Abstract
The guanosine analogs BMS-200475 and lobucavir have previously been shown to effectively suppress propagation of the human hepatitis B virus (HBV) and woodchuck hepatitis virus (WHV) in 2.2.15 liver cells and in the woodchuck animal model system, respectively. This repression was presumed to occur via inhibition of the viral polymerase (Pol) by the triphosphate (TP) forms of BMS-200475 and lobucavir which are both produced in mammalian cells. To determine the exact mode of action, BMS-200475–TP and lobucavir-TP, along with several other guanosine analog-TPs and lamivudine-TP were tested against the HBV, WHV, and duck hepatitis B virus (DHBV) polymerases in vitro. Estimates of the 50% inhibitory concentrations revealed that BMS-200475–TP and lobucavir-TP inhibited HBV, WHV, and DHBV Pol comparably and were superior to the other nucleoside-TPs tested. More importantly, both analogs blocked the three distinct phases of hepadnaviral replication: priming, reverse transcription, and DNA-dependent DNA synthesis. These data suggest that the modest potency of lobucavir in 2.2.15 cells may be the result of poor phosphorylation in vivo. Kinetic studies revealed that BMS-200475–TP and lobucavir-TP competitively inhibit HBV Pol and WHV Pol with respect to the natural dGTP substrate and that both drugs appear to bind to Pol with very high affinities. Endogenous sequencing reactions conducted in replicative HBV nucleocapsids suggested that BMS-200475–TP and lobucavir-TP are nonobligate chain terminators that stall Pol at sites that are distinct yet characteristically two to three residues downstream from dG incorporation sites.
Human hepatitis B virus (HBV), the prototype member of a small family of related hepadnaviruses, remains a major agent of liver infection and a cause of liver disease throughout the world. Although the majority of HBV infections are acute in nature and clinically resolve within 6 months, the chronic persistent HBV infection seen in some 5 to 10% of acutely infected individuals places them at greatly increased risk of liver disease, including cirrhosis and hepatocellular carcinoma (1).
Current efforts to break the cycle of persistent HBV infection have mostly centered on nucleoside analogs as inhibitors of the multifunctional viral polymerase (Pol) (14), the key enzyme in the unique hepadnaviral replication scheme. Pol converts a greater-than-genome-length, plus-strand pregenomic RNA (pgRNA) into the partially double-stranded circular DNA genome via a multistep process (11, 23, 33). The first and most remarkable step is a discrete priming reaction (45) in which a specific tyrosine residue of Pol, located in an N-terminal priming domain, acts as the acceptor for the initiating deoxynucleotide residue, usually a dGTP (20, 21, 50). Priming is templated by a bulge sequence in the stem-loop structure (epsilon or ɛ) (27, 41) on the pgRNA and results in a short DNA oligomer which is covalently linked to Pol. The next step is marked by translocation of the Pol-DNA adduct to a complementary sequence found in the DR1 element at the 3′ end of pgRNA (27, 41, 45), followed by the elongation of minus-strand DNA via RNA-dependent DNA synthesis (reverse transcription [RT]) (39). Finally, Pol also mediates DNA-dependent plus-strand DNA synthesis which is primed by an RNA primer (22) resulting from incomplete degradation of pgRNA by an RNase H activity at the carboxyl terminus of Pol (30).
BMS-200475 is a cyclopentyl guanine with an exo carbon-carbon double bond (2, 37). BMS-200475 has been shown to be an effective (50% effective concentration [EC50], 3.8 ± 1.4 nM) and selective (selectivity index, ∼8,000) inhibitor of HBV replication in cultured 2.2.15 liver cells (17). Furthermore, BMS-200475 exhibits excellent efficacy against woodchuck hepatitis virus (WHV) in chronically infected carrier woodchucks at oral dosages as low as 0.02 mg/kg of body weight/day (13). The cyclobutyl guanine (9) lobucavir, formerly designated BMS-180194, exhibits a broad antiviral spectrum including antihepadnaviral activity in 2.2.15 cells (EC50, 2.5 ± 0.85 μM) (17) as well as in WHV-infected woodchucks (4).
Inhibition of hepadnaviral replication is presumed to occur through the triphosphate (TP) forms of BMS-200475 and lobucavir, with both nucleosides being converted to their respective TP forms in mammalian cells by cellular enzymes. In this report, we used three distinct in vitro Pol assays to determine the mechanism of inhibition of HBV polymerase activity by BMS-200475–TP and lobucavir-TP. The results revealed that the TP forms of both guanosine analogs are similarly potent in inhibiting HBV, WHV, and duck hepatitis B virus (DHBV) Pol in vitro and that they effectively block all three replication steps.
MATERIALS AND METHODS
Compounds.
The following compounds were chemically synthesized at Bristol-Myers Squibb: BMS-200475, formerly SQ-34676 {[1S-(1α,3α,4β)]-2-amino- 1,9-dihydro-9-[4-hydroxy-3-(hydroxymethyl)-2-methylenecyclopentyl]-6H-purin-6-one monohydrate}; lobucavir, formerly cyclobut G or BMS-180194 {(1R)-(1α,2β,3α)-[2,3-bis(hydroxymethyl)cyclobutyl]guanine}; SQ-32829 or (±)-HHCG {(±)-(1α,2β,3α)-9-[2-hydroxy-3-(hydroxymethyl)cyclobutyl]guanine} (18, 44); acyclovir [ACV; 9-(2-hydroxyethoxymethyl)-guanine]; ganciclovir [GCV; 9-(1,3-dihydroxy-2-propoxymethyl)-guanine]; and lamivudine (3TC; (−)-β-l-2′,3′-dideoxy-3′-thiacytidine]. The TP forms of BMS-200475, ACV (43), and 3TC were synthesized chemically; lobucavir-TP, SQ-32829–TP, and GCV-TP were prepared enzymatically by using herpes simplex virus type 1 thymidine kinase essentially as described previously (44). Other deoxynucleoside-TPs (dNTPs) and 2′,3′-dideoxynucleoside-TPs (ddNTPs) were purchased from Pharmacia (Piscataway, N.J.).
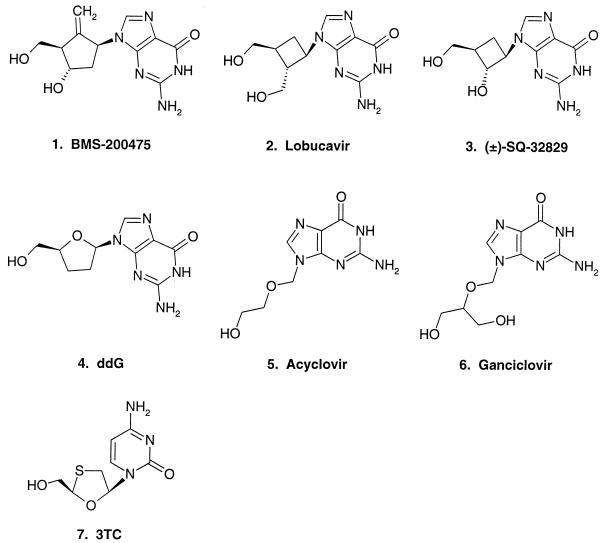
The structures of the corresponding nucleoside moieties of all seven TPs tested are depicted in Fig. 1. BMS-200475 (2) is structurally similar to 2′-deoxyguanosine except for the exo carbon-carbon double bond replacing the natural furanose oxygen and inhibits HBV replication, with an EC50 of 3.8 ± 1.4 nM in cultured 2.2.15 liver cells (17). Lobucavir (9), which bears a cyclobutyl replacement for the sugar moiety, has an EC50 of 2.5 ± 0.85 μM against HBV (17) and is also active against herpesviruses (48). The dC analog 3TC exhibits EC50s of 60 to 116 nM against HBV in cultured cells (17, 19) and is effective in the clinical setting of HBV infection (8, 28). The well-known antiherpesvirus drugs ACV and GCV are guanosine analogs with acyclic sugars. GCV reportedly inhibits DHBV replication in primary duck hepatocytes with an EC50 of 2.5 μM (3), while ACV has only modest activity against HBV in vitro (19): both compounds have been used against HBV in animal models (7, 42) and humans (12, 32). ddG is active against HBV in cultured cells, with a reported EC50 of 2.3 μM (19), and its TP form selectively inhibits DHBV Pol (16). SQ-32829 (18) has activity against herpesviruses (44) but has not been previously tested against HBV; it was included in this study because of its close structural similarity to lobucavir (they differ only in a hydroxyl group rather than a hydroxymethyl group at the 2′ position of the sugar ring), which should provide insight into the specificity of the interaction with hepadnaviral Pols.
FIG. 1.
Chemical structures of dG and dC analogs.
Pol assays and IC50 determinations.
Three in vitro assays were used to measure separately the priming, RT, and DNA-dependent DNA polymerization activities of hepadnaviral Pols. Mammalian hepadnaviral Pols were assayed by an endogenous Pol assay (EPA) that measures the Pol activity within virions or viral nucleocapsids (25). Concentrated WHV virions, partially freed of serum components, were derived from the sera of chronically infected woodchucks purchased from Marmotech (Cortland, N.Y.). The sera were thawed and clarified (20 min, 10,000 × g, 4°C), and the virus particles were pelleted through 25% (wt/vol) sucrose–TNE (10 mM Tris hydrochloride [pH 7.4], 160 mM NaCl, 1 mM EDTA) plus 0.75% Triton X-100 in a tabletop ultracentrifuge (TLA100.3 rotor; 550,000 × g, 1 h, 4°C). The virions were resuspended in TNE at 1/10 of the original serum volume. In a standard in vitro EPA (see below), these virions conduct a late-stage hepadnaviral replication reaction that reflects the synthesis of second-strand DNA (6). Recombinant human HBV nucleocapsids harboring an active HBV Pol were generated as described previously (35). Briefly, these nucleocapsids are obtained by expressing the HBV core and Pol proteins in trans in SF9 or SF21 insect cells via two distinct baculovirus vectors. Replicative nucleocapsids were purified to >80% purity by ultracentrifugation procedures following mild protease and nuclease treatment. In a standard EPA (see below), these nucleocapsids undergo mostly the earliest phases of the HBV replication reaction, namely, the priming step and the RT of first-strand HBV DNA (35).
Gel-based 50% inhibitory concentration (IC50) determinations were performed with 2-μl aliquots of 10×-concentrated WHV virions (equal to 20 μl of serum) and approximately 1 μg of immunocomplexed HBV nucleocapsids, respectively, per reaction mixture. The latter were immunoprecipitated from infected insect cell lysates as described recently (35).
For standard EPAs (35), WHV virions or immunocomplexed HBV capsids were resuspended in 50 μl of EPA buffer (50 mM Tris hydrochloride [pH 7.4], 75 mM NH4Cl, 1 mM EDTA, 20 mM MgCl2, 0.1 mM β-mercaptoethanol, 0.5% Tween 20) supplemented with 50 μM (or in some reactions 12.5 μM) unlabeled dNTPs (dGTP, dCTP, and TTP) and 33 nM [α-32P]dATP (3,000 Ci/mmol; NEN-Dupont, Boston, Mass.). Following incubation at 37°C for 12 to 16 h (WHV) and 6 h (HBV), respectively, endogenously labeled DNA products were extracted as described previously (35) and were then analyzed by gel electrophoresis. Double-stranded WHV genomes were resolved on 1% agarose gels in TBE (90 mM Tris [pH 8.0], 90 mM borate, 0.1 mM EDTA). Covalently linked minus-strand HBV RT products were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The products were visualized by autoradiography following fixation and drying of the gels. For determination of IC50s, the dried gels were scanned on a phosphorimager (Storm 860; Molecular Dynamics, Sunnyvale, Calif.) prior to data evaluation with Imagequant software (Molecular Dynamics). 35S-labeled, in vitro-translated HBV Pol was run as a reference.
The in vitro priming activity of DHBV Pol was assayed by the method of Wang and Seeger (46). DHBV Pol was expressed from a full-length DHBV pgRNA-like transcript (15) generated in a coupled in vitro transcription-translation system (TNT-SP6; Promega, Madison, Wis.). The translation reaction (12.5 μl) contained 250 ng of plasmid pSP65-D (kindly provided by Jesse Summers, Alberquerque, N.M.) and was incubated for 2 h at 30°C. Subsequently, the priming reaction was carried out for 30 min at 30°C in a 25-μl reaction volume containing 250 nM [α-32P]dGTP (3,000 Ci/mmol; NEN-Dupont) and unlabeled dCTP, dATP, and TTP each at a concentration of 220 nM; even in the presence of all four deoxynucleotides the predominant outcome of this assay is a limited priming reaction (data not shown; see Results). Aliquots (1.5 μl) were resolved on SDS–8% polyacrylamide gels. The fixed and dried gels were visualized, scanned, and quantitated as described above.
As an alternative to gel-based assays, a plate-format assay (see below) was used to obtain inhibition data for the mammalian hepadnaviral polymerase preparations. The IC50s presented in this report are expressed in the form of the ratio of drug concentration: concentration of natural substrate that is required to give 50% activity relative to the activities of the no-drug controls.
Kinetics.
For kinetic studies (34), resuspended WHV virions or HBV nucleocapsids were subjected to EPAs adapted to a 96-well plate format. All reactions were performed in triplicate. A typical assay was run for 2 h at 30°C and contained (per well) 75 mM NH4Cl, 50 mM Tris-HCl (pH 7.4), 20 mM MgCl2, 0.1% Tween 20, 100 μg of bovine serum albumin per ml, 200 μg of tRNA per ml, and either ∼500 ng of baculovirus-expressed HBV nucleocapsids or 2 μl of 10×-concentrated WHV virions (equal to 20 μl of serum) in a final volume of 20 μl. Deoxynucleotides and inhibitors were added as outlined below. Reactions were terminated with 25 μl of chilled 20% trichloroacetic acid (TCA)–2% sodium pyrophosphate. Following 15 min of incubation on ice, precipitates were collected on glass-fiber-filter plates (Unifilter 96; Packard Instruments, Meriden, Conn.), washed extensively with water and then ethanol, and quantified by liquid scintillation counting (Topcount; Packard Instruments).
For competitive inhibition studies the WHV TCA plate assay included eight dilutions of [α-33P]dGTP (125 to 5.55 nM; 2,000 Ci/mmol; NEN-Dupont); unlabeled dATP, dCTP, and TTP at concentrations of 5 μM each; and 0, 0.5, 2, 5, or 10 nM BMS-200475–TP or lobucavir-TP. The HBV TCA plate assay included eight dilutions of [α-33P]dGTP (167 to 5 nM); unlabeled dATP, dCTP, and TTP at concentrations of 5 μM each; and BMS-200475–TP or lobucavir-TP at 0, 0.5, 2, 5, or 10 nM. The Kis of BMS-200475–TP and lobucavir-TP were obtained by plotting the analog concentrations against the slopes calculated from double-reciprocal graphs of 1/V (where V is the reaction rate) versus 1/S (where S is the substrate concentration) at each drug concentration (Lineweaver-Burk plots).
Alternatively, Kis of BMS-200475–TP, lobucavir-TP, and 3TC-TP against WHV and HBV Pols were determined from independent measurements of (i) the Km of the natural substrates (dGTP and dCTP) and (ii) the IC50s of the analogs (BMS-200475–TP, lobucavir-TP, and 3TC-TP). Km determinations included eight dilutions of [α-33P]dGTP or [α-33P]dCTP (250 to 7.81 nM); the IC50 assays used eight dilutions of BMS-200475–TP (1,000 to 0.3 nM) at 33 nM [33P]dGTP, eight dilutions of lobucavir-TP (6.67 to 0.052 nM) at 2.5 nM [33P]dGTP, or eight dilutions of 3TC-TP (1000 to 1 nM) at 22.75 nM [33P]dCTP. The Ki was then calculated by the equation Ki = IC50/(1 + S/Km).
Endogenous sequencing.
Endogenous sequencing reactions were performed as described by Molnar-Kimber et al. (26), with the following modifications: purified baculovirus-expressed HBV nucleocapsids (10 μg per 100-μl reaction mixture) replaced DHBV capsids, and the reaction mixtures were incubated for 3 h at 37°C and included the four α-32P-labeled dNTPs (800 Ci/mmol; NEN-Dupont) at concentrations of 130 nM each and either one of the four ddNTPs (26 μM) or else BMS-200475–TP or lobucavir-TP at 2 and 10 μM, respectively. Unincorporated radiolabeled dNTPs were removed by pelleting the capsids through an equal volume of 25% (wt/vol) sucrose in TNE plus 0.75% Triton X-100 in a TLA100.1 fixed-angle rotor for 1.5 h at 550,000 × g and 4°C with a tabletop TL 100 ultracentrifuge (Beckman Instruments, Fullerton, Calif.). Pellets were resuspended in 10 μl of TE (Tris-EDTA)–0.1% SDS and were then digested with 0.2 mg of proteinase K (Boehringer Mannheim, Indianapolis, Ind.) per ml for 45 min at 37°C. Half of each sample was electrophoresed through a denaturing 6% polyacrylamide gel containing 8 M urea in TBE. The incorporated radioactivity was visualized by scanning the dried gel with a phosphorimager (Molecular Dynamics).
RESULTS
Inhibition of hepadnaviral Pols in vitro.
An initial set of experiments examined the inhibition of different hepadnaviral Pols and discrete hepadnaviral replication steps by the TP forms of BMS-200475 and lobucavir. For comparison purposes, this analysis also included five other nucleoside-TPs, four guanosine analogs and a single dC derivative, which were selected on the basis of either their activity against hepadnavirus Pols or structural considerations.
To determine whether the TPs of these nucleoside analogs were capable of inhibiting hepadnaviral Pols, we initially titrated the selected nucleoside analog-TPs against the HBV, WHV, and DHBV Pols in three different in vitro assays which measure the three replication steps of priming, RT, and plus-strand DNA synthesis. Because these three systems require widely different concentrations of dNTPs, the analog-TP concentration-to-natural dNTP substrate concentration ratio provides a more meaningful comparison between the different systems than does the actual inhibitor concentration.
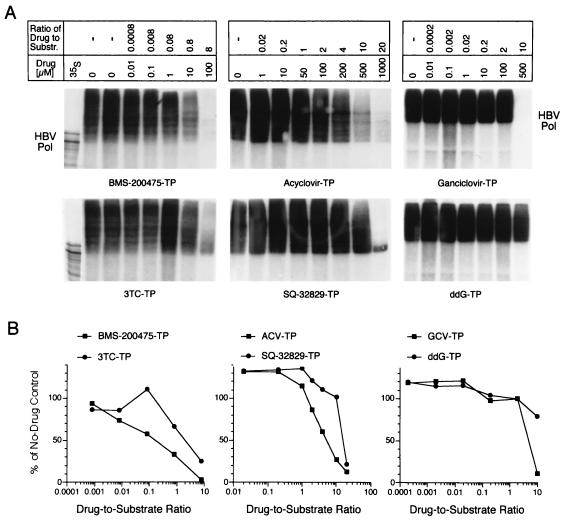
In representative drug titration experiments (Fig. 2), HBV Pol was assayed in the context of recently described baculovirus-derived replicative HBV cores (35) which yield abundant Pol-linked, 32P-labeled HBV minus-strand DNA in a standard endogenous Pol reaction. To facilitate a fair comparison between G and C analogs, [α-32P]dATP was used as the label and the drug was titrated against the unlabeled dGTP substrate for the G analogs or against dCTP for 3TC (the unlabeled substrate was typically present in the assay at 12.5 to 50 μM). The Pol-linked species were disclosed by SDS-PAGE and autoradiography (Fig. 2A) and were quantitated by phosphorimaging of the gels (Fig. 2B). BMS-200475–TP reduced the production of Pol-linked products in a dose-dependent manner, eliminating 50% of the products at a BMS-200475–TP concentration-to-dGTP substrate concentration ratio of 0.2 and essentially all products at a ratio of 8. Since lobucavir-TP is inherently difficult to synthesize, there was too little of this material to titrate in the gel-based assay; however, when it was titrated in the modified HBV TCA plate assay its inhibitory profile appeared to be comparable to that of BMS-200475–TP (see Table 1). 3TC-TP was the next most effective HBV Pol inhibitor, reducing DNA synthesis by 50% at an inhibitor concentration-to-substrate concentration ratio of ∼2, followed by ACV-TP and GCV-TP which inhibited synthesis by 50% when present at an approximately sixfold excess over the amount of substrate present. However, whereas ACV-TP and GCV-TP appeared to eliminate all Pol-linked species uniformly, some smaller-product species clearly escaped inhibition by an eightfold excess of 3TC-TP (see below). For SQ-32829–TP, a high molar excess (>10) was required to inhibit Pol activity significantly, and a 20-fold excess of drug again left a distinctive tight band of small Pol-linked DNA products; these presumably reflect species generated in the discrete priming reaction which is apparently not inhibited by certain nucleoside-TPs (see below). ddGTP was largely inactive against HBV Pol (see Table 1).
FIG. 2.
Inhibitory effects of nucleoside analogs on HBV Pol-mediated viral minus-strand DNA synthesis. Recombinant HBV nucleocapsids were subjected to endogenous Pol assays in the presence of increasing (from left to right) concentrations of dG analog TPs or the dC analog 3TC-TP and constant concentrations of cold dNTPs and [α-32P]dATP. (A) The radiolabeled Pol-linked minus-strand DNA products were extracted and analyzed through SDS–8% polyacrylamide gels. The position of HBV Pol (∼93 kDa) is indicated at the sides of the figure; an in vitro-translated 35S-labeled Pol marker is shown in lane 35S (authentic 93-kDa HBV Pol is the second largest species in this lane). Substr., substrate. (B) Inhibition curves were generated by phosphorimaging of the gels shown in panel A.
TABLE 1.
In vitro inhibition of hepadnaviral Pol activities by nucleoside TPs
| Analog-TP | IC50 (analog-TP concn/dNTP concn)
|
||
|---|---|---|---|
| HBV end. RT and priming | WHV end. Pol | DHBV priming | |
| BMS-200475a | 0.32 ± 0.28 | 0.18 ± 0.13 | 0.30 ± 0.40 |
| Lobucavira | 0.28 ± 0.12 | 0.29 ± 0.07 | 0.30 ± 0.20 |
| SQ-32829b | 12.10 | 4.12 | >400 |
| Ganciclovirb | 47.50 | 7.20 | >500 |
| Acyclovirb | 5.20 | 1.56 | >400 |
| ddGb | >17 | 3.35 | 214 |
| 3TCb | >4.7 | 7.44 | >500 |
Values are means and standard deviations derived from eight experiments.
Average values from two experiments (except for ACV which could be assayed only once).
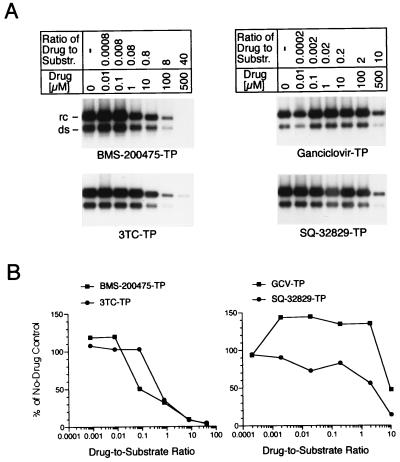
The inhibitory effects of representative analog-TPs on WHV Pol was measured via EPAs conducted with serum-derived WHV virions. Mature hepadnaviruses house largely completed linear and relaxed circular double-stranded DNA species (47). Thus, upon the addition of exogenous dNTPs in vitro, the encapsidated WHV Pol conducts only a limited DNA-dependent plus-strand DNA synthesis (6). Following deproteinization, these 32P-labeled genomic DNA species can readily be visualized by agarose gel electrophoresis and autoradiography (Fig. 3A) and can be quantitated by phosphorimaging (Fig. 3B). The titration curves revealed BMS-200475–TP to be a very potent inhibitor of WHV second-strand synthesis, with 50% inhibition attained at a very low drug concentration-to-substrate concentration ratio of ∼0.08. The inhibitory profile of lobucavir-TP against WHV Pol was very similar to that of BMS-200475–TP, as determined by the plate assay method (data not shown; see below). 3TC-TP was more active than SQ-32829–TP or GCV-TP against WHV Pol, but 3TC-TP was still some six- to sevenfold less potent than BMS-200475–TP.
FIG. 3.
Inhibitory effect of select nucleoside analogs on WHV Pol-dependent viral plus-strand DNA synthesis. (A) Partially purified virions from the serum of a woodchuck with chronic WHV infection were used to conduct endogenous Pol reactions with various amounts of guanosine-TPs or cytosine-TPs, as indicated at the top, and constant concentrations of cold dNTPs and [α-32P]dATP. The characteristic double-stranded linear (ds) and relaxed circular (rc) DNA species were isolated, resolved on 1% agarose gels, and imaged by autoradiography. Substr., substrate. (B) Inhibition curves generated by phosphorimaging analysis of the gels in panel A.
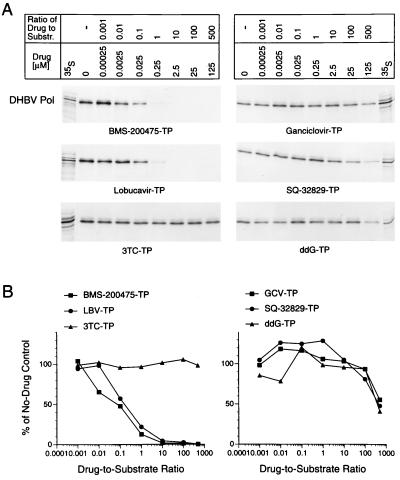
The inhibitory effects of the TP analogs against the priming reaction of DHBV Pol (Fig. 4) was monitored in an in vitro translation-priming system (46, 49, 50). Priming was conducted in the presence of comparable concentrations of all four deoxynucleotides (including the [α-32P]dGTP label) as described in Materials and Methods. DHBV Pol activity largely remained limited to a discrete priming reaction under these conditions which allow the different inhibitors to be more fairly compared. The covalently protein-linked DHBV oligonucleotide-primer products (Pol-GTAA) were analyzed by SDS–8% PAGE (Fig. 4A). Phosphorimaging of the gels (Fig. 4B) revealed that BMS-200475–TP and lobucavir-TP are similarly effective against the dGTP-based priming reaction, giving 50% inhibition at drug concentration-to-substrate concentration ratios of ∼0.1. The remaining guanosine-TP analogs were inhibitory only when present in a 100- to 500-fold excess over the amount of dGTP substrate present. 3TC-TP had no effect on a priming reaction even at a drug concentration-to-substrate concentration ratio of 500.
FIG. 4.
Effect of guanosine versus cytosine analogs on DHBV priming. In vitro-translated DHBV Pol was incubated with 250 nM [α-32P]dGTP; 220 nM unlabeled dCTP, dATP, and TTP; and increasing concentrations of the indicated analog-TPs. (A) The radiolabeled Pol-oligonucleotide adducts were analyzed by conventional SDS-PAGE and autoradiography. The migration positions of the 35S-labeled DHBV Pol (lane 35S) are indicated at the sides. Substr., substrate. (B) Titration curves were generated by phosphorimaging.
A more quantitative and comprehensive summary of the effects of the nucleoside-TPs on the three hepadnaviral Pols as well as on the three distinct replication steps is presented in Table 1. The IC50s presented are again expressed as ratios of drug-to-natural substrate (dGTP and dCTP, respectively) and represent the mean values and standard deviations obtained from eight independent BMS-200475–TP and lobucavir-TP titration experiments. The data are derived from both plate- and gel-based assays for BMS-200475–TP and plate-based assays only for lobucavir-TP; the two formats gave very comparable results (data not shown). The remaining compounds were typically assayed in at least two experiments, the results of which were averaged. It is apparent from the uniformly low IC50s of ∼0.3 seen across the first two rows of Table 1 that BMS-200475–TP and lobucavir-TP are remarkably similar (i) in inhibiting the three Pol species (HBV, WHV, DHBV) and, more importantly, (ii) in suppressing all three distinct replication reactions: priming, RT of viral minus strand, and plus-strand DNA synthesis. The fractional inhibitor concentration-to-substrate concentration ratios required to elicit a 50% reduction in Pol activity further imply that the analogs bind to Pol better than the natural substrate does, and this is supported by their low Kis (see below). In contrast, SQ-32829–TP, GCV-TP, ACV-TP, ddG-TP, and 3TC-TP showed negligible activity against DHBV priming, were 5- to 25-fold less active than BMS-200475–TP and lobucavir-TP against WHV plus-strand DNA synthesis, and were 15- to 150-fold less active against HBV Pol-mediated minus-strand DNA polymerization.
The preceding data established that BMS-200475–TP inhibits the DHBV priming reaction, while 3TC-TP does not (Fig. 4). This is consistent with the idea that dCTP cannot be incorporated into the DHBV primer (GTAA) (5, 46) or the HBV primer (GAA or TGAA) (21, 35). This difference between BMS-200475 and 3TC was further tested by conducting in vitro EPA reactions with the recombinant HBV Pol-bearing preparations that are competent for both priming and minus-strand DNA elongation. HBV nucleocapsids were exposed to no drug, to 50 μM BMS-200475–TP, or to 250 μM 3TC-TP; these high concentrations were used to ensure that both drugs exerted their maximal suppressive effects. As is apparent from the gel profile (Fig. 5), BMS-200475–TP uniformly suppressed priming and the minus-strand DNA elongation activities of HBV Pol. In contrast, the inhibitory effect of 3TC-TP appeared to be restricted to the blocking of RT, leaving the priming reaction intact, as evidenced by the tight band of Pol adducts with the approximate size of the 35S-labeled HBV Pol standard (93 kDa). 3TC appears very similar in this regard to phosphonoformic acid, a known inhibitor of hepadnavirus elongation (24) but not priming (21, 35, 46).
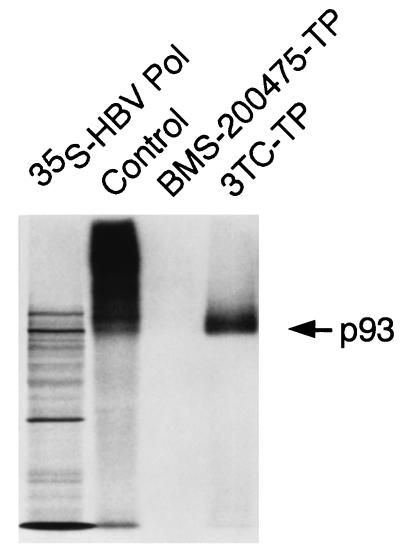
FIG. 5.
Effect of BMS-200475-TP versus 3TC-TP on the HBV priming and reverse transcription activities. Cores were subjected to endogenous Pol reactions in the presence of 50 μM BMS-200475–TP or 250 μM 3TC-TP. The product pattern generated in the absence of drug (lane Control) is shown for comparison. In vitro-translated [35S]-labeled HBV pol (p93) served as size marker.
Kinetic studies of inhibition.
To further explore the mechanism of action of lobucavir-TP and BMS-200475–TP, we next turned to some basic enzyme kinetics (34) using microtiter plate-adapted versions of both the WHV virion and replicative HBV nucleocapsid Pol assay systems (Fig. 6). This assay measures the level of incorporation of α-33P-labeled dGTP into TCA-precipitable DNA (see Materials and Methods).
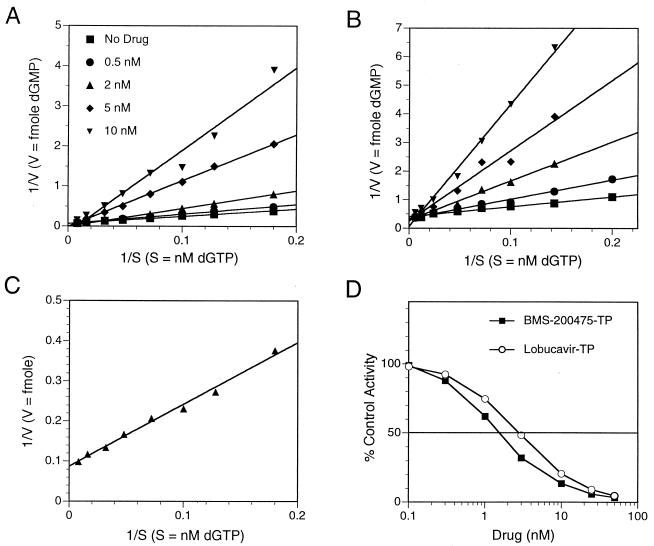
FIG. 6.
Kinetic analyses of dGTP, BMS-200475–TP, and lobucavir-TP using WHV and HBV Pols. Lineweaver-Burk plots indicate that lobucavir-TP (A) and BMS-200475-TP (B) are competitive inhibitors of dGTP for WHV Pol (A) and HBV Pol (B). The velocities of the Pol reactions were measured as femtomoles of [α-33P]dGMP incorporation in 120 min at 30°C. (C) Double-reciprocal plot to determine the Km of dGTP with HBV Pol. The reaction velocity was measured as described above. (D) IC50 titrations of BMS-200475–TP and lobucavir-TP. HBV nucleocapsids were incubated for 120 min at 30°C with dNTPs and serial dilutions of the indicated guanosine analog-TPs. The percentage of [α-33P]dGTP incorporation relative to that for the no-drug control is plotted on a semilogarithmic scale (mean of triplicate samples). The values from panels C and D were used to calculate the Kis of the guanosine analog-TPs.
We first asked whether lobucavir-TP and BMS-200475–TP are competitive inhibitors of Pol with respect to the natural substrate dGTP. Constant analog concentrations ranging from 0.5 to 10 nM were titrated against the relevant Pol enzyme at a variety of S to determine the corresponding V. Representative Lineweaver-Burk reciprocal plots of 1/V versus 1/S (Fig. 6A and B and data not shown) revealed that lobucavir-TP and BMS-200475–TP are indeed competitive inhibitors of the WHV and HBV Pol enzymes with regard to dGTP.
The apparent affinity of Pol for the natural substrate dGTP (Km) versus the guanosine analogs (Ki) was then evaluated by two methods; Kis were either determined from plots (data not shown) of the analog-TP concentrations against the slopes calculated from the double-reciprocal graphs at each drug concentration (Fig. 6A and B) or via independent measurements of (i) the Km of dGTP (Fig. 6C) and (ii) IC50 determinations from serial dilutions of BMS-200475–TP and lobucavir-TP at a constant dGTP concentration (Fig. 6D). The corresponding Ki values and Ki/Km ratios were then calculated by the formula Ki = IC50/(1 + S/Km), with the Km of dGTP determined from the double-reciprocal plot (slope × Vmax) (34).
Table 2 lists the average values that were obtained from three to four experiments performed with HBV nucleocapsids by using BMS-200475–TP and lobucavir-TP. For comparison, we also estimated the corresponding Km and Ki values for dCTP and 3TC-TP, respectively. Strikingly, the Kis of the guanosine analogs were approximately three- to fourfold lower than the Km of dGTP (3.2 versus 12 nM and 4.9 versus 13.1 nM, respectively) leading to Ki/Km ratios of 0.27 for BMS-200475–TP and 0.37 for lobucavir-TP. The opposite was observed for the dCTP and 3TC-TP pairing, for which the Ki of 3TC-TP (41.5 nM) was roughly 3.2-fold higher than the corresponding Km of dCTP (13 nM).
TABLE 2.
Kis of analogs and Kms of dGTP and dCTP substratesa
| Substrate | Km (nM) | Analog | Ki (nM) | Ki/Km |
|---|---|---|---|---|
| dGTP | 12.0 | BMS-200475–TP | 3.2 | 0.27 |
| dGTP | 13.1 | Lobucavir-TP | 4.86 | 0.37 |
| dCTP | 13.0 | 3TC-TP | 41.5 | 3.19 |
Values were calculated from four (BMS-200475-TP) and three (lobucavir-TP, 3TC-TP) independent experiments, respectively, in which baculovirus-derived HBV nucleocapsids were used as a polymerase source (35).
Comparable kinetic parameters were obtained for both guanosine analog-dGTP pairs against WHV Pol. On the basis of estimates of 1.6 nM for the IC50 of BMS-200475–TP and a Km of 17.65 nM for the natural substrate dGTP, a Ki of 1.02 nM and a Ki/Km of 0.058 were calculated for BMS-200475–TP. Lobucavir-TP, when titrated in the WHV Pol assay, yielded a Ki of 0.9 nM. The Km for dGTP in this experiment was determined to be 28.21 nM, giving a Ki/Km value of 0.032.
Chain termination of HBV DNA synthesis.
Many nucleoside analogs with antiviral activities share a common mode of action; they act as substrates for viral Pols and incorporate into viral DNA chains, an event which results in the termination of DNA synthesis. 3TC-TP has been reported to act as an obligate chain terminator of DHBV Pol (36), a result which was confirmed for HBV Pol in our pilot experiments (data not shown). Due to their structures (Fig. 1), neither BMS-200475 nor lobucavir can act as an obligate chain terminator of DNA synthesis, but it nonetheless remains possible that incorporation of BMS-200475–TP and lobucavir-TP into growing hepadnaviral DNA chains causes a subsequent failure of DNA synthesis.
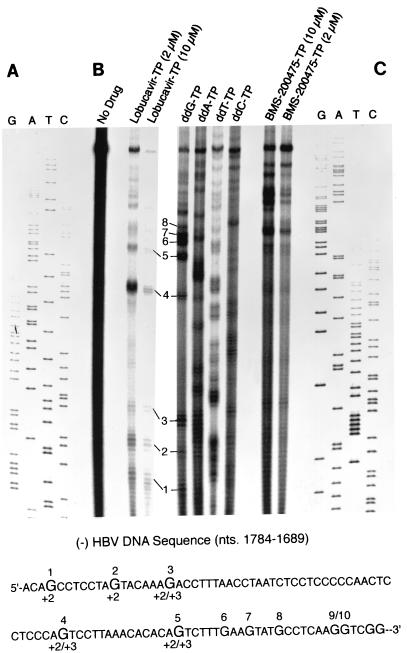
To test this possibility, we used an approach termed “endogenous sequencing” first introduced by Mason and colleagues (26). Using immature DHBV core particles from the livers of several ducks, those investigators conducted EPAs in the presence of all four α-32P-labeled dNTPs and one of each of the four ddNTP chain terminators, resulting in a nested set of randomly terminated DHBV minus-strand DNA chains. Since these DNAs arise from a discrete sequence within the replication origin, DR1, at the 3′ end of the viral pgRNA, they generate a dideoxynucleotide sequencing ladder following deproteinization and resolution on a polyacrylamide sequencing gel. Until now, this approach has not been applicable to HBV due to the difficulty of obtaining enough immature HBV cores. However, endogenous sequencing with recombinant replicative HBV cores is feasible, as seen in Fig. 7B, which also shows the influences of BMS-200475–TP and lobucavir-TP on the elongation of HBV minus-strand DNA chains. Compared to conventional dideoxysequencing ladders (31) included as size controls (Fig. 7A and C), the discrete dideoxynucleotide sequence ladder generated by replicative HBV core-associated HBV Pol in the presence of ddNTPs is hard to read, primarily due to an “echo” created by initiations occurring on either side of the main minus-strand DNA start site (35). Nevertheless, the sequence ladder can be unequivocally aligned with a region (HBV nucleotides 1784 to 1689; Fig. 7) (10) located just 45 to 140 nucleotides upstream of DR1. This alignment is as expected for nascent minus-strand DNA products arising from DR1.
FIG. 7.
(B) Endogenous sequencing of viral minus-strand DNA from replicative recombinant HBV nucleocapsids. Purified cores were incubated with 130 nM [α-32P]dNTPs and either one of the four cold ddNTPs (26 μM) or unlabeled lobucavir-TP or BMS-200475–TP (2 and 10 μM), as indicated at the top. Deproteinized reaction products were electrophoresed through a urea–6% polyacrylamide gel alongside conventional dideoxynucleotide sequencing ladders (A and C). The incorporated radioactivity was visualized by scanning the dried gel with a phosphorimager. The corresponding sequence of HBV minus-strand DNA is depicted below the figure. Major drug-induced termination sites (sites 1 to 8), are indicated, as are their alignments relative to the G residues (+2 and +3). nts., nucleotides.
Addition of BMS-200475–TP or lobucavir-TP to the EPA reaction (at either 2 or 10 μM) clearly suppressed HBV Pol activity compared to the activity of the no-drug control lane and also caused stalling at discrete sites. For lobucavir-TP, the stops characteristically occurred two or three residues beyond each and every one of the relatively few dG incorporation sites in the region of the sequence that can be read from the gel (sites 1 to 10). The earlier sites (sites 1 to 3) appeared to be preferred at higher drug concentrations. The termination pattern of BMS-200475–TP obviously differs from that of lobucavir-TP, yet the two patterns are clearly related, as is evident from the exact alignment of certain of the higher bands in Fig. 7 which correspond to sites 5 to 10 and beyond. Inspection of the sequence suggests that BMS-200475–TP is less efficient at eliciting chain termination at the proximal termination sites (which mainly comprise single dG residues) but rather effects termination at sequence elements such as closely spaced dG residues or dG doublets; again, these termination sites appear to be two to three residues beyond the normal dG incorporation sites.
DISCUSSION
In the study described in this report we have assessed the precise modes of action of BMS-200475 and lobucavir by testing the effect of their TP forms using in vitro assays which assess the WHV, DHBV, and HBV Pols as well as the three distinct steps of hepadnaviral replication (11, 23). The WHV endogenous Pol reaction uses detergent-permeabilized WHV virions which are mainly competent for late-stage WHV DNA synthesis, i.e., limited synthesis of plus-strand DNA (6). The DHBV priming reaction described by Wang and Seeger (46) mostly measures (in our experiments, almost exclusively) the priming activity of the DHBV Pol. Finally, HBV Pol activity was measured in a recently described preparation of HBV nucleocapsids that are robustly competent for at least two phases of the HBV replication reaction: the priming step and RT of minus DNA strands (35).
In terms of their effect on hepadnaviral Pol activity in vitro, the TP forms of BMS-200475 and lobucavir were essentially indistinguishable. Both were powerful inhibitors of the DHBV, WHV, and HBV Pols in vitro and appeared to be almost equipotent against the priming, minus-strand elongation, and plus-strand synthesis reactions. The comparable efficacy seen at the TP level contrasts markedly with the sharp potency difference observed for the parent nucleosides in 2.2.15 cells (3.8 nM for BMS-200475 versus 2.5 μM for lobucavir), suggesting that poor phosphorylation is the likely explanation for the weaker potency of lobucavir in cultured cells.
With respect to potency, in our hands BMS-200475–TP and lobucavir-TP were clearly superior to the TP forms of SQ-32829, GCV, ACV, ddG, and 3TC. This was true for all hepadnaviral Pols and replication steps assayed (Table 1). The best of the remaining inhibitors were probably ACV and ddG in the WHV Pol assay and ACV and 3TC in the HBV Pol assay. However, these TPs had to be present at levels 5- to 15-fold higher than the BMS-200475–TP and lobucavir-TP levels to elicit 50% inhibition of the respective assay. Penciclovir-TP was not tested in this work, but it was less effective than acyclovir-TP at inhibiting hepadnaviral minus-strand DNA elongation in a recent in vitro study (5).
The different TPs were distinguished most sharply with respect to their effects on the synthesis of the short Pol-linked DNA primer product. In the unique hepadnaviral priming reaction, Pol elaborates a short 3- or 4-base DNA oligomer by copying an RNA motif located in the bulge of the epsilon stem-loop (27, 41, 45). The DNA primer corresponds to GTAA for DHBV Pol and GAA or TGAA for HBV Pol. Our findings suggest that the DHBV and HBV Pol priming reactions were very sensitive to inhibition by BMS-200475–TP and lobucavir-TP but were largely refractory to all other TPs tested; ACV-TP and GCV-TP showed some suppression of HBV priming, but only at the highest concentrations tested. In contrast to BMS-200475–TP or lobucavir-TP, 3TC-TP essentially appeared to be inactive against the hepadnaviral priming reaction and actually appeared to enhance HBV priming, presumably reflecting a buildup in priming products caused by the ability of 3TC-TP to effectively block all replication beyond the priming step.
The findings on the priming reaction described above accord well with those obtained in some recent studies and add BMS-200475 and lobucavir to a growing list of hepadnaviral priming inhibitors. The ability of a given dNTP analog to interfere with hepadnaviral priming appears to correlate with its ability to incorporate into the primer chain. Thus, guanosine analogs such as BMS-200475–TP and lobucavir (this report), as well as penciclovir-TP, ACV-TP, and carbocyclic 2′-deoxyguanosine-TP (2′CDG-TP) (5), inhibit both HBV and DHBV priming. The TTP analogs fialuridine-TP and 2′-fluoro-β-l-arabinofuranosyluracil-TP (β-l-FMAU-TP) inhibit DHBV priming (38, 49) but have not been shown to block the HBV priming reaction. However, guanosine analogs differ markedly in their ability to suppress priming, as demonstrated by our findings and by those of Dannaoui et al. (5), who showed that penciclovir-TP and ACV-TP are comparably modest inhibitors of the DHBV priming reaction. To date, the only truly effective priming inhibitors appear to be BMS-200475–TP and lobucavir-TP (this work) and the structurally related compound 2′CDG-TP (5). It remains to be experimentally determined whether BMS-200475–TP and lobucavir-TP become covalently linked to Pol during the priming reaction, as we suspect, and whether this would have any effect on the subsequent addition of dNTPs. However, our data on HBV Pol priming (Fig. 5) suggest that BMS-200475–TP severely limits the subsequent addition of [α-32P]dATP into the HBV primer; in contrast, 2′CDG-TP reportedly only weakly inhibits addition of downstream dNTPs (5). We note that the mechanism by which any of these dG analogs inhibit priming remains to be elucidated, and mechanisms other than incorporation have not been ruled out.
Of considerable mechanistic interest were the quite different effects on the HBV Pol reaction of the two very closely related compounds SQ-32829–TP and lobucavir-TP (Fig. 1). Lobucavir-TP was a much stronger overall inhibitor of hepadnaviral Pols than SQ-32829–TP, emphasizing the exquisite selectivity with which these enzymes can distinguish even the most closely related structures. More remarkably, however, SQ-32829 ultimately suppressed all HBV Pol elongation at the highest concentrations, but without showing any inhibition of HBV priming. To our knowledge, SQ-32829 is the only guanosine analog to show a sharp discrimination between the two HBV Pol replication steps (Fig. 2). The remaining G analog-TPs generally inhibited both reactions, although the data in Table 1 suggest that some were better at inhibiting elongation versus priming. It is believed that Pol adopts different conformations for the priming and elongation reactions (40), as evidenced by the drug phosphonoformic acid, which blocks elongation but not priming (21, 24, 35, 46). The fact that a dGTP analog can discriminate between the priming and elongation activities of Pol further reinforces the idea that a restructuring of the architecture of the dNTP binding domain of HBV Pol must accompany the transition between these two states.
Our kinetic analyses of the effect of BMS-200475–TP and lobucavir-TP on the WHV and HBV Pol reactions confirm that these carbocyclic guanine compounds are very potent inhibitors of hepadnaviral Pols. Indeed, the low Ki/Km ratios (0.27 to 0.37) seen in our studies imply that Pol has a higher affinity for the two TP analogs than for the natural substrate. An earlier report by Price et al. (29) on 2′CDG-TP determined a Ki/Km for this compound to be ∼0.18 in 2.2.15-derived HBV cores, and carbovir-TP also appears to be highly active against hepadnaviral Pols (6). Carbocyclic guanine nucleoside analogs thus appear to provide a particularly potent class of HBV Pol inhibitors.
Finally, although BMS-200475 and lobucavir, like 2′CDG, are not obligate chain terminators of DNA synthesis by virtue of the OH group content of their sugar moieties, our endogenous sequencing data strongly suggest that template-dependent incorporation of either analog in place of a normal dG residue can lead to chain termination 2 or 3 nucleotides later. This finding argues that both nucleoside analogs may act as structural terminators, perhaps by introducing enough structural distortion to preclude the enzyme from optimal interaction with the 3′ end of the growing DNA chain. Surprisingly, we found different termination patterns for lobucavir-TP and BMS-200475–TP. While the former terminated chain elongation following single dGs, BMS-200475–TP caused preferential termination after dG doublets or dG-rich stretches.
In summary, this study has confirmed that the TP forms of BMS-200475 and lobucavir are potent inhibitors of the HBV, WHV, and DHBV Pols, and that they effectively suppress the priming and elongation steps of HBV replication. Both are competitive inhibitors of Pol with respect to dGTP and are apparently preferred by hepadnaviral Pols over the natural substrate. They act, at least in part, via structural, nonobligate chain termination of HBV Pol, although other modes of inhibition are not excluded. These findings further validate efforts to assess the potential of these two drugs as therapeutic agents against chronic HBV infection in humans.
ACKNOWLEDGMENTS
Our colleagues Brian Terry, Chris Cianci, Greg Bisacchi, and Bob Zahler provided TPs. Bill Mason (Fox Chase Cancer Center, Philadelphia, Pa.) shared his expertise concerning endogenous sequencing. Finally, we thank Steve Innaimo as well as Junius Clark and members of the in vivo group for encouragement and valuable discussion.
REFERENCES
- 1.Beasley R P, Hwang L Y. Overview on the epidemiology of hepatocellular carcinoma. In: Hollinger F B, Lemon S M, Margolis M, editors. Viral hepatitis and liver disease. Baltimore, Md: The Williams & Willkins Co.; 1991. pp. 532–535. [Google Scholar]
- 2.Bisacchi G S, Chao S T, Bachard C, Daris J P, Innaimo S F, Jacobs J A, Kocy O, Lapointe P, Martel A, Merchant Z, Slusarchyk W A, Sundeen J E, Young M G, Colonno R J, Zahler R. BMS-200475, a novel carbocyclic 2′-deoxyguanosine analog with potent and selective anti-hepatitis B virus activity in vitro. Bioorg Med Chem Lett. 1997;7:127–132. [Google Scholar]
- 3.Civitico G, Shaw T, Locarnini S. Interaction between ganciclovir and foscarnet as inhibitors of duck hepatitis B virus replication in vitro. Antimicrob Agents Chemother. 1996;40:1180–1185. doi: 10.1128/aac.40.5.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, J. M. 1997. Unpublished data.
- 5.Dannaoui E, Trépo C, Zoulim F. Inhibitory effect of penciclovir-triphosphate on duck hepatitis B virus reverse transcription. Antivir Chem Chemother. 1997;8:38–46. [Google Scholar]
- 6.Davis M G, Wilson J E, VanDraanen N A, Miller W H, Freeman G A, Daluge S M, Boyd F L, Aulabaugh A E, Painter G R, Boone L R. DNA polymerase activity of hepatitis B virus particles: differential inhibition by l-enantiomers of nucleotide analogs. Antivir Res. 1996;30:133–145. doi: 10.1016/0166-3542(96)00938-2. [DOI] [PubMed] [Google Scholar]
- 7.Dean J, Bowden S, Locarnini S. Reversion of duck hepatitis B virus DNA replication in vivo following cessation of treatment with the nucleoside analogue ganciclovir. Antivir Res. 1995;27:171–178. doi: 10.1016/0166-3542(94)00081-i. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag J L, Perillo R P, Schiff E R, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 9.Field A K, Tuomari A V, McGeever-Rubin B, Terry B J, Mazina K E, Haffey M L, Hagen M E, Clark J M, Braitman A, Slusarchyk W A, Young M G, Zahler R. (±)-(1 alpha,2 beta,3 alpha)-9-[2,3-Bis(hydroxymethyl)-cyclobutyl] guanine [(±)-BHCG or SQ 33,054]: a potent and selective inhibitor of herpesviruses. Antivir Res. 1990;13:41–52. doi: 10.1016/0166-3542(90)90043-7. [DOI] [PubMed] [Google Scholar]
- 10.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 11.Ganem D, Pollack J R, Tavis J. Hepatitis B virus reverse transcriptase and its many roles in hepadnavirus genomic replication. Infect Agents Dis. 1994;3:85–93. [PubMed] [Google Scholar]
- 12.Garnier J-L, Chossegros P, Daoud S, Chevallier P, Dubernard J-M, Trepo C, Touraine J-L. Treatment of hepatitis B virus replication by ganciclovir in kidney transplant patients. Transplant Proc. 1997;29:817. doi: 10.1016/s0041-1345(96)00146-7. [DOI] [PubMed] [Google Scholar]
- 13.Genovesi E V, Lamb L, Medina I, Taylor D, Seifer M, Innaimo S, Colonno R J, Standring D N, Clark J M. Efficacy of the carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob Agents Chemother. 1998;42:3209–3217. doi: 10.1128/aac.42.12.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honkoop P, deMan R A. Clinical aspects of nucleoside analogues for chronic hepatitis B. Int Antivir News. 1995;3:78–80. [Google Scholar]
- 15.Horwich A L, Furtak K, Pugh J, Summers J. Synthesis of hepadnavirus particles containing replication-defective duck hepatitis B virus genomes in cultured Huh-7 cells. J Virol. 1990;64:642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe A Y, Robins M J, Wilson J S, Tyrrell D L. Selective inhibition of the reverse transcriptase of duck hepatitis B virus by binding of 2′,3′-dideoxyguanosine 5′-triphosphate to the viral polymerase. Hepatology. 1996;23:87–96. doi: 10.1002/hep.510230113. [DOI] [PubMed] [Google Scholar]
- 17.Innaimo S F, Seifer M, Bisacchi G S, Standring D N, Zahler R, Colonno R J. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob Agents Chemother. 1997;41:1444–1448. doi: 10.1128/aac.41.7.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs G A, Tino J A, Zahler R. Synthesis of SQ-32,829, a new nucleoside antiviral agent. Tetrahedron Lett. 1989;30:6955–6958. [Google Scholar]
- 19.Korba B E, Boyd M R. Penciclovir is a selective inhibitor of hepatitis B virus replication in cultured human hepatoblastoma cells. Antimicrob Agents Chemother. 1996;40:1282–1284. doi: 10.1128/aac.40.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford R E, Notvall L, Lee H, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanford R E, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lien J M, Aldrich C E, Mason W S. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand synthesis. J Virol. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locarnini S A, Civitico G M, Newbold J E. Hepatitis B: new approaches for antiviral chemotherapy. Antivir Chem Chemother. 1996;7:53–64. [Google Scholar]
- 24.Löfgren B, Nordenfelt E, Oeberg B. Inhibition of RNA- and DNA-dependent duck hepatitis B virus DNA polymerase activity by nucleoside and pyrophosphate analogs. Antivir Res. 1989;12:301–310. doi: 10.1016/0166-3542(89)90057-0. [DOI] [PubMed] [Google Scholar]
- 25.Miller R, Marion P, Robinson W. Hepatitis B viral DNA-RNA hybrid molecules in particles from infected liver are converted to viral DNA molecules during an endogenous DNA polymerase reaction. Virology. 1984;139:64–72. doi: 10.1016/0042-6822(84)90330-1. [DOI] [PubMed] [Google Scholar]
- 26.Molnar-Kimber K L, Summers J, Taylor J M, Mason W S. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J Virol. 1983;45:165–172. doi: 10.1128/jvi.45.1.165-172.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevens F, Main J, Honkoop P, Tyrrell D L, Barber J, Sullivan M T, Fevery J, De Man R A, Thomas H C. Lamivudine therapy for chronic hepatitis B: a six-month randomized dose-ranging study. Gastroenterology. 1997;113:1258–1263. doi: 10.1053/gast.1997.v113.pm9322520. [DOI] [PubMed] [Google Scholar]
- 29.Price P M, Banerjee R, Jeffrey A M, Acs G. The mechanism of inhibition of hepatitis B virus replication by the carbocyclic analog of 2′-deoxyguanosine. Hepatology. 1992;16:8–12. doi: 10.1002/hep.1840160103. [DOI] [PubMed] [Google Scholar]
- 30.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Bio/Technology. 1992;24:104–108. [PubMed] [Google Scholar]
- 32.Schlam, S. W., R. A. Heytink, H. R. Van Buuren, and R. A. De Man. 1986. Acylovir, oral, intravenous and combined with interferon for chronic HBeAg-positive hepatitis. J. Hepatol. 3(Suppl. 2):S137–S141. [DOI] [PubMed]
- 33.Seeger C, Summers J, Mason W S. Viral DNA synthesis. Curr Top Microbiol Immunol. 1991;168:41–59. doi: 10.1007/978-3-642-76015-0_3. [DOI] [PubMed] [Google Scholar]
- 34.Segel I H. Biochemical calculations. New York, N.Y: John Wiley & Sons, Inc.; 1975. Enzymes; pp. 208–323. [Google Scholar]
- 35.Seifer M, Hamatake R K, Bifano M, Standring D N. Generation of replication-competent hepatitis B virus nucleocapsids in insect cells. J Virol. 1998;72:2765–2776. doi: 10.1128/jvi.72.4.2765-2776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severini A, Liu X Y, Wilson J S, Tyrrell D L. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-beta-L-2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1995;39:1430–1435. doi: 10.1128/aac.39.7.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slusarchyk, W. A., A. K. Field, J. A. Greytok, P. Taunk, A. V. Tooumari, M. G. Young, and R. Zahler. 1992. 4-Hydroxy-3-(hydroxymethyl)-2-methylcyclopentyl purines and pyrimidines, a novel class of anti-herpesvirus agents. Antivir. Res. 17(Suppl. 1):98. (Abstract.)
- 38.Staschke K A, Colacino J M. Priming of duck hepatitis B virus reverse transcription in vitro: premature termination of primer DNA induced by the 5′-triphosphate of fialuridine. J Virol. 1994;68:8265–8269. doi: 10.1128/jvi.68.12.8265-8269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 40.Tavis J E, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavis J E, Perri S, Ganem D E. Hepadnaviral reverse transcription initiates within a stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tencza M G, Newbold J E. Heterogeneous response for mammalian hepadnavirus infection to acyclovir: drug arrested intermediates of minus strand viral DNA are enveloped and secreted from infected cells as virion-like particles. J Med Virol. 1997;51:6–16. [PubMed] [Google Scholar]
- 43.Terry B J, Mazina K E, Tuomari A V, Haffey M L, Hagen M, Feldman A, Slusarchyk W A, Young M G, Zahler R, Field A K. Broad-spectrum antiviral activity of the acyclic guanosine phosphonate (R,S)-HPMPG. Antivir Res. 1988;10:235–252. doi: 10.1016/0166-3542(88)90034-4. [DOI] [PubMed] [Google Scholar]
- 44.Terry B J, Mazina K E, Tuomari A V, Hagen M E, Haffey M L, Jacobs G A, Zahler R, Field A K. Anti-herpetic activity of (±)-(1a,2b,3a)-9-[2-hydroxy-3-(hydroxymethyl)cyclobutyl]guanine and inhibition of HSV-1 DNA polymerase. Antivir Chem Chemother. 1990;1:263–268. [Google Scholar]
- 45.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 47.Yamada M, Ishii M, Miura M, Sato S, Kanno A, Ohori H, Toyota T. Three distinct Southern blot hybridization patterns of HBV-DNA in the sera of HBV carriers. Tohoku J Exp Med. 1993;170:219–228. doi: 10.1620/tjem.170.219. [DOI] [PubMed] [Google Scholar]
- 48.Yang H Y, Drain R L, Franco C A, Clark J M. Efficacy of BMS-180194 against experimental cytomegalovirus infections in immunocompromised mice. Antivir Res. 1996;29:233–241. doi: 10.1016/0166-3542(95)00901-9. [DOI] [PubMed] [Google Scholar]
- 49.Zoulim F. Improving hepatitis B virus therapy—new inhibitors of reverse transcriptase. Int Antivir News. 1997;5:110–112. [Google Scholar]
- 50.Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]