512 TRA and 1,433 TRB) from 1,873 cells and 43 subjects (Table S1).
For our initial experiments, we focused on Cohort 1. Approximately 90% of these subjects had high-risk DRB1*0401 HLA class II alleles, while ~ 10% had DRB1*0301 alleles. Using molecular cloning, lentiviral re-expression, and TCR functional assays, we identified multiple specific islet antigen epitopes that triggered 29/47 (~ 62%) of the TCRs tested from this cohort 12,13. These TCRs, therefore, represented IAR T cells from peripheral blood with a wide range of specificities.
For PIT cells, we utilized paired TCRs identified by Reverse Transcription PCR of islets or pancreatic tissues from organ donors provided by the Network for Pancreatic Organ Donors with Diabetes (nPOD) 18; a protocol at Vanderbilt University Medical Center/University of Pittsburgh; the Integrated Islet Distribution program (IIDP); and the Alberta Diabetes Institute Islet Core (ADI). PIT cell TCRs comprised a total of 9,798 unique TRA and TRB junctions from pancreatic tissues of autoantibody negative donors without diabetes, autoantibody positive donors representing prediabetic/preclinical stages of T1D, and T1D donors (1,785, 1,447 and 6,566, respectively). TCR junctions represented 4,706 T cells and 5,092 CD8 + T cells. PIT cell TCR sequences contained multiple perfectly matched amino acid junction sequences from preproinsulin reactive TCRs found in islets and peripheral blood 19. IAR T cell and PIT cell TCR sequences were subjected to several filtering steps before use in subsequent analyses (Materials and Methods) and are presented in Table S2. To maximize power, TCR sequences were not subset by subject group or cell type before analysis, unless otherwise indicated.
TCRs from IAR T cells in blood share TRA chain sequence identity with TCRs from PIT cells.
To test for TCR overlap, we first utilized string matches to test for perfectly matched recombined V-J or V-D-J junction sequence overlap between IAR and PIT cell TCRs. Unless otherwise noted, sequence comparisons were made at the amino acid level. We reasoned that perfectly matched junctions are most likely to reflect conserved function. This test showed multiple perfectly matched junction sequences, mainly TRA junctions compared to TRB junction matches (Fig. 1A). Because of the predominance of TRA chain matches, we focused subsequent analyses on TRA chains only. In initial experiments, we obtained essentially identical fractions of PIT-matching with IAR T in both CD4 + and CD8 + T cells. Since both cell types gave equivalent results, we pooled them to increase power in subsequent analyses. Overall, ~ 2.9% (47/1,606) of total IAR T cell TRA junctions perfectly matched PIT junctions; conversely, ~ 0.45% (44/9,798) of PIT cell TCRs matched IAR T cell TCRs. The distribution of perfect matches between different subject groups (HC, T1D and newT1D) and cell types (CD4+, CD8+) did not differ significantly from the distribution of total TCR populations from each group (p-value > 0.05, Fisher’s exact test). Thus, there were numerous perfectly matched TRA junction sequences shared between IAR T cells and PIT TCRs, even though the PIT TCRs were from different individuals than the IAR T cells.
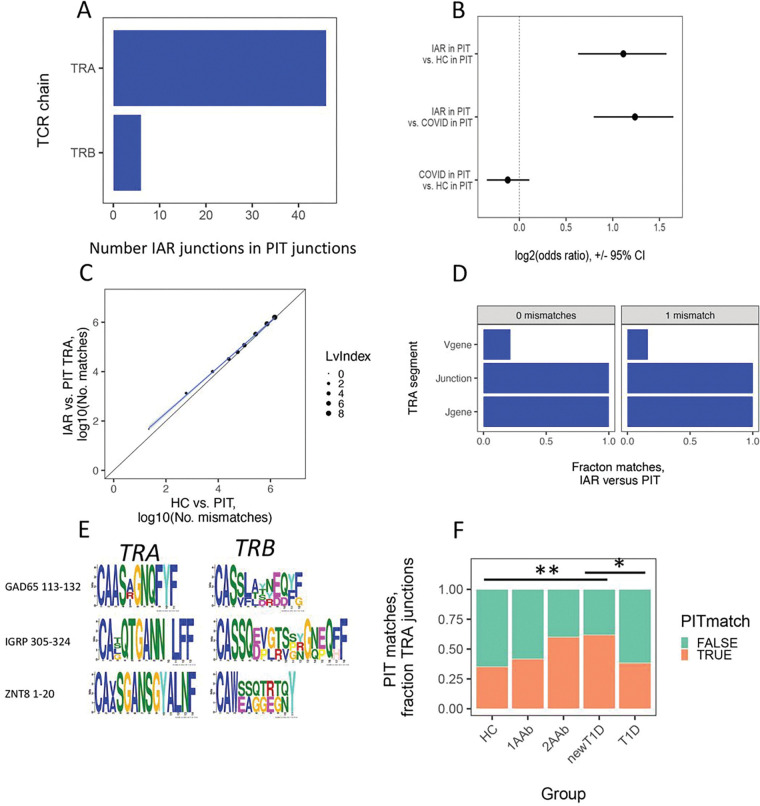
Figure 1. IAR T and PIT TCR sequence matching.
A) Numbers of perfectly matched IAR T cell TCR TRA and TRB junctions from Cohort 1 in PIT TCRs. B) Numbers of PIT-matched versus non-matched TRA junctions from Cohort 1 IAR T cell TCRs compared with HC, and COVID-19 patient TCRs 20. Presented are log2 odds ratios calculated by Fischer’s exact test. Error bars, 95% confidence intervals. Dotted vertical line, log2 odds ratios of identity. C) Numbers of PIT matches in TRA junctions from Cohort 1 IAR T versus matches from a subset of HC donors 20, randomly size-matched to Cohort 1, over a range of mismatch values. Dot sizes, number of sequence pair matches with the indicated numbers of mismatches (Levenshtein index values). Diagonal line, equivalency line. Blue line, best fit line from linear modeling. Gray shading, 95% confidence intervals. D) PIT matches by different TRA junction segments in from Cohort 1 T cells (N = 1,389 TRA junctions). Lv0 and Lv1, Levenshtein index values: 0 (N = 47) and 1 (N = 1,342), respectively. E) MEME plots representing three groups of PIT-matching IAR sequences The topmost TCR in each group recognized 13 the indicated autoantigen epitopes: GAD65 113–132 (N = 4 TCRs); IGRP 305–324, (N=2); and ZNT8 1–20 (N = 2). F) Frequencies of PIT-matched and non-matched TRA junction numbers in IAR T cells by disease group. TRA chains from combined Cohort 1 and Cohort 2 were subset to HLA-DRB1*04 donors, and further separated by expanded and non-expanded cells (467 expanded and 1347 non-expanded cells). There were 12, 15, 145, 55 and 240 cells from 13 HC, 4 1AAb,3 2AAb, 11 newT1D, and 12 T1D donors, respectively. Differences were assessed using Fisher’s exact test. **, pAdj <1e-2; *, pAdj <5e-2. Comparisons not indicated did not differ significantly.
PIT-matched TRA junctions are enriched in IAR CD4 T cells relative to unselected TCR repertoires.
We hypothesized that junction sequence matching between IAR and PIT TCRs was greater than would occur by chance in an unselected repertoire. For comparison, we selected two PBMC repertoires from Su et al. (16 HC and 129 subjects with COVID-19, comprising 2,513 and 198,753 unique junctions for HC and COVID-19 patients, respectively) 20. To compare IAR PIT-matches with those in the unselected repertoires, we first down-sampled the unselected repertoires to match the size of unique IAR TRA junctions (N = 1,606). We then used Fisher’s exact test to compare numbers of perfectly PIT-matched to non-matched TRA junctions from IAR T cells versus unselected HC and COVID-19 T cells. We found more PIT matches with IAR TRA junctions than with junctions from unselected repertoires: there were 47 perfect matches with IAR junctions (2.9%) versus 22 (1.4%) for HC junctions. For IAR T cell junctions, this comparison yielded a log2 odds ratio > 1 (or > 2-fold in linear units), far outside the 95% confidence intervals (Fig. 1B) (p-value < 9e-6, Fisher’s exact test). In contrast, the log2 odds ratio was ~ 0 (or ~ 1 in linear units), and clearly within the 95% confidence intervals, for PIT-matched to non-matched junctions from COVID-19 patients versus HC subjects (Fig. 1B) (p-value > 0.05). Thus, there was a higher fraction of PIT TCR perfect matches with IAR T cells than unselected human TCR repertoires from HC and COVID-19 subjects.
TCRs from IAR T cells with mismatched junctions may also share functional properties. While we expect that shared function would be more likely with mismatches that conserve amino acid electrochemical properties, we have ignored these properties to simplify subsequent analyses. We tested for enrichment of IAR T cell TRA chains in PIT TCRs over a range of mismatch values by calculating pairwise Levenshtein index values. (The Levenshtein index is the number of residue changes needed to transform one sequence into another.) A plot of the numbers of PIT TRA junction matches with IAR versus HC T cells at different PIT mismatch thresholds (Fig. 1C) showed significant off-diagonal skewing in the direction of IAR TCRs (slope = 0.872, p-value (that the slope was not equal to 1) = 3e-3, by linear modeling). Thus, enrichment of PIT-matched to non-matched TRA junctions versus unselected HC TCRs was not limited to perfect sequence matches. Since 95% confidence intervals for PIT junction sequence overlaps with 0 and 1 mismatches showed greatest divergence between IAR T cell and HC TCRs, we chose to define IAR T cell TRA junctions with 0 and 1 mismatches as “PIT matches” in subsequent analyses.
PIT-matched TRA junctions are paired with diverse TRB chains.
It was important to know whether PIT-matched TRA chains were paired with PIT-matched TRB chains. Although we observed a few TRB perfect matches (Fig. 1A), these were not paired with perfectly matched TRA chains. Likewise, perfectly PIT-matched TRA chains were paired with quite different TRB chain sequences (Table 1). We noted similar findings with single mismatched PIT junctions with TCRs of known specificity 13 (Table 1 and Fig. 1E). In this case, TCRs having specificity for one of several islet antigens and epitopes had TRA chains that more clearly matched PIT TRA chains than TRB chains. We conclude that PIT-matched TRA chains of TCRs with multiple antigen specificities were paired with diverse TRB chains. This diversity of TRB chains was not accounted for by different HLA specificities because most of the IAR TCRs used in this comparison shared MHC class II alleles.
Table 1.
TCRs sharing perfectly matched TRA chains between IAR and PIT T cells. A
| TRA chain | TRB chain | |||||
|---|---|---|---|---|---|---|
| Cells | V gene | Junction | J gene | V gene | Junction | J gene |
| IAR | TRAV1-2 | CAVRMNTGFQKLVF | TRAJ8 | TRBV11-2 | CASSFGGGATDTQYF | TRBJ2-3 |
| PIT | TRAV1-2 | CAVRMNTGFQKLVF | TRAJ8 | TRBV9 B | CASSVGMDPGLGYNEQFF | TRBJ2-1 |
| IAR | TRAV12-1 | CVVNDQAGTALIF | TRAJ15 | TRBV7-2 | CASSLDAGRNSPLHF | TRBJ1-6 |
| PIT | TRAV12-1 | CVVNDQAGTALIF | TRAJ15 | TRBV20-1 | CSARGYNSYEQYF | TRBJ2-7 |
| IAR | TRAV12-1 | CVVQGGSYIPTF | TRAJ6 | TRBV5-4 | CASSLVTSGENEQFF | TRBJ2-1 |
| PIT | TRAV12-1 | CVVQGGSYIPTF | TRAJ6 | ND C | ||
| IAR | TRAV12-2 | CAVNQAGTALIF | TRAJ15 | TRBV28 | CASSFGSGADYGYTF | TRBJ1-2 |
| PIT | TRAV12-2 | CAVNQAGTALIF | TRAJ15 | TRBV29-1 | CSVFDWDRGPGELFF | TRBJ2-2 |
| IAR | TRAV12-2 | CAVRSNFGNEKLTF | TRAJ48 | TRBV19 | CASGTDSY-EQYF | TRBJ2-7 |
| PIT | TRAV12-2 | CAVRSNFGNEKLTF | TRAJ48 | TRBV28 | CASRTTGGTEAFF | TRBJ1-1 |
| IAR | TRAV13-1 | CAASIGTGTASKLTF | TRAJ44 | TRBV9 | CASSVA-GGGY-EQYF | TRBJ2-7 |
| PIT | TRAV13-1 | CAASIGTGTASKLTF | TRAJ44 | TRBV24-1 | CATSDPSGGGGNEQFF | TRBJ2-1 |
| IAR | TRAV41 | CAASNTGNQFYF | TRAJ49 | TRBV28 | CAIGGRVYNEQFF | TRBJ2-1 |
| PIT | TRAV41 | CAASNTGNQFYF | TRAJ49 | TRBV5-1 | CASSGSNYGYT-F | TRBJ1-2 |
| IAR | TRAV5 | CAERGLTGGGNKLTF | TRAJ10 | TRBV9 | CASSVGGDFYNEQFF | TRBJ2-1 |
| PIT | TRAV5 | CAERGLTGGGNKLTF | TRAJ10 | TRBV12-5 | CASGLTRGSTDTQYF | TRBJ2-3 |
| IAR | TRAV8-2 | CVVSGGSNYKLTF | TRAJ53 | TRBV29-1 | CSAHGGGGT-EAFF | TRBJ1-1 |
| PIT | TRAV8-2 | CVVSGGSNYKLTF | TRAJ53 | TRBV6-1 | CASSQGTPQYNEQFF | TRBJ2-1 |
| IAR | TRAV8-3 | CAVGPTGTASKLTF | TRAJ44 | TRBV7-6 | CASSTNHQ------ETQYF | TRBJ2-5 |
| PIT | TRAV8-3 | CAVGPTGTASKLTF | TRAJ44 | TRBV3-1 | CASSGTGTGGLSPQETQYF | TRBJ2-5 |
Shown are amino acid sequence comparisons of unique IAR and PIT T cell TCRs. Dashes indicate gaps.
Bold font indicates a mismatch with the IAR reference sequence (top row of each pair).
ND, not determined
PIT TRA junction matches extend to include the J gene but not the V gene.
We hypothesized that different TCRs with greater sequence identity are more likely to share binding and functional specificity. To test the extent of sequence identity for IAR TRA chains with PIT matches, we extended the requirement for sequence matches into the V and J gene segments flanking matched junctions (Fig. 1D). Since the V and J gene segments in TCRs are much longer than junctions 21, they are less amenable to string match comparisons, prompting us to consider V and J genes as identical if they simply had the same name. For all junctions with 0 or 1 PIT mismatches, sequence identity extended towards the C terminus to include use of identical J genes (Fig. 1D). In contrast, sequence identity towards the N terminus was less marked, with only ~ 20–25% of matched junctions having identical V genes.
Since the TCR V gene contains the CDR1 and CDR2 regions that contact the MHC class I or class II molecules 22, it was possible that the low frequency of V gene identical matches reflected peptide presentation to PIT TCRs by different HLA molecules. IAR T cell TCRs were associated primarily with high-risk HLA genotypes 13, whereas PIT TCRs were from individuals having a wider variety of HLA genotypes. We therefore broke down the matching data from Fig. 1D by HLA class II DRB1 genotype. Individuals having 03:01, 04:01, 07:01 and pooled other DRB1 genotypes all showed similar frequencies of V gene identical matches (Figure S1). This led us to conclude that the low frequency of PIT matches in V gene regions was not attributable to peptide presentation by different HLA molecules.
We obtained results in Figs. 1A–1E from Cohort 1 (Materials and Methods). To validate these findings, we repeated these analyses with all samples in an independent cohort (Cohort 2). We obtained essentially identical results, demonstrating that our observations were not restricted to a single data set and therefore had a potentially broader range of islet specificities.
PIT TRA matches with IAR T cell junctions are expanded early in disease.
We speculated that PIT TRA matches were related to T cell expansion and T1D progression. As a test, we quantified levels of PIT TRA matches in IAR TCR junctions found in > 1 cell (expanded) versus non-expanded TRA sequences in a cross-sectional analysis by disease group, initially using the Cohort 1 data set. To ensure that results were not biased by unequal numbers of junctions between disease groups, we randomly down sampled to equal numbers of junctions per group. This preliminary analysis revealed that the fraction of PIT TRA junction matches with expanded IAR T cells was significantly elevated, and the fraction of PIT TRA junction non-matches reduced, in newT1D subjects, relative to HC and T1D subjects.
To extend these results more broadly along the continuum of stages during the T1D disease process 23, we repeated the analysis using Combined cohorts (Fig. 1F). Because the peptide pools we used for Cohort 1 were biased towards HLA-DRB1*04 donors, we restricted our extended analysis for the combined data sets to this HLA type. The HLA-DRB1*04 subset of Combined cohorts comprised 1,873 cells and 2,967 junctions (1,512 TRA and 1,433 TRB) from 43 donors (Tables S1 and S2). Since we were able to sample relatively few 1AAb and 2AAb HLA-DRB1*04 subjects in Cohort 2, we performed this analysis without correction for different sample sizes. In cells with expanded TCRs, we saw a progressive increase in the fraction of PIT-matched junctions in IAR T cells collected from 1AAb, 2AAb and newT1D donors, then a decrease in T1D subjects to a similar level as in HC donors (Fig. 1F). There were no significant differences in PIT-matches between groups with non-expanded junctions.
To control for p-value inflation by increased numbers of junctions from relatively few donors, we repeated this analysis at the donor level (Figure S2). We observed the same trends as with junction frequency, though the effects were weaker and less significant. This suggests that these studies were underpowered at the donor (sample) level. Despite this limitation, these results supported the conclusion that an elevation of expanded PIT-matched IAR T cell TRA chains in blood occurred prior to the onset of clinical disease.
PIT-matched TRA junctions were more germline-like and hydrophobic than non-matched chains.
We hypothesized that PIT-matched TRA junctions, like public TRA junctions 13, have different sequence and chemical properties than non-matched IAR T cell TRA junctions. As a first test, we asked whether PIT-matched TRA junctions were more likely to be shared between donors (public junctions) than non-matched junctions, as was expected by the different donor pools used to isolate IAR and PIT cells. Previously, we identified a population of IAR T cells with diverse specificities determined by TCRs with restricted TCR alpha junctions and germline-constrained antigen recognition properties 13. To determine how this population of public TCRs was related to PIT-matched/non-matched TRA junctions, we tested for junction sequence overlap. We found significant overlap between public IAR T cell and PIT-matched TRA junctions (p-value = 1.46e-14, hypergeometric distribution) but not private IAR T cell and PIT-matched TRA junctions (p-value > 0.05). Thus, there was strong overlap between cell populations with public and PIT-matching TCRs.
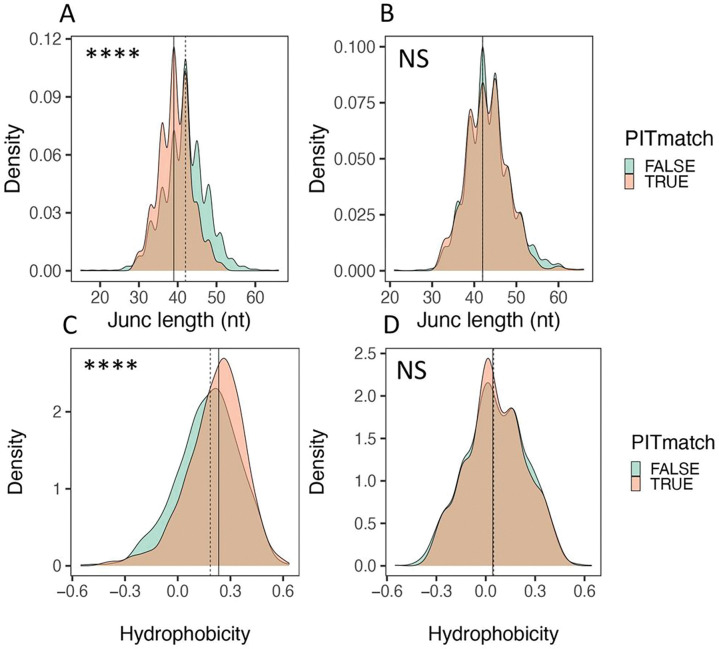
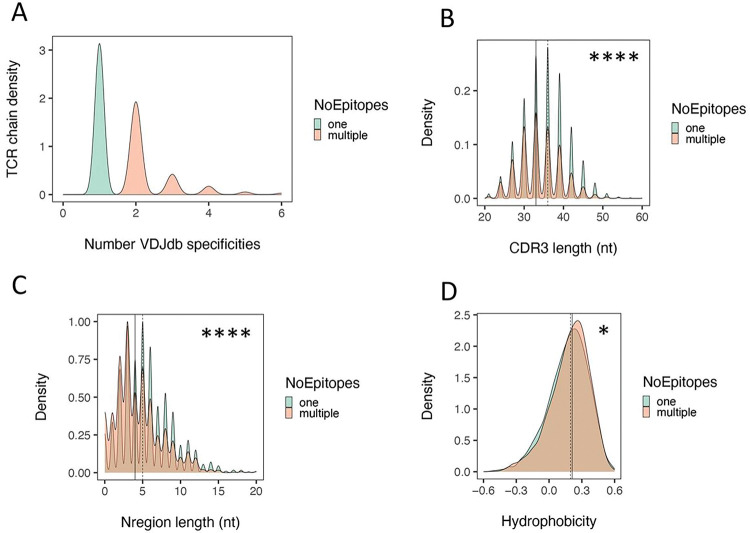
As another test of differences between PIT-matched and non-matched junctions, we compared sequence features that have been correlated with TCR autoreactivity 13,24,25.(Fig. 2). We utilized density plots for these comparisons to elucidate the range of TCR junction lengths. We found that the distribution of TRA junctions was significantly left shifted (shorter) in PIT-matched junctions than in non-matched junctions (p-value < 1e-4, Kolmogorov–Smirnov test) (Fig. 2A). PIT-matched TRA junctions had a median length of ~ 39 nucleotides (nt), versus a median length of ~ 42 nt for PIT-non-matched junctions (i.e., 3 nt or 1 AA residue difference) (Fig. 2A). We did not see a significant difference with TRB junctions paired with PIT-matched and non-matched junctions; TRB junctions from both PIT-matched and non-matched TRA junctions had identical median length values of ~ 42 nt (Fig. 2B). Furthermore, TRA junctions (Fig. 2C), but not TRB junctions (Fig. 2D), were more hydrophobic in PIT-matched versus non-matched TCRs (p-value < 1e-4). PIT-matched and non-matched TRA junctions had median hydrophobicity values of 0.23 and 0.18, respectively, on the Eisenberg hydrophobicity scale 26, falling between values for proline and tyrosine (0.12 and 0.26, respectively). Thus, PIT-matched TRA junctions, but not their paired TRB junctions, were shorter, and more hydrophobic than PIT-non-matched TRA chains.
Figure 2. PIT-matched TRA junctions are shorter and more hydrophobic than PIT non-matched junctions.
Distributions of TRA and TRB junction lengths (A and B) and hydrophobicity (C and D) calculated from amino acid sequences of TRA (A and C) and TRB junctions (B and D). The significance of differences was assessed using Kolmogorov-Smirnov tests. ****, pAdj <1e-4; **, pAdj <1e-2; NS, not significant. Solid vertical line, median value from PIT-matched junctions; dashed vertical line, median value from PIT-non-matched junctions.
Length differences between PIT-matched and non-matched TRA junctions map to peptide-contact regions.
We speculated that sequence features of PIT-matching TRA junctions were keys to the function(s) of IAR T cells having these TCRs. To further elucidate these sequence features, we used IMGT/HighV-QUEST 27 to characterize individual TCRs from their nucleotide (nt) sequences. We first determined the positions of single AA mismatches between PIT and IAR T cell TRA junctions. Since both V and J gene regions are germline-encoded, we predicted that PIT mismatches would not localize to these regions. Instead, we predicted that mismatches would instead occur primarily in the V-J recombination region, which is non-germline-encoded and varies in sequence between different TCRs. We found that mismatched residues were distributed symmetrically around amino acid position 4 (Figure S3). By comparison, the C terminal AA from V genes (3’ end) and the N terminal AA of J genes (5’ end), which mark the boundaries of germline-encoded residues in the respective gene segments, were centered at AA residues 3 and 5, respectively (Figure S3). Quantitatively, ~ 88% (1,178/1,342) of PIT-matched TRA junction mismatches were located between the 3’ends of V genes and the 5’ends of J genes. Normalizing the site of mismatches for junction length reduced variability but did not change the essential features of the results. Thus, PIT mismatches occurred primarily in the non-germline-encoded V-J recombination region.
We next focused our comparisons on regions important for TCR binding and function. Analogous to immunoglobulin molecules, TCR complementary determining regions (CDRs) convey the specificity for antigen and major histocompatibility complex molecules 28. A comparison of CDR1 region lengths identified using IMGT/HighV-QUEST 27 showed that the median CDR1 length was identical in PIT-matched and non-matched junctions, but the distribution was left-shifted significantly (i.e., CDR1 regions were shorter) for PIT-matched junctions (Figure S4A). CDR2 regions showed no length differences between PIT-matched and non-matched TRA chains (Figure S4B), whereas CDR3 regions, as expected since they comprise most of the junction sequences compared in Fig. 2, were shorter in in PIT-matched TRA chains (Figure S4C), by ~ 3 nt (1 AA). In other analyses, we found that framework (FR) regions FR1, FR2, FR3 and FR4 did not differ in length between PIT-matched and non-matched TRA chains, showing selectivity of the differences in CDR1 and CDR3 lengths.
We wished to determine the source(s) of junction length differences between PIT-matched and non-matched TRA chains. Since sequence variability of CDR3 regions mostly comes from random addition of nucleotides not encoded in the genome (N region), we compared N region lengths between the two groups of TRA chains. This comparison showed that PIT-matched junctions had median N region lengths of ~ 3 nt (1 AA) whereas PIT-non-matched junctions had median N region lengths of ~ 5 nt (1–2 AA) (Figure S4D). We also quantified contributions of the V gene and J gene segments adjacent to the N region. The 3’ end of the V region was slightly shorter in PIT-matched than non-matched TRA junctions (10 versus 11 nt) (Figure S4E) whereas the 5’ end of the J region was slightly longer (27 versus 25 nt) (Figure S4F). Thus, the difference in CDR3 lengths was a complex product of genome- and non-genome-encoded regions in PIT-matched junctions. Together, these analyses showed that PIT-matched and non-matched TRA junctions differed in V gene regions CDR1 and CDR3 that are important for peptide binding.
The difference in CDR1 region lengths was an unexpected result, since CDR1 regions are germline-encoded and not considered variable. We hypothesized that the basis for the shift in CDR1 length of PIT-matched TRA chains was enrichment for TRA V gene segments with shorter CDR1 regions. To test this, we compared enrichment of V gene segments in PIT-matched versus non-matched TRA chains (Figure S5). This showed that TRAV12–2*01 and TRAV41*01 V genes were significantly over-represented in PIT-matched IAR T cell TRA chains, whereas TRAV4*01 and TRAV26–2*01 were under-represented (or over-represented in PIT non-matched TRA chains). Over-represented V genes, TRAV12–2*01 and TRAV41*01, had CDR1 region lengths of 15 and 18 nt (5–6 AA). In contrast, under-represented V genes, TRAV4*01 and TRAV26–2*01, had 21 nt (7 AA) CDR1 regions. Thus, selective utilization of V genes with shorter CDR1 lengths was a likely explanation for enrichment for TRA V gene segments having shorter CDR1 regions in PIT-matched junctions.
Determining predicted peptide binding contacts for PIT-matching and non-matching TRA chains.
CDR1 and CDR3 regions are involved in binding to peptides presented by MHC molecules 22. Based on this fact, we hypothesized that the observed length variation of these regions suggested altered peptide binding properties for PIT-matched TRA chains. As a test, we utilized molecular modeling to predict peptide binding residues in tri-molecular models of PIT-matched and PIT-non-matched TCRs complexed with peptide-class II MHC complexes. For modeling, we used TCRmodel2, which is based on the AI system, AlphaFold v2.3 29, a newly described method that predicts protein structure with high accuracy at unmatched scale 30. TCRmodel2 uses focused databases of TCR and MHC sequences to expedite multiple sequence alignment feature building; optimization of the TCR template selection; and utilization of peptide–MHC complex structures as templates to improve modeling accuracy. For model input, we used paired TRA and TRB sequences from a set of 30 IAR TCRs with known specificity (16 PIT-matched and 14 PIT-non-matched TCRs), together with their cognate peptides 13; and sequences of HLA DRA*01:01 and DRB1*0401 MHC class II subunits 31.
From best fit models for each TCR, we identified likely peptide contact residues (TCR residues < 5 Å in distance from the bound peptide chain) and mapped these to TRA and TRB sequence features. We first visually compared TCR-peptide contact residues in representations of PIT-matched and PIT-non-matched TCRs with extremes in TRA junction length (Fig. 3A–B). This comparison showed that Clone 604 (10 AA, PIT-matched) had more predicted peptide contacts that mapped to CDR1, and fewer that mapped to CDR3, than Clone 2253 (17 AA, PIT-non-matched). To extend these observations to the larger data set, we compared the numbers of TCR contacts of each chain from the set of 30 IAR TCRs (Table S3). We displayed the numbers of contacts in different sequence features in both chains from each TCR as a function of TRA and TRB junction lengths (Figure S6). As expected, the TRA and TRB CDR3 regions contributed the most peptide contacts. There was a significant positive relationship between the number of peptide contacts mapping to the TRA CDR3 region and TRA junction length, and a weaker negative relationship with the TRB CDR3 region (Figure S6). While the number of contacts was low, there was also a negative relationship between the TRA CDR1 region and TRA junction length.
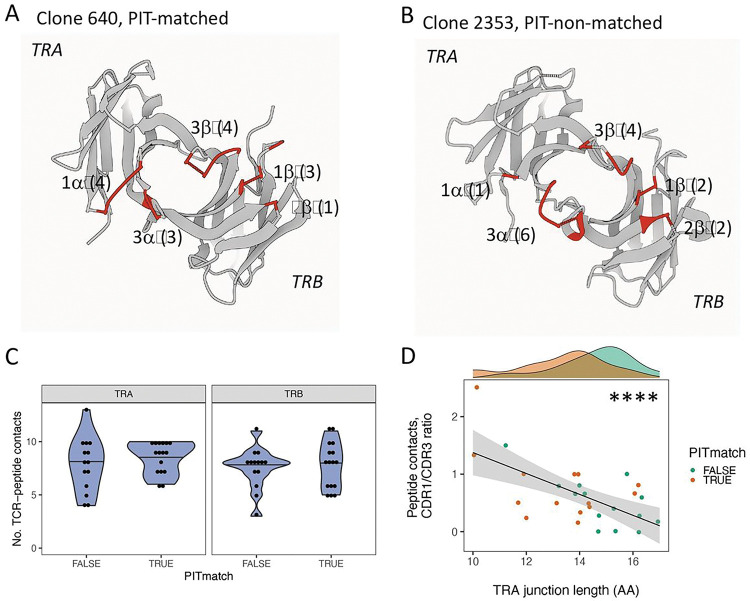
Figure 3. Increased ratios of germline-encoded to recombined peptide contacts in PIT-matched TCRs.
A) Gaussian Surface representation of a model of Clone 640, a PIT-matching TCR with a 10 AA TRA junction (7 CDR contacts) paired with a 12 AA TRB junction (8 CDR contacts), that binds the GAD65 113–132 peptide 56 presented by the HLA DRB1*0401 class II molecule 13. The viewing plane is from the interface with the MHC molecules, which together with the peptide, have been removed from the representation for clarity. Labels indicate the CDR loops, with letters denoting the chain (α or β); and numbers, the CDR loop. Numbers in parentheses are the number of predicted peptide contacts with the CDR loop. B) A model of Clone 2353, a PIT-non-matching TCR with a 17AA TRA junction (7 CDR contacts) paired with a 14 AA TRB junction (8 CDR contacts), that binds the IGRP305–324 peptide presented by the HLA DRB1*0401 class II molecule. C) The number of total peptide contacts for either the TRA or TRB chains did not differ significantly between PIT-matched and PIT-non-matched TCRs (p-value >0.05, Wilcox test). D) Ratios of TRA CDR1 to CDR3 peptide contacts decreased significantly with increasing TRA junction length. ****, p-value < 1e-4 for a slope of 0, calculated using linear modeling.
Increased dependence on germline-encoded residues for binding of shorter and PIT-matching TCRs.
The variation in contact residues by junction length (and PIT match) could indicate either differences in overall number of contacts, or different ratios of contacts in different regions. To test these possibilities, we first compared the overall number of TCR contacts by TCR chain in PIT-matched versus PIT-non-matched TCRs. This comparison showed no significant difference in overall peptide contacts in either TRA or TRB chains between the two groups (Fig. 3C). In contrast, the ratio of TRA CDR1 to CDR3 contacts varied significantly with TRA junction length (p-value < 1e-4 for line slope, by linear modeling), while maintaining the separation in length between PIT-matched and PIT-non-matched junctions (Fig. 3D). Thus, peptide recognition by PIT-matched TCRs shows a trend towards greater reliance on the germline-encoded residues in the TRA CDR1 region than do PIT-non-matched TCRs, which in turn show greater reliance on CDR3 residues generated by V(D)J recombination.
Concomitant with the increase in the ratio of CDR1 to CDR3 contacts, PIT-matched TRA chains also showed a decrease in CDR1 region length (Figure S4A). This incongruity could suggest a change in physical properties of the shorter CDR1 regions to make them more amenable to close peptide contacts. Thus, it may be important that PIT-matched CDR1 regions were weakly more hydrophobic than PIT-non-matched regions (p-value = 0.049, one-sided Kolmogorov–Smirnov test).
TCRs with PIT-matching TRA junctions show evidence of multi-specificity.
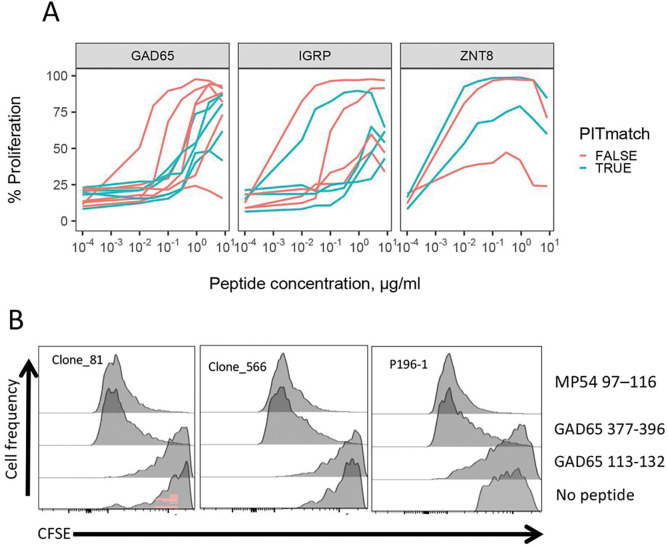
We hypothesized that different peptide binding modes of PIT-matched and non-matched TRA junctions would lead to altered peptide recognition properties, such as strength and/or specificity of binding. As a measure of binding strength, we compared the functional avidities of TCRs having PIT-matched and non-matched TRA junctions specific for different peptide epitopes as described previously 13. We plotted cell proliferation, as measured by CFSE 13, versus peptide concentration (Fig. 4A) for the subset of TCRs having PIT-matching or non-matching TRA junctions (Table 2). To increase power, we aggregated results with different peptides from each of three different islet antigens (GAD, IGRP and ZNT8). These results showed wide variability in range of dose responses, but no consistent differences between TCRs with PIT-matched and non-matched TRA junctions. EC50 values 13 for aggregated GAD65- and IGRP-specific TCRs with PIT-matched and non-matched TRA junctions did not differ significantly by unpaired Wilcox tests; since there were only two ZNT8 TCRs, a p-value could not be calculated. We obtained similar overall results when individual peptides were considered. These data do not support there being large differences in functional avidity between TCRs with PIT-matched and non-matched TRA junctions.
Figure 4. Functional properties of selected PIT-matching and non-matching TRA chains of known specificity.
A) Dose response curves for TCRs recognizing GAD65, IGRP and ZNT8 epitopes 13. Curves for multiple epitopes per target antigen were pooled to increase power; examination of curves for individual epitopes yielded conclusions consistent with the pooled data. B) Cross-reactivity of TCRs with PIT-matching TRA junctions (Table 3) for autoimmune (GAD65) and viral (influenza M protein) epitopes in a CFSE proliferation assay.
Table 2.
TCRs sharing single mismatched TRA chains between PIT and IAR T cells of known specificity. A
| TRA chain | TRB chain | |||||
|---|---|---|---|---|---|---|
| specificity | V gene | Junction | J gene | V gene | Junction | J gene |
| GADp15 | TRAV41 | CAAA-GNQFYF | TRAJ49 | TRBV12-4 | CASSFT-YNEQFF | TRBJ2-1 |
| TRAV29/DV5 B | CAAR-GNQFYF | TRAJ49 | TRBV6-2 | CASSLLNLDNEQFF | TRBJ2-1 | |
| TRAV21 | CAAI-GNQFYF | TRAJ49 | TRBV29-1 | CSVLRDRASYEQYF | TRBJ2-7 | |
| TRAV29/DV5 | CAASAGNQFYF | TRAJ49 | TRBV4-1 | CASSLAATRDDYGYTF | TRBJ1-2 | |
| IGRP39 | TRAV25 | CAGQTGANNLFF | TRAJ36 | TRBV4-3 | CASSQEVGTVPNQPQHF- | TRBJ1-5 |
| TRAV16 | CALQTGANNLFF | TRAJ36 | ||||
| TRAV13-1 | CATQTGANNLFF | TRAJ36 | TRBV9 | CASSVGR-SSYNEQFF | TRBJ2-1 | |
| TRAV24 | CASQTGANNLFF | TRAJ36 | TRBV4-1 | CASSQDPLTSGRGNEQFF | TRBJ2-1 | |
| ZNP1 | TRAV13-1 | CAASGANSGYALNF | TRAJ41 | TRBV30 | CAWSAQGETQYF | TRBJ2-5 |
| TRAV8-4 | CAVSGANSGYALNF | TRAJ41 | TRBV30 | CAWESGTRGNYGYTF | TRBJ1-2 | |
Shown are amino acid sequence comparisons of TCRs having single mismatched TRA junctions between IAR and PIT TCRs. IAR TCRs of the indicated specificities were used as reference sequences.
Bold font indicates a mismatch with the IAR reference sequence (top row of each pair).
Based on these results, we next hypothesized that TCRs with PIT-matched and non-matched TRA junctions differed in peptide binding specificity. This was supported by our previous finding of multi-specificity of some public IAR TCRs 13. Though limited in number (n = 3), the frequency of these multi-specific clones was higher in public compared with private clones 13. In the present studies, we noted that TRA chains from all three of these multi-specific TCRs were PIT-matching. Indeed, multi-specific TRA junctions were modestly more frequent in PIT-matching than non-matching TRA junctions (one-sided p-value = 0.030, Fisher’s exact test).
Two of these PIT-matching multi-specific TCRs (Clones 81 and 566) recognized non-overlapping GAD65 epitopes 13. In parallel and independent experiments, we unexpectedly found that the TRA chain of a TCR (P196–1) from Influenza A/MP54- reactive CD4 + T cells perfectly matched the TRA chain from Clones 81 and 566 (Table 3). Furthermore, P196–1 had TRB chains that differed from Clones 81 and 566 at only a single AA position (Table 3). This sequence similarity led us to reason that closely related Clone 81, Clone 566 and P196–1 were all multi-specific. As a test, we compared the ability of activating peptides (MP54 97–116 and GAD65 377–396), a non-activating peptide (GAD65 113–132) to trigger proliferation of TCR-transduced primary CD4 + T cells (Fig. 4B). We found that both MP54 97–116 and GAD65 377–396 peptides identically activated Clone 81, Clone 566, and P196–1 TCRs. Thus, these TCRs with PIT-matched TRA junctions were all multi-specific, supporting our hypothesis that TCR sequence features, including shorter and more hydrophobic TRA junctions, are linked to an inherent tendency for multi-specificity.
Table 3.
Mismatched TRB chains from multi-specific TCRs with perfectly matched TRA chains. A
| Clone ID | Specificity | Epitope | TRBV-gene | Junction | TRBJ-gene |
|---|---|---|---|---|---|
| Clone_81 | Islet | GAD65 377-396 | TRBV12-4 | CASSPQGGNTEAFF | TRBJ1-1 |
| Clone_566 | Islet | GAD65 377-396 | TRBV12-4 | CASSVQGGNTEAFF | TRBJ1-1 |
| P196-1 | Influenza | MP54 97-116 | TRBV12-4 | CASSLQGGNTEAFF | TRBJ1-1 |
Shown are mismatched TRB chain amino acid sequence comparisons of multi-specific TCRs that recognized both GAD65 and MP54 peptides. All TCRs had identical TRA chains (TRAV29DV5-CAASRYSGGGADGLTF-TRAJ45). Sequences of GAD65 377-396 54 (HKWKLSGVERANSVTWNPHK, where bold font denotes core sequence complexed with the TCR) and MP74 97-116 55 (VKLYRKLKREITFHGAKEIS) peptides showed minimal sequence similarity by multiple sequence alignment.
Bold font indicates mismatches between the three TRB chains.
To elucidate possible mechanisms of cross-reactivity of the Clone 81 TCR for the MP54 97–116 and GAD65 377–396 peptides, we constructed molecular models 32 of the Clone 81 TCR 13 and complexes of these peptides together with HLA-DRA1*0101/DRB1*0401 molecules 31. The cognate GAD65 377–396 (HKWKLSGVERANSVTWNPHK, where bold font denotes core sequence complexed with the TCR) and MP74 97–116 (VKLYRKLKREITFHGAKEIS) peptides did not show compelling sequence similarity by multiple sequence alignment. They also showed no evidence of structure 33 or aromatic side chain 34 conservation characterizing “hotspots” of molecular mimicry used by some crossreactive TCRs. Despite their low degree of peptide sequence similarity, molecular models of Clone 81 TCR with both peptides yielded identical scores of 0.88. In addition, TRA and TRB chains in both models showed similar predicted topography in their interactions with peptide-MHC class II complexes (Figure S7A-B). Overlapping but non-identical residues in the modeled TCR TRA and TRB chain CDR1, CDR2 and CDR3 regions contacted the different peptides (Figure S7C). Alignment of predicted structures of the TRA (Figure S7D) and TRB (Figure S7E) chains from models made with the different peptides showed nearly perfect superposition except in the CDR3 regions. This suggests that the PIT-matched Clone 81 TCR accommodates quite different peptide sequences through interactions involving conformationally conserved CDR1 (and CDR2 chains for TRB) regions, together with more variable CDR3 regions.
Multi-specific TCRs shared sequence features with PIT-matched TCRs.
We wished to test in grerater depth our hypothesis that PIT-matched and non-matched TRA junctions differed in peptide binding specificity. Recognizing that the number of known multi-specific TCRs from IAR T cells was too small at present to enable firm population-based conclusions about multi-specificity, we took an alternative approach to test our hypothesis. In other experiments, we observed that VDJdb, a curated public database of TCRs with known specificities 35, contains TCR sequences that recognize both single and multiple peptide epitopes. While we recognized that these designations were likely biased, with some TCRs being designated as single specificity because they were understudied, and others being designated as multi-specific on the basis of reactivity with closely related peptides. We reasoned, however that these errors would tend to offset each other, and that the size and scope of this database would provide a more comprehensive source of specific and multi-specific TCRs, which could be used to test their junction sequence features. There were N = 17,826 unique TRA chains in VDJdb that recognized single epitopes and N = 1,669 TRA chains that recognized multiple epitopes (Fig. 5A). Since nt sequences used to identify junction sequence features are not readily available for VDJdb TCRs, we utilized the software package, Stitchr, to produce complete TCR cDNAs from V/J/CDR3 AA sequences 36. We then used IMGT/HighV-QUEST 27 to identify TCR sequence features from the predicted cDNA sequences. We found that TRA CDR3 regions (Fig. 5B) from TCRs that recognized multiple epitopes were significantly shorter (by ~ 3 nt in median length, or ~ 1 AA) than junctions from TCRs that recognized single epitopes. We found equivalent differences in junction AA sequences, indicating that the results were independent of Stitchr. Likewise, TRA chains from multi-specific TCRs contained fewer non-templated (N region) nucleotides (~ 1 nt in median length) than TCRs with single specificity (Fig. 5C). Finally, TRA junction amino acid sequences from multi-specific TCRs were more hydrophobic in TCRs that recognized multiple epitopes (Fig. 5D). None of these differences were seen with TRB chains (Figure S8 A-D). Thus, multi-specific TCRs from VDJdb shared multiple sequence features with PIT-matched TCRs, including shorter TRA but not TRB chains, that had fewer N region nt, and encoded more hydrophobic AA sequences.
Figure 5. TRA junctions from cross-reactive TCRs in the VDJdb database shared junction sequence features with PIT-matched TRA junctions.
A) Density of TRA chains from TCRs with one versus multiple specificities in VDJdb. There were N = 17,826 unique TRA chains from TCRs with single specificity and N = 1,664 with multiple specificities. B) Distribution of TRA CDR3 nt lengths from TCRs with one versus multiple specificities; C) Distribution of TRA N region nt lengths. D) Distribution of TRA junction AA hydrophobicity. Solid vertical lines, median values from TRA chains from multi-specific TCRs; dashed vertical lines, median values from TCRs with single reported specificities. Significance of differences in distributions were assessed using Kolmogorov-Smirnov tests. ****, pAdj <1e-4; *; pAdj <0.05.
Discussion
It has long been unclear whether and how rare autoreactive cells in peripheral blood represent autoimmunity in in target organs. In other words, are these cells drivers or passengers in autoimmune processes? To help resolve this question, we show here that a significant fraction of IAR TCRs from peripheral blood share matching TRA chains with PIT TCRs, and vice versa. Thus, a subset of IAR T cells shares a TCR chain with TCRs present in the pancreas. Since there are no familial or genetic relationships between the Cohort 1 and Cohort 2 blood donors and the nPOD organ donors, the TCR chain sharing we demonstrate involves public sequences. We also show that frequencies of PIT-matching TRA chains in blood increase prior to the time of diagnosis, suggesting a temporal linkage of levels of PIT-matching TRA chains in blood with disease progression. Shared TRB chains between peripheral blood and insulin-reactive TCRs from the pancreas were reported recently 19. These combined studies place autoreactive TCR chains at the scene of disease (the pancreas) at the right time (early in the disease process) to influence disease development.
However, we must emphasize that, at present, we do not know how many of these autoreactive TCR chains are associated with functional autoreactivity versus β-cells. At present, there is no way to predict with certainty that TCRs share specificity unless they show complete identity of both chains. In the present case, PIT-matching TRA chains shared quite divergent TRB chains, and it remains unknown what fraction of these TCRs recognizes autoantigens. Nonetheless, our demonstration of TRA chain sharing strengthens the link between IAR T cells in blood and autoimmune destruction of the pancreas.
T cell specificity is key to cellular immunity 37. Paradoxically, TCR cross-reactivity is necessary because the diversity of TCRs, while huge, is dwarfed by the vast array of potential foreign peptide-MHC complexes 38,39. Cross-reactivity expands the potential range of a given repertoire towards foreign antigens, but it comes at the expense of greater potential for increased self-reactivity or autoimmunity. These considerations emphasize the importance of understanding of sequence and structural features determining TCR cross-reactivity. Unfortunately, at present such an understanding remains incomplete 37,40. Our data show that cross-reactive (multi-specific) TCRs share sequence features with PIT-matched TCRs, namely shorter and more hydrophobic TRA junctions and diverse TRB chains, reminiscent of a previous report in mouse 41. Supporting this conclusion, the few islet-specific TCRs for which we have demonstrated cross-reactivity are PIT-matched TCRs. Earlier work demonstrated shortened 25 and more hydrophobic 24 TRB chains in bulk T cell subsets from T1D subjects, and suggested that these features are important in development of self-reactive T cells.
TCR CDR3 regions form most of the primary contacts with peptide epitopes presented by MHC molecules 22. For most TCRs, TRB CDR3 contacts are most numerous 42, although there are multiple examples of MHC class I-restricted TCRs that predominantly utilize germline-like TRA CDR3 regions 43–47. Less is known about such TRA-centric, MHC class II-restricted TCRs, such as those we have observed. One of the V genes enriched in PIT-matched TRA chains (Figure S5), TRAV12–2, is also a feature of multiple MHC class I-restricted TCRs 43,44,46,47. The other V gene enriched in PIT-matched TRA chains was TRAV21–2, which is associated with celiac disease, while paired with TRBV9 chains Our findings suggest that utilization of TRA-centric binding is a previously undescribed but widespread feature of IAR TCRs, and perhaps MHC class II-restricted TCRs in general.
Molecular modeling showed that germline-encoded CDR1, rather than CDR3, residues from TRAV12–2 contributed critically to binding of an immunodominant TCR to Yellow fever virus 46. This feature may contribute to the high precursor frequency and immunodominance of this TCR 46. PIT-matching IAR T cell TRA chains also employed less non-templated and more germline-like (innate) mode(s) of epitope engagement. TRA CDR3 regions of PIT-matching IAR varied from the genome by relatively few N region nucleotide sequences. In addition, PIT-matching IAR T cell TRA chains showed shorter CDR3 regions and longer TRAJ segments than non-matching IAR T cell TRA chains. TCR CDR1 regions contact both peptide epitope and MHC residues, while CDR2 regions generally contact only MHC residues 22. Thus, TCR regions important for peptide binding were shortened in PIT-matching TRA chains and had altered peptide binding properties. These PIT-matching TRA chains were also more hydrophobic at the amino acid level, suggesting partner agnostic interactioins via the hydrophobic effect versus more residue specific effects such as hydrogen bonding. Our results also suggest that these shorter and more hydrophobic, germline-like, TRA chains are a feature of many cross-reactive TCRs. While some TCRs that predominantly utilizing germline-like TRA CDR3 regions are cross-reactive 49, this has not been previously demonstrated on the global level we show here.
One caveat to our study is that we have addressed only sharing of public TCRs. This was by necessity, since TCR sequences from IAR and PIT T cells from the same individuals were not available. While this feature enhances broadness of potential translational applications, it also limits broad mechanistic conclusions. Future studies may be able to address this weakness by testing of additional nPOD tissues 18, including spleen and pancreas draining lymph nodes, from the same donors used for PIT TCR identification. Another weakness in our studies is that it is underpowered with respect to numbers of subjects studied. This is apparent in our examination of individual donor frequencies between PIT-matched and non-matched TRA junctions (compare Fig. 1F versus Figure S2). Our study is also underpowered for autoreactive TCRs of known specificity needed to directly test our hypothesis that PIT-matched TCRs tend to be multi-specific. Such a test is not possible at present because the numbers of IAR T cell TCRs with demonstrated specificity remains small. While we used VDJdb sequences of different specificities as an indirect test, a. more comprehensive collection of autoreactive TCRs with known specificity will be needed to further clarify the situation.
Our studies have potential translational implications. Typically, studies on TCRs as biomarkers have taken a reductionist approach by focusing on one or a few TCRs to monitor disease progression and/or response to therapy 19,50,51. In contrast to this approach, our broader-based studies place many TCRs with different islet antigen specificities and distinct TRA sequence features in both the pancreas and blood in early stages of T1D progression. Together, these new findings suggest that an alternative to the reductionist approach for translational studies is to consider autoreactivity as a web of T cell specificities and potential cross reactivities. Our findings provide a basis for expansion of future biomarker studies to include a broader network of autoreactive and potentially multi-specific TCRs.
Materials and Methods
Repository information.
Code and data for generating Figures, as well as pdb files of molecular models, are available at https://github.com/BenaroyaResearch/. Profiles yielding TCRs were deposited in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE182870 (Cohort 1) and GSEXXXXX (Cohort 2).
Study approval.
Protocols for these studies were approved by the Institutional Review Board of Benaroya Research Institute (IRB7109–332). Protocols for the clinical studies were approved under the auspices of NCT00129259 for the AbATE trial 52 and NCT00515099 for the START study.53
Experimental methods
Methods are available as Supplemental Methods.
Acknowledgments
We acknowledge Vivian Gersuk, Kimberly O’Brien, Jeffry Yaplee, Thao Huynh, and Quynh-Anh Nguyen for molecular profiling assistance; the BRI TrialNet Clinical Center led by Dr. Carla Greenbaum and Dr. Sandra Lord for collecting samples from subjects with and at-risk for T1D; and Mario Rosasco for assistance with TCR analysis. This research was performed with the support from the Immune Tolerance Network (5UM1AI109565 to Gerald T Nepom), the American Diabetes Association (1-19-ICTS-006 to KC), and the Network for Pancreatic Organ donors with Diabetes (nPOD; RRID:SCR_014641), a collaborative type 1 diabetes research project supported by JDRF (nPOD: 5-SRA-2018-557-Q-R) and The Leona M. & Harry B. Helmsley Charitable Trust (Grant#2018PG-T1D053, G-2108-04793). The content and views expressed are the responsibility of the authors and do not necessarily reflect the official view of the ADA or nPOD. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded through a cooperative agreement by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases (NIAID), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and JDRF.
Footnotes
Additional Declarations: There is NO Competing Interest.
Contributor Information
Peter Linsley, Benaroya Research Institute at Virginia Mason.
Maki Nakayama, Barbara Davis Center for Childhood Diabetes.
Elisa Balmas, Benaroya Research Institute.
Janice Chen, Benaroya Research Institute.
Fariba Pour, Benaroya Research Institute.
Shubham Bansal, Benaroya Research Institute.
Elisavet Serti, The Henry Jackson Foundation.
Cate Speake, Benaroya Research Institute.
Alberto Pugliese, City of Hope.
Karen Cerosaletti, Benaroya Research Institute, Seattle, WA.
References
- 1.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009. Apr 16;360(16):1646–1654. [DOI] [PubMed] [Google Scholar]
- 2.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005. May 12;435(7039):224–228. [DOI] [PubMed] [Google Scholar]
- 3.Pathiraja V, Kuehlich JP, Campbell PD, Krishnamurthy B, Loudovaris T, Coates PTH, Brodnicki TC, O’Connell PJ, Kedzierska K, Rodda C, Bergman P, Hill E, Purcell AW, Dudek NL, Thomas HE, Kay TWH, Mannering SI. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes. 2015. Jan;64(1):172–182. [DOI] [PubMed] [Google Scholar]
- 4.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009. Feb;155(2):173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennon GP, Bettini M, Burton AR, Vincent E, Arnold PY, Santamaria P, Vignali DAA. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity. 2009. Oct 16;31(4):643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigby MR, Ehlers MR. Targeted immune interventions for type 1 diabetes: not as easy as it looks! Curr Opin Endocrinol Diabetes Obes. 2014. Aug;21(4):271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culina S, Lalanne AI, Afonso G, Cerosaletti K, Pinto S, Sebastiani G, Kuranda K, Nigi L, Eugster A, Østerbye T, Maugein A, McLaren JE, Ladell K, Larger E, Beressi JP, Lissina A, Appay V, Davidson HW, Buus S, Price DA, Kuhn M, Bonifacio E, Battaglia M, Caillat-Zucman S, Dotta F, Scharfmann R, Kyewski B, Mallone R, ImMaDiab Study Group. Islet-reactive cd8+ t cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol. 2018. 02;3(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danke NA, Koelle DM, Yee C, Beheray S, Kwok WW. Autoreactive T cells in healthy individuals. J Immunol Baltim Md 1950. 2004. May 15;172(10):5967–5972. [DOI] [PubMed] [Google Scholar]
- 9.Danke NA, Yang J, Greenbaum C, Kwok WW. Comparative study of GAD65-specific CD4+ T cells in healthy and type 1 diabetic subjects. J Autoimmun. 2005. Dec;25(4):303–311. [DOI] [PubMed] [Google Scholar]
- 10.Skowera A, Ladell K, McLaren JE, Dolton G, Matthews KK, Gostick E, Kronenberg-Versteeg D, Eichmann M, Knight RR, Heck S, Powrie J, Bingley PJ, Dayan CM, Miles JJ, Sewell AK, Price DA, Peakman M. β-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes. 2015. Mar;64(3):916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janeway CA Jr, Travers P, Walpert M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. [Google Scholar]
- 12.Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, Murray SA, Israelsson E, Speake C, Gersuk VH, Eddy JA, Reijonen H, Greenbaum CJ, Kwok WW, Wambre E, Prlic M, Gottardo R, Nepom GT, Linsley PS. Single-Cell RNA Sequencing Reveals Expanded Clones of Islet Antigen-Reactive CD4(+) T Cells in Peripheral Blood of Subjects with Type 1 Diabetes. J Immunol Baltim Md 1950. 2017. May 31; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linsley PS, Barahmand-Pour-Whitman F, Balmas E, DeBerg HA, Flynn KJ, Hu AK, Rosasco MG, Chen J, O’Rourke C, Serti E, Gersuk VH, Motwani K, Seay HR, Brusko TM, Kwok WW, Speake C, Greenbaum CJ, Nepom GT, Cerosaletti K. Autoreactive T cell receptors with shared germline-like α chains in type 1 diabetes. JCI Insight. 2021. Nov 22;6(22):e151349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landry LG, Anderson AM, Russ HA, Yu L, Kent SC, Atkinson MA, Mathews CE, Michels AW, Nakayama M. Proinsulin-Reactive CD4 T Cells in the Islets of Type 1 Diabetes Organ Donors. Front Endocrinol. 2021;12:622647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama M, Michels AW. Determining Antigen Specificity of Human Islet Infiltrating T Cells in Type 1 Diabetes. Front Immunol. 2019. Mar 8;10:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babon JAB, DeNicola ME, Blodgett DM, Crèvecoeur I, Buttrick TS, Maehr R, Bottino R, Naji A, Kaddis J, Elyaman W, James EA, Haliyur R, Brissova M, Overbergh L, Mathieu C, Delong T, Haskins K, Pugliese A, Campbell-Thompson M, Mathews C, Atkinson MA, Powers AC, Harlan DM, Kent SC. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med. 2016. Dec;22(12):1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauken KE, Lagattuta KA, Lu BY, Lucca LE, Daud AI, Hafler DA, Kluger HM, Raychaudhuri S, Sharpe AH. TCR-sequencing in cancer and autoimmunity: barcodes and beyond. Trends Immunol [Internet]. Elsevier; 2022. Jan 25 [cited 2022 Jan 26];0(0). Available from: https://www.cell.com/trends/immunology/abstract/S1471-4906(22)00002-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell-Thompson M, Wasserfall C, Kaddis J, Albanese-O’Neill A, Staeva T, Nierras C, Moraski J, Rowe P, Gianani R, Eisenbarth G, Crawford J, Schatz D, Pugliese A, Atkinson M. Network for Pancreatic Organ Donors with Diabetes (nPOD): developing a tissue biobank for type 1 diabetes. Diabetes Metab Res Rev. 2012. Oct;28(7):608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell AM, Baschal EE, McDaniel KA, Simmons KM, Pyle L, Waugh K, Steck AK, Yu L, Gottlieb PA, Rewers MJ, Nakayama M, Michels AW. Temporal development of T cell receptor repertoires during childhood in health and disease. JCI Insight. 2022. Sep 22;7(18):e161885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, Voillet V, Duvvuri VR, Scherler K, Troisch P, Baloni P, Qin G, Smith B, Kornilov SA, Rostomily C, Xu A, Li J, Dong S, Rothchild A, Zhou J, Murray K, Edmark R, Hong S, Heath JE, Earls J, Zhang R, Xie J, Li S, Roper R, Jones L, Zhou Y, Rowen L, Liu R, Mackay S, O’Mahony DS, Dale CR, Wallick JA, Algren HA, Zager MA, ISB-Swedish COVID19 Biobanking Unit, Wei W, Price ND, Huang S, Subramanian N, Wang K, Magis AT, Hadlock JJ, Hood L, Aderem A, Bluestone JA, Lanier LL, Greenberg PD, Gottardo R, Davis MM, Goldman JD, Heath JR. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell. 2020. Dec 10;183(6):1479–1495.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008. Jul 1;36(Web Server issue):W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennecke J, Wiley DC. T Cell Receptor–MHC Interactions up Close. Cell. Elsevier; 2001. Jan 12;104(1):1–4. [DOI] [PubMed] [Google Scholar]
- 23.Battaglia M, Anderson MS, Buckner JH, Geyer SM, Gottlieb PA, Kay TWH, Lernmark Å, Muller S, Pugliese A, Roep BO, Greenbaum CJ, Peakman M. Understanding and preventing type 1 diabetes through the unique working model of TrialNet. Diabetologia. 2017. Nov;60(11):2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadinski BD, Shekhar K, Gómez-Touriño I, Jung J, Sasaki K, Sewell AK, Peakman M, Chakraborty AK, Huseby ES. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol. 2016. Aug;17(8):946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Tourino I, Kamra Y, Baptista R, Lorenc A, Peakman M. T cell receptor β-chains display abnormal shortening and repertoire sharing in type 1 diabetes. Nat Commun. 2017. 27;8(1):1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. [DOI] [PubMed] [Google Scholar]
- 27.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol Clifton NJ. 2012;882:569–604. [DOI] [PubMed] [Google Scholar]
- 28.Katayama CD, Eidelman FJ, Duncan A, Hooshmand F, Hedrick SM. Predicted complementarity determining regions of the T cell antigen receptor determine antigen specificity. EMBO J. 1995. Mar 1;14(5):927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021. Aug;596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoline LMF, Lima AN, Krieger JE, Teixeira SK. Before and after AlphaFold2: An overview of protein structure prediction. Front Bioinforma [Internet]. 2023. [cited 2023 Aug 2];3. Available from: 10.3389/fbinf.2023.1120370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-Ray Crystal Structure of HLA-DR4 (DRA*0101, DRB1*0401) Complexed with a Peptide from Human Collagen II. Immunity. Elsevier; 1997. Oct 1;7(4):473–481. [DOI] [PubMed] [Google Scholar]
- 32.Yin R, Ribeiro-Filho HV, Lin V, Gowthaman R, Cheung M, Pierce BG. TCRmodel2: high-resolution modeling of T cell receptor recognition using deep learning. Nucleic Acids Res. 2023. Jul 5;51(W1):W569–W576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harkiolaki M, Holmes SL, Svendsen P, Gregersen JW, Jensen LT, McMahon R, Friese MA, van Boxel G, Etzensperger R, Tzartos JS, Kranc K, Sainsbury S, Harlos K, Mellins ED, Palace J, Esiri MM, van der Merwe PA, Jones EY, Fugger L. T Cell-Mediated Autoimmune Disease Due to Low-Affinity Crossreactivity to Common Microbial Peptides. Immunity. 2009. Mar 20;30(3):348–357. [DOI] [PubMed] [Google Scholar]
- 34.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MCJ, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J. T cell allorecognition via molecular mimicry. Immunity. 2009. Dec 18;31(6):897–908. [DOI] [PubMed] [Google Scholar]
- 35.Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G, Komech EA, Sycheva AL, Koneva AE, Egorov ES, Eliseev AV, Van Dyk E, Dash P, Attaf M, Rius C, Ladell K, McLaren JE, Matthews KK, Clemens EB, Douek DC, Luciani F, van Baarle D, Kedzierska K, Kesmir C, Thomas PG, Price DA, Sewell AK, Chudakov DM. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 2018. 04;46(D1):D419–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heather JM, Spindler MJ, Alonso MH, Shui YI, Millar DG, Johnson DS, Cobbold M, Hata AN. Stitchr: stitching coding TCR nucleotide sequences from V/J/CDR3 information. Nucleic Acids Res. 2022. Jul 8;50(12):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh NK, Riley TP, Baker SCB, Borrman T, Weng Z, Baker BM. Emerging concepts in T cell receptor specificity: rationalizing and (maybe) predicting outcomes. J Immunol Baltim Md 1950. 2017. Oct 1;199(7):2203–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewell AK. Why must T cells be cross-reactive? Nat Rev Immunol. 2012. Sep;12(9):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998. Sep;19(9):395–404. [DOI] [PubMed] [Google Scholar]
- 40.Antunes DA, Rigo MM, Freitas MV, Mendes MFA, Sinigaglia M, Lizée G, Kavraki LE, Selin LK, Cornberg M, Vieira GF. Interpreting T-Cell Cross-reactivity through Structure: Implications for TCR-Based Cancer Immunotherapy. Front Immunol [Internet]. 2017. [cited 2023 Feb 4];8. Available from: 10.3389/fimmu.2017.01210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Jasinski JM, Kobayashi M, Davenport B, Johnson K, Davidson H, Nakayama M, Haskins K, Eisenbarth GS. Analysis of T cell receptor beta chains that combine with dominant conserved TRAV5D-4*04 anti-insulin B:9–23 alpha chains. J Autoimmun. 2009. Aug;33(1):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer I, Tickotsky N, Louzoun Y. Contribution of T Cell Receptor Alpha and Beta CDR3, MHC Typing, V and J Genes to Peptide Binding Prediction. Front Immunol [Internet]. 2021. [cited 2023 Oct 5];12. Available from: 10.3389/fimmu.2021.664514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gil A, Kamga L, Chirravuri-Venkata R, Aslan N, Clark F, Ghersi D, Luzuriaga K, Selin LK. Epstein-Barr Virus Epitope-Major Histocompatibility Complex Interaction Combined with Convergent Recombination Drives Selection of Diverse T Cell Receptor α and β Repertoires. mBio. 2020. Mar 17;11(2):e00250–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J Biol Chem. 2009. Oct 2;284(40):27281–27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamga L, Gil A, Song I, Brody R, Ghersi D, Aslan N, Stern LJ, Selin LK, Luzuriaga K. CDR3α drives selection of the immunodominant Epstein Barr virus (EBV) BRLF1-specific CD8 T cell receptor repertoire in primary infection. PLoS Pathog. 2019. Nov;15(11):e1008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bovay A, Zoete V, Dolton G, Bulek AM, Cole DK, Rizkallah PJ, Fuller A, Beck K, Michielin O, Speiser DE, Sewell AK, Fuertes Marraco SA. T cell receptor alpha variable 12–2 bias in the immunodominant response to Yellow fever virus. Eur J Immunol. 2018. Feb;48(2):258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996. Nov 14;384(6605):134–141. [DOI] [PubMed] [Google Scholar]
- 48.Broughton SE, Petersen J, Theodossis A, Scally SW, Loh KL, Thompson A, van Bergen J, Kooy-Winkelaar Y, Henderson KN, Beddoe T, Tye-Din JA, Mannering SI, Purcell AW, McCluskey J, Anderson RP, Koning F, Reid HH, Rossjohn J. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity. 2012. Oct 19;37(4):611–621. [DOI] [PubMed] [Google Scholar]
- 49.Dolton G, Rius C, Wall A, Szomolay B, Bianchi V, Galloway SAE, Hasan MS, Morin T, Caillaud ME, Thomas HL, Theaker S, Tan LR, Fuller A, Topley K, Legut M, Attaf M, Hopkins JR, Behiry E, Zabkiewicz J, Alvares C, Lloyd A, Rogers A, Henley P, Fegan C, Ottmann O, Man S, Crowther MD, Donia M, Svane IM, Cole DK, Brown PE, Rizkallah P, Sewell AK. Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy. Cell. 2023. Aug 3;186(16):3333–3349.e27. [DOI] [PubMed] [Google Scholar]
- 50.Jacobsen LM, Posgai A, Seay HR, Haller MJ, Brusko TM. T Cell Receptor Profiling in Type 1 Diabetes. Curr Diab Rep. 2017. Oct 11;17(11):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seay HR, Yusko E, Rothweiler SJ, Zhang L, Posgai AL, Campbell-Thompson M, Vignali M, Emerson RO, Kaddis JS, Ko D, Nakayama M, Smith MJ, Cambier JC, Pugliese A, Atkinson MA, Robins HS, Brusko TM. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight. 2016. 08;1(20):e88242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herold KC, Gitelman SE, Ehlers MR, Gottlieb PA, Greenbaum CJ, Hagopian W, Boyle KD, Keyes-Elstein L, Aggarwal S, Phippard D, Sayre PH, McNamara J, Bluestone JA, the AbATE Study Team. Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: Metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013. Jul 8; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gitelman SE, Gottlieb PA, Felner EI, Willi SM, Fisher LK, Moran A, Gottschalk M, Moore WV, Pinckney A, Keyes-Elstein L, Harris KM, Kanaparthi S, Phippard D, Ding L, Bluestone JA, Ehlers MR, ITN START Study Team. Antithymocyte globulin therapy for patients with recent-onset type 1 diabetes: 2 year results of a randomised trial. Diabetologia. 2016. Jun;59(6):1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, James EA, Sanda S, Greenbaum C, Kwok WW. CD4+ T cells recognize diverse epitopes within GAD65: implications for repertoire development and diabetes monitoring. Immunology. 2013. Mar;138(3):269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ettinger RA, Buitinga M, Vandamme C, Afonso G, Gomez R, Arribas-Layton D, Bissenova S, Speake C, Reijonen H, Kinnunen T, Overbergh L, Mallone R, Kwok WW, James EA. Technical Validation and Utility of an HLA Class II Tetramer Assay for Type 1 Diabetes: A Multicenter Study. J Clin Endocrinol Metab. 2023. Jul 20;dgad434. [DOI] [PubMed] [Google Scholar]
- 56.Linsley PS, Barahmand-Pour-Whitman F, Balmas E, DeBerg HA, Flynn KJ, Hu AK, Rosasco MG, Chen J, O’Rourke C, Serti E, Gersuk VH, Motwani K, Seay HR, Brusko TM, Kwok WW, Speake C, Greenbaum CJ, Nepom GT, Cerosaletti K. Autoreactive T cell receptors with shared germline-like α chains in type 1 diabetes. JCI Insight. 2021. Nov 22;6(22):e151349. [DOI] [PMC free article] [PubMed] [Google Scholar]