Abstract
Modular cloning has become a benchmark technology in synthetic biology. However, a notable disparity exists between its remarkable development and the need for standardization to facilitate seamless interoperability among systems. The field is thus impeded by an overwhelming proliferation of organism-specific systems that frequently lack compatibility. To overcome these issues, we present Golden Standard (GS), a Type IIS assembly method underpinned by the Standard European Vector Architecture. GS unlocks modular cloning applications for most bacteria, and delivers combinatorial multi-part assembly to create genetic circuits of up to twenty transcription units (TUs). Reliance on MoClo syntax renders GS fully compatible with many existing tools and it sets the path towards efficient reusability of available part libraries and assembled TUs. GS was validated in terms of DNA assembly, portability, interoperability and phenotype engineering in α-, β-, γ- and δ-proteobacteria. Furthermore, we provide a computational pipeline for parts characterization that was used to assess the performance of GS parts. To promote community-driven development of GS, we provide a dedicated web-portal including a repository of parts, vectors, and Wizard and Setup tools that guide users in designing constructs. Overall, GS establishes an open, standardized framework propelling the progress of synthetic biology as a whole.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Synthetic biology is a research field in continuous expansion and increased opportunities for new industrial processes (1). Its focuses on the creation of new genetic solutions to control cellular processes, which is achieved through rational design of transcription units (TUs) and, by extension, complex biological pathways and genetic circuits (2). The modular and hierarchical nature of biological designs reveals new possibilities for the development of rational and standardized mechanisms in order to improve the engineering process of specific biological solutions.
DNA assembly cloning is a widely used method to build synthetic genetic circuits. Traditionally, these cloning strategies were performed by digestion and ligation of DNA fragments (from genomic, plasmid or PCR origin), using BioBricks (3) or Gibson assembly (4). However, these approaches require specific designs for each step that can hamper complete standardization and parts reuse (3,4). Golden Gate cloning is a methodology based on Type IIS restriction enzymes and has emerged as a powerful tool for standardizing the assembly of genetic parts (5). A popular and standardized Golden Gate method, called Modular Cloning (MoClo) was first developed to work with plants, supporting fractionation of TUs into their basic parts (or level 0 parts): promoters, ribosome binding sites (RBS), coding sequences (CDS), terminators, etc. These modular elements can be further assembled into synthetic TUs (level 1 constructs), and more complex structures, such as genetic circuits (level 2 constructs) (6). To accomplish this, MoClo assembly combines Type IIS restriction enzymes and T4 DNA ligase in a one-pot reaction, thus making it possible to digest and assemble several DNA fragments simultaneously. Overall, Type IIS restriction enzymes are capable of cleaving any sequence outside of their recognition site, which opens up the possibility of designing compatible overhang ends (fusion sites) (7). Therefore, by placing the Type IIS enzyme recognition sites flanking level 0 parts it is possible to digest multiple DNA fragments while simultaneously assembling new DNA fragments in a directional, defined orientation to form level 1 TUs (8). The main DNA manipulation which is required when using MoClo technology is known as ‘domestication’ of DNA parts and host vectors through the elimination of relevant Type IIS restriction enzyme sites. This simple process can be performed through PCR or by using chemically synthesized DNA parts.
The efficiency of MoClo technology as a cloning method for engineering genetic constructs has been demonstrated in a variety of diverse organisms, including bacteria (9–14), yeasts (15), plants (8,16) and mammalian cells (17) (Table 1). In addition, the use of MoClo as a cloning system has seen an increase in the iGEM competition (13). Most of the MoClo systems use Type IIS enzymes that generate 4-nucleotide overhang fusion sites. Based on the fusion sites for level 0 parts described by Engler (8), BsaI has been established as the reference restriction enzyme for level 0. This common syntax and structure facilitates the construction of reusable and interchangeable part libraries and the development of standard protocols (18). This standardization effort has not prevented the appearance of alternative fusion site designs, thus increasing the variability of MoClo solutions. However, this increased scope has precluded full compatibility of the available libraries of parts and TUs categorized as level 0 and level 1, respectively. This incompatibility has also been augmented in some cases by the introduction of Type IIS enzymes that generate 3-nt overhangs (such as SapI) in the DNA parts, during the construction of TUs (11), and synthetic circuits (16).
Table 1.
MoClo systems and microorganisms when have been experimentally tested
| Name | Organism | Compatible with SEVA | Parts compatible with GS | Max. parts assembled in TUs | Max. TUs | Ref. |
|---|---|---|---|---|---|---|
| MoClo | Plants | No | Yes | 8 (Prom, RBS, N-tag x2, CDS, C-tag x2, Ter) | 17 | (6,8,19) |
| Golden Braid | Plants, Fungi, Animalia | No | Yes | 11 (Prom, RBS, N-tag, CDS, C-tag, linker x5, Ter) | N.D.a | (20,21) |
| CIDAR | E. coli | No | Yes | 4 (Prom, RBS, CDS, Ter) | 4 | (9) |
| EcoFlex | E. coli | No | No | 5 (Prom, RBS, N-tag, CDS, Ter) | 20 | (10) |
| Mobius assembly | E. coli | No | Yes | 10 (Prom, RBS, N-tag, CDS, C-tag, linker x4, Ter) | N.D.a | (22) |
| Midas | Fungi | No | Yes | 3 (Prom, CDS, Ter) | N.D.a | (15) |
| Start-Stop assembly | E. coli | No | No | 4 (Prom, RBS, CDS, Ter) | 15 | (11) |
| Cyano Gate | Cyanobacteria | No | Yes | 7 (UpNeutral sites x2, Prom, CDS, Ter, DownNeutral sites x2) | 4 | (12) |
| Multi-Kingdom | Fungi, Eubacteria, Protista, Plants, Animalia | Yes | No | 6 (Prom, RBS, N-tag, CDS, Ter, linker) | 5 | (23) |
| uLoop | Diatoms, yeast, plants, bacteria | No | Yes | 4 (Prom, RBS, CDS, Ter) | N.D.a | (24) |
| JUMP | E. coli and Pseudomonas | Yes | Yes | 6 (Prom, RBS, N-tag, CDS, C-tag, Ter) | N.D.a | (25) |
| The Marburg Collection | V. natriegens | No | Yes | 6 (Prom, RBS, N-tag, CDS, C-tag, Ter) | 25 | (13) |
| SEVAtile | Pseudomonas | Yes | No | 6 (Prom, RBS, CDS, Ter, RBS, Ter) | 1 | (14) |
| Golden Standard | Proteobacteria and Actinomyces | Yes | N.A.b | 8 (Prom, RBS, N-tag x2, CDS, C-tag x2, Ter) | 20 | This work, (26) |
Abbreviations: Prom, promoter; RBS, ribosome binding site; N-tag, N-terminal tag; CDS, coding sequence; C-tag, C-terminal tag; Ter, terminator.
aN.D.: not defined.
bN.A: not applied.
Despite most of the available Golden Gate systems have been specifically designed to engineer single species, new systems targeting more than one cellular host have recently been developed. In this sense, it is worth highlighting the JUMP (25), Multi-Kingdom (23) and SEVAtile (14) assembly systems (Table 1). JUMP assembly introduces a MoClo system for prokaryotic cells based partially on the architecture of SEVA vectors, thus increasing its portability. Multi-Kingdom proposes a universal system that supports construction of synthetic expression systems for both prokaryotes and eukaryotes. However, despite the fact that it uses a digestion/assembly system based on the alternation of Type IIS BsaI and BpiI enzymes, it is designed to use a unique syntax (e.g. exclusive fusion sites), thus reducing its operational standardization by limiting the possibility to directly exchange parts between different MoClo systems. SEVAtile assembly also proposes a MoClo system based on the architecture of SEVA vectors. However, besides only being able to construct a single transcription unit with no possibility of including tags, it is limited by the impossibility to share parts due to the unique syntax used by this system.
Overall, a recurrent limitation and unsolved bottleneck in available MoClo systems is the absence of a meaningful community-guided development effort supporting individual users’ contributions to expand the universe of available DNA parts, receptor vectors or even novel applications. Furthermore, there is also a lack of intuitive computational frameworks to implement assembly of target TUs while providing the necessary rational, standardized and quantitative support to build complex metabolic pathways. Plant and cyanobacteria-based systems such as GoldenBraid (20) and CyanoGate (12) were designed with an eye on partially fulfilling these goals. However, they are clade or phyla-specific systems. Further efforts are needed to develop similar systems that are able to cover a broad range of organisms.
Despite these significant progress done in term of standardization of the physical assembly of genetic devices, important challenges arise when it comes to characterizing biological parts (such as promoters, RBSs, terminators) and complex devices (such as inducible expression systems and intricate circuits). This is because the behaviour of genetic parts ultimately depends on multiple extrinsic (environmental) and intrinsic (host) variables that influence their performance (27). Extrinsic variables, such as temperature and/or carbon source cause differences regarding absolute activities while maintaining its relative behaviour (28). However host-dependent variables are much more unpredictable (29). Compounding the problem, there is a lack of widely accepted methods for parts characterization, and often the methods employed are inaccessible, highly subjective, and have low reproducibility. As a result, the behaviour of a single part can vary from one laboratory to another, greatly hindering predictability. Consequently, there is a need for concerted efforts towards standardizing and improving the accessibility of part characterization methods.
Here, we present Golden Standard (GS), a standardized modular cloning technology that unifies and harmonizes available methods by extending the MoClo approach to a wide range of bacteria. This allows portability of the resulting final vectors for use in diverse microorganisms. Golden Standard adheres to SEVA standards, allowing not only modularity in the assembly process, but also modularization of the host vectors, thus resulting in a significant scope expansion. We provide additionally an objective and user-friendly computational workflow for the proper characterization of DNA parts. In order to facilitate widespread adoption and future community-driven development of GS, a web portal featuring the GS database, protocols and tutorials is available to make GS accessible to non-experts in modular cloning (http://sysbiol.cnb.csic.es/GoldenStandard/home.php).
MATERIALS AND METHODS
Additional materials and methods are available in the Supplementary Information SI pdf file.
Bacterial strains and plasmid construction
Escherichia coli DH5α and DH10B were used for all DNA assembly, unless otherwise stated while E. coli W (ATCC 9637) was used for GS characterization. E. coli strains were routinely grown on LB agar or in LB broth with shaking at 170 rpm at 37°C. Pseudomonas putida KT2440, Agrobacterium tumefaciens DSM 30150, Sphingobium sp. SYK-6, Cupriavidus metallidurans CH34 and Parazoarcus communis were grown on LB agar or in LB broth with shaking at 170 rpm at 30°C. Geobacter sulfurreducens DSM 12127 was grown anaerobically in MC medium (30) supplemented with fumarate (40 mM), acetate (15 mM), sodium selenate (100 μM), yeast extract (0.001%) and cysteine (5 mM) at 30°C. Media was supplemented with ampicillin (100 μg·ml−1), kanamycin (50 μg·ml−1, 250 μg·ml−1 for Geobacter sulfurreducens and 1250 μg·ml−1 for Cupriavidus metallidurans CH34), gentamicin (10 μg·ml−1) or streptomycin (75 μg·ml−1) as required. For PHB production in E. coli, LB was supplemented with glucose (10 g·L−1) and inoculated at OD600 0.3 from an overnight culture of OD600 ∼3. These cultures were grown for 24 h at 37°C and 200 rpm. Growth was monitored by measuring optical density at 600 nm (OD600) using a portable spectrophotometer (ThermoFisher Scientific). The level 1 host vectors were constructed by standard cloning methods (Supplementary Table S1). Briefly, MoClo fragments were amplified by PCR using primers with BpiI and BsaI sites and using the reporter LacZα as DNA template (Supplementary Table S4). The PCR product was blunt ligated into pSEVA281, pSEVA23g1 and pSEVA221 plasmids digested with SmaI to divide the multicloning site into a prefix (PacI-SmaI) and a suffix (BamHI-SpeI). To remove a BpiI site from the pBBR1 origin of replication, pBBR1 was amplified in two fragments using the oligo pairs 5pBBR1_1–3pBBR1_1 and 5pBBR1_2–3pBBR1_2 that include overlapping regions with a single nucleotide mutation to eliminate the BpiI site. The two PCR products then served as a template for overlap extension PCR to generate a fragment containing the domesticated origin of replication (3 g oriC in SEVA nomenclature). This fragment was used as pBBR1 origin of replication. Level 2 and level 3 host vectors were constructed following the same method used for the level 1 vectors (Supplementary Table S1).
Construction of level 0 parts
Fragments were amplified by PCR, oligonucleotide annealing or by chemical synthesis while introducing silent mutations (when appropriate) to eliminate restriction enzyme sites used in the assembly system. If subsequent traditional restriction enzyme cloning is planned, restriction sites present in the SEVA backbone were also removed. Further details can be found in the supplementary materials and methods.
Modular assembly protocol
Assembly reactions were set up using 40 fmol of each donor plasmid part, 200 U T4 ligase (NEB), 10 U BsaI-HFv2 (NEB) for assembly into levels 1 and 3 or 10 U BpiI (Thermo Scientific) for assembly into levels 2 in 20 μl 1× T4 ligase buffer (NEB). The reaction condition was: 28 cycles of 37°C 1.5 min plus 16°C 3 min, then 50°C 5 min, 37°C 5 min and 80°C 10 min. Finally, E. coli DH5α or DH10B were transformed with the mix reaction and plated on LB agar with antibiotic plus 40 μg/ml X-Gal and incubated at 37°C overnight. White colonies were selected and screened by DNA sequencing.
Proteobacteria transformation by electroporation
2 mm gap electroporation cuvette and 200 ng of reporter plasmids were used in all the electroporations. Sphingobium sp. strain SYK-6, Agrobacterium tumefaciens, Parazoarcus communis, Escherichia coli W and Pseudomonas putida were transformed following the Choi et al. protocol (31). Cupriavidus metallidurans CH34, Geobacter sulfurreducens were transformed following the Coppi et al. protocol (32) with modifications. Briefly, cells were washed 3 times with electroporation buffer (1 mM HEPES [pH 7], 1 mM MgCl2, 175 mM sucrose) and resuspended in the same buffer with 100 μl. After applying a pulse (2.5 kV on an Eppendorf Eporator). Cupriavidus cells were incubated for 2 h while Geobacter cells were incubated for 5 h to recovering.
Polyhydroxybutyrate (PHB) quantification and monomer composition
PHB monomer composition and PHB content were determined by gas chromatography–mass spectrometry (GC–MS) of the methanolysed polyester (33) (see supplementary material and methods for details).
Analysis of expression of level 1 N- and C-tagged OleD glucosyltransferase
E. coli were electroporated with a series of level 1 OleD GT vectors with different N-terminal purification tags and/or C-terminal GFP (see supplementary information, Table S2). Isolated clones were used to inoculate 1 ml (mixed assembly) or 25 ml (individual assemblies) of LB broth supplemented with kanamycin and cultured overnight at 37°C with shaking at 120 rpm. Cultures were subsequently harvested at 4000 × g for 30 min and cell pellets were resuspended in 0.5 ml (mix) or 5 ml (individual) of lysis buffer (30 mM Na phosphate pH 8.0, 300 μg/ml lysozyme), sonicated (2.5 min, amplitude 80%, 5 s pulse and 5 s pause) and centrifuged 20 000 × g for 30 min at 4°C. The resulting supernatant was directly used in further analysis of protein concentration (individual assemblies), fluorescence, glucosyltransferase activity, SDS-PAGE (individual assemblies) and protein purification (individual assemblies, N-His6x version only). Protein concentration was evaluated by Bradford assay using BSA as the reference for calibration (34). Fluorescence was measured using a Cary Eclipse Varian fluorimeter with unmodified OleD cell free extract (CFE) as a reference. Glucosyltransferase relative activity was evaluated via quick HPLC assay (for details of sample preparation, product and analytical procedure see supplementary information), using 100 μL of CFE in a sodium phosphate buffer (pH 8.0, 30 mM) supplemented with 0.05 mM xanthohumol (10 μl of DMSO stock solution) as glucose acceptor and 0.1 mM UDP-Glucose as glucose donor in a final reaction volume of 1 ml for 20 min at 30°C and 800 rpm using a Thermoshaker (Eppendorf). CFE used to test individual assemblies were diluted to an exact protein concentration of 50 μg/ml. IMAC protein purification was performed on a GE Healthcare His SpinTrap column following the manufacturer's instructions. SDS-PAGE was performed using standard conditions with Laemmli denaturing buffer, precast TGX gels (Bio-Rad) and Broad Range Protein Marker (New England Biolabs).
Expression and purification of level 1 2xN- and 2xC-tagged protein
P. putida was electroporated with Lv1 pSEVA23g19g1-His-RFP-GST-HRV3C-GFP-Strep vector (see supplementary information, Table S3). One ‘red’ isolated clone was used to inoculate 300 ml of LB broth supplemented with kanamycin and cultured for 24 h at 30°C with shaking at 120 rpm. Culture was harvested at 4000 × g for 30 min and cell pellet was resuspended in 30 ml of standard His-tag binding buffer with lysozyme (300 μg/ml lysozyme), sonicated (2.5 min, amplitude 80%, 5 s pulse and 5 s pause) on ice and centrifuged 20 000 × g for 30 min at 4°C. The supernatant was directly used for IMAC protein purification using 5 ml His-trap column (GE Healthcare, UK) following the standard product protocol. 10 ul of HRV-3C protease (Sigma-Aldrich) was added to 12 ml of combined eluted fractions that ‘glow’ under UV light, and left overnight at 4°C for digestion. Resulting mixture was directly used in another round of protein purification using 5 ml Strep-Trap column (GE Healthcare, UK) following the manufacturer's instructions. SDS-PAGE was performed using standard conditions with Laemmli denaturing buffer, 12% polyacrylamide gel, Coomassie staining and Precision Plus Protein Marker (Bio-Rad).
Parts characterization and analysis
Plasmids used are listed in Supplementary Table S3. Each plasmid was introduced into E. coli W and P. putida by electroporation except for the inducible promoter plasmids, which were introduced in P. putida by triparental mating (using E. coli HB101 harbouring pRK600 plasmid as the mating helper) and selected on cetrimide-agar. Recombinant strains were grown in LB agar plates, followed by overnight precultures of 10 ml M9 minimal medium (35) supplemented with 34 nM EDTA and using 0.2 % (w/v) glucose as carbon source (M9 + Glu). Overnight precultures were diluted to 0.2 OD600 and grown in 20 ml of M9 + Glu until mid-exponential phase (0.6–0.8 OD600). Cells were collected, washed with saline solution (NaCl 0.85%) and adjusted to 0.2 OD600 in fresh M9 media with 0.2 % (w/v) succinate as C-source (M9 + Suc). 200 μl of these cultures were grown in 96-well plates (black, clear flat-bottom from Greiner Bio-One) for 24 h (P. putida) or 48 h (E. coli W) in a Victor NiVo plate reader (PerkinElmer, MA, USA) recording OD600 and fluorescence each 30 min with an orbital shaking step each 15 min. 580 and 625 nm filters were used for excitation and emission of mRFP1, respectively, with 500 ms exposure. 480 and 530 nm filters were used for GFPmut3, with 100 ms exposure.
For inducible promoters, 100 μl of adjusted 0.4 OD600 cultures were diluted with another 100 μl of M9 + Suc supplemented with adequate concentration of inducers. Stock solutions of the inducers Isopropyl ß-d-1-thiogalactopyranoside (IPTG), l-rhamnose and m-toluic acid were prepared in H2O at concentrations of 1 M or 0.5 M for m-toluic acid.
Part activity was computed using Python (v3.8.5) custom scripts available at (https://github.com/SBGlab/GoldenStandardDataset) together with the datasets obtained. Briefly, for each experiment raw absorbance data was normalized by subtracting the mean of at least 3 wells containing media only. Then, the experimental data points defining the time window of two doubling times at maximal growth rate were computed. Part activity was defined as the average of (dRFP/dt)/OD within the calculated window. The average activity resulting from control strains was subtracted for background auto-fluorescence normalization. For terminators, the ratio between the activity obtained for GFP and RFP is reported. For inducible promoters, obtained activities were fit to Hill equation using GraphPad Prism software v7.04 (San Diego, CA, USA). A detailed description of part activity computations can be found in the supplementary materials and methods.
RESULTS
Golden Standard features, transcription units and circuit assemblies
GS is a flexible synthetic biology tool for modular cloning based on PhytoBricks parts (6). In addition, GS adopts the standardized vector structure provided by SEVA (36). As with other Golden Gate-based DNA assembly systems, GS is based on the hierarchical assembly of functional DNA parts into increasingly complex genetic circuits using receptor vector levels 0 to 3.
To build the basic level 0 parts (promoters, RBS, CDS, terminators, N-, and C-terminal tags), DNA fragments can be created by PCR amplification, oligonucleotide annealing, or by chemical synthesis. Irrespective of the method used to generate the DNA fragment, the sequence must be domesticated in order to eliminate BsaI and BpiI recognition sites. It is also strongly recommended to eliminate the restriction sites that define the different modules of the SEVA vectors (37) if further traditional cloning steps are envisioned. GS level 0 parts were cloned into the acceptor vector pSEVA182 linearized with SmaI to facilitate blue-white selection of positive clones. Furthermore, parts from other MoClo systems with the same fusion site syntaxes can be used directly as GS parts, including level 0 parts from MoClo (6,8), CIDAR (9), Mobius (22), CyanoGate (12) and Marburg Collection (13) (Table 1), see below.
GS level 0 parts are flanked by BsaI restriction sites with unique 4-nucleotide fusion sites, labelled A to I depending on their category (Figure 1). By concatenating compatible fusion sites, multiple parts can be assembled in order to create customized TUs (Figure 1). By using a one-pot restriction and ligation reaction, correctly assembled parts lose the BsaI site while re-ligated fragments are re-cleaved again in each new cycle. Once the level 0 parts are assembled into TUs, no BsaI sites remain in the final level 1 construct, thus achieving highly efficient assembly (≥95%, Figure S1). Basic functional GS level 1 TUs are made up of four level 0 parts: promoter, ribosomal binding site (RBS), coding sequence (CDS) and terminator, assembled within a level 1 host vector between fusion sites A to I, and using the combination of fusion sites A-B, B-D, D-G and G-I (Figure 1). To assemble more complex TUs requiring additional functional parts, such as signal peptides, anchors or tags, there are four additional positions available: two for N-terminal (C and E) and two for C- terminal (F and H) additions that can be used to deliver the final construct between the fusion sites A to I (Figure 1). Overall, up to eight level 0 parts can be combined and assembled into different level 1 receptor vectors, see below.
Figure 1.
Golden Standard fusion sites. Modules can be combinatorically assembled into a level 1 host vectors to make a transcription unit.
Depending on the future position of the TUs in more complex circuits, there are six level 1 receptor vectors available (Figure 2). Additionally, we also created special level 1 host vectors with inverted I to A fusion sites to construct TUs with the opposite transcription orientation. We initially included three sets of level 1 host vectors covering a large range of origins of replication, RK2, pBBR1 and pUC for low, medium and high copy number plasmids, respectively (Supplementary Table S1). The broad range of origins of replication (RK2 and pBBR1) provide compatibility within proteobacteria, thus providing a significant advantage with respect to other MoClo systems (9–11,22). Level 1 host vectors contain a kanamycin resistance cassette. Flanking the BsaI sites in level 1 constructs there are additional type IIS BpiI sites to allow subsequent assembly of multiple TUs in level 2 host vectors (Figures 2 and 3).
Figure 2.
Structure and nomenclature of GS pSEVA vectors (Supplementary Table S1). Following SEVA nomenclature, GS pSEVA vectors are named by 3 digits where the first number denotes the antibiotic resistance marker (blue), the second number identifies the origin of replication (orange), and the third number identifies the cargo (green). Golden Standard cargo is denoted with the number 19. In addition, vector level 1 (Lv1) receptors, level 2 (Lv2) receptors and level 3 (Lv3) receptor are named with the letter g followed of numbers, capital letters and Greek letters, respectively. The position of the fusion sites (FSs) of each specific vector and the basic structure of the cargos of each type of receptor vector (Lv1, Lv2 and Lv3) is indicated. Sequences of the BsaI FSs (blue) and BpiI FSs (red) are shown in the right tables.
Figure 3.
Hierarchy and graphical description of Golden Standard assembly. Hierarchy of Golden Standard assembly ranges from basic DNA parts (level 0), which are assembled to generate transcription units (level 1), which in turn can be assembled into more complex genetic circuits (level 2 and level 3). A Golden Standard reaction consists of a series of restriction/ligation cycles, whose operation is the same, regardless of the level under construction. Restriction steps release the genetic features flanked by the corresponding Type IIS enzyme (BsaI or BpiI), and removes the lacZ gene from the receptor vector. Ligation steps assemble the genetic features in a pre-defined order into the receptor vector.
Five level 2 vectors are available, thus supporting assembly of up to six TUs. The assembly of level 2 constructs is driven in a fixed and directional order through the fusion sites generated by BpiI digestion numbered 1 to 7 (Figures 2 and 3). The choice of the final level 2 host vector will depend on the number of transcription units to be assembled in the final construct. In contrast to other MoClo systems with a single level 2 receptor vector that require linker parts to fill empty positions, the library of level 2 vectors provided by GS significantly increases assembly efficiency by avoiding the use of extra adapters. The level 2 host vector collection includes plasmids with RK2, pBBR1 and pUC origins of replication and the gentamycin resistance cassette in order to facilitate selection (Supplementary Table S1). These vectors also contain BsaI sites external to the BpiI sites in order to allow fusion of multiple transcription units into level 3 host vectors (Figures 2 and 3). We constructed four level 3 host vectors containing BsaI fusion sites to assemble between seven and twenty TUs using the appropriate linkers, all of them featuring the kanamycin resistance cassette (Figure 2).
In addition, all receptor vectors (levels 1–3) include lacZ-alpha fragment gene for easy selection in E. coli cloning strains, and the SEVA multicloning site divided into a prefix (PacI to SmaI sites) and suffix (BamHI to SpeI sites) flanking the MoClo cargo, thus allowing further pathway/circuit modifications by traditional restriction enzyme cloning (Figure 2). GS cargo has been assigned as number 19 following SEVA nomenclature (Figure 2) (36).
To increase the flexibility and reusability of GS, we additionally created linker DNA parts (short non-coding DNA sequences) that can be used to de-functionalize any level 0 part, level 1 TU and level 2 circuits. The linkers are available as level 0/2 (BsaI and their corresponding fusion sites) and level 1 (BpiI and their corresponding fusion sites). The inclusion of multiple linkers allows the construction of polycistronic circuits (by replacing promoters and/or terminators) and the generation of genetic circuits of up to twenty transcription units, among other applications (a detailed protocol describing the construction of these complex circuits is provided in the supplementary materials and methods, Figure S2).
GS is initially launched with a library of: (i) six constitutive and four inducible promoters; (ii) four mono and two bicistronic RBSs; (iii) seven N-terminal and five C-terminal tags; (iv) two widely used CDS such as fluorescent reporter genes and a protease target sequence peptide; (v) six terminators; (vi) 15 linkers for construction of TUs and assembly of level 2 and 3 circuits; (vii) three preassembled transcription factors in order to allow the use of inducible promoters and (viii) a collection of 41 host vectors. The list of 96 available plasmids for distribution (including receptor plasmids and parts) can be found in Supplementary Table S1. To validate the feasibility and functionality of GS we successfully assembled in the 20 TUs level 3 pSEVA23g19[gδ] vector a complex and functional circuit expressing the gfp and rfp genes under the control of rhaSR/PrhaBAD and XylS/Pm, respectively (Supplementary Figure S3).
Expanding vector portability, bacterial hosts and gene expression control through Golden Standard assembly
Portability is the capability to exchange biological parts across organisms regardless of the origin of the DNA sequence. In the frame of synthetic biology and DNA assembly, portability is one of the key features allowing part reusability (38). Among other benefits, portability allows saving the vast effort required to generate and characterize parts for each individual organism. Despite the large effort to make highly portable libraries of DNA parts in the context of MoClo systems, libraries of receptor vectors suitable for use in a broad range of bacterial hosts are just starting to be developed (23,39).
By adopting SEVA standards GS supports, at least theoretically, the expansion of MoClo approaches to most of Gram negative bacteria including the whole group of Proteobacteria (37). This is especially important as many of these bacteria currently lack efficient and standardized genetic tools (40,41). To validate the portability of the GS system, we constructed a TU that expressed the reporter gene gfp under the control of the constitutive Pem7 promoter. This TU was then integrated into receptor vectors with varying origins of replication, including RK2, pBBR1, and pUC, which allowed for an increase in the copy number of the assembled TU. We then tested the performance of these expression plasmids monitoring the fluorescence in proteobacteria including Alpha- (i.e. Agrobacterium tumefaciens and Sphingobium sp. Strain SYK-6), Beta- (i.e. Cupriavidus metallidurans CH34 and Parazoarcus communis), Gamma- (i.e. E. coli W and Pseudomonas putida) and Deltaproteobacteria (i.e. Geobacter sulfurreducens) (Figure 4). As expected, the constructed broad spectrum plasmids were found to be interchangeable among all the tested strains. In the majority of the cases the expression profiles aligned with the anticipated number of copies conferred by each origin of replication (42). An interesting exception was observed in C. metallidurans, where both RK2 and pBBR1 displayed similar fluorescence levels suggesting similar copy numbers. Significantly, we observed that the absolute fluorescence levels were largely influenced by the host organism. Specifically, A. tumefaciens exhibited the highest fluorescence levels, while P. communis showed considerably lower fluorescence. These results clearly demonstrate the host-dependent expression of GFP, regardless of the plasmid used, emphasizing the influence of the biological context on the performance of genetic devices, as previously demonstrated (29). Consequently, precise monitoring of the performance of specific parts, TUs, and/or circuits in the desired bacterial chassis necessitates accurate measurements. Critically, GS largely facilitates this important task in synthetic biology.
Figure 4.
Portability of Golden Standard assembly in Proteobacteria. GFP fluorescence of cells carrying plasmids with RK2 (red), pBBR1 (blue) or pUC (green) origins of replication. Negative control is shown in black.
Golden Standard offers a set of well-characterized DNA parts
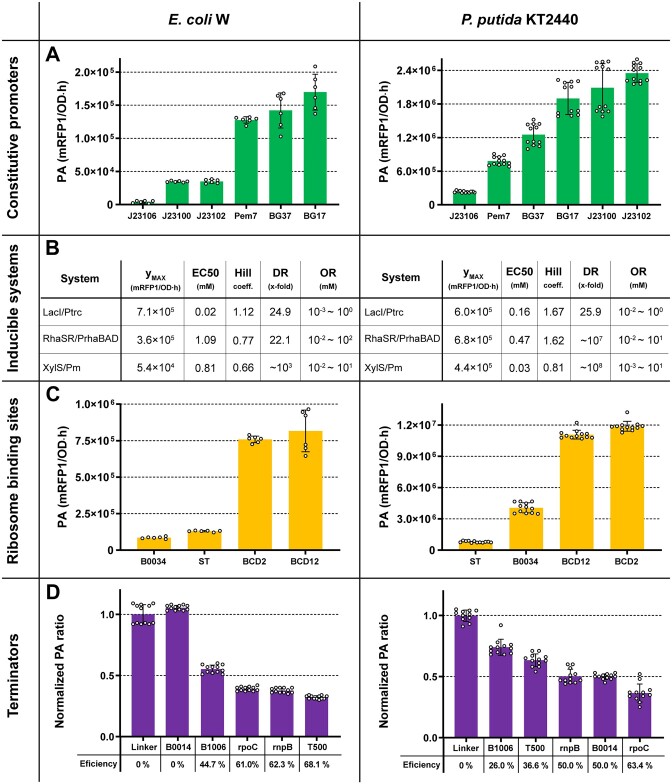
As above mentioned, a major cornerstone in synthetic biology is the lack of a widely accepted and accessible method for parts characterization which limit predictability. Golden Standard addresses this is issue by offering a complete catalogue of well-characterized parts using a highly reproducible and unbiased computational workflow under FAIR principles. To highlight the context-dependency, we characterized GS parts in model (E. coli) and non-model (P. putida) bacteria (Supplementary Figure S4). Regarding constitutive promoters, GS combines selected promoters from two well-known libraries: Anderson (J23100, J23102, J23106) (9) and Zobel's (Pem7, BG17, BG37) (43) collections. We found an evenly stepped 42-fold activity range between the promoters tested in E. coli W while in the case of P. putida, 10-fold activity range were obtained (Figure 5A). Interestingly, we found that Anderson's promoters exhibited higher activity in P. putida, whereas Zobel's promoters showed greater activity in E. coli. These results evidence that while relative activities are preserved among promoter families, they can operate at completely different scales in a host-dependent manner from different collections.
Figure 5.
Characterization of GS parts in E. coli W and P. putida KT2440. GS constitutive promoters (A), inducible expression systems (B), RBS (C) and terminators (D) characterization. For promoters and RBS, absolute part activity values are given. Inducible expression systems parameters were computed using Hill's equation, where ymax is maximum activity, EC50 is half maximal effective concentration, DR is dynamic range, and OR is operational range. For terminators, efficiency was measured by computing the ratio of GFPmut3/mRFP1 and normalizing it to a non-terminator linker (0% efficiency). Each point represents a single replicate well from two independent experiments. Error bars indicate standard deviation.
Regarding the inducible systems characterization, we analysed the dynamic behaviour of three well-known inducible expression systems included in GS: LacI/Ptrc, RhaSR/PrhaBAD, and XylS/Pm (Figure 5B). We screened the inducer effect spanning six orders of magnitude (from 1 to 100 mM) and the resulting activities data were used to fit a Hill equation function. Every construction was assembled using Golden Standard's set of RK2 oriC low-copy vectors to avoid metabolic burden. We noticed a growth deficiency when adding the highest concentration of IPTG, as well as for 10 and 100 mM of m-toluic acid in P. putida, and so was the case for 100 mM of m-toluic acid in of E. coli W. Similarly, we observed a significant host-dependent performance in terms of maximal expression, EC50 values, dynamic range, and operational range.
We next evaluated the performance of mono- and bicistronic RBSs in both strains. As it could be anticipated, the bicistronic BCD2 and BCD12 RBSs exhibited significantly higher activities. Noteworthy, we observed more homogeneous ranges of activities across hosts, with approximately 15- and 9.5-fold differences observed for P. putida and E. coli W, respectively (Figure 5C). Finally, we asset the performance of synthetic (T500, B1006, B0014) and natural (rnpB, rpoC) terminators as part of GS. We also found a large host-dependence. The maximum termination efficiency for P. putida was achieved the by rpoC terminator, which showed a 63.4% termination efficiency. For E. coli W, the synthetic terminator T500 reached a 68.1% termination efficiency, however reaching only 36.6% in P. putida (Figure 5D).
Overall, our results evidence how the same genetic elements completely change their performance depending on the microbial chassis, highlighting the importance of characterizing parts in different biological contexts. In this regard, GS provides an unprecedented toolset for conducting such analyses across a wide variety of bacterial species.
Golden Standard provides high interoperability with available MoClo systems
In addition to portability, interoperability and compatibility with existing systems becomes an essential feature of any genetic device when it comes to fostering the community-driven development of increasingly complete platforms. These features play a crucial role in promoting the reusability of parts and plasmids, enabling researchers to build upon existing resources and expand the capabilities of synthetic biology platforms. In order to validate the compatibility of GS with a well-known Golden Gate system based on standard syntax of fusion sites, we constructed a complex biosynthetic pathway using the repertoire of parts and level 1 vectors from the original MoClo system (6) and the library of GS level 2 vectors which provide extra flexibility in terms of receptor vectors. Specifically, we addressed the production of polyhydroxybutyrate (PHB) in E. coli. PHB is a biotechnologically useful natural polyester produced by many microorganisms under nutritional imbalances which has similar properties to conventional plastics. Production of PHB in model strains such as Cupriavidus necator H16 requires the action of (i) 3-ketothiolase (PhaA) to condense two acetyl-CoA molecules into acetoacetyl-CoA, (ii) acetoacetyl-CoA reductase (PhaB1) to reduce acetoacetyl-CoA to 3-hydroxybutyryl-CoA and iii) PHB synthase (PhaC1). We built the required level 0 plasmids following the procedures described by Weber (6), where: i) the Synpro16 promoter containing the AGGGGG RBS sequence (43,44) was cloned into an A-D level 0 vector; ii) genes phaA, phaB1, and phaC1 from C. necator H16 were domesticated, E. coli codon usage optimized and cloned into D-G level 0 vectors; and iii) three different terminators (rnpB_T1, rpoC and T500) were cloned into G-I level 0 vectors (Figure 6A). Subsequently, we assembled these level 0 parts into three level 1 plasmids using MoClo level 1 expressing phaA, phaB1 and phaC1 from positions 1, 2 and 3, respectively (see Supplementary Material, Table S3).
Figure 6.
(A) PHB operon construction combining Golden Standard with MoClo. Level 0 parts for PHB production previously constructed with the MoClo toolkit were assembled in level 1 MoClo vectors to obtain three TUs. Afterwards, these Tus were assembled in three level 2 GS pSEVA plasmids with RK2, pBBR1 or pUC origins of replication. (B) PHB granules present in E. coli DH10B cells harbouring plasmids pSEVA6219[gB] PHB (8% PHB), pSEVA63g19[gB] PHB (13% PHB) and pSEVA6819[gB] PHB (50% PHB). Image captured using phase contrast microscopy.
These level 1 plasmids were finally assembled into level 2 GS vectors featuring different origins of replication: pBBR1, RK2 or pUC (Figure 6A). The performance of these constructs was monitored following the production of PHB in E. coli DH10B during 24 h by GC-MS. Considering the expected number of copies provided by each origin of replication (42), we found a positive correlation between the expected number of plasmid copies and the amount of PHB produced, i.e. 0.9931 fit (Supplementary Figure S5, Table S5). Thus, the E. coli strain harbouring the level 2 pUC plasmid yielded the highest PHB levels, >50% cell dry weight (CDW) followed by the strains harbouring the pBBR1 and RK2 plasmids ∼13% and ∼8% respectively (Figure 6B). Overall, we demonstrated that the GS system is able to incorporate level 0 parts constructed using homologous systems like CIDAR or MoClo while providing a compatible platform for the effective reuse and further optimization of available level 1 plasmids in the context of a broad scope of level 2 and 3 vectors with variable copy numbers.
Golden Standard allows efficient and robust optimization of complex synthetic pathway expression using combinatorial assembly
One of the main features of Golden Gate technology is the possibility to optimize the performance of a given synthetic pathway by using combinatorial assembly (45,46). This paves the way to the construction of libraries of a given metabolic pathway under a variety of expression outputs, thus facilitating systematic exploration of expression space limited by the available parts. To accomplish this, each individual TU involved in the metabolic pathway (level 1) can be built using a set of regulatory parts, e.g. promoters, RBSs, etc. The library of individual TUs can be further combined to construct a collection of synthetic gene circuits and/or metabolic pathways (level 2). Following this approach and using a high-throughput screening method, e.g. a specific biosensor, the expression of an entire pathway can be optimized in a single step. Thus, the GS system can be used to engineer optimized gene clusters comprising several TUs. In order to validate this utility, we used GS to optimize the biosynthetic pathway for zeaxanthin production in E. coli (47–49). The biosynthesis of this carotenoid in E. coli involves five enzymatic steps: i) Synthesis of geranylgeranyl diphosphate (GGPP) from farnesyl pyrophosphate (FPP) and isopentyl diphosphate (IPP) catalysed by geranylgeranyl synthase (CrtE); ii) Condensation of two GGPP to one phytoene molecule catalysed by phytoene synthase (CrtB); iii) Desaturation of phytoene to lycopene by phytoene desaturase (CrtI); iv) Cyclization of lycopene by lycopene cyclase (CrtY); and v) Hydroxylation of β-carotene by β-carotene hydroxylase (CrtZ) (Figure 7A). The yellow colour of zeaxanthin not only simplifies the identification of colonies successfully expressing the whole pathway, but it also allows for visual recognition of relative colouration intensities between colonies. In this way, colonies expressing the optimal GS construct can be selected (11,22,50).
Figure 7.
Combinatorial assembly of the zeaxanthin biosynthetic pathway using the GS system. (A) Scheme of the heterologous zeaxanthin biosynthetic pathway in E. coli from glyceraldehyde 3-phosphate (G3P) and pyruvate using enzymes from P. agglomerans: FPP, farnesyl pyrophosphate; IPP, isopentyl diphosphate; GGPP, geranylgeranyl diphosphate; crtE, geranygeranyl synthase; crtB, phytoene synthase; crtI, phytoene desaturase; crtY, lycopene cyclase; crtZ, b-carotene hydroxylase. (B) Scheme of the assembly of level 0 parts and level 1 TUs for zeaxanthin production with multiple level 0 Anderson promoter parts (http://parts.igem.org/Promoters/Catalog/Anderson) into a polycistronic level 2 genetic device using Golden Standard assembly. (C) LB petri dish plate image showing colonies of E. coli cells transformed with the mix of level 2 parts for zeaxanthin production. Yellow colonies are positive colonies producing zeaxanthin, while the blue colonies are negative colonies carrying the empty vector. (D) Restreaking of different intensity yellow colonies selected by eyesight for subsequent analysis of the promoters used (1 to 5), 0, negative control colony. (E). Heat map showing the combination of Anderson's promoters for each CDS part in the five selected colonies.
Following the procedure to construct basic level 0 parts, the five CDSs from Pantoea agglomerans (48) encoding the enzymes catalysing the biosynthetic pathway of zeaxanthin (crtE, crtB, crtI, crtY and crtZ), were E. coli-codon-usage-optimized, properly domesticated removing internal BsaI and BpiI sites and flanked by the proper D and G -based fusion sites. Subsequently, they were cloned in the SmaI site of pSEVA182. We further engineered libraries of transcription units (level 1) expressing crtE, crtB, crtY and crtZ by combinatorial assembly using an equimolar mixture of selected Anderson's promoters (J23100, J23106, J23107 and J23116) -see http://parts.igem.org/Promoters/Catalog/Anderson, obtained from the CIDAR MoClo kit (Addgene Kit #1000000059). To avoid the known toxicity of crtI overexpression (51), this gene was kept at a constant low expression level using the weak promoter J23103 (Figure 7B). Twenty positive clones (white kanamycin-resistant colonies) from each TU assembly reaction were selected, mixed and grown together. Following plasmid extractions, we generated libraries of transcription units’ level 1 plasmids for crtE, crtB, crtY and crtZ which were used to construct the synthetic level 2 cluster using a pUC-based multicopy receptor plasmid. No yellow colonies were obtained and only a few blue colonies were observed (data not shown). However, when a pBBR-based level 2 receptor plasmid was used instead, we achieved a high number of positive yellow colonies (∼95%), with a transformation efficiency of 2 × 104 colonies/μg of receptor plasmid (Figure 7C). Five positive colonies displaying different intensities of yellow colouration were selected for further analysis (Figure 7D). After sequencing the promoter regions, we observed a significant predominance of the medium strength promoters J23106 and J23107 (Figure 7E). It was noteworthy that colony 2, which had the most intense yellow colony colour, expressed the crtB gene under the control of the high strength J23100 promoter, strongly suggesting that high expression of this gene is important for a high level of zeaxanthin production. Despite the few clones analysed, it was interesting that there was an overall lack of the strong promoter J23100, which could be indicative of the toxicity of this pathway when overexpressed in E. coli (22,52,53). In fact, successful strategies for the production of carotenoids in E. coli generally have relied on the use of inducible promoters and/or stabilization of the pathway through chromosome integration (49,52,53). In this regard, the constructs for production of zeaxanthin in E. coli we obtained using GS combinatorial assembly, in the context of alternative copy number plasmids, highlight the usefulness of this technology in optimized synthetic metabolic pathway engineering, both in terms of improving gene expression and reducing the metabolic burden to avoid cell growth impairment and plasmid instability (54).
Golden standard for optimizing expression of recombinant proteins
The main applications of MoClo assembly lie in synthetic biology and focus on construction and optimization of complex metabolic pathways and/or genetic circuits featuring multiple transcription units. Applications targeting the optimization of recombinant protein production or purification are uncommon. In this context, despite MoClo having been used in modular construction of proteins including N- and/or C- terminal tags, to our knowledge this interesting feature is underexploited and lacks direct applications. Therefore, we present here a straightforward application of GS to modular construction of proteins via introduction of solubility/purification and selection/quantification tags (Figure 8). We used Streptomyces antibioticus’ oleandomycin glucosyltransferase (OleD) as the model protein in our assays. First, native OleD and a fusion protein with GFP as a C-terminal tag were constructed confirming that this modification did not hinder OleD enzymatic activity (Supplementary Figure S6, Table S6). Finally, we evaluated the influence of purification/solubility tags on OleD expression and the activity of the resulting fusion proteins. To this end, we selected four commonly used N-terminal purification/solubility tags (His6x, strep, glutathione S-transferase with an HRV 3C protease cleavage site and maltose binding protein). Four GS assembly setups were prepared, one for each of the individual N-tags (Figure 8A, see Supplementary Figure S6 for details). Plasmid sequencing and SDS-PAGE analyses (Supplementary Figure S7) confirmed proper assembly of each of the N-terminal tags used.
Figure 8.
Schematic diagram and results of N- and C-terminal tag assemblies. (A) Graphical representation of individual MoClo assemblies of the OleD variants tested. (B) Combined plotted results (conversion of xanthohumol to 4′-O-β-d-glucoside of xanthohumol versus fluorescence of GFP-tag) in cell-free extracts from random clones obtained using mixed MoClo assembly with different N-terminal tags. (C) Graphical representation of MoClo assembly of 2xN- and 2xC protein with calculated MW masses of proteins; (D) SDS-PAGE analysis – on each well 1 ug of proteins was resolved. L – protein ladder; 1 – P. putida crude protein extact; 2 – eluted fraction after IMAC purification; 3 – HRV-3C digestion reaction; 4 – proteins not bound to Strep-Tactin column; 5 – proteins eluted from Strep-Tactin column. Photograph of fractions 3–5 irradiated with UV light are shown.
Analysis of both fluorescence and enzymatic activity is established as common practice in high-throughput screening of protein engineering libraries (55,56). In this sense, instead of performing laborious dilutions to standardize protein concentration by means of fluorescence and further activity assays, we calculated the ratio of activity and fluorescence from the same volume of CFE. Taking this ratio as a key parameter, we posited that GS could be used to optimize the expression/activity of heterologously produced proteins by constructing a combinatorial library of fusion proteins. Therefore, the four N-terminal tags were mixed stoichiometrically in the MoClo reaction and, after transformation, 30 positive clones were selected and analysed (Supplementary Figure S8). As expected, the mixed assembly resulted in 4 populations that shared similar values of fluorescence and activity of cell-free extracts (Figure 8B). Two clones from each population were sequenced to identify the tag responsible for these different behaviours. A near linear correlation between fluorescence and activity could be drawn as a logical consequence of enzyme concentration (Figure 8B).
The versatility of the GS in protein engineering lies also in double tag assembly enabling design and production of even more complex fusion proteins (Figure 1). To present the application of double N- and C-terminal tag assembly, we have decided to create a complex fluorescent protein with two independent purification tags, two fluorescent proteins and core GST protein with protease cleavage site that allows separation of fluorescent proteins after digestion (Figure 8C). Constructed lv 1 vector (pBBR1) was used in transformation of P. putida and resulted in red colonies as a consequence of application of strong constitutive promoter. Randomly chosen colony, verified by sequencing, was used in overnight expression. The resulting CFE was further purified using IMAC, and directly digested using HRV-3C protease, then the reaction products were separated using Strep-tactin column. Successful expression and purification was confirmed by SDS-PAGE and visualized by direct UV-light irradiation of obtained fractions (Figure 8C, D). This experiment clearly demonstrates simplicity and ease of application of GS in protein engineering and might serve as the standard for any combinatorial protein design and discovery.
In summary, we proved that GS can be successfully applied to screen optimal recombinant protein expression and perform complex target protein modifications. Although we present data for the constitutive expression of recombinant proteins, the same procedure could be applied to inducible promoter expression systems using a randomised library of level 1 vectors with PT7, Pm, PrhaBAD or any other promoter. It is worth pointing out that a similar approach could be applied to serial, standardised experiments using, among others, protein linker selection, protease cleavage site selection, anchor, secretory or signal peptide selection, and could be tested in hosts within the proteobacteria phylum and beyond.
Web portal for community-driven developed of Golden Standard
We created a website http://sysbiol.cnb.csic.es/GoldenStandard/home.php to host the Golden Standard constructions, metadata, and necessary tools to design new assemblies. This is divided into multiple interconnected modules to provide flexibility for future growth. The modules are named Golden Standard DB, Golden Standard Hub and Wizard.
The Golden Standard DB module is the Golden Standard database interface. It features tools to search and identify DNA parts and constructions stored in the database.
The Golden Standard Hub module serves as a SynBioHub server (57). The data therein follows the well-established SBOL (Synthetic Biology Open Language) syntax to aid in sharing constructs. This module is linked internally to the Golden Standard DB module to provide it with the sequences and other annotations. This module cannot be directly accessed by the user, but it is required to interconnect with external tools.
The Wizard module includes a computational framework to support in silico assembly of constructs. This interactive tool simplifies design and assembly by allowing the user to make multiple selections of components stored in the Golden Standard Hub (levels 0, 1 or 2) and using them to simulate the cloning steps for assembly. It also supports the addition of custom elements to handle specific parts not included in the Golden Standard Hub. The user only has to provide a given sequence and its concentration to launch the simulation. The server is able to handle constructions with up to 6 level 1 elements. However, it also supports higher levels of complexity in terms of level through the use of an iterative procedure. Another key tool included in this module is the ‘Setup’ tool, which generates a detailed report with the assembly protocol that is specific to the simulated constructs.
A step-by-step user guide to the GS Hub and Wizard is provided with the supplementary information.
DISCUSSION AND OUTLOOK
Our use and characterization of Golden Standard assembly demonstrates that it is a robust modular cloning system capable of constructing complex expression genetic devices in a wide range of bacteria. Although our focus has primarily been on proteobacteria, it is important to note that GS assembly is not limited to this group of bacteria, as it has been successfully expanded and employed even beyond Gram-bacteria. As interesting proof-of-concept, GS has been recently expanded to Streptomyces (26). This highlights the potential of GS to become a standard system within the Golden Gate technology framework, further promoting its widespread adoption and utility in the synthetic biology community. GS assemblies with varying total numbers of fragments, size and DNA composition were performed to demonstrate that it is a high performance (≥95%), routine assembly method. GS system takes advantage of: (i) a common syntax for DNA parts that is compatible with existing Golden Gate systems, thus allowing exchangeability and reusability of parts by researchers (Table 1), (ii) a straightforward assembly scheme, from a single transcription unit up to genetic circuits, with a set of vectors that supports construction of up to up twenty TUs for rapid and high-throughput combinatorial assemblies, (iii) a streamlined protocol supporting a logical framework and parts reusability for hierarchical assembly, (iv) the ability to continually make improvements to the collection of building blocks (parts and modules) in order to simplify current and future engineering processes. Therefore, GS fulfils, to a great extent, the recommendations for standardization recently highlighted in the White Book on standardization in synthetic biology (58).
In order to facilitate the design of constructs using modular assembly, we developed a set of web-based tools that provide protocols and assistance for in silico design. We anticipate that these will enable rapid adoption of GS by non-specialist users of modular cloning. The implementation of the web server supports easy design of target synthetic pathways by: (i) facilitating user-friendly selection of promoters, RBSs, and plasmid origins of replication and (ii) providing detailed visualization of the final construct. Additionally, the custom-part-handling feature supports assembly procedures involving new elements and paves the way to increasing the parts database, which is expected to become a high-value asset to the scientific community.
Another advantage of the GS platform is that plasmids for genetic circuits feature a range of origins of replication to avoid undesired effects on cells. For instance, it may be necessary to use a low-to-medium copy plasmid when high expression of heterologous genetic constructs leads to an excessive metabolic burden (59) or to avoid toxic side effects on the chassis cell when membrane proteins are expressed at a high level. The comprehensive collection of parts available in the toolkit supports customized levels of gene expression.
GS is underpinned by a strong will to inspire the necessary cross-community effort to develop a universal, highly portable and standardized MoClo system. It has been designed to undergo constant updates in order to increase its functionality and scope. Following on from successful expansion of SEVA plasmids by multiple synthetic biology labs, we encourage this community to expand GS system by using the SEVA standards together with the GS syntax. Therefore, we envision a continuous community-guided GS expansion focussed in new organisms, the creation of new parts and modules (i.e. CRISPRi, multiplexing, genome editing, etc). Similarly, we anticipate the incorporation of quantitative outputs of parts and circuits, not only in the context of the large array of receptor vectors available, but also considering the increasing number of host environments and chassis. This feature is expected to unlock rational design and construction of complex metabolic pathways highly customized to the host chassis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Fabián Moreno, Alejandro Ronco, and Darwin Carranza for helping in the construction of some Golden Standard plasmids. We acknowledge and appreciate plasmid donations from Till Tiso and Lars Blank (RWTH Aachen University, Aachen, Germany); Nick Wierckx (Forschungszentrum Jülich, Jülich, Germany); and Sylvestre Marillonnet (Leibniz Institute of Plant Biochemistry, Halle, Germany). We thanks Sofía Fraile for providing Cupriavidus metallidurans CH34, and Daniela Torruella and Abraham Esteve for providing Geobacter sulfurreducens. The authors are especially grateful to Esteban Martínez-García and Víctor de Lorenzo (Centro Nacional de Biotecnología, CSIC, Spain) for their helpful, guidance and assistance adopting SEVA formalisms and critical reading of the manuscript. Finally, the authors wish to thank Clive A. Dove for proofreading of the manuscript.
Authors’ contributions: B.B., D.S.L., J.T.B., A.P. and J.N. conceived and designed the study. B.B., A.G.L., J.T.B., I.M., R.K., S.G., J.P., S.S., A.W. performed and analysed the experiments. D.S.L. constructed GS web server. B.B., I.M., J.T.B., A.G.L., D.S.L., J.P. and S.S. drafted the paper. E.H., A.P. and J.N.: Review, editing and funding acquisition. J.N. is orresponding author. All authors read and approved the final manuscript.
Contributor Information
Blas Blázquez, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain; Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain.
David San León, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain; Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain.
Jesús Torres-Bacete, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain.
Álvaro Gómez-Luengo, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain; Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain.
Ryan Kniewel, Microbial and Plant Biotechnology Department, Biological Research Center-Margarita Salas, CSIC, Madrid, Spain.
Igor Martínez, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain.
Sandra Sordon, Wrocław University of Environmental and Life Sciences, Department of Food Chemistry and Biocatalysis, Norwida 25, 50-375, Wrocław, Poland.
Aleksandra Wilczak, Wrocław University of Environmental and Life Sciences, Department of Food Chemistry and Biocatalysis, Norwida 25, 50-375, Wrocław, Poland.
Sergio Salgado, Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain; Microbial and Plant Biotechnology Department, Biological Research Center-Margarita Salas, CSIC, Madrid, Spain.
Ewa Huszcza, Wrocław University of Environmental and Life Sciences, Department of Food Chemistry and Biocatalysis, Norwida 25, 50-375, Wrocław, Poland.
Jarosław Popłoński, Wrocław University of Environmental and Life Sciences, Department of Food Chemistry and Biocatalysis, Norwida 25, 50-375, Wrocław, Poland.
Auxiliadora Prieto, Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain; Microbial and Plant Biotechnology Department, Biological Research Center-Margarita Salas, CSIC, Madrid, Spain.
Juan Nogales, Department of Systems Biology, Centro Nacional de Biotecnología, CSIC, Madrid, Spain; Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain.
DATA AVAILABILITY
The plasmids listed in Table S1 have been deposited at SEVA database (http://seva-plasmids.com/) and they are available under request. Data and tools for hierarchical assembly are available through the Golden Standard portal http://sysbiol.cnb.csic.es/GoldenStandard/home.php. Python custom scripts for part characterization and data are available at https://zenodo.org/record/8159259.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union's Horizon 2020 research and innovation program [814650 (Synbio4Flav), 633962 (P4SB), 814418 (SinFonia), 870294 (MixUp)]; Spanish Ministry of Science and Innovation for the RobExplode project PID2019‐108458RB‐I00 [AEI /10.13039/501100011033], BIO2017-83448-R and PID2020-112766RB-C21, and CSIC’s Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy+ (PTI-SusPlast+); A.G.L. was funded by ‘Fondo Social Europeo’ and ‘Iniciativa de Empleo Juvenil (YEI)’ [PEJ-2020-AI/BIO-18028] and S.S. by a FPU (Ayuda para la formación de profesorado universitario) fellowship [FPU17/03978] from the Spanish Ministry of Universities. Funding for open access charge: Consejo Superior de Investigaciones Científicas (CSIC) [82582328].
Conflict of interest statement. None declared.
REFERENCES
- 1. Freemont P.S. Synthetic biology industry: data-driven design is creating new opportunities in biotechnology. Emerg. Top. Life Sci. 2019; 3:651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradley R.W., Buck M., Wang B.. Tools and principles for microbial gene circuit engineering. J. Mol. Biol. 2016; 428:862–888. [DOI] [PubMed] [Google Scholar]
- 3. Shetty R.P., Endy D., Knight T.F.. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008; 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 5. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S.. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011; 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szybalski W., Kim S.C., Hasan N., Podhajska A.J.. Class-IIS restriction enzymes - a review. Gene. 1991; 100:13–26. [DOI] [PubMed] [Google Scholar]
- 8. Engler C., Youles M., Gruetzner R., Ehnert T.M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S.. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 2014; 3:839–843. [DOI] [PubMed] [Google Scholar]
- 9. Iverson S.V., Haddock T.L., Beal J., Densmore D.M.. CIDAR MoClo: improved MoClo assembly standard and new E. coli part library enable rapid combinatorial design for synthetic and traditional biology. ACS Synth. Biol. 2016; 5:99–103. [DOI] [PubMed] [Google Scholar]
- 10. Moore S.J., Lai H.E., Kelwick R.J.R., Chee S.M., Bell D.J., Polizzi K.M., Freemont P.S.. EcoFlex: a multifunctional MoClo Kit for E. coli synthetic biology. ACS Synth. Biol. 2016; 5:1059–1069. [DOI] [PubMed] [Google Scholar]
- 11. Taylor G.M., Mordaka P.M., Heap J.T.. Start-Stop Assembly: a functionally scarless DNA assembly system optimized for metabolic engineering. Nucleic Acids Res. 2019; 47:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vasudevan R., Gale G.A.R., Schiavon A.A., Puzorjov A., Malin J., Gillespie M.D., Vavitsas K., Zulkower V., Wang B., Howe C.J.et al.. Cyanogate: a modular cloning suite for engineering cyanobacteria based on the plant moclo syntax. Plant Physiol. 2019; 180:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stukenberg D., Hensel T., Hoff J., Daniel B., Inckemann R., Tedeschi J.N., Nousch F., Fritz G.. The marburg collection: a Golden Gate DNA assembly framework for synthetic biology applications in Vibrio natriegens. ACS Synth. Biol. 2021; 10:1904–1919. [DOI] [PubMed] [Google Scholar]
- 14. Lammens E.M., Boon M., Grimon D., Briers Y., Lavigne R.. SEVAtile: a standardised DNA assembly method optimised for Pseudomonas. Microb. Biotechnol. 2021; 15:370–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Dolleweerd C.J., Kessans S.A., Van De Bittner K.C., Bustamante L.Y., Bundela R., Scott B., Nicholson M.J., Parker E.J.. MIDAS: a modular DNA assembly system for synthetic biology. ACS Synth. Biol. 2018; 7:1018–1029. [DOI] [PubMed] [Google Scholar]
- 16. Pollak B., Cerda A., Delmans M., Álamos S., Moyano T., West A., Gutiérrez R.A., Patron N.J., Federici F., Haseloff J.. Loop assembly: a simple and open system for recursive fabrication of DNA circuits. New Phytol. 2019; 222:628–640. [DOI] [PubMed] [Google Scholar]
- 17. Martella A., Matjusaitis M., Auxillos J., Pollard S.M., Cai Y.. EMMA: an extensible mammalian modular assembly toolkit for the rapid design and production of diverse expression vectors. ACS Synth. Biol. 2017; 6:1380–1392. [DOI] [PubMed] [Google Scholar]
- 18. Patron N.J., Orzaez D., Marillonnet S., Warzecha H., Matthewman C., Youles M., Raitskin O., Leveau A., Farré G., Rogers C.et al.. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol. 2015; 208:13–19. [DOI] [PubMed] [Google Scholar]
- 19. Werner S., Engler C., Weber E., Gruetzner R., Marillonnet S.. Fast track assembly of multigene constructs using golden gate cloning and the MoClo system. Bioeng. Bugs. 2012; 3:38–43. [DOI] [PubMed] [Google Scholar]
- 20. Sarrion-Perdigones A., Vazquez-Vilar M., Palací J., Castelijns B., Forment J., Ziarsolo P., Blanca J., Granell A., Orzaez D. Goldenbraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013; 162:1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kundert P., Sarrion-Perdigones A., Gonzalez Y., Katoh-Kurasawa M., Hirose S., Lehmann P., Venken K.J.T., Shaulsky G.. A GoldenBraid cloning system for synthetic biology in social amoebae. Nucleic Acids Res. 2020; 48:4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andreou A.I., Nakayama N.. Mobius assembly: a versatile golden-gate framework towards universal DNA assembly. PLoS One. 2018; 13:e0189892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiasson D., Giménez-Oya V., Bircheneder M., Bachmaier S., Studtrucker T., Ryan J., Sollweck K., Leonhardt H., Boshart M., Dietrich P.et al.. A unified multi-kingdom Golden Gate cloning platform. Sci. Rep. 2019; 9:10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pollak B., Matute T., Nuñez I., Cerda A., Lopez C., Vargas V., Kan A., Bielinski V., Von Dassow P., Dupont C.L.et al.. Universal loop assembly: open, efficient and cross-kingdom DNA fabrication. Synth. Biol. 2020; 5:ysaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valenzuela-Ortega M., French C.E.. Joint universal modular plasmids: a flexible platform for golden gate assembly in any microbial host. Methods in Molecular Biology. 2020; 2205:Humana Press Inc; 255–273. [DOI] [PubMed] [Google Scholar]
- 26. Magadán-Corpas P., Ye S., Pérez-Valero Á., McAlpine P.L., Valdés-Chiara P., Torres-Bacete J., Nogales J., Villar C.J., Lombó F.. Optimized de novo eriodictyol biosynthesis in Streptomyces albidoflavus using an expansion of the Golden Standard toolkit for its use in Actinomycetes. Int. J. Mol. Sci. 2023; 24:8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cardinale S., Arkin A.P.. Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 2012; 7:856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keren L., Zackay O., Lotan-Pompan M., Barenholz U., Dekel E., Sasson V., Aidelberg G., Bren A., Zeevi D., Weinberger A.et al.. Promoters maintain their relative activity levels under different growth conditions. Mol. Syst. Biol. 2013; 9:701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tas H., Grozinger L., Stoof R., de Lorenzo V., Goñi-Moreno Á.. Contextual dependencies expand the re-usability of genetic inverters. Nat. Commun. 2021; 12:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blázquez B., Carmona M., Díaz E.. Transcriptional regulation of the peripheral pathway for the anaerobic catabolism of toluene and m-xylene in Azoarcus sp. CIB. Front. Microbiol. 2018; 9:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi K.H., Kumar A., Schweizer H.P.. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods. 2006; 64:391–397. [DOI] [PubMed] [Google Scholar]
- 32. Coppi M.V., Leang C., Sandler S.J., Lovley D.R.. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 2001; 67:3180–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Revelles O., Tarazona N., García J.L., Prieto M.A.. Carbon roadmap from syngas to polyhydroxyalkanoates in Rhodospirillum rubrum. Environ. Microbiol. 2016; 18:708–720. [DOI] [PubMed] [Google Scholar]
- 34. Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 35. Abril M.A., Michan C., Timmis K.N., Ramos J.L.. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 1989; 171:6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martínez-García E., Fraile S., Algar E., Aparicio T., Velázquez E., Calles B., Tas H., Blázquez B., Martín B., Prieto C.et al.. SEVA 4.0: an update of the Standard European Vector Architecture database for advanced analysis and programming of bacterial phenotypes. Nucleic Acids Res. 2023; 51:D1558–D1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., De Las Heras A., Páez-Espino A.D., Durante-Rodríguez G., Kim J., Nikel P.I.et al.. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013; 41:D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casini A., Storch M., Baldwin G.S., Ellis T.. Bricks and blueprints: methods and standards for DNA assembly. Nat. Rev. Mol. Cell Biol. 2015; 16:568–576. [DOI] [PubMed] [Google Scholar]
- 39. Valenzuela-Ortega M., French C.. Joint universal modular plasmids (JUMP): a flexible vector platform for synthetic biology. Synth. Biol. 2021; 6:ysab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nora L.C., Westmann C.A., Martins-Santana L., Alves L.d.F., Monteiro L.M.O., Guazzaroni M.E., Silva-Rocha R.. The art of vector engineering: towards the construction of next-generation genetic tools. Microb. Biotechnol. 2019; 12:125–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turco F., Garavaglia M., Van Houdt R., Hill P., Rawson F.J., Kovacs K.. Synthetic biology toolbox, including a single-plasmid CRISPR-Cas9 system to biologically engineer the electrogenic, metal-resistant bacterium Cupriavidus metallidurans CH34. ACS Synth. Biol. 2022; 11:3617–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jahn M., Vorpahl C., Hübschmann T., Harms H., Müller S.. Copy number variability of expression plasmids determined by cell sorting and droplet digital PCR. Microb. Cell Fact. 2016; 15:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zobel S., Benedetti I., Eisenbach L., De Lorenzo V., Wierckx N., Blank L.M.. Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth. Biol. 2015; 4:1341–1351. [DOI] [PubMed] [Google Scholar]
- 44. Tiso T., Sabelhaus P., Behrens B., Wittgens A., Rosenau F., Hayen H., Blank L.M.. Creating metabolic demand as an engineering strategy in Pseudomonas putida – Rhamnolipid synthesis as an example. Metab. Eng. Commun. 2016; 3:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smanski M.J., Bhatia S., Zhao D., Park Y.J., Woodruff L.B.A., Giannoukos G., Ciulla D., Busby M., Calderon J., Nicol R.et al.. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014; 32:1241–1249. [DOI] [PubMed] [Google Scholar]
- 46. Coussement P., Bauwens D., Maertens J., De Mey M.. Direct combinatorial pathway optimization. ACS Synth. Biol. 2017; 6:224–232. [DOI] [PubMed] [Google Scholar]
- 47. Sajilata M.G., Singhal R.S., Kamat M.Y.. The carotenoid pigment zeaxanthin—a review. Compr. Rev. Food Sci. Food Saf. 2008; 7:29–49. [Google Scholar]
- 48. Cunningham F.X., Gantt E.. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: adonis aestivalis as a case study. Photosynth. Res. 2007; 92:245–259. [DOI] [PubMed] [Google Scholar]
- 49. Ruther A., Misawa N., Böger P., Sandmann G.. Production of zeaxanthin in Escherichia coli transformed with different carotenogenic plasmids. Appl. Microbiol. Biotechnol. 1997; 48:162–167. [DOI] [PubMed] [Google Scholar]
- 50. Wang C., Zhao S., Shao X., Park J.B., Jeong S.H., Park H.J., Kwak W.J., Wei G., Kim S.W. Challenges and tackles in metabolic engineering for microbial production of carotenoids. Microb. Cell Fact. 2019; 18:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Albermann C. High versus low level expression of the lycopene biosynthesis genes from Pantoea ananatis in Escherichia coli. Biotechnol. Lett. 2011; 33:313–319. [DOI] [PubMed] [Google Scholar]
- 52. Chen Y.Y., Shen H.J., Cui Y.Y., Chen S.G., Weng Z.M., Zhao M., Liu J.Z.. Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biotechnol. 2013; 13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li X.R., Tian G.Q., Shen H.J., Liu J.Z.. Metabolic engineering of Escherichia coli to produce zeaxanthin. J. Ind. Microbiol. Biotechnol. 2015; 42:627–636. [DOI] [PubMed] [Google Scholar]
- 54. Silva F., Queiroz J.A., Domingues F.C.. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 2012; 30:691–708. [DOI] [PubMed] [Google Scholar]
- 55. Listwan P., Terwilliger T.C., Waldo G.S.. Automated, high-throughput platform for protein solubility screening using a split-GFP system. J. Struct. Funct. Genomics. 2009; 10:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santos-Aberturas J., Dörr M., Waldo G.S., Bornscheuer U.T.. In-depth high-throughput screening of protein engineering libraries by split-GFP direct crude cell extract data normalization. Chem. Biol. 2015; 22:1406–1414. [DOI] [PubMed] [Google Scholar]
- 57. McLaughlin J.A., Myers C.J., Zundel Z., Mlslrll G., Zhang M., Ofiteru I.D., Goñi-Moreno A., Wipat A.. SynBioHub: a Standards-Enabled Design Repository for Synthetic Biology. ACS Synth. Biol. 2018; 7:682–688. [DOI] [PubMed] [Google Scholar]
- 58. Ordozgoiti E., Porcar M., Baldwin G., de Lorenzo V., Ríos L., Elfick A., Schyfter P., Maria Delgado A., Schmidt M., Garfinkel M.et al.. Standardisation in Synthetic Biology: a white book state-of-the-art and recommendations for policy makers. 2021; [Google Scholar]
- 59. Ceroni F., Boo A., Furini S., Gorochowski T.E., Borkowski O., Ladak Y.N., Awan A.R., Gilbert C., Stan G.B., Ellis T.. Burden-driven feedback control of gene expression. Nat. Methods. 2018; 15:387–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plasmids listed in Table S1 have been deposited at SEVA database (http://seva-plasmids.com/) and they are available under request. Data and tools for hierarchical assembly are available through the Golden Standard portal http://sysbiol.cnb.csic.es/GoldenStandard/home.php. Python custom scripts for part characterization and data are available at https://zenodo.org/record/8159259.