Abstract
As essential components of the protein synthesis machinery, tRNAs undergo a tightly controlled biogenesis process, which include the incorporation of numerous posttranscriptional modifications. Defects in these tRNA maturation steps may lead to the degradation of hypomodified tRNAs by the rapid tRNA decay (RTD) and nuclear surveillance pathways. We previously identified m1A58 as a late modification introduced after modifications Ψ55 and T54 in yeast elongator tRNAPhe. However, previous reports suggested that m1A58 is introduced early during the tRNA modification process, in particular on primary transcripts of initiator tRNAiMet, which prevents its degradation by RNA decay pathways. Here, aiming to reconcile this apparent inconsistency on the temporality of m1A58 incorporation, we examined its introduction into yeast elongator and initiator tRNAs. We used specifically modified tRNAs to report on the molecular aspects controlling the Ψ55 → T54 → m1A58 modification circuit in elongator tRNAs. We also show that m1A58 is efficiently introduced on unmodified tRNAiMet, and does not depend on prior modifications. Finally, we show that m1A58 has major effects on the structural properties of initiator tRNAiMet, so that the tRNA elbow structure is only properly assembled when this modification is present. This observation provides a structural explanation for the degradation of hypomodified tRNAiMet lacking m1A58 by the nuclear surveillance and RTD pathways.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Transfer RNAs (tRNAs) are essential components of the cellular protein synthesis machinery, but also serve additional functions outside translation (1–4). To achieve their wide range of functions within cells, tRNAs undergo a tightly controlled biogenesis process leading to the formation of mature tRNAs (5–8). The biogenesis of tRNAs typically includes the removal of the 5′-leader and 3′-trailer sequences from the precursor-tRNA transcripts, the addition of the 3′-CCA amino-acid accepting sequence, and the incorporation of a large number of posttranscriptional chemical modifications. These modifications occur at specific sites in a tightly controlled manner, which ensures that the tRNA biogenesis process effectively leads to the formation of functional tRNAs (9–13). All the cellular functions of tRNAs are, to various extents, affected by modifications. In particular, modifications in and around the anticodon are implicated in the decoding process (9,14–17), whereas modifications found in the tRNA core are collectively implicated in the folding and stability of tRNAs (18–21). Posttranscriptional modifications are thus central to tRNA biology. Maturation defects, resulting in lack of modifications in the tRNA core, may result in alternative folding (22,23), and often reduce tRNA stability, leading to the degradation of hypomodified tRNAs by the rapid tRNA decay (RTD) pathway (24–26) and the nuclear surveillance pathway (27–29).
Although modifications are typically introduced in tRNAs independently of each other, several modification circuits have been identified in which one or more modifications stimulate or repress the incorporation of another modification (11,30,31). This obviously drives a defined sequential order in the tRNA modification process. Most of the reported examples of an ordered modification process occur in the tRNA anticodon loop region (32–36), but modification circuits in the tRNA core have also been reported (37–39).
One such circuit in the tRNA core involves modifications in the T-loop of yeast tRNAs. Using NMR spectroscopy to monitor the maturation of tRNAs in a time-resolved fashion in yeast extracts (40), we previously identified a sequential order in the introduction of T54, Ψ55 and m1A58 in yeast tRNAPhe, with Ψ55 being introduced first, then T54 and finally m1A58 (39). Using specific deletion strains, we uncovered a cross-talk between these three modifications, with the m1A58 modification strongly dependent on the two others. In a pus4Δ strain, lacking Ψ55, we indeed observed a severe slow-down in the introduction of both T54 and m1A58. Similarly, in a trm2Δ strain, lacking T54, we observed a slow-down in the introduction of m1A58 (39). In addition, we showed, using liquid-chromatography coupled with tandem mass spectrometry (LC-MS/MS), that levels of m1A58 and T54 are affected in the pus4Δ and trm2Δ strains, in both yeast tRNAPhe and in total yeast tRNAs, in a manner compatible with the cross-talk observed with NMR spectroscopy in yeast extracts. This demonstrated that these cross-talks in the T-loop are manifest not only in tRNAPhe but also in other yeast tRNAs. Overall, the slow-down in the incorporation of modifications and the corresponding decrease in the modification levels observed in the absence of a specific enzyme, namely in the pus4Δ and trm2Δ strains, was interpreted as a positive effect of the corresponding modification on the introduction of the other ones. We thus concluded that two modification circuits exist in the T-loop of yeast tRNAs, the long-branch Ψ55  T54
T54 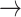 m1A58 circuit and the direct-branch Ψ55
m1A58 circuit and the direct-branch Ψ55  m1A58 circuit, without being able to conclude on the direct or indirect nature of the effect of Ψ55 on m1A58 (39).
m1A58 circuit, without being able to conclude on the direct or indirect nature of the effect of Ψ55 on m1A58 (39).
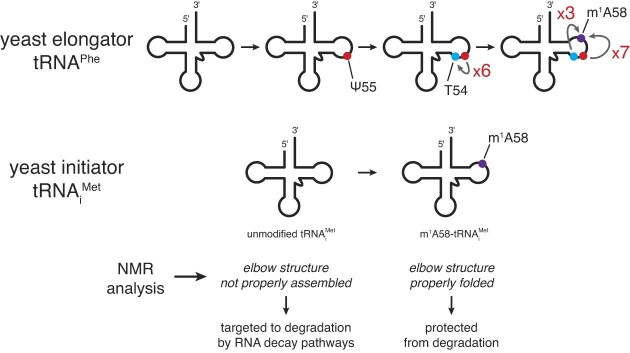
Overall, this report on yeast tRNAPhe identified m1A58 as a late modification, introduced after earlier modifications such as Ψ55, T54 and m7G46 (39). However, previous reports suggested that m1A58 is introduced early along the tRNA modification process in yeast, with m1A58 being introduced on initial pre-tRNA transcripts (5). Yeast initiator pre-tRNAiMet lacking m1A58, but containing the 5′-leader and part of the 3′-trailer sequences, is targeted by the nuclear surveillance and RTD pathways (27,28,41,42). In yeast tRNAiMet, the m1A58 modification is part of an unusual tRNA elbow structure involving non-canonical nucleotides A20, A54 and A60. This unusual substructure is assembled via an intricate network of interactions between the D- and T-loops and is likely conserved in eukaryotic initiator tRNAs (43). Altogether, these reports led to the model that m1A58 is introduced on pre-tRNAiMet transcripts, which stabilizes the tRNAiMet unique substructure, thereby preventing its degradation. In addition, degradation of tRNAiMet lacking m1A58 by the RTD pathway was recently shown to be conserved in the phylogenetically distant yeast species S. pombe and S. cerevisiae (42), suggesting that throughout eukaryotes the m1A58 modification is crucial to tRNAiMet biology.
Here, aiming to reconcile the apparent inconsistency regarding the incorporation of m1A58 in yeast tRNAs, namely as a late modification in elongator tRNAPhe and as an early modification in initiator tRNAiMet, we decided to examine the m1A58 modification pathways in yeast elongator and initiator tRNAs (see Supplementary Figure S1 for the sequence and modifications of yeast tRNAPhe and tRNAiMet). On the elongator tRNAPhe, we aimed at characterizing the molecular details related to the modification circuits present in the T-loop and involving Ψ55, T54 and m1A58, in order, in particular, to untangle direct from indirect effects. On the initiator tRNAiMet, we sought to investigate the introduction of m1A58 and its dependence on other modifications. In addition, we aimed at investigating the impact of the m1A58 modification on the structural properties of the tRNAiMet elbow region. Understanding the maturation process of initiator tRNAiMet, and in particular the m1A58 incorporation, which has consequences on its stability and quality control, is indeed crucial considering the central role of tRNAiMet in translation initiation and hence gene expression.
For that, we first implemented a generic approach enabling the preparation of tRNAs containing specific modifications. We then used these specifically modified tRNAs to demonstrate that the incorporation of T54 in tRNAPhe is directly stimulated by Ψ55, and that the incorporation of m1A58 in tRNAPhe is directly and individually stimulated by Ψ55 and T54, with a remarkable cumulative effect when they are present together, thereby reporting in detail the molecular mechanisms controlling the Ψ55  T54
T54  m1A58 modification circuit in yeast elongator tRNAs. We also show that m1A58 is efficiently introduced on unmodified tRNAiMet, and does not strictly need any prior modification, although m5C48,49 have a slight stimulatory effect on m1A58 incorporation. Finally, we show that the m1A58 single modification has major effects on the structural properties of yeast tRNAiMet, with the tRNA elbow structure being properly assembled only when this modification is present. This provides a structural basis to the degradation of hypomodified tRNAiMet lacking m1A58 by the nuclear surveillance and RTD pathways.
m1A58 modification circuit in yeast elongator tRNAs. We also show that m1A58 is efficiently introduced on unmodified tRNAiMet, and does not strictly need any prior modification, although m5C48,49 have a slight stimulatory effect on m1A58 incorporation. Finally, we show that the m1A58 single modification has major effects on the structural properties of yeast tRNAiMet, with the tRNA elbow structure being properly assembled only when this modification is present. This provides a structural basis to the degradation of hypomodified tRNAiMet lacking m1A58 by the nuclear surveillance and RTD pathways.
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this study are listed in Supplementary Table S1. The wild-type S. cerevisiae BY4741 strain and the YKO collection kanMX strains carrying deletions of the genes for modification enzymes Trm1, Trm2, Trm4, Trm8, Trm10, Trm11, Pus4, Dus1, Dus3 and Rit1, were obtained from Euroscarf and used for tRNA preparations for MS analysis. The proteinase-deficient S. cerevisiae strain c13-ABYS-86 and the derived strain c13-ABYS-86-trm4Δ were used for the preparation of yeast extracts for NMR experiments. All strain constructions were verified by PCR using appropriate oligonucleotides (listed in Supplementary Table S2).
E. coli strains
E. coli strains used in this study are listed in Supplementary Table S1. The E. coli BL21(DE3) CodonPlus-RIL yggH::kan (trmB) strain was constructed by transferring the yggH::kan cassette from the appropriate K-12 strain of the Keio collection (44) to a BL21(DE3) CodonPlus-RIL strain (Agilent) by phage P1 vir-mediated transduction (45) (Supplementary Table S1). Deletion of the yggH gene and its replacement by the kanamycin resistance cassette in the BL21(DE3) CodonPlus-RIL strain was checked with PCR using appropriate sets of primers (Supplementary Table S2).
Modification enzymes cloning
The gene encoding the full-length yeast Pus4 (M1 to V403 – Uniprot entry P48567) was cloned from BY4741 genomic DNA between the EcoRI and NotI sites of a modified pET28a vector (Novagen) encoding an N-terminal His6-tag cleavable with TEV protease (pET28-Pus4). The gene encoding the full-length yeast Trm2 (M1 to I639 – Uniprot entry P33753) was initially cloned from BY4741 genomic DNA between the EcoRI and NotI sites of a pGEX-6p-1 vector (pGEX-Trm2). However, this construct was insoluble and poorly expressed in E. coli BL21(DE3) CodonPlus-RIL cells. Since the N-terminal part of Trm2 contains highly hydrophobic stretches of amino-acids, and does not correspond to the catalytic domain of the protein, a second construct corresponding to V116 to I639 was cloned between the BamHI and XhoI sites of a pRSFDuet-Smt3 vector leading to an N-terminal His6-SUMO- fusion of Trm2 (pSUMO-Trm2). The naturally present BamHI site in the yeast trm2 gene was first removed by a silent mutation of the codon encoding for D564 from GAT to GAC with site directed mutagenesis. The genes encoding yeast Trm6/Trm61 heterodimer (Trm6: M1 to I478 – Uniprot entry P41814; Trm61: M1 to K383 – Uniprot entry P46959) were cloned from BY4741 genomic DNA between the BamHI and NotI sites for Trm6 and NdeI and XhoI sites for Trm61 of a pETDuet-1 vector (Novagen) thereby encoding an N-terminal His6-tag on Trm6 (pETDuet-Trm6/Trm61).
Modification enzymes purification
Pus4, Trm2 and Trm4 were overexpressed in E. coli BL21(DE3) CodonPlus-RIL cells (Agilent) in LB media. Trm6/Trm61 heterodimer was overexpressed in E. coli BL21(DE3) CodonPlus-RIL yggh::kan cells lacking the E. coli enzyme catalyzing m7G46 modifications in tRNAs, namely TrmB since initial expression and purification in E. coli BL21(DE3) CodonPlus-RIL cells lead to a Trm6/Trm61 heterodimer contaminated with an m7G46 modification activity (see Supplementary Figure S2). The cells were grown at 37°C to OD600 ∼0.4, cooled down to 18–30°C and induced at OD600 ∼0.6 by adding (IPTG) to a final concentration of 0.4–0.5 mM. Cells were harvested 6–22 h after induction by centrifugation. Cell pellets were resuspended in the corresponding lysis buffer supplemented with an EDTA-free antiprotease tablet (Roche) and lysed by sonication. Cell lysates were centrifuged for 30 min at 35 000 g. All column chromatography purifications were performed on a ÄKTA Pure purification system (Cytiva) at 4°C. The cell lysate supernatant was loaded on a Ni-NTA column and the protein of interest was eluted with an imidazole gradient. Fractions containing the protein were pooled, concentrated with an Amicon 50 000 MWCO (Millipore) and further purified with a combination of hydrophobic and size exclusion chromatography depending on the protein. Purified protein samples loaded on size exclusion chromatography were eluted in the corresponding protein storage buffer, confirmed for purity using SDS-PAGE (Supplementary Figure S3), concentrated with an Amicon (Millipore) to ∼5–10 mg/ml and stored at –20°C. The protein concentrations were determined by absorbance at 280 nm using the corresponding mM extinction coefficient (See Supplementary Table S3 for specific details on the purifications).
RNA sample preparation for NMR and enzyme activity assays
Unmodified yeast tRNAPhe-WT, tRNAiMet-WT, tRNAPhe-ΔU17, tRNAPhe-A20A60, tRNAPhe-A54, tRNAPhe-UAAA, tRNAiMet-U17, tRNAiMet-G20C60, tRNAiMet-U54 and tRNAiMet-UGCU were prepared by standard in vitro transcription following previously published procedures, either with unlabelled NTPs or 15N-labelled Us and Gs (39,46). We replaced the first Watson Crick base pair A1-U72 of tRNAiMet with a G1-C72 base pair in order to improve in vitro transcription efficiency. To prepare the single modified Ψ55-tRNAPhe, 112 μM of refolded tRNAPhe was incubated with 3.3 μM of purified Pus4 for 40 min at 30°C in an 800 μl reaction mix. To prepare T54-tRNAPhe, 80 μM of refolded tRNAPhe was incubated with 12 μM of purified Trm2 and a ∼6–8-times excess of S-adenosyl-L-methionine (SAM) in an 800 μl reaction mix for 14 h at 30°C. To prepare the double modified Ψ55-T54-tRNAPhe, 80 μM of Ψ55-tRNAPhe was incubated with 8 μM Trm2 and a ∼6–8-times excess of SAM in an 800 μl reaction mix for 4 h at 30°C. To prepare m5C48,49-tRNAiMet, 146 μM of refolded unmodified-tRNAiMet was incubated with 23 μM of purified Trm4 for 17 h at 30°C in a 500 μl reaction mix. All reactions were performed in the following maturation buffer (MB): 100 mM NaH2PO4/K2HPO4 pH 7.0, 5 mM NH4Cl, 2 mM DTT and 0.1 mM EDTA. The tRNA reaction products were then purified by ion exchange chromatography (MonoQ, Cytiva), dialyzed extensively against 1 mM Na-phosphate pH 6.5, and refolded by heating at 95°C for 5 min and cooling down slowly at room temperature. Buffer was added to place the tRNAs in the NMR buffer (10 mM Na-phosphate pH 6.5, 10 mM MgCl2), and the samples were concentrated using Amicon 10000 MWCO (Millipore) to ∼80 μM for further use in kinetic assays, or ∼1.4–1.5 mM for the NMR study of tRNAiMet maturation in yeast extracts.
Trm2 and Trm6/Trm61 kinetic assays on different substrates
To measure initial velocities of m1A58 and T54 formation, 10 μM of unmodified tRNAPhe, Ψ55-tRNAPhe, T54-tRNAPhe, Ψ55-T54-tRNAPhe, unmodified tRNAiMet and m5C48,49-tRNAiMet were incubated each in a 300 μl reaction with enzyme concentrations varying from 50 to 300 nM depending on enzyme and substrate type, 18 μM non-radioactive SAM and 50 nM of radioactive [3H]-SAM (see Supplementary Table S4 for details on the reaction mixes). Reactions were performed in the MB buffer and were incubated at 30°C for 30 min except for the reaction with Trm6/Trm61 and the unmodified tRNAPhe that was incubated for 96 min. Aliquots of 50 μl were taken of each reaction at 6, 12, 18, 24 and 30 min (for the 30 min reactions) and at 24, 48, 72 and 96 min (for the 96 min reaction) and the samples were quenched by adding 5% (v/v) cold trichloracetic acid (TCA). Quenched samples were filtered through Whatman glass microfibers disks pre-soaked with 5% (v/v) TCA, washed four times with 5% (v/v) TCA and one final time with ethanol. The filter disks were dried, then 5 ml Optiphase ‘HISAFE’ 2 scintillation cocktail (PerkinElmer) were added, and the counts per minute (CPM) equivalent to the incorporated [3H]-methyl were determined by scintillation counting. Then CPM values were converted to concentrations of modified tRNAs using [3H]-SAM/CPM calibration standards. Enzymatic reactions were performed in triplicates or quadruplicates. Since Trm6/Trm61 and Trm2 activity turned out to vary greatly between different substrates, different enzyme concentrations were used to perform the kinetic assays. Therefore, we normalized the quantities of modified tRNAs to an equivalent of 50 nM of enzyme. Initial velocities (Vi) were determined by linear regression using Prism7 (GraphPad), i.e. data were fitted to a single linear function: y = Vi .x while forcing the curve to pass through the origin, and standard errors (SE) on the Vi were determined by taking into account the data spread.
Trm6/Trm61 activity assays on yeast tRNAPhe and tRNAiMet variants
To measure m1A58 formation, 10 μM of unmodified yeast tRNAPhe-WT, tRNAPhe-ΔU17, tRNAPhe-A20A60, tRNAPhe-A54, tRNAPhe-UAAA, tRNAiMet-WT, tRNAiMet-U17, tRNAiMet-G20C60, tRNAiMet-U54 and tRNAiMet-UGCU were incubated each in a 100 μl reaction with 600 nM of purified Trm6/Trm61, 18 μM non-radioactive SAM and 100 nM of radioactive [3H]-SAM. Reactions were performed in the MB buffer at 30°C and concentrations of modified tRNAs were measured at t = 60 min. Samples were then treated as described above. Enzymatic reactions were performed in six replicates (N = 6). Standard deviations were relatively uniform across the different tRNA substrates and corresponded to 20–33% of the average value for tRNAPhe variants and to 17–20% for tRNAiMet variants. CPM values were converted to concentrations of modified tRNAs using [3H]-SAM/CPM calibration standards.
NMR spectroscopy
All NMR spectra of yeast tRNAPhe and tRNAiMet were measured at 38°C on a Bruker AVIII-HD 700 MHz spectrometer equipped with TCI 5-mm cryoprobe with 5-mm Shigemi tubes in the NMR buffer (10 mM Na-phosphate pH 6.5, 10 mM MgCl2) supplemented with 5% (v/v) D2O. To verify that the desired modifications were incorporated quantitatively in yeast tRNAPhe, 1D jump-and-return-echo NMR spectra (47,48) of the different tRNAs were measured and compared to previously characterized samples (39,49). To analyse the effect of nucleotide swapping on the structural properties of yeast tRNAPhe and yeast tRNAiMet, 2D (1H,15N)-BEST-TROSY spectra of unmodified yeast tRNAPhe-WT, tRNAPhe-ΔU17, tRNAPhe-A20A60, tRNAPhe-A54, tRNAPhe-UAAA, tRNAiMet-WT, tRNAiMet-U17, tRNAiMet-G20C60, tRNAiMet-U54 and tRNAiMet-UGCU were measured at 38°C in the NMR buffer. In addition, to evaluate the effect of specific modifications on the structural properties of yeast tRNAiMet, 2D (1H,15N)-BEST-TROSY spectra of unmodified tRNAiMet, m5C48,49-tRNAiMet and m1A58-tRNAiMet were measured at 38°C in the NMR buffer. Imino resonances of the m1A58-tRNAiMet were assigned using 2D jump-and-return-echo (1H,1H)-NOESY (47,48) and 2D (1H,15N)-BEST-TROSY (50) experiments. For monitoring the maturation of tRNAiMet in yeast extract, wild-type and trm4Δ yeast extracts were prepared in the c13-ABYS-86 background, as previously described (40). NMR spectra were measured at 30°C with unmodified 15N-[U/G]-labelled tRNAiMet at 40 μM in yeast extracts supplemented with NaH2PO4/K2HPO4 pH 6.5 150 mM, NH4Cl 5 mM, MgCl2 5 mM, DTT 2 mM, EDTA 0.1 mM, SAM 4 mM, ATP 4 mM, NADPH 4 mM and D2O 5% (v/v) (51). Each 2D (1H,15N)-BEST-TROSY experiment of the series was measured with a recycling delay of 200 ms, a SW(15N) of 26 ppm and 96 increments for a total experimental time of 120 min. The data were processed using TOPSPIN 3.6 (Bruker) and analysed with Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Total tRNA samples from yeast for mass spectrometry
Total tRNA from S. cerevisiae BY4741 wild-type or mutant strains used for mass spectrometry analysis were prepared as described previously (39). For each strain, all cultures and tRNA preparations were performed in triplicate for statistical analysis. Yeast tRNAiMet was isolated from ∼1 μg total tRNA samples with a first step of SEC and a subsequent purification using T1 Dynabeads (Thermo Fisher Scientific, Product no. 65801D) and a DNA probe specific to tRNAiMet ([Btn]- AAA-TCG-GTT-TCG-ATC-CGA-GGA-CAT-CAG-GGT-TAT-GA, Sigma-Aldrich, Munich, Germany) as previously reported (39,52,53).
Digestion of tRNAs to nucleosides and quantification by mass spectrometry
Purified tRNAiMet samples were digested to single nucleosides following previously published procedures (39) and stable isotope-labelled internal standard (SILIS, 0.1 volume of 10X solution) from yeast was added for absolute quantification (54). Quantification of the m1A modification in tRNAiMet was performed with an Agilent 1290 Infinity II equipped with a DAD combined with an Agilent Technologies G6470A Triple Quad system and electro-spray ionization (ESI-MS, Agilent Jetstream) following previously published procedures (39,54). Absolute abundance of m1A from wild-type yeast corresponded to 0.69 ± 0.04 m1A per tRNAiMet. The absolute quantities of m1A in the deleted strains were normalized to that of the wild-type strain to determine abundance relative to wild-type. Analyses of the variations compared to the wild-type strain were conducted from the determination of the confidence intervals at 95% (CI 95%) using Prism7 (GraphPad).
RESULTS
A generic approach to prepare tRNAs with specific modifications
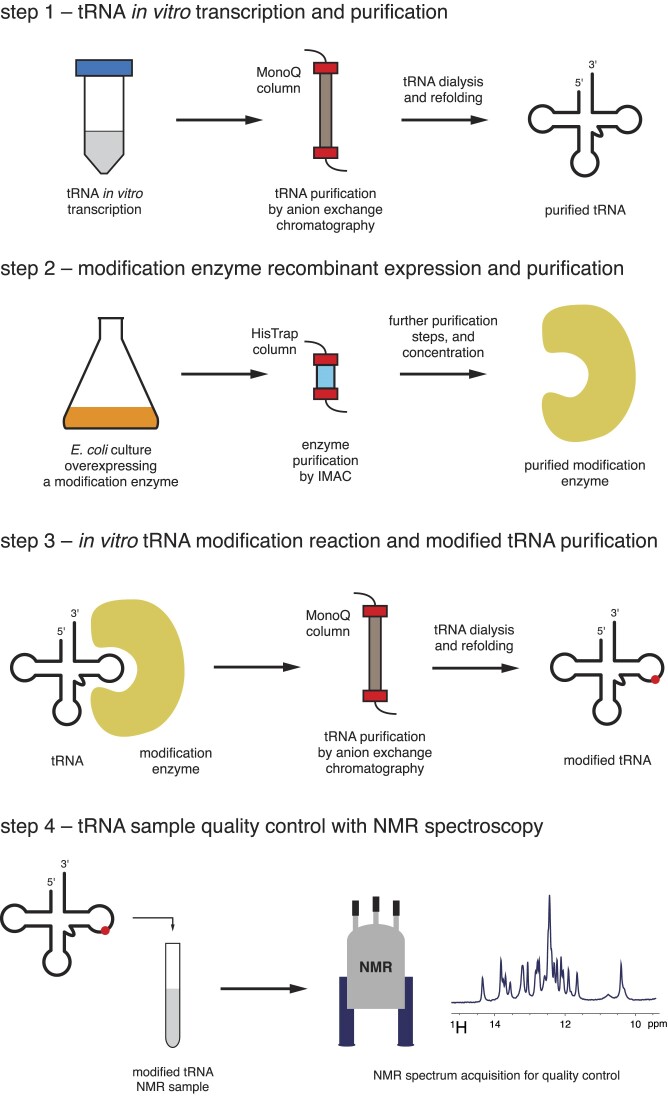
In order to evaluate the effect of pre-existing modifications on the introduction of further ones, we have implemented a generic method for preparing tRNA samples with a single or a specific set of modifications (Figure 1). Our approach is divided into four successive steps, (1) tRNA in vitro transcription and purification, (2) modification enzyme expression and purification, (3) in vitro tRNA modification reaction and modified tRNA purification and (4) tRNA sample quality control by NMR spectroscopy (Figure 1). To introduce several modifications on a tRNA, steps 3 and 4 can be reiterated on a tRNA sample already carrying modification(s).
Figure 1.
A generic approach to prepare tRNAs with specific modifications. (Step 1) The unmodified tRNA is transcribed in vitro and purified by anion exchange chromatography. (Step 2) The desired modification enzyme is overexpressed in E. coli and purified by immobilized metal affinity chromatography (IMAC) and further purification steps if needed. (Step 3) The unmodified tRNA is modified in vitro with the purified modification enzyme in presence of cofactors and subsequently purified by anion exchange chromatography. (Step 4) A quality control step is performed by 1D 1H NMR in order to establish that the desired modifications were fully incorporated in the tRNA population.
For the present study on yeast tRNAPhe and tRNAiMet, in addition to the unmodified tRNAPhe and tRNAiMet, we applied our methodology to produce: tRNAPhe samples carrying single modifications (Ψ55-tRNAPhe and T54-tRNAPhe), or double modifications (Ψ55-T54-tRNAPhe), and tRNAiMet samples carrying m5C48,49 or m1A58 modifications (m5C48,49-tRNAiMet and m1A58-tRNAiMet). For this purpose, we first transcribed and purified, using anion exchange chromatography, the yeast unmodified tRNAPhe and tRNAiMet (Figure 1, step 1). We then overexpressed and purified the yeast enzymes Pus4 that introduces Ψ55, Trm2 that adds T54 (or m5U54), Trm6/Trm61 that adds m1A58 and Trm4 that introduces m5C48 and m5C49 (see Materials and Methods and Supplementary Figure S1; Figure 1, step 2). Next, preliminary activity tests with these different enzymes allowed us to estimate the enzyme to tRNA ratios and the incubation times needed to introduce the desired modifications quantitatively. We thus incubated the unmodified tRNAs with the appropriate enzymes and cofactors for the required duration, and then purified the in vitro modified tRNAs using anion exchange chromatography (Figure 1, step 3). Finally, we verified that the desired modifications were introduced quantitatively by performing a quality control of our samples with NMR spectroscopy (Figure 1, step 4).
The introduction of T54 by Trm2 to the yeast tRNAPhe is stimulated by Ψ55
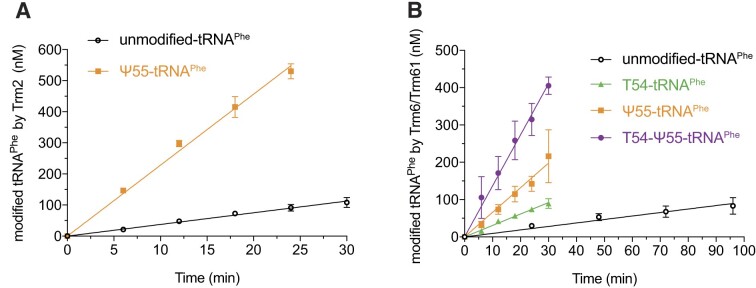
The fact that we observed a slower incorporation of T54 in tRNAPhe in the pus4Δ yeast extract, and that the amount of T54 in tRNAPhe as well as in the total tRNA population is drastically reduced in the pus4Δ strain, suggested that the Ψ55 modification had a positive effect on the introduction of T54 by Trm2 (39). However, we could not exclude that the defect in T54 incorporation was due to a negative effect of other modification(s) that only become apparent in the absence of Ψ55 or that the genetic expression of Trm2 was affected in the pus4Δ strain. Here, in order to unambiguously determine whether the introduction of T54 on the yeast tRNAPhe by Trm2 is directly dependent on the presence of Ψ55, we conducted activity assays with Trm2 on unmodified tRNAPhe and Ψ55-tRNAPhe (produced as described above). Trm2 was incubated with each of the tRNAs in the presence of the methyl-donor cofactor SAM carrying a radioactive methyl group (S-adenosyl-l-methionine [methyl-3H]), and aliquots were taken at different time points to determine the initial velocities (Vi) of the methylation reactions (Figure 2A, Table 1, and Supplementary Figure S4). These activity assays clearly demonstrated that the methylation reaction catalysed by Trm2 is about 6 times faster on the Ψ55-tRNAPhe when compared to the unmodified tRNAPhe (Table 2). This shows that the catalytic efficiency of Trm2 introducing T54 to tRNAPhe directly depends on the prior presence of Ψ55 and establishes the direct positive link between Ψ55 and the introduction of T54 by Trm2.
Figure 2.
Influence of pre-existing modifications on Trm2 and Trm6/Trm61 activities on tRNAPhe. (A) Time course of the introduction of T54 in tRNAPhe depending on the prior presence (orange) or absence (black) of the Ψ55 modification. (B) Time course of the introduction of m1A58 in tRNAPhe depending on pre-existing modifications: unmodified tRNAPhe (black), single modified T54-tRNAPhe (green) and Ψ55-tRNAPhe (orange), and double modified T54-Ψ55-tRNAPhe (purple). Modified tRNA quantities were measured for 4 or 5 time points in at least three independent experiments (N = 3 or 4), and initial velocities (Vi) were determined by linear regression (see Tables 1 and 2).
Table 1.
Initial velocities (Vi) of Trm2 and Trm6/Trm61 acting on yeast tRNAs presenting different modification profiles. Initial velocities were determined by linear regression and normalized to an equivalent of 50 nM of enzyme. The reported errors correspond to the standard error (SE) of the slope determination (see material and methods)
| Enzyme | Trm2 | Trm6/Trm61 | ||||||
| Yeast tRNAs | tRNAPhe | Ψ55-tRNAPhe | tRNAPhe | T54-tRNAPhe | Ψ55-tRNAPhe | Ψ55-T54-tRNAPhe | tRNAiMet | m5C-tRNAiMet |
| Vi (nM/min) | 3.8 ± 0.1 | 22.9 ± 0.5 | 0.93 ± 0.05 | 3.1 ± 0.1 | 6.6 ± 0.3 | 13.7 ± 0.3 | 10.7 ± 0.3 | 15.2 ± 0.4 |
Table 2.
Ratios of initial velocities (Vi) showing enzyme efficiency depending on the presence of pre-existing modifications on the yeast tRNAPhe and tRNAiMet. The reported errors of the ratios were calculated by taking into account the propagation of uncertainties
| Enzyme | Trm2 | Trm6/Trm61 | ||||||
| Yeast tRNAs | Ψ55-tRNAPhe/ tRNAPhe | T54-tRNAPhe/ tRNAPhe | Ψ55-tRNAPhe/ tRNAPhe | Ψ55-T54-tRNAPhe/ tRNAPhe | m5C-tRNAiMet/ tRNAiMet | tRNAiMet/ tRNAPhe | tRNAiMet/ Ψ55-T54-tRNAPhe | m5C-tRNAiMet/ Ψ55-T54-tRNAPhe |
| Vi ratio | 6.1 ± 0.2 | 3.3 ± 0.2 | 7.1 ± 0.5 | 15 ± 0.8 | 1.4 ± 0.1 | 11.5 ± 0.7 | 0.8 ± 0.03 | 1.1 ± 0.04 |
The introduction of m1A58 by Trm6/Trm61 to the yeast tRNAPhe is stimulated by Ψ55 and T54
Likewise, our previous work suggested a positive effect of the Ψ55 and T54 modifications on the introduction of m1A58 by the Trm6/Trm61 complex (39). However, as explained above for Trm2, we could not exclude that the observed behaviours were due to alternative effects. In addition, considering the above-mentioned effect of Ψ55 on T54, it was not possible to distinguish a direct effect of Ψ55 on m1A58 from an indirect effect via T54. To definitely establish whether the introduction of m1A58 on the yeast tRNAPhe is directly dependent on the presence of Ψ55 and T54, we conducted activity assays with the Trm6/Trm61 complex on unmodified tRNAPhe, Ψ55-tRNAPhe, T54-tRNAPhe and Ψ55-T54-tRNAPhe. The Trm6/Trm61 complex was incubated with each of the tRNAs in the presence of a radioactive [methyl-3H]-SAM cofactor, and aliquots were taken at different time points to derive the initial velocities (Figure 2B, Table 1, and Supplementary Figure S5a–d). With these activity assays, we observed that the introduction of m1A58 by Trm6/Trm61 is 3.3 times more efficient when the T54 modification is present compared to the unmodified tRNAPhe, 7.1 times more efficient in the presence of Ψ55 and 15 times more efficient if both T54 and Ψ55 are present in the yeast tRNAPhe (Table 2). This demonstrates that T54 and Ψ55 have individually a positive effect on the introduction of m1A58, as well as a cumulative positive effect if they are both simultaneously present. Therefore, the catalytic activity of Trm6/Trm61 directly depends on the presence of both the T54 and Ψ55 modifications. Additionally, our measurements indicate that Ψ55 stimulates the introduction of m1A58 about two times more efficiently than T54.
The m1A58 modification is efficiently introduced on an unmodified tRNAiMet
The results presented above showed that efficient introduction of m1A58 in tRNAPhe, strongly depends on the prior presence of Ψ55 and T54 in the T-loop. In addition, since the levels of modifications observed for total yeast tRNAs and tRNAPhe, are similarly affected in the pus4Δ and trm2Δ strains, the stimulation effect of Ψ55 and T54 on the introduction of m1A58 is certainly a common feature of several yeast tRNAs (39). At first sight, it might seem paradoxical that the efficiency of an enzyme encoded by two essential genes, i.e. trm6/trm61 (5,55,56), is highly dependent on the prior presence of modifications encoded by non-essential genes, i.e. pus4 and trm2. However, the origin of the essentiality of the m1A58 modification has been studied in detail in yeast and has been shown to be related to its importance for the maturation of initiator tRNAiMet (55). Hypomodified initiator tRNAiMet lacking m1A58 are indeed targeted to degradation by RNA decay pathways (27,28,42). As a possible explanation to this paradox, we noted that yeast tRNAiMet does not carry T54 and Ψ55 in the T-loop, but contains unmodified A54 and U55 (Supplementary Figure S1). Altogether, we anticipated that the initiator tRNAiMet would have its own pathway of modification in the T-arm, in which the m1A58 modification did not depend on pre-existing modifications. More generally, since tRNAiMet transcripts lacking m1A58 are degraded by RNA decay pathways, it seems reasonable that levels of m1A58 in tRNAiMet should not be altered in different strains or growth conditions. Modification levels should indeed reflect the requirement of m1A58 for tRNAiMet stability.
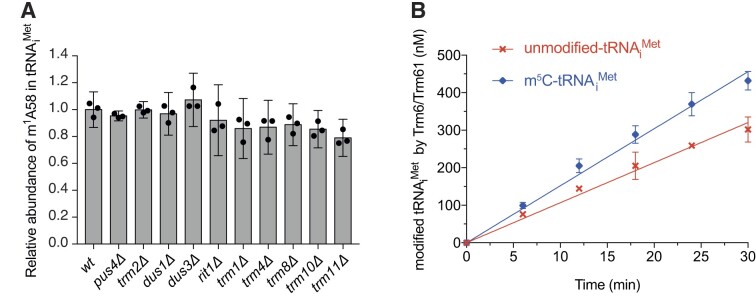
To examine these points, we measured, using LC–MS/MS, the levels of m1A in tRNAiMet from pus4Δ and trm2Δ strains, and from dus1Δ, dus3Δ, rit1Δ, trm1Δ, trm4Δ, trm8Δ, trm10Δ and trm11Δ strains, involved in the introduction of modifications D16, D47, Ar(p)64, m22G26, m5C48,49, m7G46, m1G9 and m2G10, respectively. These levels were compared with the levels of m1A in tRNAiMet from wild-type yeast cultured under the same experimental conditions. As expected, we observed no substantial changes in the amount of m1A in any of these deleted strains compared with the wild-type level (Figure 3A). The slight variations observed between some deleted strains and the wild-type could reflect a certain stability of m1A58-depleted tRNAiMet and/or small variations in the degree of purity of tRNAiMet recovered from the total tRNA population in the purification procedure. In any case, our data do not allow to conclude that these slight variations are significant. Overall, this shows that the lack of any other single modification does not prevent the formation of mature tRNAiMet carrying m1A58, and suggests that m1A58 can be correctly introduced on unmodified tRNAiMet.
Figure 3.
Influence of pre-existing modifications on m1A58 abundance and on Trm6/Trm61 activity on tRNAiMet. (A) Quantitative analysis of nucleoside modifications in yeast tRNAiMet with LC–MS/MS. Histograms showing the relative abundance of m1A58 modification in purified yeast tRNAiMet prepared from modification-enzyme-deleted strains using the wild-type levels as reference. Black dots represent individual measurements, data heights represent the mean of the biological replicates. Error bars correspond to the confidence interval at 95% (CI 95%). Modifications were quantified in three independent biological replicates (N = 3). (B) Time course of the introduction of m1A58 in tRNAiMet depending on the prior presence (blue) or absence (red) of the m5C48,49 modifications. Modified tRNA quantities were measured for 5 time points in three independent experiments (N = 3), and initial velocities (Vi) were determined by linear regression (see Tables 1 and 2).
To evaluate the efficiency of m1A58 modification on an unmodified tRNAiMet, we conducted activity assays with Trm6/Trm61 on unmodified tRNAiMet produced by in vitro transcription as described for tRNAPhe (Figure 3B, Table 1, and Supplementary Figure S5e). We observed that the introduction of m1A58 by Trm6/Trm61 is 11.5 times more efficient on the unmodified tRNAiMet than on the unmodified tRNAPhe (Table 2). This rate corresponds to an efficiency of about 0.8 times that measured on the doubly-modified Ψ55-T54-tRNAPhe (Table 2). Our data therefore establish that, on the contrary to its introduction on unmodified tRNAPhe, m1A58 is efficiently introduced on unmodified tRNAiMet, with an efficiency that is comparable to that observed for an optimally modified tRNAPhe bearing both Ψ55 and T54.
A54 is required for an efficient incorporation of m1A58 on unmodified tRNAiMet
Aiming to identify the sequence elements and associated structural properties implicated in the differences observed for m1A58 incorporation in tRNAPhe and tRNAiMet, we designed a set of tRNA variants with the objective of transfering elongator sequence elements and associated structural properties to initiator tRNAiMet, and vice versa. Since m1A58 is part of the specific initiator elbow structure (43), residues involved in this unique substructure, namely A20, A54 and A60, were primarily targeted for mutations. In addition, since the absence of nucleotide U17 is also a characteristic of initiator tRNAiMet, we chose to add it in a tRNAiMet variant. All tRNA variants, with their specific mutation or set of mutations are schematically summarized on Supplementary Figure S6.
First, in order to evaluate the effect of the nucleotide swapping between tRNAPhe and tRNAiMet from a structural point of view, we conducted NMR analysis on each tRNA variant. The comparison of the NMR fingerprint of unmodified tRNAPhe and tRNAiMet revealed clear differences (Supplementary Figure S6). The NMR spectrum of tRNAPhe-WT displays sharp and uniform NMR signals, characteristic of a stable, homogeneously folded tRNA. On the contrary, the NMR spectrum of tRNAiMet-WT exhibits a heterogeneous NMR signal profile with both weak and strong signals, as well as signals with atypic line shapes. These classic exchange-broaden signals reflect a less homogeneous folding for unmodified tRNAiMet that probably exchanges between several folding states, an exchange occurring in the intermediate regime relative to the NMR chemical shift time scale. The NMR fingerprints of the different variants revealed that for tRNAPhe, important structural changes are taking place in the tRNAPhe-A54 and tRNAPhe-UAAA variants, which tend to acquire a heterogeneous NMR spectrum profile (Supplementary Figure S6a). Conversely, for tRNAiMet, structural changes are apparent mostly for the tRNAiMet-U54 and tRNAiMet-UGCU variants, which exhibit slightly less heterogeneous or atypic NMR line shapes (Supplementary Figure S6b).
Next, we conducted activity assays with Trm6/Trm61 on unmodified tRNAPhe and tRNAiMet variants. On one hand, we observed that Trm6/Trm61 is ∼5.5 times less efficient on tRNAiMet-U54, where A54 is replaced by U54, compared to tRNAiMet-WT (Figure 4). This shows that A54 is required for an efficient incorporation of m1A58 by Trm6/Trm61 on unmodified tRNAiMet. We do not observe any other significant changes in the efficiency of m1A58 introduction on other tRNAiMet variants and in particular on tRNAiMet-UGCU (Figure 4). This is quite puzzling since tRNAiMet-UGCU also lacks the A54 residue. The additional mutations U17, G20 and C60 seem to neutralize the negative effect of the lack of A54. This demonstrates the inherent complexity and challenge associated with comprehending how nucleotides collaborate to establish intricate networks of interactions that shape the structure of tRNAs. On the other hand, the tRNAPhe-A54 variant, in which U54 is replaced by A54, does not show an increased efficiency of m1A58 incorporation (Figure 4). This is a good illustration that converting a good substrate into a poor substrate through the removal of a single key element is considerably simpler compared to the transformation of a poor substrate into a good one by introducing the same key element. In addition, we do not observe any changes in the efficiency of m1A58 introduction on tRNAPhe-ΔU17 and tRNAPhe-A20A60 variants compared to tRNAPhe-WT. This shows that neither adding nor removing U17, A20 and A60 residues to either tRNAiMet or tRNAPhe has any effect on m1A58 formation. Although determining the sequence and structural elements that govern Trm6/Trm61 activity appeared complicated, we identified nucleotide A54, which interacts with the Hoogsteen face of the target A58, as a key element for the efficient introduction of m1A58 into tRNAiMet.
Figure 4.
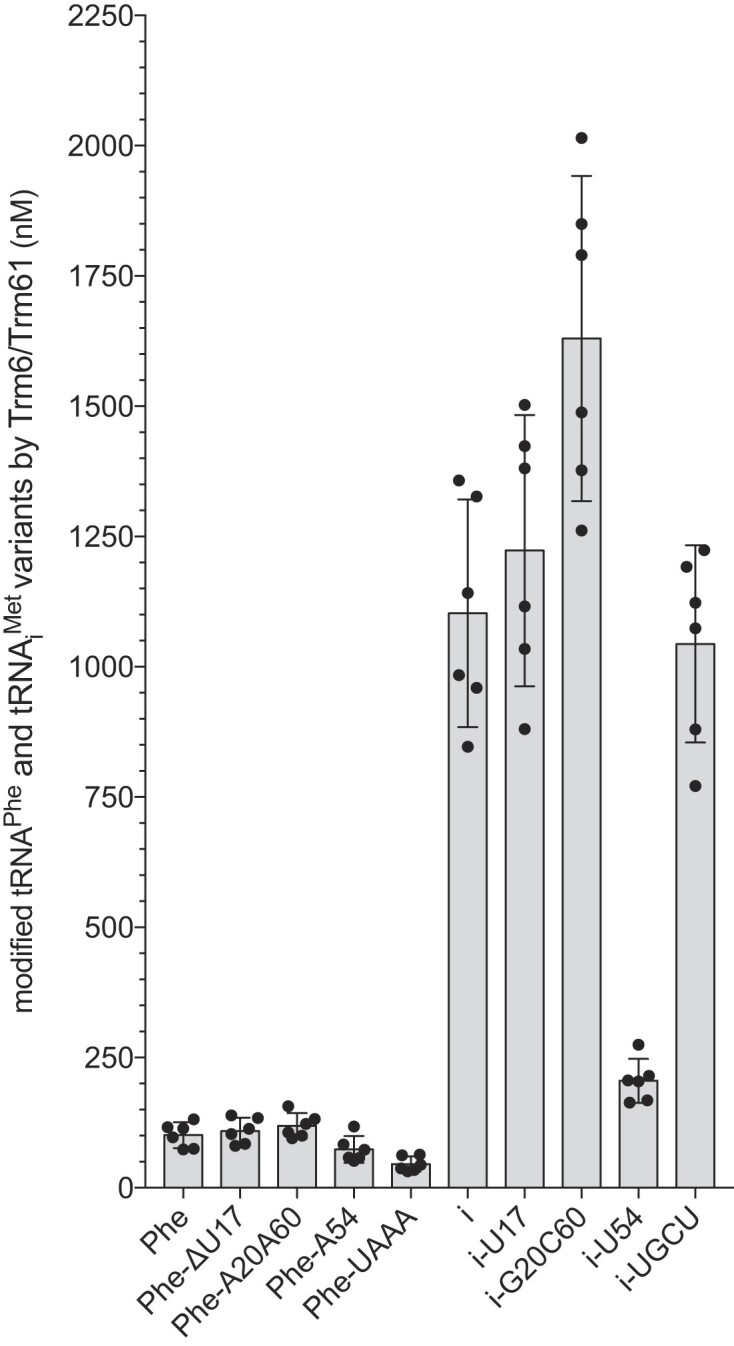
Influence of specific nucleotide swapping between tRNAPhe and tRNAiMet on Trm6/Trm61 activity. Histogram comparing the quantity of m1A58 introduced by Trm6/Trm61 in tRNAPhe and tRNAiMet variants (in nM). Names of the tRNAs are indicated below the graph and correspond to a specific nomenclature (see Supplementary Figure S6 for correspondence and details). Black dots represent individual measurements. Modified tRNA quantities were measured for 1 time point at t = 1 h in six independent experiments (N = 6). Data heights represent the mean of the replicates. Error bars correspond to the confidence interval at 95% (CI 95%).
The introduction of m1A58 by Trm6/Trm61 to the yeast tRNAiMet is slightly stimulated by m5C48,49
After studying the structural effects on the introduction of m1A58 by mutating nucleotides implicated in the unique tRNAiMet elbow structure, we wondered whether more subtle alterations that may also affect the tRNAiMet local structure could modulate m1A58 incorporation. In particular, posttranscriptional modifications that are close in space to m1A58, and that participate in the tRNAiMet tertiary interactions, could be considered as prime targets. Among tRNAiMet modifications, m5C48 meets these criteria. Indeed, m5C48 is relatively close to m1A58 in the tRNAiMet structure (<10 Å), and m5C48 and m1A58 are together implicated in the particular tRNA elbow structure of tRNAiMet involving the previously mentioned non-canonical nucleotides A20, A54 and A60 (43). More precisely, m5C48 is involved in an intricate network of interactions with G15, A20 and A59, with A59 and A20 forming a relay with another network involving A60 and m1A58 (Supplementary Figure S7). Another aspect prompted us to examine the link between m5C modifications and m1A58. Indeed, since the yeast trm4Δ mutant has been implicated in the RTD pathway in combinations with several other mutations (24–26), and since hypomodified tRNAiMet is targeted by the nuclear surveillance pathway and the RTD pathway, we wondered whether the modifications introduced by Trm4 could have an impact on the introduction of m1A58 by Trm6/Trm61, thereby affecting tRNAiMet stability.
For these reasons, we investigated whether m5Cs have any effect on the introduction of m1A58 in tRNAiMet. Note that in yeast initiator tRNAiMet, Trm4 introduces m5Cs at two positions, namely m5C48 and m5C49 (Supplementary Figure S1). We therefore conducted activity assays with Trm6/Trm61 on m5C48,49-tRNAiMet. We observed that Trm6/Trm61 is about 40% more efficient in the presence of m5C48,49 as compared with the unmodified tRNAiMet (Figure 3B, Table 1, and Supplementary Figure S5f). This corresponds to an efficiency of about 1.1 times the one measured on the doubly-modified Ψ55-T54-tRNAPhe (Table 2). Thus, even though the m5C48,49 modifications are not strictly required for m1A58 introduction by Trm6/Trm61, their presence enhances the efficiency of m1A58 introduction in tRNAiMetin vitro.
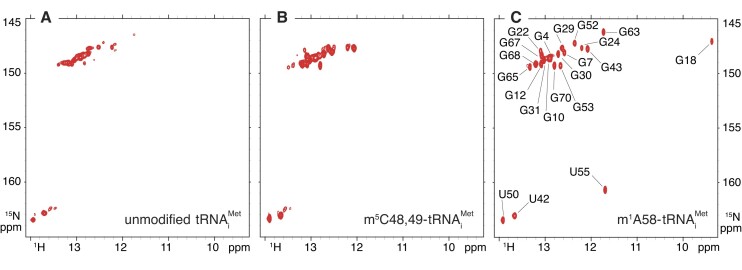
In order to get a clearer idea of the origin of m5Cs positive effects on m1A58 introduction, we analysed m5C-containing tRNAiMet with NMR spectroscopy. We produced an m5C48,49-tRNAiMet sample 15N-labelled on its imino groups, thereby allowing for the measurements of 2D 1H–15N NMR spectrum, which corresponds to its NMR-fingerprint and reflects folding homogeneity and structural integrity, as explained previously for the tRNAPhe and tRNAiMet variants. The comparison of the 1H–15N BEST-TROSY experiments of unmodified and m5C48,49-tRNAiMet samples revealed marked differences (Figure 5A, B). Additional signals appear on the spectrum of the m5C48,49-tRNAiMet, and a decrease in signal line broadening is observed. In addition, the signal heterogeneity present in the unmodified tRNAiMet, with weak and strong signals coexisting, is less pronounced in the m5C48,49-tRNAiMet spectrum, which shows a more homogeneous signal profile, with overall stronger signals than in the unmodified tRNAiMet (Figure 5A, B). As previously explained, NMR signals of RNA imino groups are only observed on condition that the imino protons are protected from exchange with the solvent by hydrogen bonding in any type of base pairing. The decrease in signal heterogeneity in the m5C48,49-tRNAiMet therefore reflects more stable base pairs, as well as a less dynamic and more homogeneous folding of this tRNA. The introduction of m5Cs by Trm4 therefore induces local and/or global changes in the folding of tRNAiMet, which could explain the increased efficiency of m1A58 incorporation (Figure 3B, Tables 1 and 2).
Figure 5.
Effect of m5C48,49 and m1A58 on the structural properties of yeast tRNAiMetimino (1H,15N) correlation spectra of 15N-labelled tRNAiMet with different modification status measured at 38°C. (A) unmodified tRNAiMet, (B) m5C48,49-tRNAiMet and (C) m1A58-tRNAiMet. The assignment of the imino resonances of the m1A58-tRNAiMet was obtained following standard methods.
Effect of m1A58 on the structural properties of yeast tRNAiMet
Since m1A58 is involved in the particular tRNA elbow structure of tRNAiMet (see (43) and text above), and since the m1A58 modification is essential for tRNAiMet stability and prevents its degradation by the nuclear surveillance and RTD pathways (27,28,42), we examined the effect of this single modification on the structural properties of tRNAiMet. We thus produced a 15N-labelled m1A58-containing tRNAiMet sample following our generic approach, and measured its NMR-fingerprint (Figure 5C). The comparison of the 1H-15N BEST-TROSY spectra of the unmodified and of the m1A58-tRNAiMet (Figure 5A–C) revealed considerable changes in the structural properties of tRNAiMet upon m1A58 modification. The pronounced signal heterogeneity present in unmodified tRNAiMet (Figure 5A) is completely absent in m1A58-tRNAiMet (Figure 5C), the NMR spectra of which display the characteristics of a stable and homogeneously folded tRNA. Thus, a single modification has major effects on the structural properties of yeast tRNAiMet, which can likely explain why hypomodified tRNAiMet lacking m1A58 is targeted by degradation pathways.
To get a deeper understanding of the structural changes arising upon m1A58 introduction, we performed the assignment of the imino resonances of the m1A58-tRNAiMet following standard methods (Figure 5C), as previously described for other tRNAs (49). With this assignment at hand, we noticed that the imino signals of G18 and U55 are only visible in the spectrum of the m1A58-tRNAiMet (Figure 5A–C). These nucleotides, and their respective imino groups, are engaged in universally conserved tertiary interactions at the level of the elbow region of tRNAs, with the imino group of U55 forming a hydrogen bond with a non-bridging oxygen of the phosphate backbone of A58, and that of G18 forming a hydrogen bond with an exocyclic carbonyl group of U55 (57). The detection of these imino groups in the NMR spectra of m1A58-tRNAiMet attests that their imino protons are protected from an exchange with the solvent, thereby demonstrating that the tRNA elbow structure is well-assembled. The imino signals of G18 and U55 can be considered as a signature of a properly folded tRNA with a well-assembled elbow structure. Conversely, their absence in the NMR-fingerprint of the unmodified tRNAiMet and the m5C48,49-tRNAiMet (Figure 5A, B) indicate that the tRNA elbow structure is not properly assembled in these tRNAs.
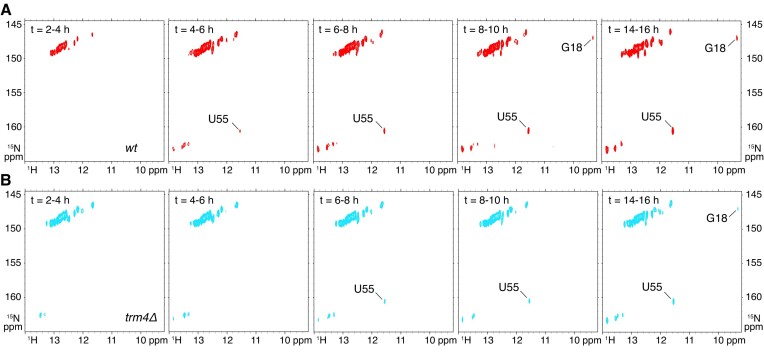
Time-resolved NMR monitoring of m1A58 introduction in tRNAiMet in yeast extract
The existence of a positive effect of m5Cs on m1A58 introduction in tRNAiMetin vitro (Figure 3B), does not necessarily imply that this effect occurs in a cellular context. For example, if m1A58 is introduced before m5Cs, no effect of m5Cs on the introduction of m1A58 can possibly be observed. In order to investigate whether this positive effect persists in a cellular context, we applied our recently developed methodology (39,40) to the monitoring of the introduction of m1A58 into tRNAiMet in yeast extracts. As seen above, the imino signals of G18 and U55 constitute an NMR signature of a properly assembled elbow structure, and therefore can be regarded as an indirect marker of m1A58 introduction in the case of tRNAiMet. We made use of this marker to monitor the introduction of m1A58 in tRNAiMet in wild-type and in trm4Δ yeast extracts using time-resolved NMR. For that, 15N-labelled unmodified tRNAiMet was incubated at 30°C in yeast extracts supplemented with the modification enzymes cofactors, SAM and NADPH. A series of 1H-15N BEST-TROSY experiments were measured for wild-type and trm4Δ yeast extracts (Figure 6). The observation of the imino signals of G18 and U55 along the tRNAiMet maturation routes revealed that m1A58 is introduced slightly faster in the wild-type extract than in an extract depleted of Trm4. This shows that lack of m5C48,49 has a negative effect on m1A58 introduction by Trm6/Trm61 in yeast tRNAiMet, which is perfectly consistent with the in vitro kinetic assays on tRNAiMet (Figure 3B). Overall, our data show that m5C modifications have a positive effect on m1A58 introduction in tRNAiMet both in vitro and in a cellular context.
Figure 6.
Time-resolved NMR monitoring of m1A58 introduction in tRNAiMet in yeast extracts. (A) Imino (1H,15N) correlation spectra of a 15N-labelled tRNAiMet measured in a time-resolved fashion during a continuous incubation at 30°C in yeast wild-type extract over 16 h. (B) Imino (1H,15N) correlation spectra of a 15N-labelled tRNAiMet measured in a time-resolved fashion during a continuous incubation at 30°C in yeast trm4Δ extract over 16 h. Each NMR spectrum measurement spreads over a 2 h time period, as indicated.
DISCUSSION
In this study, we implemented a generic approach for the preparation of specifically modified tRNAs in order to pursue a thorough investigation of the cross-talk between modifications Ψ55, T54 and m1A58 in yeast tRNAPhe. We demonstrated a direct positive and cumulative effect of modifications Ψ55 and T54 on the incorporation of m1A58 in this elongator tRNA. Conversely, we report that m1A58 is efficiently introduced on unmodified initiator tRNAiMet without the need of any prior modification, revealing distinct pathways for m1A58 incorporation in yeast elongator and initiator tRNAs. Finally, we show that the m1A58 single modification has a considerable impact on the structural properties of yeast tRNAiMet. This provides an explanation with structural basis for the degradation of hypomodified tRNAiMet lacking m1A58 by the nuclear surveillance and RTD pathways. Our study has important implications for understanding tRNA modification pathways and in particular for the investigation of modification circuits. These aspects are discussed below.
Genetic approaches are very effective strategies for identifying cross-talk between different genes, and genes encoding modification enzymes are no exception (30). These are however most effective when used in conjunction with biochemical approaches, allowing for a detailed characterization of the molecular aspects contributing to the observed phenotypes. Using specific deletion strains, we previously identified an interdependence between the Ψ55, T54 and m1A58 modifications in yeast tRNAPhe from the observation of a slow-down in the incorporation of certain modifications in the absence of other specific enzymes (39). With a biochemical approach, we now establish that the incorporation of T54 is directly stimulated by Ψ55, and that the incorporation of m1A58 is directly and individually stimulated by Ψ55 and T54, with a notable cumulative effect when they are both present, thus reporting that the effects of the modifications are direct and not the result of other indirect effects. These modification circuits in the T-arm of yeast elongator tRNAs concern modifications T54, Ψ55 and m1A58, which are among the most conserved modified nucleotides in all sequenced tRNAs (58,59). These modifications participate in maintaining the universal tRNA tertiary fold, more precisely at the level of the elbow region, assembled via conserved contacts between the T- and D-loops (57,60). The characterization of this circuit involving modifications of the tRNA core is therefore of general interest for understanding the relation between modifications and structure in tRNAs.
Simple chemical modifications, namely an isomerisation in case of Ψ55, and a methylation in case of T54, can thus render a given tRNA a substantially better substrate for subsequent modification enzymes. The Ψ55  T54
T54  m1A58 and Ψ55
m1A58 and Ψ55  m1A58 modification circuits reported here are robust circuits with highly pronounced effects, with for instance an initial velocity of m1A58 incorporation that is increased by a factor 15 in presence of both Ψ55 and T54 (Table 2). The presence of Ψ55 alone also greatly stimulates the activity of Trm6/Trm61, with a positive effect on m1A58 incorporation that is about two-times larger than the positive effect of T54 (Table 2). This marked effect of Ψ55 leads to undetectable levels of m1A58 along the maturation route of tRNAPhe in pus4Δ yeast extracts monitored by NMR spectroscopy (39). In addition, previous time-resolved NMR study of tRNAPhe in pus4Δ and trm2Δ yeast extracts pointed towards the m1A58 incorporation being more affected by Ψ55 than by T54, which is perfectly in agreement with the kinetic data reported here. This indicates that the time-resolved NMR approach we have developed in cellular extracts (40), is not only reliable to identify cross-talks between modifications, but also to discriminate between weak and strong dependencies.
m1A58 modification circuits reported here are robust circuits with highly pronounced effects, with for instance an initial velocity of m1A58 incorporation that is increased by a factor 15 in presence of both Ψ55 and T54 (Table 2). The presence of Ψ55 alone also greatly stimulates the activity of Trm6/Trm61, with a positive effect on m1A58 incorporation that is about two-times larger than the positive effect of T54 (Table 2). This marked effect of Ψ55 leads to undetectable levels of m1A58 along the maturation route of tRNAPhe in pus4Δ yeast extracts monitored by NMR spectroscopy (39). In addition, previous time-resolved NMR study of tRNAPhe in pus4Δ and trm2Δ yeast extracts pointed towards the m1A58 incorporation being more affected by Ψ55 than by T54, which is perfectly in agreement with the kinetic data reported here. This indicates that the time-resolved NMR approach we have developed in cellular extracts (40), is not only reliable to identify cross-talks between modifications, but also to discriminate between weak and strong dependencies.
The question remains of the molecular origin of the differences in the catalytic efficiencies of Trm6/Trm61 regarding tRNAPhe and tRNAiMet, as well as of the molecular basis of such ordered modification circuit in tRNAPhe. In a circuit of modifications, the observed effect of the initial modification on the subsequent enzyme is reflected in an increased turnover rate, meaning either a better substrate binding, or a better catalytic efficiency, or a better product release, depending on the enzyme considered (61). For RNA modification enzymes, the rate-determining step of the reaction has been reported to be the catalytic step (62,63), the product release (64,65), or conformational changes of both the RNA and protein, most probably to accommodate the target nucleotide into the active site (66). Since yeast Trm6/Trm61 exhibits high structural similarity with its human homolog (67), the structure of human Trm6/Trm61 in complex with tRNALys(UUU) can be examined to understand the tRNA recognition and modification mechanism of Trm6/Trm61 (68). This structure reveals that unfolding of the tRNA tertiary structure is required to allow access to the methylation target A58. In particular, the interactions between the T- and D-loops are disrupted and the D-arm is moved away from its position as a result of interactions with the N-terminal β-barrel domain of Trm61. Nucleotides 55–60 in the T-loop also change their conformation to accommodate the A58 target into the Trm61 active site (68). This structure suggests that a weak interaction between the D- and T-arms would probably lead to a more favorable substrate accommodation for m1A58 modification by Trm6/Trm61. This could explain why unmodified tRNAiMet, where the tRNA elbow is not properly assembled without the m1A58 modification (Figure 5), is efficiently modified by Trm6/Trm61. On the contrary, given the structural properties of the unmodified tRNAPhe (Supplementary Figure S6), this substrate would be less favourably recognized by Trm6/Trm61. Furthermore, in the case of elongator tRNAPhe, the stabilization of the T-arm structure via the modifications T54 and Ψ55 has a positive effect on m1A58 incorporation by Trm6/Trm61 (Figure 2 and Table 1). The comparison of the NMR spectra of Ψ55- and T54-containing tRNAPhe with that of the unmodified tRNAPhe, shows very limited chemical shift variations, suggesting that these modifications do not induce large global rearrangements in the structure of tRNAPhe (39,49). However, since nucleotides 55–60 in the T-loop largely change their conformation upon accommodation of the A58 target into the active site of Trm61 (68), modifications Ψ55 and T54 may directly affect this step of T-loop reorganization. These modifications could indeed lead to local and/or global changes in the dynamic properties of the tRNA substrate. Such a mechanism has recently been reported in the case of E. coli tRNAfMet, in which conformational fluctuations on the local level are increased in the modified tRNA (69). Modifications could thus help reach otherwise inaccessible structural conformations that are more suited to substrate accommodation by the next modification enzyme, which would explain the increased efficiency of m1A58 incorporation in presence of T54 and Ψ55.
Even though modification circuits are widespread and have been reported in several organisms, including S. cerevisiae, S. pombe, E. coli, T. thermophilus, drosophila, human and plants (32–38,70–74), the role of such ordered circuits of modifications remains an open question. For modification circuits in the anticodon-loop region, however, it has been recently proposed that modifications introduced first act as additional recognition elements for the subsequent enzyme. This would provide the means for adding modifications with considerable variation in the anticodon-loop region (31). This hypothesis is quite convincing for modifications in the anticodon-loop region, but cannot explain the actual modification circuit in the T-loop of yeast elongator tRNAs. This modification circuit in the tRNA core indeed involves modifications that are highly conserved. Until recently, modification circuits in the tRNA core region have been only reported in the case of the extremely-thermophilic bacterium T. thermophilus (37,38), and are most likely not implicated in sequential orders of modification incorporation, but rather in a fine tuning of modification levels in relation to an adaptation to variations in growth temperature (75). The Ψ55  T54
T54  m1A58 modification circuit in yeast elongator tRNAs therefore constitutes the first description of an ordered circuit of modification involving modifications from the tRNA core region. Since their identification remains difficult, particularly because real-time monitoring of tRNA maturation at a single nucleotide level is technically challenging (76), we are convinced that modification circuits in the tRNA core region are certainly more widespread than currently thought. Such circuits are likely to be identified in the near future through the use of nanopore sequencing technologies applied to tRNAs (77,78).
m1A58 modification circuit in yeast elongator tRNAs therefore constitutes the first description of an ordered circuit of modification involving modifications from the tRNA core region. Since their identification remains difficult, particularly because real-time monitoring of tRNA maturation at a single nucleotide level is technically challenging (76), we are convinced that modification circuits in the tRNA core region are certainly more widespread than currently thought. Such circuits are likely to be identified in the near future through the use of nanopore sequencing technologies applied to tRNAs (77,78).
One of the most striking features of our study concerns the changes in the structural properties of tRNAiMet upon m1A58 modification. NMR-fingerprints of unmodified tRNAiMet and m1A58-tRNAiMet indeed revealed important structural rearrangements upon addition of a single methyl group. Even though the NMR spectra of unmodified tRNAiMet indicate a certain dynamic that probably leads to intermediate exchange on the NMR chemical shift time scale, the presence at the almost exact same chemical shifts of the imino groups of U42, U50, G68, G70, G12, G24 and G30 in the NMR-fingerprints of unmodified- and m1A58-tRNAiMet attest to the proper secondary structure assembly of this tRNA (Figure 5). All RNA helices, namely the T-, D-, anticodon- and the acceptor-stems, are thus likely correctly assembled. However, the three-dimensional structure of the tRNA is not properly formed as demonstrated by the lack of signals attesting to a properly assembled tRNA elbow structure, namely imino groups of U55 and G18 (Figure 5). These structural rearrangements of yeast tRNAiMet upon m1A58 modification are most probably at the origin of the specific degradation of hypomodified tRNAiMet lacking m1A58 by the nuclear surveillance and RTD pathways (27,28,41,42), while the properly folded m1A58-tRNAiMet is protected from degradation. It is noteworthy that other hypomodified tRNAs lacking at least one tRNA core modification are targeted to degradation by the RTD pathway. In all reported cases, this degradation in the absence of one or two modifications is tRNA specific, meaning that only specific tRNAs are targeted to degradation. For instance, tRNAVal(AAC) lacking m7G46 and m5C49 is rapidly degraded by the RTD pathway in S. cerevisiae (24); tRNASer(CGA) and tRNASer(UGA) lacking either Um44 and ac4C12 or m2,2G26 and m5C48 are also rapidly degraded by the RTD pathway in S. cerevisiae (26,79); tRNATyr(GUA) and tRNAPro(AGG) lacking m7G46 are rapidly degraded by the RTD pathway in S. pombe (80). Comparing these reports with the case of tRNAiMet lacking m1A58, it is tempting to speculate that the modifications involved might be responsible for large structural effects and stabilize the tRNA tertiary structure in these particular cases. The same modifications would, in comparison, not much alter much the structure of non-targeted tRNAs, a hypothesis that would need to be tested experimentally in future structural work.
Another important point revealed by the monitoring of the m1A58 introduction in tRNAiMet in yeast extracts resides in the fact that our NMR-based methodology for monitoring tRNA maturation in cell extracts has the ability to report both on the introduction of chemical modifications, and on structural changes occurring during maturation. This point was not fully appreciated in the NMR study of yeast tRNAPhe, since this tRNA is, to a certain extent, properly folded without modifications (39). Changes in the NMR spectra of tRNAPhe upon modification are modest (39,49), and mainly reflect the incorporation of new chemical groups, with probably also some minor structural rearrangements. The example of tRNAiMet has highlighted that NMR spectroscopy is an ideal method that can report, in a time-resolved fashion, on how the modification process affects tRNA structural properties.
In this work, we have described different modification pathways for m1A58 incorporation in yeast elongator and initiator tRNAs. Unmodified elongator tRNAPhe is an intrinsically poor substrate of Trm6/Trm61, whereas unmodified tRNAiMet is an intrinsically good substrate of the same enzyme. This raises the general question of what makes a good versus a poor substrate for a modification enzyme? To look into this matter, it is important to bear in mind that modifications may not necessarily have the same beneficial effect on all tRNAs (81). For instance, a certain modification may be particularly important for a certain tRNA, which constitutes the evolutionary pressure for retaining this modification enzyme, but might be much less important, if significant at all, in other tRNAs. In this context, dealing with good and poor substrates represents an ordinary challenge faced by modification enzymes. Indeed, tRNAs are to some extent sufficiently similar to be recognized and employed by the translation machinery, but need at the same time to be sufficiently different to be uniquely recognized by their cognate aminoacyl-tRNA synthetases. The modification enzymes therefore should handle a population of highly similar but unique tRNAs and the tRNA modification patterns can be regarded as the result of millions of years of coevolution of modification enzymes with the tRNA population (11,82). In this context, we believe that a potential role of modification circuits could be to allow the modification of both good and poor tRNA substrates. In the case of yeast elongator and initiator tRNAs, which must have sufficiently different structural properties to be recognized by elongation or initiation factors, the existence of the Ψ55  T54
T54  m1A58 modification circuit enables the incorporation of m1A58 in certain elongator tRNAs, such as tRNAPhe, which are poor intrinsic substrate of Trm6/Trm61. In conclusion, modification circuits might be a solution found to deal with the problem of having at the same time poor and good tRNA substrates that all require to be eventually modified.
m1A58 modification circuit enables the incorporation of m1A58 in certain elongator tRNAs, such as tRNAPhe, which are poor intrinsic substrate of Trm6/Trm61. In conclusion, modification circuits might be a solution found to deal with the problem of having at the same time poor and good tRNA substrates that all require to be eventually modified.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Josette Banroques for providing the yeast YKO kanMX strains, Grégory Boël for providing the E. coli K-12 yggH::kan, Julien Henri for providing the pRSFDuet-Smt3 vector, Sylvie Auxilien and Yuri Motorin for providing the pET28-Trm4 expression plasmid, Alexandre Gato for preliminary data acquisition, Christel Le Bon for ensuring the best performance of the NMR infrastructure at the IBPC, and Jacqueline Plumbridge for careful reading of the manuscript. The authors acknowledge access to the biomolecular NMR platform of the IBPC that is supported by the CNRS, the Labex DYNAMO (ANR-11-LABX-0011), the Equipex CACSICE (ANR-11-EQPX-0008) and the Conseil Régional d'Île-de-France (SESAME grant). M.-J.Y. is supported by a Q-Life PhD fellowship (Q-life ANR-17-CONV-0005). In addition, this work was funded by the ANR JCJC CiMoDyMo (ANR-19-CE44-0013), the Deutsche Forschungsgemeinschaft (255344185-SPP 1784) and the Horizon 2020 program (ID-952373).
Author contributions: P.B. conceived the study. P.B. cloned the constructs for Pus4, Trm2 and Trm6/Trm61 overexpression and constructed the E. coli BL21 yggH::kan strain. M.-J.Y. purified all modification enzymes with support from M.C. and under the supervision of P.B. and C.T.; M.-J.Y. prepared tRNA samples for NMR and kinetic assays; M.-J.Y. performed and analyzed the enzymatic assays; M.-J.Y. prepared total tRNA samples for MS; Y.Y. isolated and digested tRNAiMet for MS; Y.Y. measured and analyzed MS data under the supervision of S.K.; M.-J.Y. and P.B. measured and analyzed the NMR spectra. M.-J.Y. and P.B. wrote the manuscript.
Contributor Information
Marcel-Joseph Yared, Expression génétique microbienne, Université Paris Cité, CNRS, Institut de biologie physico-chimique, Paris, France.
Yasemin Yoluç, Department of Chemistry, Ludwig Maximilians University, Munich, Germany.
Marjorie Catala, Expression génétique microbienne, Université Paris Cité, CNRS, Institut de biologie physico-chimique, Paris, France.
Carine Tisné, Expression génétique microbienne, Université Paris Cité, CNRS, Institut de biologie physico-chimique, Paris, France.
Stefanie Kaiser, Department of Chemistry, Ludwig Maximilians University, Munich, Germany; Institute of Pharmaceutical Chemistry, Goethe-University, Frankfurt, Germany.
Pierre Barraud, Expression génétique microbienne, Université Paris Cité, CNRS, Institut de biologie physico-chimique, Paris, France.
DATA AVAILABILITY
The data underlying this article will be shared on reasonable request to the corresponding author.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Conseil Régional d'Île-de-France; Deutsche Forschungsgemeinschaft [255344185-SPP]; Horizon 2020 Framework Programme [952373]; Centre National de la Recherche Scientifique; Agence Nationale de la Recherche [ANR-11-EQPX-0008, ANR-11-LABX-0011, ANR-17-CONV-0005, ANR-19-CE44-0013]. Funding for open access charge: ANR grant [ANR-19-CE44-0013].
Conflict of interest statement. None declared.
REFERENCES
- 1. Katz A., Elgamal S., Rajkovic A., Ibba M.. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol. Microbiol. 2016; 101:545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fields R.N., Roy H.. Deciphering the tRNA-dependent lipid aminoacylation systems in bacteria: novel components and structural advances. RNA Biol. 2018; 15:480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oberbauer V., Schaefer M.R.. tRNA-derived small rnas: biogenesis, modification, function and potential impact on Human disease development. Genes (Basel). 2018; 9:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su Z., Wilson B., Kumar P., Dutta A.. Noncanonical roles of tRNAs: TRNA fragments and beyond. Annu. Rev. Genet. 2020; 54:47–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hopper A.K. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013; 194:43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shepherd J., Ibba M.. Bacterial transfer rnas. FEMS Microbiol. Rev. 2015; 39:280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jarrous N., Mani D., Ramanathan A.. 2022) Coordination of transcription and processing of tRNA. FEBS J. 289:3630–3641. [DOI] [PubMed] [Google Scholar]
- 8. Phizicky E.M., Hopper A.K.. The life and times of a tRNA. RNA. 2023; 29:898–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Yacoubi B., Bailly M., de Crécy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer rnas. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 10. Jackman J.E., Alfonzo J.D.. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA. 2013; 4:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barraud P., Tisné C.. To be or not to be modified: miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life. 2019; 71:1126–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Crécy-Lagard V., Jaroch M.. Functions of bacterial tRNA modifications: from ubiquity to diversity. Trends Microbiol. 2021; 29:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boccaletto P., Stefaniak F., Ray A., Cappannini A., Mukherjee S., Purta E., Kurkowska M., Shirvanizadeh N., Destefanis E., Groza P.et al.. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022; 50:D231–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tuorto F., Lyko F.. Genome recoding by tRNA modifications. Open Biol. 2016; 6:160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helm M., Alfonzo J.D.. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014; 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ranjan N., Leidel S.A.. The epitranscriptome in translation regulation: MRNA and tRNA modifications as the two sides of the same coin?. FEBS Lett. 2019; 593:1483–1493. [DOI] [PubMed] [Google Scholar]
- 18. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 19. Väre V.Y.P., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F.. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lorenz C., Lünse C.E., Mörl M.. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biela A., Hammermeister A., Kaczmarczyk I., Walczak M., Koziej L., Lin T.-Y., Glatt S.. The diverse structural modes of tRNA binding and recognition. J. Biol. Chem. 2023; 299:104966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Helm M., Giegé R., Florentz C.. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999; 38:13338–13346. [DOI] [PubMed] [Google Scholar]
- 23. Urbonavicius J., Armengaud J., Grosjean H.. Identity elements required for enzymatic formation of N2,N2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J. Mol. Biol. 2006; 357:387–399. [DOI] [PubMed] [Google Scholar]
- 24. Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M.. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006; 21:87–96. [DOI] [PubMed] [Google Scholar]
- 25. Chernyakov I., Whipple J.M., Kotelawala L., Grayhack E.J., Phizicky E.M.. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev. 2008; 22:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dewe J.M., Whipple J.M., Chernyakov I., Jaramillo L.N., Phizicky E.M.. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012; 18:1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kadaba S., Krueger A., Trice T., Krecic A.M., Hinnebusch A.G., Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004; 18:1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kadaba S., Wang X., Anderson J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006; 12:508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gudipati R.K., Xu Z., Lebreton A., Séraphin B., Steinmetz L.M., Jacquier A., Libri D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell. 2012; 48:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sokołowski M., Klassen R., Bruch A., Schaffrath R., Glatt S.. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 2018; 1861:409–418. [DOI] [PubMed] [Google Scholar]
- 31. Han L., Phizicky E.M.. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018; 24:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou M., Long T., Fang Z.-P., Zhou X.-L., Liu R.-J., Wang E.-D.. Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2'-O-methyltransferase. RNA Biol. 2015; 12:900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guy M.P., Phizicky E.M.. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA. 2015; 21:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johannsson S., Neumann P., Wulf A., Welp L.M., Gerber H.-D., Krull M., Diederichsen U., Urlaub H., Ficner R.. Structural insights into the stimulation of S. pombe Dnmt2 catalytic efficiency by the tRNA nucleoside queuosine. Sci. Rep. 2018; 8:8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han L., Marcus E., D'Silva S., Phizicky E.M. S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA. 2017; 23:406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J., Wang Y.-N., Xu B.-S., Liu Y.-P., Zhou M., Long T., Li H., Dong H., Nie Y., Chen P.R.et al.. Intellectual disability-associated gene ftsj1 is responsible for 2'-O-methylation of specific tRNAs. EMBO Rep. 2020; 21:e50095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomikawa C., Yokogawa T., Kanai T., Hori H.. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010; 38:942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., Iwashita C., Hori H.. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011; 39:2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barraud P., Gato A., Heiss M., Catala M., Kellner S., Tisné C.. Time-resolved NMR monitoring of tRNA maturation. Nat. Commun. 2019; 10:3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gato A., Catala M., Tisné C., Barraud P.. A method to monitor the introduction of posttranscriptional modifications in tRNAs with NMR spectroscopy. Methods Mol. Biol. 2021; 2298:307–323. [DOI] [PubMed] [Google Scholar]
- 41. Ozanick S.G., Wang X., Costanzo M., Brost R.L., Boone C., Anderson J.T. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate rnas in Saccharomyces cerevisiae. Nucleic Acids Res. 2009; 37:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tasak M., Phizicky E.M.. Initiator tRNA lacking 1-methyladenosine is targeted by the rapid tRNA decay pathway in evolutionarily distant yeast species. PLoS Genet. 2022; 18:e1010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basavappa R., Sigler P.B.. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991; 10:3105–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomason L.C., Costantino N., Court D.L.. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007; 79:1.17.1–1.17.8. [DOI] [PubMed] [Google Scholar]
- 46. Catala M., Gato A., Tisné C., Barraud P.. Preparation of yeast tRNA sample for NMR spectroscopy. Bio Protoc. 2020; 10:e3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Plateau P., Gueron M.. Exchangeable proton NMR without base-line distorsion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982; 104:7310–7311. [Google Scholar]
- 48. Sklenar V., Bax A.. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J Mag. Res. 1987; 74:469–479. [Google Scholar]
- 49. Catala M., Gato A., Tisné C., Barraud P.. 1H, 15N chemical shift assignments of the imino groups of yeast tRNAPhe: influence of the post-transcriptional modifications. Biomol. NMR Assign. 2020; 14:169–174. [DOI] [PubMed] [Google Scholar]
- 50. Farjon J., Boisbouvier J., Schanda P., Pardi A., Simorre J.-P., Brutscher B.. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J. Am. Chem. Soc. 2009; 131:8571–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freund J., Kalbitzer H.R.. Physiological buffers for NMR spectroscopy. J. Biomol. NMR. 1995; 5:321–322. [DOI] [PubMed] [Google Scholar]
- 52. Chionh Y.H., Ho C.-H., Pruksakorn D., Babu R., I. N., C. S., Hia F., McBee M.E., Su D., Pang Y.L.J.et al.. A multidimensional platform for the purification of non-coding RNA species. Nucleic Acids Res. 2013; 41:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson J., Phan L., Hinnebusch A.G.. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Borland K., Diesend J., Ito-Kureha T., Heissmeyer V., Hammann C., Buck A.H., Michalakis S., Kellner S.. Production and application of stable isotope-labeled internal standards for RNA modification analysis. Genes (Basel). 2019; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anderson J., Phan L., Cuesta R., Carlson B.A., Pak M., Asano K., Björk G.R., Tamame M., Hinnebusch A.G.. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998; 12:3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Calvo O., Cuesta R., Anderson J., Gutiérrez N., García-Barrio M.T., Hinnebusch A.G., Tamame M.. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J., Ferré-D’Amaré A.R.. The tRNA elbow in structure, recognition and evolution. Life (Basel). 2016; 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grosjean H., Sprinzl M., Steinberg S.. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995; 77:139–141. [DOI] [PubMed] [Google Scholar]
- 59. Machnicka M.A., Olchowik A., Grosjean H., Bujnicki J.M.. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014; 11:1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roovers M., Droogmans L., Grosjean H.. Post-transcriptional modifications of conserved nucleotides in the T-loop of tRNA: a tale of functional convergent evolution. Genes (Basel). 2021; 12:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boschi-Muller S., Motorin Y.. Chemistry enters nucleic acids biology: enzymatic mechanisms of RNA modification. Biochemistry. 2013; 78:1392–1404. [DOI] [PubMed] [Google Scholar]
- 62. Wright J.R., Keffer-Wilkes L.C., Dobing S.R., Kothe U.. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA. 2011; 17:2074–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schultz S.K., Meadows K., Kothe U.. Molecular mechanism of tRNA binding by the Escherichia coli N7 guanosine methyltransferase TrmB. J. Biol. Chem. 2023; 299–104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Christian T., Lahoud G., Liu C., Hou Y.-M.. Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J. Mol. Biol. 2010; 400:204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garcia G.A., Chervin S.M., Kittendorf J.D.. Identification of the rate-determining step of tRNA-guanine transglycosylase from Escherichia coli. Biochemistry. 2009; 48:11243–11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krishnamohan A., Dodbele S., Jackman J.E.. Insights into catalytic and tRNA recognition mechanism of the dual-specific tRNA methyltransferase from Thermococcus kodakarensis. Genes (Basel). 2019; 10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang M., Zhu Y., Wang C., Fan X., Jiang X., Ebrahimi M., Qiao Z., Niu L., Teng M., Li X.. Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae. Sci. Rep. 2016; 6:32562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Finer-Moore J., Czudnochowski N., O’Connell 3rd, J.D., Wang A.L., Stroud R.M.. Crystal structure of the Human tRNA m(1)A58 methyltransferase-tRNA(3)(Lys) complex: refolding of substrate tRNA allows access to the methylation target. J. Mol. Biol. 2015; 427:3862–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Biedenbänder T., de Jesus V., Schmidt-Dengler M., Helm M., Corzilius B., Fürtig B.. RNA modifications stabilize the tertiary structure of tRNAfMet by locally increasing conformational dynamics. Nucleic Acids Res. 2022; 50:2334–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arimbasseri A.G., Iben J., Wei F.-Y., Rijal K., Tomizawa K., Hafner M., Maraia R.J.. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA. 2016; 22:1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Benítez-Páez A., Villarroya M., Douthwaite S., Gabaldón T., Armengod M.-E.. YibK is the 2'-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA.(Leu) isoacceptors. RNA. 2010; 16:2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Müller M., Hartmann M., Schuster I., Bender S., Thüring K.L., Helm M., Katze J.R., Nellen W., Lyko F., Ehrenhofer-Murray A.E.. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015; 43:10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guy M.P., Shaw M., Weiner C.L., Hobson L., Stark Z., Rose K., Kalscheuer V.M., Gecz J., Phizicky E.M.. Defects in tRNA anticodon loop 2'-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 2015; 36:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.-L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H.. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006; 311:395–398. [DOI] [PubMed] [Google Scholar]
- 75. Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014; 5:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Helm M., Motorin Y.. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017; 18:275–291. [DOI] [PubMed] [Google Scholar]
- 77. Thomas N.K., Poodari V.C., Jain M., Olsen H.E., Akeson M., Abu-Shumays R.L.. Direct nanopore sequencing of individual full length tRNA strands. ACS Nano. 2021; 15:16642–16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lucas M.C., Pryszcz L.P., Medina R., Milenkovic I., Camacho N., Marchand V., Motorin Y., Ribas de Pouplana L., Novoa E.M.. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 2023; 10.1038/s41587-023-01743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kotelawala L., Grayhack E.J., Phizicky E.M.. Identification of yeast tRNA um(44) 2'-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNA(Ser) species. RNA. 2008; 14:158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. De Zoysa T., Phizicky E.M.. Hypomodified tRNA in evolutionarily distant yeasts can trigger rapid tRNA decay to activate the general amino acid control response, but with different consequences. PLoS Genet. 2020; 16:e1008893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Phizicky E.M., Alfonzo J.D.. Do all modifications benefit all tRNAs?. FEBS Lett. 2010; 584:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grosjean H., de Crécy-Lagard V., Marck C.. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010; 584:252–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.