Abstract
Two highly potent dihydroalkoxybenzyloxopyrimidine (DABO) derivatives targeting the nonnucleoside inhibitor (NNI) binding site of human immunodeficiency virus (HIV) reverse transcriptase (RT) have been designed based on the structure of the NNI binding pocket and tested for anti-HIV activity. Our lead DABO derivative, 5-isopropyl-2-[(methylthiomethyl)thio]-6-(benzyl)-pyrimidin-4-(1H)-one, elicited potent inhibitory activity against purified recombinant HIV RT and abrogated HIV replication in peripheral blood mononuclear cells at nanomolar concentrations (50% inhibitory concentration, <1 nM) but showed no detectable cytotoxicity at concentrations as high as 100 μM.
The nonnucleoside inhibitor (NNI) binding site of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) has been studied extensively (20, 21, 29, 32, 40, 41, 44, 46). Several crystal structures of HIV-1 RT complexed with NNIs have been determined and have provided important structural information about the NNI binding site. These RT-NNI crystal structures have shown distinct properties of the NNI binding pocket which can be utilized for structure-based rational drug design (20, 21, 29, 32, 40, 41, 44, 46). However, each reported structure revealed a unique binding pattern, indicating that rational drug design efforts should not rely on one particular crystal structure. Therefore, we have used the NNI binding site coordinates of nine RT-NNI structures to generate a composite binding pocket revealing a molecular surface which defines a larger-than-presumed NNI binding pocket and serves to summarize critical features unique to the NNI binding site (35, 48). The composite binding pocket allowed a clear identification of features which could be successfully exploited in new inhibitor designs. In the present study, we have utilized this composite pocket, together with a computer docking procedure and a structure-based semiempirical score function, for the design of novel derivatives of dihydroalkoxybenzyloxopyrimidine (DABO) as potent NNIs of HIV-1 RT. Here, we report two highly potent DABO derivatives which were found to inhibit HIV RT. Our most potent DABO derivative, 5-isopropyl-2-[(methylthiomethyl)thio]-6-benzyl)pyrimidin-4-(1H)-one (3c), showed potent inhibitory activity against purified recombinant HIV RT and abrogated HIV replication in peripheral blood mononuclear cells (PBMNC) (50% inhibitory concentration at nanomolar concentrations [IC50], <1 nM) but showed no detectable cytotoxicity at concentrations as high as 100 μM.
MATERIALS AND METHODS
Construction of the NNI composite binding pocket.
Modeling studies required the construction of a binding pocket which encompassed the superimposed crystal structure coordinates of all known RT-NNI complexes, including nine different structures of RT complexed with 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT), 6-benzyl-2-(ethoxymethyl)-5-isopropyluracil (MKC), 6-benzyl-1-[(benzyloxy)methyl]-5-isopropyluracil (TNK), α-anilinophenylacetamide (APA), nevirapine (dipyridodiazepinone derivative), N-ethyl nevirapine derivative, 9-Cl-tetrahydrobenzodiazepine derivative (9-Cl TIBO) (41), 9-Cl TIBO (17), and 8-Cl-TIBO (protein data bank [PDB] access codes, 1rti, 1rt1, 1rt2, 1hni, 1vrt, 1rth, 1rev, 1tvr, and 1hnv, respectively). The “thumb” region of RT complexes is relatively variable compared with the “palm” region. Therefore, a total of 117 Cα atoms of residues 97 to 213, which cover part of the NNI binding site and the palm region, were used for a least-squares superimposing procedure within program O (30). The RMS values for the superimposed coordinates were 1.00, 0.98, 0.99, 0.62, 0.80, 0.87, 0.94, and 0.65 Å for HEPT, MKC, TNK, APA, cyclopropanyl nevirapine, N-ethyl nevirapine derivative, and the two 9-Cl TIBO complexes, respectively. The coordinates of the corresponding inhibitor molecules were then transformed according to the same matrices derived from the superimposition. Lastly, the overlaid coordinates of all inhibitors were read into the program GRASP (38), from which an overall molecular surface was generated and provided a binding pocket encompassing all inhibitors. The molecular surface was used to better visualize the available space within the NNI binding pocket of RT and to more easily identify potentially usable space in a qualitative way. The generated binding pocket, referred to as the composite binding pocket, was used as a basis for the analysis of inhibitor binding.
Molecular docking and Ki prediction.
Fixed docking in the Affinity program within InsightII (37) was used for docking of small molecules into the NNI binding site, which was taken from a crystal structure (PDB code, 2rt1; RT-MKC complex) (17, 29). As the modeling calculations progressed, the residues within a defined radius (5 Å) of the NNI molecule were allowed to move in accordance with energy minimization. Ten final docking positions were initially chosen for each inhibitor modeling calculation but failed to reveal more than two promising positions. Later, only two calculated positions were set for the search target. The limited number of suitable docked positions is consistent with the observation that the NNI binding site of RT is sufficiently enclosed so that inhibitors can only bind in a small number of specific orientations, unlike other proteins, where the binding site is relatively open and molecules can bind in a number of orientations. Binding calculations were carried out on an SGI INDIGO2 by using the CVFF force field in the Discover Program and a Monte Carlo Search Strategy in Affinity (33). No solvation procedures were used. Since the total number of movable atoms exceeded 200, conjugated gradient minimization was used instead of the Newton minimization method to conserve computation time. The initial coordinates of the compounds were generated by using the Sketcher module within InsightII, which calculates an energy-minimized conformation for the molecule. InsightII was also used to calculate intermolecular energy (van der Waals and Coulombic interaction energy) for some compounds to compare energy differences between different conformers. The largest conformational differences can result from a rotation around the bond joining the two rings in DABO compounds. For our study, several conformers were generated which differed in the orientation of the phenyl ring around this bond and were used to calculate an energy score for each conformer. Calculated interaction scores for inhibitor conformations were considered which differed from the conformation we eventually used for docking. Our chosen conformation was based on the RT-HEPT and RT-MCK442 crystal structures and was found to be one of the lowest-energy conformers, which probably represents the most reasonable conformation for binding. This conformation was then compared with our small-molecule crystal structure of the compound, which was found to be conformationally similar. The CVFF force field was used to obtain a low-energy conformation of a DABO compound representing a local minimum. After the docking run was completed, by using the chosen conformer, each final position was then evaluated by an interaction score function using the Ludi module. Therefore, a final docked position was chosen for each DABO compound which was based on a combination of both the CVFF energy minimum and the best Ludi score (the final position of the DABO compound in the RT active site was first minimized and evaluated by using the CVFF force field and then scored based on the Ludi scoring function). The top-scoring model was then compared with the constructed binding pocket and the known crystal structures of similar compounds and used for further analyses, which involved an evaluation of the compound in the composite binding pocket. A qualitative assessment was made of how well the compound fit the binding site relative to other DABO compounds or other NNIs. This was accomplished by superimposing the compounds into the binding site and visually inspecting unoccupied regions, favorable interactions with RT, and unfavorable interactions. This analysis was done to see if the final Ludi scores were reasonable in light of biological data or known binding modes of NNIs based on crystal structures.
The calculated inhibitory constants (Ki values) of the positioned DABO compounds were evaluated by using the Ludi score function (6, 7). (The InsightII program used for docking and Ludi scoring employed a calibration procedure during its establishment which involved calculation of the Ki values of 45 protein-ligand complexes having known Ki values and known crystal structures and comparing the calculated Ki values to the experimentally determined Ki values. The Ki value has a correlation with the free energy of binding, ΔG, where ΔG = −RTlnKi.) We imposed several modifications during the calculation. First, the molecular surface areas were directly calculated from the coordinates of the compounds in docked conformations by using the MS program (13). (The molecular surface area is used to calculate a Lipo score, which is an important component of binding affinity in the Ludi score function.) Second, we reevaluated the number of rotatable bonds, which was assessed inaccurately by InsightII. Third, we assumed that the conserved hydrogen bond with RT did not deviate significantly from the ideal geometry (the backbone carbonyl of RT residue 101 interacts with a hydrogen bond acceptor on the inhibitor, as observed in the RT-HEPT, RT-MKC442, RT-TIBO, and RT-APA crystal structures). This assumption was supported by the fact that in the known crystal structures of RT complexes, all hydrogen bonds between NNIs and RT are near ideal in geometry. Consequently, the Ki values for our modeled compounds were more predictable than they would be otherwise without such constraints (6, 7).
In the Ludi scoring function used to evaluate the binding of NNIs to RT, an ideal hydrogen bond corresponds to a maximum hydrogen bond score of 85. Our analysis showed that all of the RT-NNI crystal structures studied can be assigned a hydrogen bond score of 50 or greater. Six of nine complexes showed a hydrogen bond score of 80 or greater, indicating nearly ideal hydrogen bond geometry for these complexes, except in the case of the RT-APA structure. For modeling studies, we used the same definitions for ideal hydrogen bonds that are used in the Ludi program, which are briefly summarized in Table 1 (6, 7).
TABLE 1.
Hydrogen bond geometry definitions used in Ludi program
| Hydrogen bonding group | Distance from H to acceptor (Å) | Hydrogen bond angle between H and A–X (°) |
|---|---|---|
| C⩵O | 1.9 | 110–180 |
| NH or OH | 1.9 | 150–180 |
| NH (charged) | 1.8 | 150–180 |
| COO− | 1.8 | 100–140 |
| ⩵N— | 1.9 | 150–180 |
| R-O-R (sp2) | 1.9 | 100–140 |
| R-O-R (sp3) | 1.9 | 90–130 |
Biological assays. (i) Purified RT assays for anti-HIV activity.
5-Methyl-2-[(methylthiomethyl)thio]-6-benzylpyrimidin-4-1H-one (compound 3a), 5-ethyl-2-[methylthiomethyl)thio]-6-benzylpyrimidin-4-1H-one (compound 3b), 5-isopropyl-2-[(methylthiomethyl)thio]-6-benzyl-pyrimidin-4-1H-one (compound 3c), and 5-isopropyl-2-[(methylthiomethyl)thio]-6-(3,5-dimethylbenzyl)-pyrimidin-4-1H-one (compound 3d) were tested for RT inhibitory activity (IC50[rRT]) against purified HIV recombinant RT (rRT) by using the cell-free Quan-T-RT system (Amersham, Arlington Heights, Ill.), which utilizes the scintillation proximity assay (SPA) principle (8). In the assay, a DNA-RNA template is bound to SPA beads via a biotin-strepavidin linkage. The primer DNA is a 16-mer oligod(T) which has been annealed to a poly(rA) template. The primer-template is bound to a strepavidin-coated SPA bead. [3H]TTP is incorporated into the primer by reverse transcription. In brief, [3H]TTP, at a final concentration of 0.5 μCi/sample, was diluted in RT assay buffer (49.5 mM Tris-Cl [pH 8.0], 80 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 2.5 mM EGTA, 0.05% Nonidet P-40) and added to annealed DNA-RNA bound to SPA beads. The compound being tested was added to the reaction mixture at 0.001 to 100 μM concentrations. Addition of 10 mU of HIV rRT and incubation at 37°C for 1 h resulted in extension of the primer by incorporation of [3H]TTP. The reaction was stopped by adding of 0.2 ml of 120 mM EDTA. The samples were counted in an open window by using a Beckman LS 7600 instrument, and IC50s were calculated by comparing the measurements to untreated samples.
(ii) p24 assays for anti-HIV activity.
Normal human PBMNC from HIV-negative donors were cultured for 72 h in RPMI 1640 medium supplemented with 20% (vol/vol) heat-inactivated fetal bovine serum, 3% interleukin-2, 2 mM l-glutamine, 25 mM HEPES, 2-g/liter NaHCO3, 50-μg/ml gentamicin, and 4-μg/ml phytohemagglutinin prior to exposure to HIV-1 at a multiplicity of infection of 0.1 during a 1-h adsorption period at 37°C in a humidified 5% CO2 atmosphere. Subsequently, cells were cultured in 96-well microtiter plates (100 μl/well; 2 × 106 cells/ml, triplicate wells) in the presence of various inhibitor concentrations, and aliquots of culture supernatants were removed from the wells on day 7 after infection for antigen p24 enzyme immunoassays (EIA), as previously described (22, 47, 49). The applied p24 EIA was the unmodified kinetic assay commercially available from Coulter Corporation/Immunotech, Inc. (Westbrooke, Maine), which utilizes a murine monoclonal antibody to HIV core protein used to coat microwell strips to which the antigen present in the test culture supernatant samples binds. Percent inhibition of viral replication was calculated by comparing the p24 values from the test substance-treated infected cells with p24 values from untreated infected cells (i.e., virus controls). In parallel, the effects of various treatments on cell viability were also examined as previously described (22, 49). In brief, noninfected PBMNC were treated with each compound for 7 days under identical experimental conditions. A microculture tetrazolium assay (MTA), using 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)-carbonyl]-2H-tetrazolium hydroxide, was performed to quantitate cellular proliferation.
X-ray crystallography.
Yellow rectangular plates of 3b and colorless plates of 3c were grown from tetrahydrofuran in separate experiments by slow evaporation at room temperature. The crystals were mounted on glass fibers by using epoxy, and X-ray diffraction data for a crystal (0.5 by 0.2 by 0.08 mm) of 3b and a crystal (0.3 by 0.2 by 0.1 mm) of 3c were collected at 22°C by using a SMART charge-coupled device X-ray detector (Bruker Analytical X-Ray Systems, Madison, Wis.). Structure solution and refinement were performed by using the SHELXTL suite of programs (Bruker Analytical X-Ray Systems). All nonhydrogen atoms were refined by using anisotropic displacement parameters. Hydrogen atoms were placed at ideal positions and refined as riding atoms with relative isotropic displacement parameters.
Chemical synthesis.
All chemicals were purchased from Aldrich Chemical Company (Milwaukee, Wis.) and used as received. Anhydrous solvents were obtained from Aldrich in Sure Seal bottles and transferred to reaction vessels via cannula under nitrogen. All reactions were carried out under nitrogen. Nuclear magnetic resonance (NMR) spectra were recorded on a Varian (Palo Alto, Calif.) 300-MHz instrument, and chemical shifts (δ) are reported in parts per million (ppm) relative to tetramethylsilane, which was the internal standard. 13C NMR spectra were recorded in CDCl3 on the same instrument by using the proton decoupling technique. The chemical shifts reported for 13C NMR spectra are referenced to chloroform at 77.0 ppm. Melting points were obtained by using a Fisher-Johns melting apparatus and left uncorrected. Mass spectral analyses were conducted by using a Finnigan (Madison, Wis.) MAT 95 mass spectrometer. Column chromatography was performed by using EM Science silica gel 60.
The 5-alkyl-2-[(methylthiomethyl)thio]-6-(benzyl)-pyrimidin-4-(1H)-one derivatives 3a to 3d were prepared as shown in Fig. 4. Ethyl-2-alkyl-4-(phenyl)-3-oxobutyrates 1a to 1d were obtained from commercially available phenylacetonitrile by a method previously described (15, 16, 34). The β-ketoesters were condensed with thiourea in the presence of sodium ethoxide to furnish the corresponding thiouracils 2a to 2d. Subsequent reaction of thiouracil with methylthiomethyl chloride in N,N-dimethylformamide (DMF) in the presence of potassium carbonate afforded compounds 3a to 3d in moderate yields.
FIG. 4.
Preparation of 5-alkyl-2-[(methylthiomethyl)thio]-6-(benzyl)-pyrimidin-4-(1H)-one derivatives 3a to 3d. Reagents and conditions: a, R2CHBrCOOEt/Zn/tetrahydrofuran; b, HCl (aqueous); c, (H2N)2CS-Na-ethanol; d, DMF, K2CO3, and chloromethyl methyl sulfide; 15 h. Me, methyl; Et, ethyl; i-Pr, isopropyl.
General procedure for synthesis of compounds 3a to 3d.
A mixture of thiouracil 2 (1 mmol), methylchloromethylsulfide (1 mmol), and potassium carbonate (1 mmol) in anhydrous DMF (5 ml) was stirred overnight at room temperature. After treatment with water (50 ml), the solution was extracted with ethyl acetate (3 × 50 ml). The combined extracts were washed with saturated NaCl (2 × 50 ml), dried (MgSO4), filtered, and concentrated in vacuo to give the crude products 3a to 3d, which were purified by column chromatography (hexane-ethyl acetate eluent). Selected analytical data for compounds 3a to 3d are shown as follows. Compound 3a yield, 62%; mp, 148 to 149°C; 1H NMR (CDCl3) δ 2.10 (s, 3H), 2.14 (s, 3H), 3.91 (s, 2H), 4.29 (s, 2H), 7.29–7.26 (m, 5H), 12.20 (s, 1H); 13C NMR (CDCl3) δ 10.7 (CH3), 15.5 (SCH3), 36.6 (CH2Ph), 41.0 (SCH2), 116.7 (C-5), 137.6 to 126.4 (Ph), 155.2 (C-6), 162.0 (C-4), 165.1 (C-2); CI-MS (chemical ionization mass spectrometry), 293.1 (M+1). Compound 3b yield, 65%; mp, 124 to 126°C; 1H NMR (CDCl3) δ 1.08 (t, 3H), 2.12 (s, 3H), 2.58 (q, 2H), 3.91 (s, 2H), 4.26 (s, 2H), 7.28-7.26 (m, 5H), 12.30 (s, 1H); 13C NMR (CDCl3) δ 13.1 (CH3), 15.4 (SCH3), 18.7 (CH2), 36.4 (CH2Ph), 40.3 (SCH2), 122.4 (C-5), 138.0 to 126.3 (Ph), 155.4 (C-6), 161.5 (C-4), 165.2 (C-2); CI-MS, 307.1 (M+1). Compound 3c yield, 57%; mp, 116 to 117°C; 1H NMR (CDCl3) δ 1.22 (d, 6H), 2.07 (s, 3H), 3.03 (q, 1H), 3.88 (s, 2H), 4.21 (s, 2H), 7.24-7.13 (m, 5H), 12.43 (s, 1H); 13C NMR (CDCl3) δ 15.4 (SCH3), 19.6 (CH3), 28.0 (CH), 36.3 (CH2Ph), 40.9 (SCH2), 125.3 (C-5), 138.3 to 126.3 (Ph), 155.5 (C-6), 161.1 (C-4), 164.5 (C-2); CI-MS, 321.1 (M+1). Compound 3d yield, 67%; mp, 116 to 120°C; 1H NMR (CDCl3) δ 1.28 (d, 6H), 2.15 (s, 3H), 2.27 (s, 6H), 3.10 (q, 1H), 3.88 (s, 2H), 4.31 (s, 2H), 6.84 (s, 3H), 12.42 (s, 1H); 13C NMR (CDCl3) δ 15.3 (SCH3), 19.6 (CH3), 21.2 (CH3), 28.0 (CH), 36.3 (CH2Ph), 40.8 (SCH2), 125.2 (C-5), 138.0 to 126.5 (Ph), 155.4 (C-6), 161.3 (C-4), 164.7 (C-2); CI-MS; 349.2 (M+1).
RESULTS
Modeling and design.
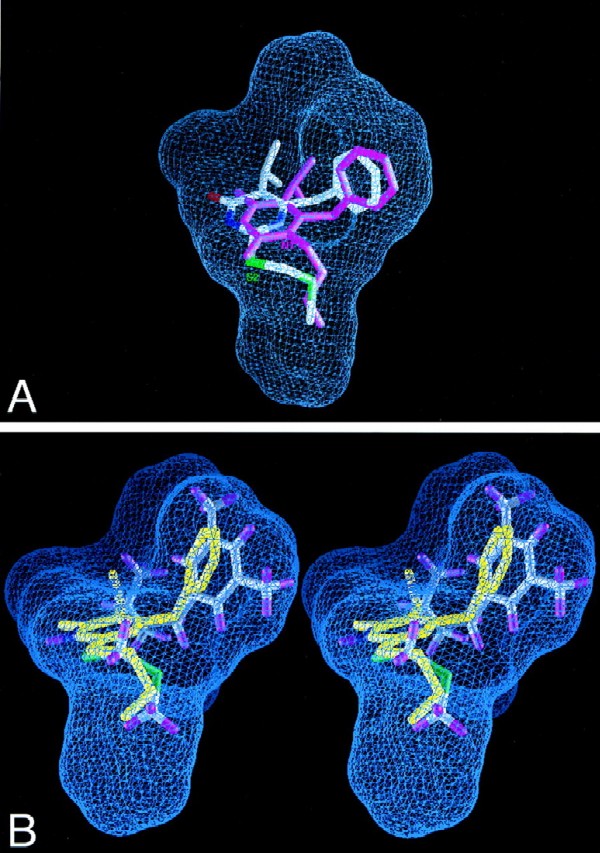
Our detailed analysis of HEPT and the HEPT derivative MKC442, both of which are active NNIs of HIV RT, revealed that the N1 substituents of HEPT derivatives occupy the same region of the binding site as the thio (S2) substituents of DABO compounds (Fig. 2A). We therefore designed new DABO derivatives and modeled their binding into the NNI site of RT by using the crystal structure coordinates of the RT-MKC complex (PDB access code, 1rt1) and a molecular docking procedure in the Affinity module of the InsightII program (37) (Table 2). We then superimposed the final coordinates of the docked molecules on the composite binding pocket to evaluate the fit within the RT NNI pocket. Notably, multiple sterically allowed unoccupied spatial gaps in the binding site were identified from the docking studies which could be filled by strategically designed functional groups (Fig. 2B). The docked DABO molecule (compound 3a) showed significant space surrounding the 6-benzyl ring and the fifth position of the thymine ring, which led to our design and synthesis of compounds 3b, 3c, and 3d. The inhibition constants of the docked molecules were calculated based on an interaction score function and are listed in Table 2. The trend of the calculated Ki values predicted that compounds having a slightly larger R2 group would show stronger inhibition of RT; this general trend was also observed for the measured IC50s (Table 2).
FIG. 2.
View of composite NNI binding pocket of HIV-1 RT. Blue grid lines represent the collective van der Waals surface of nine different inhibitor crystal structures superimposed on the active site and highlight the space available for binding (inhibitor structures included HEPT, MKC, TNK, APA, nevirapine, N-ethyl nevirapine derivative, 8-Cl TIBO, and two 9-Cl TIBO compounds with PDB access codes 1rti, 1rt1, 1rt2, 1hni, 1vrt, 1rth, 1hnv, 1rev, and 1tvr, respectively). (A) Compound 3c superimposed on the composite NNI binding site of the crystal structure of the RT-MKC442 complex (hydrogen atoms are not shown; PDB access code, 1rt1). MKC442 (from crystal structure) is shown in pink, and compound 3c (from docking calculations) is multicolored. Compound 3c was docked into the active site of the RT-MKC442 complex and then superimposed onto the composite NNI binding pocket based on the matrix used in pocket construction. The S2 substituent of DABO analog 3c occupies the same region of the binding pocket as the N1 substituent of HEPT analog MKC442. (B) X-ray crystal structure of compound 3b (yellow) superimposed on the docked model of compound 3d in the composite NNI binding pocket of RT, demonstrating their similar conformations.
TABLE 2.
Interaction scores, calculated Ki values, and biological assay data for inhibitors of HIV RT
| Compound | R1 | R2 | MSa (Å2) | BSb (%) | Lipo score | LudicKi (μM) | IC50[rRT]d (μM) | IC50[p24]d (μM) | CC50[MTA]e (μM) | Minimum SIf |
|---|---|---|---|---|---|---|---|---|---|---|
| 3a | H | Meg | 275 | 88 | 709 | 3.3 | 28.8 ± 10.0 | 3.4 ± 1.1 | >100 | 32 ± 10 |
| 3b | H | Eth | 283 | 88 | 730 | 2.0 | 12.0 ± 2.3 | 0.8 ± 0.2 | >100 | 146 ± 38 |
| 3c | H | i-Pri | 301 | 89 | 785 | 0.56 | 5.6 ± 2.4 | <0.001 ± 0.000j | >100 | 100,000 ± 0 |
| 3d | Me | i-Pr | 329 | 89 | 875 | 0.05 | 7.0 ± 2.2 | 0.016 ± 0.011 | >100 | 31,520 ± 31,310 |
| MKC442 | NAk | NA | NDl | ND | ND | ND | 0.8 ± 0.1 | 0.004 ± 0.001 | >100 | 25,830 ± 4,383 |
| Delavirdine | NA | NA | ND | ND | ND | ND | 2.3 ± 1.0 | ND | ND | ND |
| Nevirapine | NA | NA | ND | ND | ND | ND | 19.8 ± 5.5 | ND | ND | ND |
MS, molecular surface area calculated by using Connolly’s MS program (13), is defined as the boundary of the volume within any probe sphere (meant to represent a water molecule) with a given radius sharing no volume with the hard sphere atoms which make up the molecule. The values are slightly lower than those approximated by the Ludi program.
BS, the buried surface, is the percentage of the molecular surface in contact with protein calculated by Ludi relative to docked positions. Based on published crystal structures of RT complexes, our calculation shows that these values can be as low as 77% (in an RT-HEPT complex) and as high as 90% (in an RT-APA complex), but most of them, including that for RT-MKC, average around 84%.
Ludi Ki values were calculated based on the empirical score function in the Ludi program (6, 37). Ideal hydrogen bond distances and angles between compounds and protein are assumed in all cases for Ludi Ki and Ludi score calculations. In published crystal structures of RT complexes, hydrogen bond geometries are indeed close to ideal; the amide carbonyl of residue A101 on a loop demonstrates substantial flexibility, which can accommodate the best geometry for hydrogen bonding. The number of rotatable bonds (two) is used in the Ludi calculation to reflect the loss of binding energy due to freezing of internal degrees of freedom.
Mean ± standard error of three or four independent experiments (two experiments for 3a), each performed in triplicate.
Even at an inhibitor concentration of 100 μM, the viability of cells was minimally affected, with 50% cytotoxic concentration (CC50[MTA]) values of >100 μM in each of three or four independent experiments.
SI, the selectivity index, is equal to the ratio of CC50[MTA] to IC50[p24]. Since the CC50 values for the inhibitors tested were >100 μM, only the minimum selectivity index estimates could be determined for these noncytotoxic agents.
Me, methyl.
Et, ethyl.
i-Pr, isopropyl.
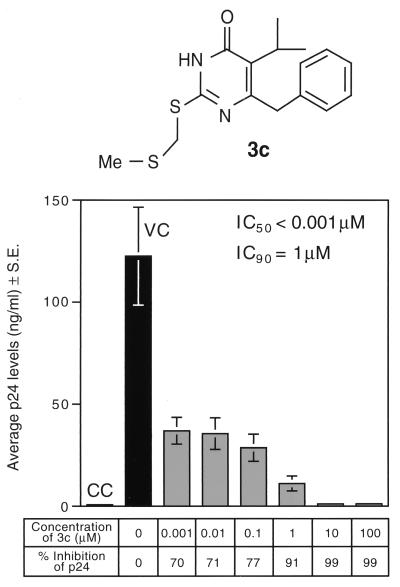
In each of four independent experiments, at a 1 nM concentration of 3c, the percent inhibition of p24 production was >50%, with a mean value of 67% ± 4%. In a dose-response study using picomolar and nanomolar concentrations of compound 3c, the subnanomolar IC50 was determined to be 130 pM.
NA, not applicable.
ND, not determined.
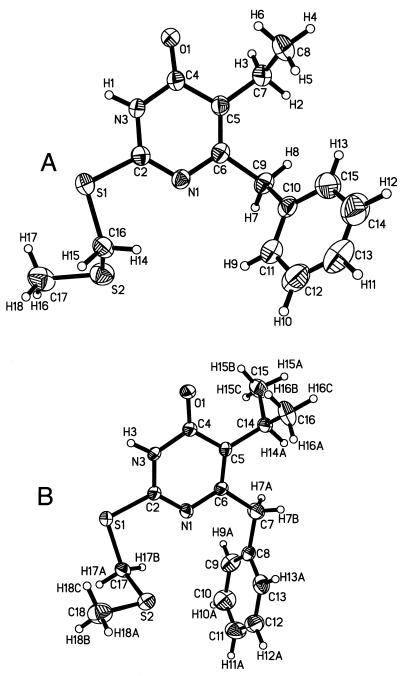
The refined small-molecule X-ray crystal structures of compounds 3b and 3c are shown in Fig. 3. Table 3 lists crystal data and structure refinement statistics, and Tables 4 and 5 list atomic coordinates for 3b and 3c. The molecular coordinates of DABO compounds which were energy minimized and docked into the NNI binding site adopted a conformation remarkably similar to that of the crystal structure of compound 3b. Fig. 2B shows the model coordinates of 3d superimposed on the crystal structure coordinates of 3b and illustrates their conformational similarity. Compound 3c adopted a different conformation in the crystal structure relative to the molecular model of 3c used for docking. The conformational differences of 3c include a 174° rotation of the isopropyl group around the C5-C4 bond (Fig. 3B) (roughly the equivalent of twofold rotation) and a 130° rotation of the sulfur tail around the S1-C17 bond (the equivalent of a mirror reflection through the S1-C17-S2 plane). This suggests that the two different molecular conformations did not differ significantly in energy, and both represent low-energy conformers. The main difference is due to a 45° rotation of the phenyl ring around the C6-C7 bond, which in our model can be stabilized by favorable contacts with binding site residues.
FIG. 3.
X-ray crystal structures of DABO analogs 3b (A) and 3c (B) (30% ellipsoids; temperature, 22°C).
TABLE 3.
Crystal data and refinement statistics for X-ray crystal structures of compounds 3b and 3ca
| Parameter | 3b (SD) | 3c (SD) |
|---|---|---|
| Unit cell | ||
| a (Å) | 8.9531 (9) | 4.7893 (4) |
| b (Å) | 23.374 (2) | 10.8709 (10) |
| c (Å) | 8.862 (9) | 30.040 (3) |
| α (°) | 90 | 90 |
| β (°) | 112.271 (2) | 92.474 (2) |
| γ (°) | 90 | 90 |
| Space group | P21/c | P21/c |
| Unit cell vol (Å3) | 1,717.4 (3) | 1,562.5 (2) |
| Z | 4 | 4 |
| θ range for data collection (°) | 1.74 to 24.99 | 1.36 to 28.27 |
| Limiting indices | −11 ≤ h ≤11 | −6 ≤ h ≤ 6 |
| −17 ≤ k ≤ 31 | −8 ≤ k ≤ 14 | |
| −11 ≤ l ≤ 11 | −39 ≤ l ≤ 37 | |
| Reflections collected | 8,167 | 8,744 |
| Independent reflections (Rint) | 2,990 (0.0676) | 3,507 (0.0486) |
| Data/restraints/parameters | 2,990/0/194 | 3,507/0/183 |
| Goodness of fit on F2 | 1.029 | 1.095 |
| Final R indices [I>2ς(I)] | R1, 0.0578; wR2, 0.0856 | R1, 0.0666; wR2, 0.1384 |
| R indices (all data) | R1, 0.1207; wR2, 0.1028 | R1, 0.1114; wR2, 0.1569 |
| Absorption coefficient (mm−1) | 0.310 | 0.338 |
| Max, min transmission | 0.4894, 0.4456 | 0.8356, 0.6542 |
| Extinction coefficient | 0.0023 (6) | 0.0004 (11) |
| Largest difference peaks (eÅ−3)b | 0.227, −0.204 | 0.279, −0.211 |
Data were collected at 22°C (λ = 0.71073 Å), refined by using full-matrix least-squares refinement on F2, and corrected for absorption by using semiempirical psi-scan data. Rint = Σ|Fo2 − <Fo2|/Σ|Fo2|, F1 = Σ‖Fo| − |Fc‖/Σ|Fo|. R1 = Σ‖Fo| − |Fc‖/Σ|Fo|. wR3 = {Σ[w(Fo2 − Fc2)2]/Σ[w(Fo2)2]}1/2. GooF = S = {Σ[w(Fo2 − FFc2)2]/(n − p)}1/2, where n is reflections and p is parameters.
eÅ, −3, electrons/Å3.
TABLE 4.
Atomic coordinates and equivalent isotropic displacement parameters for 3b based on its X-ray crystal structure at 22°C
| Atom | Atomic coordinate (104) and equivalent isotropic displacement parameter (Å, 103)b
|
|||
|---|---|---|---|---|
| x | y | z | U(eq)a | |
| S1 | 8,080 (2) | 2,998 (1) | 532 (1) | 60 (1) |
| S2 | 5,831 (2) | 5,424 (1) | 871 (1) | 79 (1) |
| O1 | 2,787 (5) | −801 (2) | 347 (1) | 74 (1) |
| N1 | 4,538 (5) | 2,146 (2) | 1,133 (1) | 49 (1) |
| N3 | 5,010 (5) | 1,011 (2) | 481 (1) | 53 (1) |
| C2 | 5,613 (6) | 1,980 (3) | 748 (1) | 47 (1) |
| C4 | 3,231 (7) | 84 (3) | 601 (1) | 55 (1) |
| C5 | 2,015 (6) | 238 (3) | 1,029 (1) | 52 (1) |
| C6 | 2,693 (6) | 1,264 (3) | 1,268 (1) | 50 (1) |
| C7 | 113 (8) | −794 (3) | 1,164 (1) | 69 (1) |
| C8 | 1,773 (10) | −1,884 (4) | 1,341 (2) | 93 (1) |
| C9 | 1,436 (7) | 1,572 (3) | 1,708 (1) | 60 (1) |
| C10 | 3,421 (7) | 1,473 (3) | 2,104 (1) | 56 (1) |
| C11 | 5,121 (8) | 2,421 (4) | 2,234 (1) | 76 (1) |
| C12 | 6,938 (10) | 2,331 (5) | 2,609 (1) | 96 (1) |
| C13 | 7,025 (11) | 1,282 (6) | 2,850 (2) | 106 (2) |
| C14 | 5,312 (13) | 345 (5) | 2,730 (2) | 117 (2) |
| C15 | 3,557 (10) | 432 (4) | 2,355 (1) | 91 (1) |
| C16 | 8,199 (7) | 4,179 (3) | 952 (1) | 62 (1) |
| C17 | 7,394 (10) | 6,198 (4) | 420 (1) | 93 (1) |
| H1 | 5,764 (5) | 969 (2) | 227 (1) | 64 |
| H2 | −1,100 (8) | −501 (3) | 1,392 (1) | 83 |
| H3 | −1,054 (8) | −1,048 (3) | 909 (1) | 83 |
| H4 | 515 (10) | −2,524 (4) | 1,425 (2) | 139 |
| H5 | 2,906 (10) | −1,637 (4) | 1,597 (2) | 139 |
| H6 | 2,952 (10) | −2,184 (4) | 1,115 (2) | 139 |
| H7 | 713 (7) | 2,406 (3) | 1,692 (1) | 72 |
| H8 | −131 (7) | 1,026 (3) | 1,751 (1) | 72 |
| H9 | 5,072 (8) | 3,146 (4) | 2,069 (1) | 91 |
| H10 | 8,083 (10) | 2,991 (5) | 2,692 (1) | 115 |
| H11 | 8,254 (11) | 1,208 (6) | 3,097 (2) | 127 |
| H12 | 5,314 (13) | −368 (5) | 2,901 (2) | 140 |
| H13 | 2,441 (10) | −237 (4) | 2,272 (1) | 110 |
| H14 | 7,847 (7) | 3,799 (3) | 1,236 (1) | 74 |
| H15 | 10,080 (7) | 4,511 (3) | 973 (1) | 74 |
| H16 | 6,399 (42) | 6,948 (15) | 355 (7) | 140 |
| H17 | 7,313 (60) | 5,678 (12) | 161 (3) | 140 |
| H18 | 9,309 (21) | 6,383 (26) | 501 (4) | 140 |
U(eq) is defined as one-third of the trace of the orthogonalized Uij tensor.
Estimated standard deviations are given in parentheses.
TABLE 5.
Atomic coordinates and equivalent isotropic displacement parameters for 3c based on its X-ray crystal structure at 22°C
| Atom | Atomic coordinate (104) and equivalent isotopic displacement parameter (Å, 103)b
|
|||
|---|---|---|---|---|
| x | y | z | U(eq)a | |
| S1 | 2,787 (1) | 1,450 (1) | 4,913 (1) | 51 (1) |
| S2 | 2,938 (1) | 2,175 (1) | 7,812 (1) | 65 (1) |
| N1 | 1,819 (3) | 759 (1) | 6,769 (3) | 47 (1) |
| N3 | 3,649 (3) | 383 (1) | 5,748 (3) | 43 (1) |
| O1 | 4,643 (3) | −515 (1) | 6,250 (3) | 58 (1) |
| C2 | 2,724 (4) | 808 (1) | 5,931 (4) | 39 (1) |
| C4 | 3,739 (4) | −147 (1) | 6,458 (4) | 40 (1) |
| C5 | 2,741 (4) | −220 (1) | 7,391 (4) | 37 (1) |
| C6 | 1,849 (4) | 239 (1) | 7,500 (4) | 43 (1) |
| C7 | 777 (5) | 213 (2) | 8,499 (5) | 78 (1) |
| C8 | 24 (5) | 766 (2) | 8,675 (5) | 50 (1) |
| C9 | −1,424 (6) | 938 (2) | 7,542 (5) | 75 (1) |
| C10 | −2,116 (5) | 1,445 (2) | 7,721 (6) | 90 (2) |
| C11 | −1,367 (6) | 1,786 (2) | 9,039 (7) | 81 (1) |
| C12 | 51 (6) | 1,617 (2) | 10,179 (5) | 70 (1) |
| C13 | 740 (4) | 1,110 (2) | 10,006 (5) | 59 (1) |
| C14 | 2,770 (4) | −789 (1) | 8,215 (4) | 44 (1) |
| C15 | 2,224 (5) | −1,278 (2) | 7,015 (5) | 70 (1) |
| C16 | 4,395 (5) | −907 (2) | 9,555 (5) | 80 (1) |
| C17 | 1,768 (4) | 1,924 (1) | 5,816 (4) | 49 (1) |
| C18 | 4,318 (5) | 2,638 (2) | 7,388 (5) | 80 (1) |
| H3 | 4,210 (3) | 445 (1) | 5,163 (3) | 52 |
| H7A | −77 (5) | −63 (2) | 7,992 (5) | 93 |
| H7B | 1,421 (5) | 71 (2) | 9,579 (5) | 93 |
| H9A | −1,949 (6) | 709 (2) | 6,637 (5) | 89 |
| H10A | −3,101 (5) | 1,557 (2) | 6,937 (6) | 108 |
| H11A | −1,830 (6) | 2,131 (2) | 9,150 (7) | 98 |
| H12A | 564 (6) | 1,845 (2) | 11,088 (5) | 84 |
| H13A | 1,714 (4) | 997 (2) | 10,809 (5) | 71 |
| H14A | 1,987 (4) | −761 (1) | 8,740 (4) | 53 |
| H15A | 2,142 (28) | −1,621 (3) | 7,576 (7) | 104 |
| H15B | 2,995 (15) | −1,335 (7) | 6,515 (22) | 104 |
| H15C | 1,190 (14) | −1,190 (5) | 6,191 (17) | 104 |
| H16A | 4,660 (15) | −603 (6) | 10,340 (17) | 120 |
| H16B | 5,206 (7) | −931 (11) | 9,094 (6) | 120 |
| H16C | 4,349 (11) | −1,262 (6) | 10,079 (22) | 120 |
| H17A | 1,384 (4) | 2,253 (1) | 5,107 (4) | 58 |
| H17B | 828 (4) | 1,727 (1) | 5,853 (4) | 58 |
| H18A | 5,064 (20) | 2,796 (9) | 8,389 (6) | 120 |
| H18B | 3,731 (5) | 2,942 (6) | 6,685 (26) | 120 |
| H18C | 4,900 (23) | 2,424 (3) | 6,862 (29) | 120 |
U(eq) is defined as one-third of the trace of the orthogonalized Uij tensor.
Estimated standard deviations are given in parentheses.
Predictable activities.
Compounds 3a to 3d were tested for RT inhibitory activity in cell-free assays using purified HIV RT (listed as IC50[rRT] in Table 2), as well as by in vitro assays of anti-HIV activity in human T-cell leukemia virus IIIB (HTLV-IIIB)-infected PBMNC (22, 47, 49) (IC50[p24] in Table 2). Compound 3a, which showed the lowest activity and the worst interaction scores, inhibited rRT in two independent experiments with IC50s of 38.8 and 18.8 μM (mean IC50[rRT], 28.8 ± 10.0 μM), whereas our lead compound, 3c, inhibited rRT in three independent experiments with IC50s of 6.1, 1.3, and 9.5 μM (mean IC50[rRT], 5.6 ± 2.4 μM). As shown in Table 2, the IC50[p24]s for the inhibition of HIV-infected cells were significantly lower than the IC50[rRT]s, which measure the inhibition of rRT. Compound 3a inhibited p24 production at micromolar concentrations with a mean IC50 of 3.4 ± 1.1 μM from three independent experiments, whereas the lead compound had an IC50 of <0.001 μM in each of four independent experiments (percent inhibition at 1 nM in the four experiments, 58.3, 76.5, 63.3, and 70.0%). Thus, the lead compound had consistently and significantly higher potency than compound 3a in both cell-free and cellular RT inhibition assays. By comparison, MKC442 inhibited p24 production consistently at slightly higher concentrations than our lead compound 3c. The individual IC50s of MKC442 in the four independent experiments were 0.006, 0.005, 0.003, and 0.003 μM, with a mean value of 0.004 ± 0.001 μM. Larger compounds, which better fill the composite binding pocket and have lower calculated Ki values, showed better IC50[rRT]s. This is reflected by the enhancement of the inhibitory activity with the addition of progressively larger groups, such as methyl (3a), ethyl (3b), and isopropyl (3c) groups, at the C-5 position of the thymine ring (Table 2). The same general trend was also observed for IC50[p24]s. Compound 3d, which differs from compound 3c by the addition of two methyl groups to the 6-benzyl ring, provides more hydrophobic contact with the NNI binding pocket but showed potency similar to that of 3c in enzymatic assays. As shown in Table 2, compound 3d, despite its comparable IC50[rRT] in cell-free assays, failed to inhibit HIV replication in HTLV-IIIB-infected cells as effectively as compound 3c, contrary to our predictions. Our calculations indicate that compounds 3a to 3d have progressively larger molecular surface areas but still maintain approximately the same percentage of the molecular surface area in contact with the protein residues. Consequently, the calculated contact surface area between the protein and the compound increases in the order 3a, 3b, 3c, and 3d. This increased surface area, in turn, dictates a decrease in calculated Ki values, with 3a having the worst value and 3d the best. The trend of calculated Ki values for compounds 3a to 3c were predictive of the general trend of their measured IC50[p24]s (Table 2).
DISCUSSION
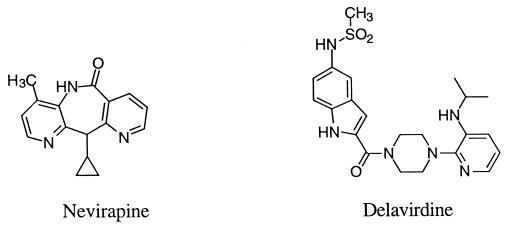
Recent translational research efforts against AIDS have focused on the development of potent inhibitors of HIV RT (18, 26, 36). Two NNIs of HIV RT that have been approved by the U.S. Food and Drug Administration for licensing and sale in the United States are nevirapine (27) and delavirdine [bis (heteroaryl)piperazine derivative U-90152] (23, 42, 43) (Fig. 1). Other promising NNI derivatives that have been studied include DABO derivatives (14–16, 34), HEPT derivatives (2, 14, 39, 45), TIBO (20), 2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2′-dioxide)pyrimidine (3, 4), oxathiin carboxanilide derivatives (9–11), quinoxaline derivatives (24, 31), thiadiazole derivatives (25), and phenethylthiazolylthiourea (PETT) derivatives (1, 5, 12, 28). NNIs have been found to bind to a specific allosteric site of HIV-1 RT near the polymerase site and interfere with reverse transcription by altering either the conformation or the mobility of RT, thereby leading to noncompetitive inhibition of the enzyme (20, 21, 32, 40, 41, 44).
FIG. 1.
Structures of nevirapine and delavirdine.
The crystal structure of RT was analyzed to determine how certain mutations might affect the binding of DABO compounds. Observed RT mutations such as L100I, V106A, and Y181C (46) involve substituting a slightly smaller residue at the active site. Our model suggests that DABO compounds 3a to 3d would not be prohibited from binding to these mutant RTs. The mutations can accommodate each compound’s slightly larger molecular volume (larger than that of the parent DABO compound and, in some cases, MKC442). Inhibition would probably not be as effective as for wild-type RT, however, due to loss of hydrophobic contacts. In this context, it is noteworthy that a novel PETT derivative, designed by using our composite HIV RT binding pocket, exhibited potent activity against the MKC442-resistant V106A RT mutant strain of HIV (data not shown). The activity of compound 3c against various NNI-resistant HIV-1 strains will be examined in future studies. In a clinical setting, the currently applied approach to handling the problem of drug resistance by RT is to use a combination of drugs which have different mechanisms of action (viz., nucleoside inhibitors, NNIs, and protease inhibitors). Another strategy is to use a sufficiently high concentration of a single NNI to eradicate the virus in the host before mutations can occur (19). The more potent the drug, the more effective this approach would be, if the drug has sufficiently low toxicity.
Our lead DABO derivative, compound 3c, elicited significant anti-HIV activity with an IC50 of less than 1 nM for inhibition of HIV replication (measured by p24 production in HIV-infected human PBMNC) and showed no detectable cytotoxicity (inhibition of cellular proliferation occurred at >100 μM, as measured by MTA) (Table 2 and Fig. 5). In contrast to all previously published data for DABO and S-DABO derivatives, which were less active than zidovudine and MKC442 (15, 16, 34) and showed selectivity indices of <1,000, compound 3c was more than fourfold more active than zidovudine and MKC442 and abrogated HIV replication in PBMNC at nanomolar concentrations with an unprecedented selectivity index (CC50[MTA]/IC50[p24]) of >100,000.
FIG. 5.
Anti-HIV activity of compound 3c. The anti-HIV activity of DABO derivative 3c was evaluated in four independent experiments (each performed in triplicate) by measuring its ability to inhibit HIV replication in HTLV-IIIB-infected PBMNC by using the p24 EIA as previously reported (22, 47, 49). The results depicted are from one representative experiment. CC, uninfected control cell cultures; VC, HTLV-IIIB-infected cultures not exposed to compound 3c; S.E., standard error. Me, methyl.
Our modeling studies showed that the trend of calculated Ki values for compounds 3a to 3c were predictive of the general trend of their measured IC50[p24]s (Table 2). However, 3d was predicted to be significantly more potent than 3c (based on calculated Ki values), which was not observed experimentally. The IC50[rRT]s of 3c and 3d were, in fact, similar, and the IC50[p24] of 3c was >100-fold better than of 3d. Compound 3c was not predicted to be quite so potent; the calculated Ki value for compound 3c differs from the measured IC50 by several orders of magnitude. This is in contrast to the other, less active, compounds in the series, which exhibit reasonably predictable Ki values. These results prompt the hypothesis that the binding interactions taken into consideration in our modeling studies may not fully account for the superior anti-HIV activity of our lead compound, 3c. For example, one of the methyl groups on the phenyl ring of 3d is in a position to contact a region of the binding pocket which is at an interface between a polar region and a nonpolar region. If the actual binding position of 3d allows the methyl group to contact the polar region rather than the nonpolar region predicted by docking, this less favorable interaction would lead to weaker RT binding than was estimated. We have described similar observations for halogen-substituted PETT derivatives, which also contact this interfacial region of the NNI binding pocket of HIV RT (48).
An additional consideration for NNI binding is the Tyr183 residue of the HIV RT located in the catalytic region which has a conserved YMDD motif characteristic of RTs. The displacement of this conserved tyrosine residue can interfere with catalysis and render the HIV-1 RT protein inactive. It has been suggested that bulky substituents at the fifth position of the thymine ring could indirectly accomplish such inactivation by displacing Tyr181, which is near Tyr183 (29). Our composite binding pocket shows sufficient room for at least a three-carbon group at the fifth position. Modeling shows that the addition of a methyl, ethyl, or isopropyl group at the fifth position of the thymine ring would lead to higher affinity for the relatively hydrophobic environment at this location of the binding pocket. The favorable hydrophobic contact increases as the hydrophobic group at the fifth position gets bulkier. As it binds to the site, the ethyl or isopropyl group can also cause the nearby Tyr181 residue to rotate away from the inhibitor. Our modeling showed that this change in conformation, in turn, affects the positions of neighboring Tyr183 and Tyr188, which may contribute to the inactivation of HIV-1 RT. The 6-benzyl ring of compounds 3a to 3d is located near a region surrounded by the hydrophobic ring planes of residues Trp229, Pro95, Y188, and Y181. The analysis of compounds 3a to 3c in the composite binding pocket suggests that the 6-benzyl ring would be located on the boundary of the pocket, near residue Y188. The para position of the ring is situated perpendicular to the ring plane of nearby Trp229, within van der Waals contact, and thus would be an unsuitable location for large-group substitution. However, there is significant unfilled space between the compound and Pro95. With slight conformational rotation of the 6-benzyl ring, compound 3d, with the addition of two methyl groups, was found to better fill the composite binding pocket (Fig. 2B). Such observations suggest that further modification of the 6-benzyl ring could lead to even more potent inhibitors.
The goal of our modeling studies was to understand the interactions between RT and its inhibitors so that better inhibitors could be designed. The results of our studies are useful for a more precise design of inhibitors. A qualitative design choice may be possible without any modeling but would probably result in significantly fewer highly effective inhibitors. Quantitating how well a design might interact with the binding site is our greater goal and is more useful for designing a new generation of compounds highly effective against HIV RT. It is important to note, however, that computer modeling of NNIs, while providing valuable guidance to direct the chemical synthesis of new inhibitors, can offer only an “educated guess” regarding the biologic activity of new designs until experimentally verified. In future studies, we will continue to use our composite binding pocket geometry, in conjunction with biological testing, for the purpose of structure-based design of other potent NNIs of HIV RT.
REFERENCES
- 1.Ahgren C, Backro K, Bell F W, Cantrell A S, Clemens M, Colacino J M, Deeter J B, Engelhardt J A, M. H, Jaskunas S R, Johansson N G, Jordan C L, Kasher J S, Kinnick M D, Lind P, Lopez C, Morin J M J, Muesing M A, Noreen R, Oberg B, Paget C J, Palkowitz J A, Parrish C A, Pranc P, Rippy M K, Rydergard C, Sahlberg C, Swanson S, Ternansky R J, Unge T, Vasileff R T, Vrang L, West S J, Zhang H, Zhou X X. The PETT series, a new class of potent nonnucleoside inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:1329–1335. doi: 10.1128/aac.39.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba M, Shigeta S, Tanaka H, Miyasaka T, Ubasawa M, Umezu K, Walker R T, Pauwels R, De Clercq E. Highly potent and selective inhibition of HIV-1 replication by 6-phenylthiouracil derivatives. Antiviral Res. 1992;17:245–264. doi: 10.1016/0166-3542(92)90021-v. [DOI] [PubMed] [Google Scholar]
- 3.Balzarini J. Novel concepts in the treatment of human immunodeficiency virus type 1 (HIV-1) infections by HIV-1-specific reverse transcriptase inhibitors. Verh K Acad Geneeskd Belg. 1995;57:575–600. [PubMed] [Google Scholar]
- 4.Balzarini J, Perez Perez M J, San Felix A, Schols D, Perno C F, Vandamme A M, Camarasa M J, De Clercq E. 2′,5′-Bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2′-dioxide) pyrimidine (TSAO) nucleoside analogues: highly selective inhibitors of human immunodeficiency virus type 1 that are targeted at the viral reverse transcriptase. Proc Natl Acad Sci USA. 1992;89:4392–4396. doi: 10.1073/pnas.89.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell F W, Cantrell A S, Hogberg M, Jaskunas S R, Johansson N G, Jordan C L, Kinnick M D, Lind P, Morin J M, Jr, Noreen R, Oberg B, Palkowitz J A, Parrish C A, Pranc P, Sahlberg C, Ternansky R T, Vasileff R T, Vrang L, West S J, Zhang H, Zhou X X. Phenethylthiazolethiourea (PETT) compounds, a new class of HIV-1 reverse transcriptase inhibitors. 1. Synthesis and basic structure-activity relationship studies of PETT analogs. J Med Chem. 1995;38:4929–4936. doi: 10.1021/jm00025a010. [DOI] [PubMed] [Google Scholar]
- 6.Bohm H J. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J Comput-Aided Mol Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 7.Bohm H J. LUDI: rule-based automatic design of new substituents for enzyme inhibitor leads. J Comput-Aided Mol Des. 1992;6:593–606. doi: 10.1007/BF00126217. [DOI] [PubMed] [Google Scholar]
- 8.Bosworth N, Towers P. Scintillation proximity assay. Nature. 1989;341:167–168. doi: 10.1038/341167a0. [DOI] [PubMed] [Google Scholar]
- 9.Buckheit R W, Jr, Kinjerski T L, Fliakas-Boltz V, Russell J D, Stup T L, Pallansch L A, Brouwer W G, Dao D C, Harrison W A, Schultz R J, Bader J P, Yang S S. Structure-activity and cross-resistance evaluations of a series of human immunodeficiency virus type 1-specific compounds related to oxathiin carboxanilide. Antimicrob Agents Chemother. 1996;39:2718–2727. doi: 10.1128/aac.39.12.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckheit R W, Jr, Hollingshead M, Stinson S, Fliakas-Boltz V, Pallansch L A, Roberson J, Decker W, Elder C, Borgel S, Bonomi C, Shores R, Siford T, Malspeis L, Bader J P. Efficacy, pharmacokinetics, and in vivo antiviral activity of UC781, a highly potent, orally bioavailable nonnucleoside reverse transcriptase inhibitor of HIV type 1. AIDS Res Hum Retroviruses. 1997;13:789–796. doi: 10.1089/aid.1997.13.789. [DOI] [PubMed] [Google Scholar]
- 11.Buckheit R W, Jr, Snow M J, Fliakas-Boltz V, Kinjerski T L, Russell J D, Pallansch L A, Brouwer W G, Yang S S. Highly potent oxathiin carboxanilide derivatives with efficacy against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus isolates. Antimicrob Agents Chemother. 1997;41:831–837. doi: 10.1128/aac.41.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantrell A S, Engelhardt P, Hogberg M, Jaskunas S R, Johansson N G, Jordan C L, Kangasmetsa J, Kinnick M D, Lind P, Morin J M, Jr, Muesing M A, Noreen R, Oberg B, Pranc P, Sahlberg C, Ternansky R J, Vasileff R T, Vrang L, West S J, Zhang H. Phenethylthiazolylthiourea (PETT) compounds as a new class of HIV-1 reverse transcriptase inhibitors. 2. Synthesis and further structure-activity relationship studies of PETT analogs. J Med Chem. 1996;39:4261–4274. doi: 10.1021/jm950639r. [DOI] [PubMed] [Google Scholar]
- 13.Connolly M L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 14.Danel K, Larsen E, Pedersen E B, Vestergaard B F, Nielsen C. Synthesis and potent anti-HIV-1 activity of novel 6-benzyluracil analogues of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine. J Med Chem. 1996;39:2427–2431. doi: 10.1021/jm9600499. [DOI] [PubMed] [Google Scholar]
- 15.Danel K, Nielsen C, Pedersen E B. Anti-HIV active naphthyl analogues of HEPT and DABO. Acta Chem Scand. 1997;51:426–430. doi: 10.3891/acta.chem.scand.51-0426. [DOI] [PubMed] [Google Scholar]
- 16.Danel K, Pedersen E B, Nielsen C. Synthesis and anti-HIV-1 activity of novel 2,3-dihydro-7H-thiazolo[3,2-a]pyrimidin-7-ones. J Med Chem. 1998;41:191–198. doi: 10.1021/jm970443m. [DOI] [PubMed] [Google Scholar]
- 17.Das K, Ding J, Hsiou Y, Clark A D, Jr, Moereels H, Koymans L, Andries K, Pauwels R, Janssen P A, Boyer P L, Clark P, Smith R H, Jr, Kroeger Smith M B, Michejda C J, Hughes S H, Arnold E. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 18.De Clercq E. Inhibitors targeted at the reverse transcriptase. J Acquired Immune Defic Syndr Res Hum Retroviruses. 1992;8:119–134. doi: 10.1089/aid.1992.8.119. [DOI] [PubMed] [Google Scholar]
- 19.De Clercq E. Toward improved anti-HIV chemotherapy: therapeutic strategies for intervention with HIV infections. J Med Chem. 1995;38:2491–2517. doi: 10.1021/jm00014a001. [DOI] [PubMed] [Google Scholar]
- 20.Ding J, Das K, Moereels H, Koymans L, Andries K, Janssen P A, Hughes S H, Arnold E. Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 21.Ding J, Das K, Tantillo C, Zhang W, Clark A D, Jr, Jessen S, Lu X, Hsiou Y, Jacobo Molina A, Andries K, Pauwels R, Moereels H, Koymans L, Janssen P A J, Smith R H J, Kroeger Koepke R, Michejda C J, Hughes S H, Arnold E. Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution. Structure. 1995;3:365–379. doi: 10.1016/s0969-2126(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 22.Erice A, Balfour H H, Jr, Myers D E, Leske V L, Sannerud K J, Kuebelbeck V, Irvin J D, Uckun F M. Anti-human immunodeficiency virus type 1 activity of an anti-CD4 immunoconjugate containing pokeweed antiviral protein. Antimicrob Agents Chemother. 1993;37:835–838. doi: 10.1128/aac.37.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esnouf R M, Ren J, Hopkins A L, Ross C K, Jones E Y, Stammers D K, Stuart D I. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc Natl Acad Sci USA. 1997;94:3984–3989. doi: 10.1073/pnas.94.8.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Font M, Monge A, Alvarez E, Cuartero A, Losa M J, Fidalgo M J, SanMartin C, Nadal E, Ruiz I, Merino I, Martinez-Irujo J J, Alberdi E, Santiago E, Prieto I, Lasarte J J, Sarobe P, Borras F. Synthesis and evaluation of new Reissert analogs as HIV-1 reverse transcriptase inhibitors. 1. Quinoline and quinoxaline derivatives. Drug Des Discovery. 1997;14:305–332. [PubMed] [Google Scholar]
- 25.Fujiwara M, Ijichi K, Hanasaki Y, Ide T, Katsuura K, Takayama H, Aimi N, Shigeta S, Konno K, Yokota T, Baba M. Thiadiazole derivatives: highly potent and selective inhibitors of human immunodeficiency virus type 1 (HIV-1) replications in vitro. Microbiol Immunol. 1997;41:301–308. doi: 10.1111/j.1348-0421.1997.tb01205.x. [DOI] [PubMed] [Google Scholar]
- 26.Greene W C. The molecular biology of human immunodeficiency virus type 1 infections. N Engl J Med. 1991;324:308–317. doi: 10.1056/NEJM199101313240506. [DOI] [PubMed] [Google Scholar]
- 27.Grob P M, Wu J C, Cohen K A, Ingraham R H, Shih C K, Hargrave K D, McTague T L, Merluzzi V J. Nonnucleoside inhibitors of HIV-1 reverse transcriptase: nevirapine as a prototype drug. AIDS Res Hum Retroviruses. 1992;8:145–152. doi: 10.1089/aid.1992.8.145. [DOI] [PubMed] [Google Scholar]
- 28.Heinisch G, Matuszczak B, Pachler S, Rakowitz D. The inhibitory activity of diazinyl-substituted thiourea derivatives on human immunodeficiency virus type 1 reverse transcriptase. Antiviral Chem Chemother. 1997;8:443–446. [Google Scholar]
- 29.Hopkins A L, Ren J, Esnouf R M, Willcox B E, Jones E Y, Ross C, Miyasaka T, Walker R T, Tanaka H, Stammers D K, Stuart D I. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J Med Chem. 1996;39:1589–1600. doi: 10.1021/jm960056x. [DOI] [PubMed] [Google Scholar]
- 30.Jones T A, Zou J Y, Cowan S W, Kjeldgaard G. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Kleim J P, Rosner M, Winkler I, Paessens A, Kirsch R, Hsiou Y, Arnold E, Riess G. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 33.Luty B A, Wasserman P F, Stouten P F, Hodge C N, Zacharias M, McCammon J A. A molecular mechanics/grid method for evaluation of ligand-receptor interactions. J Comp Chem. 1995;16:454–464. [Google Scholar]
- 34.Mai A, Artica M, Sbardella G, Quartarone S, Massa S, Loi A G, Montis A D, Scintu F, Putzolu M, La Colla P. Dihydro(alkylthio)(naphthylmethyl)oxopyrimidines: novel non-nucleoside reverse transcriptase inhibitors of the S-DABO series. J Med Chem. 1997;40:1447–1454. doi: 10.1021/jm960802y. [DOI] [PubMed] [Google Scholar]
- 35.Mao C, Vig R, Venkatachalam T K, Sudbeck E A, Uckun F M. Structure-based design of N-[2-(1-piperidinylethyl)]-N′-[2-(5-bromopyridyl)]-thiourea and N-[2-(1-piperazinylethyl)]-N′-[2-(5-bromopyridyl)]-thiourea as potent non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg Med Chem Lett. 1998;8:2213–2218. doi: 10.1016/s0960-894x(98)00384-9. [DOI] [PubMed] [Google Scholar]
- 36.Mitsuya H, Yarchoan R, Broder S. Molecular targets for AIDS therapy. Science. 1990;249:1533–1544. doi: 10.1126/science.1699273. [DOI] [PubMed] [Google Scholar]
- 37.Molecular Simulations Inc. InsightII. San Diego, Calif: Molecular Simulations Inc.; 1996. [Google Scholar]
- 38.Nicholls A. GRASP graphical representation and analysis of surface properties. 1992. New York, N.Y. [Google Scholar]
- 39.Pontikis R, Benhida R, Aubertin A M, Grierson D S, Monneret C. Synthesis and anti-HIV activity of novel N-1 side chain-modified analogs of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) J Med Chem. 1997;40:1845–1854. doi: 10.1021/jm960765a. [DOI] [PubMed] [Google Scholar]
- 40.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, Stammers D. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 41.Ren J, Esnouf R, Hopkins A, Ross C, Jones Y, Stammers D, Stuart D. The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design. Structure. 1995;3:915–926. doi: 10.1016/S0969-2126(01)00226-X. [DOI] [PubMed] [Google Scholar]
- 42.Romero D L, Morge R A, Genin M J, Biles C, Busso M, Resnick L, Althaus I W, Reusser F, Thomas R C, Tarpley W G. Bis(heteroaryl)piperazine (BHAP) reverse transcriptase inhibitors: structure-activity relationships of novel substituted indole analogues and the identification of 1-[(5-methanesulfonamido-1H-indol-2-yl)-carbonyl]-4-[3-[(1-methylethyl)amino]-pyridinyl]piperazine monomethanesulfonate (U-90152S), a second-generation clinical candidate. J Med Chem. 1993;36:1505–1508. doi: 10.1021/jm00062a027. [DOI] [PubMed] [Google Scholar]
- 43.Romero D L, Olmsted R A, Poel T J, Morge R A, Biles C, Keiser B J, Kopta L A, Friis J M, Hosley J D, Stefanski K J, Wishka D G, Evans D B, Morris J, Stehle R G, Sharma S K, Yagi Y, Voorman R L, Adams W J, Tarpley W G, Thomas R C. Targeting delavirdine/atevirdine resistant HIV-1: identification of (alkylamino)piperidine-containing bis(heteroaryl)piperazines as broad spectrum HIV-1 reverse transcriptase inhibitors. J Med Chem. 1996;39:3769–3789. doi: 10.1021/jm960158n. [DOI] [PubMed] [Google Scholar]
- 44.Smerdon S J, Jager J, Wang J, Kohlstaedt L A, Chirino A J, Friedman J M, Rice P A, Steitz T A. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:3911–3915. doi: 10.1073/pnas.91.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka H, Baba M, Hayakawa H, Sakamaki T, Miyasaka T, Ubasawa M, Takashima H, Sekiya K, Nitta I, Shigeta S, Walker R T, Balzarini J, De Clercq E. A new class of HIV-1-specific 6-substituted acyclouridine derivatives: synthesis and anti-HIV-1 activity of 5- or 6-substituted analogues of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) J Med Chem. 1991;34:349–357. doi: 10.1021/jm00105a055. [DOI] [PubMed] [Google Scholar]
- 46.Tantillo C, Ding J, Jacobo Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 47.Uckun F M, Chelstrom L M, Tuel-Ahlgren L, Dibirdik I, Irvin J D, Chandan-Langlie M, Myers D E. TXU (anti-CD7)-pokeweed antiviral protein as a potent inhibitor of human immunodeficiency virus. Antimicrob Agents Chemother. 1998;42:383–388. doi: 10.1128/aac.42.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vig R, Mao C, Venkatachalan T K, Tuel-Ahlgren L, Sudbeck E A, Uckun F M. Rational design and synthesis of phenethyl-5-bromopyridyl thiourea derivatives as potent non-nucleoside inhibitors of HIV reverse transcriptase. Bioorg Med Chem. 1998;6:1–9. doi: 10.1016/s0968-0896(98)00108-4. [DOI] [PubMed] [Google Scholar]
- 49.Zarling J M, Moran P A, Haffar O, Sias J, Richman D D, Spina C A, Myers D E, Kuebelbeck V, Ledbetter J A, Uckun F M. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+ cells by monoclonal antibodies. Nature. 1990;347:92–95. doi: 10.1038/347092a0. [DOI] [PubMed] [Google Scholar]