Abstract
Enhancer reprogramming has been proposed as a key source of transcriptional dysregulation during tumorigenesis, but the molecular mechanisms underlying this process remain unclear. Here, we identify an enhancer cluster required for normal development that is aberrantly activated in breast and lung adenocarcinoma. Deletion of the SRR124–134 cluster disrupts expression of the SOX2 oncogene, dysregulates genome-wide transcription and chromatin accessibility and reduces the ability of cancer cells to form colonies in vitro. Analysis of primary tumors reveals a correlation between chromatin accessibility at this cluster and SOX2 overexpression in breast and lung cancer patients. We demonstrate that FOXA1 is an activator and NFIB is a repressor of SRR124–134 activity and SOX2 transcription in cancer cells, revealing a co-opting of the regulatory mechanisms involved in early development. Notably, we show that the conserved SRR124 and SRR134 regions are essential during mouse development, where homozygous deletion results in the lethal failure of esophageal–tracheal separation. These findings provide insights into how developmental enhancers can be reprogrammed during tumorigenesis and underscore the importance of understanding enhancer dynamics during development and disease.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Developmental enhancers are commissioned during early embryogenesis, as transcription factors progressively restrict the epigenome through the repression of regulatory regions associated with pluripotency (1,2) and the activation of enhancers that control the expression of lineage-specific developmental genes (3–5). This establishes a cell type-specific epigenetic regulatory ‘memory’ that maintains cell lineage commitment and reinforces transcriptional programs (6). As cells mature and development ends, developmental-associated enhancers are decommissioned, and the enhancer landscape becomes highly restrictive and developmentally stable (6). This landscape, however, becomes profoundly disturbed during tumorigenesis, as cancer cells aberrantly acquire euchromatin features at regions near oncogenes (7,8) that are often associated with earlier stages of cell lineage specification (6). This ‘enhancer reprogramming’ has been proposed to result in a dysfunctional state that causes widespread abnormal gene expression and cellular plasticity (9–13). Although the misactivation of enhancers has been suggested as a major source of transcriptional dysregulation (reviewed in 14,15), it remains largely unclear how this mechanism unfolds during the progression of cancer. To study this process, we evaluated cis-regulatory elements involved in driving transcription during normal development and disease.
SRY-box transcription factor 2 (SOX2) is a pioneer transcription factor required for pluripotency maintenance in embryonic stem cells (16,17), involved in reprogramming differentiated cells to induced pluripotent stem cells in mammals (18–20), and acts as an oncogene in several different types of cancer (reviewed in 21,22). During later development, SOX2 is also required for tissue morphogenesis and homeostasis of the brain (23), eyes (24), esophagus (25), inner ear (26), lungs (27), skin (28), stomach (29), taste buds (30) and trachea (31) in both human and mouse. In these tissues, SOX2 expression is regulated precisely in space and time at critical stages of development, although in most cases the cis-regulatory regions that mediate this precision remain unknown. For example, proper levels of SOX2 expression are required during early development for the complete separation of the anterior foregut into the esophagus and trachea in mice (25,32,33) and in humans (34–36), as the disruption of SOX2 expression leads to an abnormal developmental condition known as esophageal atresia with distal tracheoesophageal fistula (EA/TEF) (reviewed in 37,38). After the anterior foregut is properly separated in mice, Sox2 expression ranges from the esophagus to the stomach in the gut (25,29), and throughout the trachea, bronchi and upper portion of the lungs in the developing airways (31). Proper branching morphogenesis at the tip of the lungs, however, requires temporary down-regulation of Sox2, followed by reactivation after lung bud establishment (27). Sox2 also retains an essential function in multiple mature epithelial tissues, where it is highly expressed in proliferative and self-renewing adult stem cells necessary for replacing terminally differentiated cells within the epithelium of the brain, bronchi, esophagus, stomach and trachea (29,31,39,40). The expression of Sox2, however, becomes repressed as stem cells differentiate in these tissues (39).
As an oncogene, overexpression of SOX2 is linked to increased cellular replication rates, aggressive tumor grades and poor patient outcomes in breast carcinoma (BRCA) (41–45), colon adenocarcinoma (COAD) (46–49), glioblastoma (GBM) (50–53), liver hepatocellular carcinoma (LIHC) (54), lung adenocarcinoma (LUAD) (55–57) and lung squamous cell carcinoma (LUSC) (58,59). These clinical and molecular characteristics arise from the participation of SOX2 in the formation and maintenance of tumor-initiating cells that resemble tissue progenitor cells, as evidenced by BRCA (45,60,61), GBM (52,62–64), LUAD (65) and LUSC (66) studies. SOX2 knockdown, on the other hand, often results in diminished levels of cell replication, invasion and treatment resistance in these tumor types (41,42,45,55,57,58,67–69). Despite the involvement of SOX2 in the progression of multiple types of cancer, little is known about the mechanisms that cause SOX2 overexpression during tumorigenesis. Two proximal enhancers were once deemed crucial for driving Sox2 expression during early development: Sox2 Regulatory Region 1 (SRR1) and SRR2 (23,70,71). Deletion of SRR1 and SRR2, however, has no effect on Sox2 expression in mouse embryonic stem cells (72). In contrast, deletion of a distal Sox2 Control Region (SCR), 106 kb downstream of the Sox2 promoter, causes a profound loss of Sox2 expression in mouse embryonic stem cells (72,73) and in blastocysts, where SCR deletion causes peri-implantation lethality (33). The contribution of these regulatory regions in driving SOX2 expression during tumorigenesis, however, remains poorly defined.
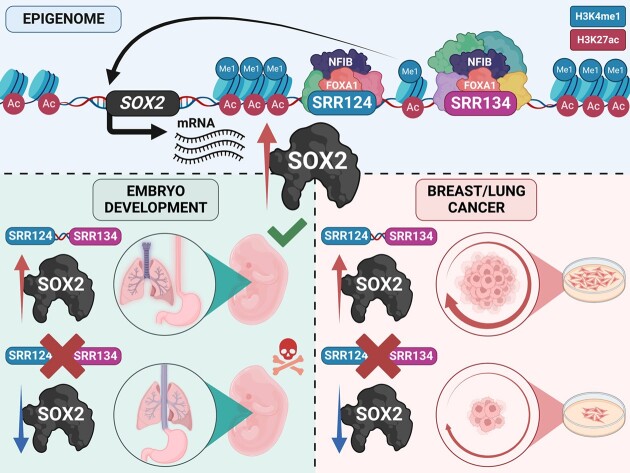
Here, we investigated the mechanisms underlying SOX2 overexpression in cancer. We found that, in breast and lung adenocarcinoma, SOX2 is driven by a novel developmental enhancer cluster we termed SRR124–134, rather than the previously identified SRR1, SRR2 or the SCR. This novel distal cluster contains two regions located 124 and 134 kb downstream of the SOX2 promoter that drive transcription in breast and lung adenocarcinoma cells. Deletion of this cluster results in significant SOX2 down-regulation, leading to genome-wide changes in chromatin accessibility and a globally disrupted transcriptome. The SRR124–134 cluster is highly accessible in most breast and lung patient tumors, where chromatin accessibility at these regions is correlated with SOX2 overexpression and is regulated positively by FOXA1 and negatively by NFIB. Finally, we found that both SRR124 and SRR134 are highly conserved in the mouse and are essential for postnatal survival, as homozygous deletion of their homologous regions results in lethal EA/TEF. These findings serve as a prime example of how different types of cancer cells reprogram enhancers that were decommissioned during development to drive the expression of oncogenes during tumorigenesis.
MATERIALS AND METHODS.
Cell culture
MCF-7 cells were obtained from Eldad Zacksenhaus (Toronto General Hospital Research Institute, Toronto, ON, Canada). H520 (HTB-182) and T47D (HTB-133) cells were acquired from the ATCC. PC-9 (90071810) cells were obtained from Sigma. Cell line identities were confirmed by short tandem repeat profiling. MCF-7 and T47D cells were grown in phenol red-free Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Gibco), 10% fetal bovine serum (FBS) (Gibco), 1× Glutamax (Gibco), 1× sodium pyruvate (Gibco), 1× penicillin–streptomycin (Gibco), 1× non-essential amino acids (Gibco), 25 mM HEPES (Gibco) and 0.01 mg/ml insulin (Sigma). H520 and PC-9 cells were grown in phenol red-free RPMI-1640 (Gibco), 10% FBS (Gibco), 1× Glutamax (Gibco), 1× sodium pyruvate (Gibco), 1× penicillin–streptomycin (Gibco), 1× non-essential amino acids (Gibco) and 25 mM HEPES (Gibco). Cells were either passaged or had their medium replenished every 3 days.
Genome editing
Guide RNA (gRNA) sequences were designed using Benchling. We minimized the possibility of unwanted off-target mutations by strictly selecting gRNA with no off-target sites with <3 bp mismatches. Pairs of gRNA plasmids were constructed by inserting a 20 bp target sequence (Supplementary Table S1) into an empty gRNA cloning vector (a gift from George Church; Addgene plasmid #41824) (74) containing either miRFP670 (Addgene plasmid #163748) or tagBFP (Addgene plasmid #163747) fluorescent markers. Plasmids were sequenced to confirm correct insertion. Both gRNA (1 μg each) vectors were co-transfected with 3 μg of pCas9_GFP (a gift from Kiran Musunuru; Addgene plasmid #44719) (75) using Neon electroporation (Life Technologies). After 72 h of transfection, cells were sorted by fluorescence-activated cell sorting (FACS) to select clones that contained all three plasmids. Sorted tagBFP+/GFP+/miRFP670+ cells were grown in a bulk population and serially diluted into individual wells to generate isogenic populations. Once fully grown, each well was screened by polymerase chain reaction (PCR) to confirm the deletion (Supplementary Table S2). Enhancer-deleted cells are available to the research community upon request.
Gene tagging
SOX2 was tagged with a P2A-tagBFP sequence in both alleles using clustered regularly interspaced palindromic repeats (CRISPR)-mediated homology-directed repair (HDR) (76). This strategy results in the expression of a single transcript that is further translated into two separate proteins due to ribosomal skipping (77). In summary, we designed a gRNA that targets the 3′ end of the SOX2 stop codon (Supplementary Table S1, Addgene plasmid #163752). We then amplified ∼800 bp homology arms upstream and downstream of the gRNA target sequence using high-fidelity Phusion Polymerase. We purposely avoided amplification of the SOX2 promoter sequence to reduce the likelihood of random integrations in the genome. Both homology arms were then joined at each end of a P2A-tagBFP sequence using Gibson assembly. Flanking primers containing the gRNA target sequence were used to reamplify SOX2-P2A-tagBFP and add gRNA targets at both ends of the fragment; this approach allows excision of the HDR sequence from the backbone plasmid once inside the cell (78). Finally, the full HDR sequence was inserted into a pJET1.2 (Thermo Scientific) backbone, midiprepped and sequenced (Addgene #163751). A 3 μg aliquot of HDR template was then co-transfected with 1 μg of hCas9 (a gift from George Church; Addgene plasmid #41815) (74) and 1 μg of gRNA plasmid using Neon electroporation (Life Technologies). A week after transfection, tagBFP+ cells were FACS sorted as a bulk population. Sorted cells were further grown for 2 weeks, and single tagBFP+ cells were isolated to generate isogenic populations. Once fully grown, each clone was screened by PCR and sequenced to confirm homozygous integration of P2A-tagBFP into the SOX2 locus (Supplementary Table S2). MCF-7 SOX2-P2A-tagBFP cells are available to the research community upon request.
Luciferase assay
Luciferase activity was measured using the dual-luciferase reporter assay (Promega #E1960) that relies on the co-transfection of two plasmids: pGL4.23 (firefly luciferase, luc2) and pGL4.75 (Renilla luciferase). Assayed plasmids were constructed by subcloning the empty pGL4.23 vector containing a minimal promoter (minP). SRR124, SRR134, SRR1, SRR2 and hSCR were PCR amplified (primers are given in Supplementary Table S3) from MCF-7 genomic DNA using high-fidelity Phusion Polymerase and inserted in the forward position downstream of the luc2 gene at the NotI restriction site. Constructs were sequenced to confirm correct insertions.
JASPAR2022 (79) was used to detect FOXA1 (GTAAACA) and NFIB (TGGCAnnnnGCCAA) motifs in the SRR134 sequence. Only motifs with a score of ≥80% were further analyzed. Bases within each motif sequence were mutated until the score was reduced below 80% without affecting co-occurring motifs or creating novel binding sites. In total, four FOXA1 motifs and two NFIB motifs were mutated (Supplementary Table S4). Engineered sequences were ordered as gene blocks (Eurofins) and inserted into pGL4.23 in the forward position. Constructs were sequenced to confirm correct insertions.
Cells were plated in 96-well plates with four technical replicates at 2 × 104 cells per well. After 24 h, a 200 ng 50:1 mixture of enhancer vector and pGL4.75 was transfected using Lipofectamine 3000 (0.05 μl of Lipofectamine:1 μl of Opti-mem). For transcription factor overexpression analysis, a 200 ng 50:10:1 mixture of enhancer vector, expression plasmid and pGL4.75 was transfected. After 48 h of transfection, cells were lysed in 1× Passive Lysis Buffer and stored at –80°C until all five biological replicates were completed. Luciferase activity was measured in the Fluoroskan Ascent FL plate reader. Enhancer activity was calculated by normalizing the firefly signal from pGL4.23 to the Renilla signal from pGL4.75.
Colony formation assay
MCF-7 and PC-9 cells were seeded at low density (2,000 cells/well) into 6-well plates in triplicate for each cell line. Culture medium was renewed every 3 days. After 12 days, cells were fixed with 3.7% paraformaldehyde for 10 min and stained with 0.5% crystal violet for 20 min to quantify the number of colonies formed. Crystal violet staining was then eluted with 10% acetic acid and absorbance was measured at 570 nm to evaluate cell proliferation. Each 6-well plate was considered one biological replicate and the experiment was repeated five times for each cell line (n = 5).
FACS analysis
For analyzing the effects of FOXA1 and NFIB overexpression, 2 × 106 SOX2-P2A-tagBFP cells were transfected with 50 nM of plasmid expressing either miRFP670 (a gift from Vladislav Verkhusha; Addgene plasmid #79987), FOXA1-T2A-miRFP670 (Addgene plasmid #182335) or NFIB-T2A-miRFP670 (Addgene plasmid #187222) in five replicates. Five days after transfection, miRFP670, tagBFP and propidium iodide (PI) (live/dead stain) signals were acquired using FACS; the amount of tagBFP signal from miRFP670+/PI– cells was compared between each treatment across all replicates.
FlowJo's chi-squared T(x) test was used to compare the effects of each treatment on tagBFP expression; T(x) scores >1000 were considered ‘strongly significant’ (***), whereas T(x) scores <100 were considered ‘non-significant’.
Transcriptome analysis
Total RNA was isolated from wild-type (WT; ΔENH+/+) and enhancer-deleted (ΔENH–/–) cell lines using the RNeasy kit. Genomic DNA was digested by Turbo DNase. A 500–2,000 ng aliquot of total RNA was used in a reverse transcription reaction with random primers. cDNA was diluted in H2O and amplified in a quantitative PCR (qPCR) using SYBR Select Mix (primers are given in Supplementary Table S5). Amplicons were sequenced to confirm primer specificity. Gene expression was normalized to PUM1 (80–82).
Total RNA was sent to The Centre for Applied Genomics (TCAG) for paired-end rRNA-depleted total RNA-seq (Illumina 2500, 125 bp). Read quality was checked by fastQC, trimmed using fastP (83) and mapped to the human genome (GRCh38/hg38) using STAR 2.7 (84). Normal breast epithelium RNA-seq was obtained from ENCODE (Supplementary Table S6) (85,86). Mapped reads were quantified using featureCounts (87) and imported into DESeq2 (88) for normalization and differential expression analysis. Genes with a |log2 fold change (FC)| > 1 and false discovery rate (FDR)-adjusted Q < 0.01 were considered significantly changed. Differential gene expression was plotted using the EnhancedVolcano package. Correlation and clustering heatmaps were plotted using the pheatmap R package (https://cran.r-project.org/web/packages/pheatmap/index.html). A signal enrichment plot was prepared using NGS.plot (89).
Cancer patient transcriptome data were obtained from TCGA (90) using the TCGAbiolinks package (91). The overall survival KM-plot (92) was calculated using clinical information from TCGA (93). Tumor transcriptome data were compared with normal tissue using DESeq2. RNA-seq reads were normalized to library size using DESeq2 (88) and transformed to a log2 scale [log2 counts]. Differential gene expression was considered significant if |log2 FC| > 1 and Q < 0.01.
Gene set enrichment analysis (GSEA) was performed by ranking genes according to their log2 FC in ΔENH–/– versus ΔENH+/+ MCF-7 cells. The ranking was then analyzed using the GSEA function from the clusterProfiler package (94) with a threshold of FDR-adjusted Q < 0.05 using the MSigDB GO term database (C5).
Chromatin accessibility analysis
Cells were grown in three separate wells (n = 3) and 50,000 cells were sent to the Princess Margaret Genomics Centre for ATAC-seq library preparation using the Omni-ATAC protocol (95). ATAC-seq libraries were sequenced using 50 bp paired-ended parameters in the Illumina Novaseq 6000 platform. Read quality was checked by fastQC, trimmed using fastP and mapped to the human genome (GRCh38/hg38) using STAR 2.7. Narrow peaks were called using Genrich (https://github.com/jsh58/Genrich). Differential chromatin accessibility analysis was performed using diffBind (96). ATAC-seq peaks with a |log2 FC| > 1 and FDR-adjusted Q < 0.01 were considered significantly changed. Correlation heatmaps were generated using diffBind. A signal enrichment plot was prepared using NGS.plot (89). Genes were separated into three categories according to their expression levels in our ΔENH+/+ MCF-7 RNA-seq data.
Transcription factor footprint analysis was performed using TOBIAS (97) with standard settings. Motifs with a |log2 FC| > 0.1 and FDR-adjusted Q < 0.01 were considered significantly enriched in each condition. Replicates (n = 3) were merged into a single BAM file for each condition. Motif enrichment at differential ATAC-seq peaks was performed using HOMER (98). ATAC-seq peaks were assigned to their closest gene within ± 1 Mb distance from their promoter using ChIPpeakAnno (99).
Cancer patient ATAC-seq data were obtained from TCGA (100). DNase-seq data from human developing tissues were obtained from ENCODE (Supplementary Table S6) (85,86). Read quantification was calculated at the RAB7a (pRAB7a), OR5K1 (pOR5K1) and SOX2 (pSOX2) promoters, together with SRR1, SRR2, SRR124, SRR134, hSCR and desert regions with a 1500 bp window centered at the core of each region (genomic coordinates of each region are given in Supplementary Table S7). Reads were normalized to library size [reads per million (RPM)] and transformed to a log2 scale (log2 RPM) using a custom script (https://github.com/luisabatti/BAMquantify). Each region's average log2 RPM was compared with that of the OR5K1 promoter for differential analysis using Dunn's test with Holm correction. Correlations were calculated using Pearson's correlation test and considered significant if FDR-adjusted Q < 0.05. Chromatin accessibility at SRR124 and SRR134 regions was considered low if log2 RPM < –1, medium if –1 ≤ log2 RPM ≤ 1 or high if log2 RPM > 1.
ATAC-seq data from developing mouse lung and stomach tissues were obtained from ENCODE (Supplementary Table S6) (85) and others (101). Conserved mouse regulatory regions were lifted from the human build (GRCh38/hg38) to the mouse build (GRCm38/mm10) using UCSC liftOver (102). The number of mapped reads was calculated at the Egf (pEgf), Olfr266 (pOlfr266) and Sox2 (pSox2) promoters, together with the mouse mSRR1, mSRR2, mSRR96, mSRR102, mSCR and desert regions with a 1500 bp window at each location (genomic coordinates are given in Supplementary Table S8). Each log2-transformed region's RPM (log2 RPM) was compared with that of the negative Olfr266 promoter control for differential analysis using Dunn's test with Holm correction.
Conservation analysis
Cross-species evolutionary conservation was obtained using phyloP (103). Pairwise comparisons between human SRR124 and SRR134 (GRCh38/hg38) and mouse mSRR96 and mSRR102 (GRCm38/mm10) sequences were aligned using Clustal Omega (104) and plotted using FlexiDot (105) with an 80% conservation threshold.
ChIP-seq analysis
ChIP-seq data for transcription factor and histone modifications were obtained from ENCODE (85) (Supplementary Table S6) and others (106–108) (Supplementary Table S9). H3K4me1 and H3K27ac tracks were normalized to input and library size (log2 RPM). Histone modification ChIP-seq tracks and transcription factor ChIP-seq peaks were uploaded to the UCSC browser (102) for visualization. Normalized H3K4me1 and H3K27ac reads were quantified and the difference in normalized signal was calculated using diffBind. Peaks with a |log2 FC| > 1 and Q < 0.01 were considered significantly changed.
Overlapping ChIP-seq and ATAC-seq peaks were analyzed using ChIPpeakAnno (99). The hypergeometric test was performed by comparing the number of overlapping peaks with the total size of the genome divided by the median peak size.
Mouse line construction
Our mSRR96–102 knockout mouse line (C57BL/6J; Chr3_SRR124-SRR134_del) was ordered from and generated by The Centre for Phenogenomics (TCP) model production core in Toronto, ON. The protocol for the generation of the mouse line has been previously described (109). Briefly, C57BL/6J zygotes were collected from superovulated, mated and plugged female mice at 0.5 days post-coitum. Zygotes were electroporated with CRISPR-associated protein 9 (Cas9) ribonucleoprotein (RNP) complexes (gRNA sequences are given in Supplementary Table S1) and transferred into pseudopregnant female recipients within 3–4 hours of electroporation. Newborn pups (potential founders) were screened by endpoint PCR and sequenced to confirm allelic mSRR96–102 deletions (Supplementary Table S2). One heterozygous mSRR96–102 founder (ΔmENH+/–) was then backcrossed twice to the parental strain to reduce the probability of off-target mutation segregation and to confirm germline transmission. Off-target mutagenesis by Cas9 is rare in mouse embryos using this protocol (110). Neither of the two gRNAs used for the mSRR96–102 deletion had any predicted off-target sites with <3 bp mismatches. Furthermore, no off-target hits were found within exonic regions on chromosome 3, where Sox2 is located. Potential changes in chromosomal copy numbers were also ruled out by real-time PCR.
Once the mouse line was established and the mSRR96–102 deletion was fully confirmed and sequenced in the N1 offspring, ΔmENH+/– mice were crossed and the number of live pups from each genotype (ΔmENH+/+, ΔmENH+/–, ΔmENH–/–) was assessed at weaning (P21). The obtained number of live pups from each genotype was then compared with the expected Mendelian ratio of 1:2:1 (ΔmENH+/+:ΔmENH+/–:ΔmENH–/–) using a chi-squared test. Once the lethality of the homozygous deletion was confirmed at weaning, E18.5 littermate embryos generated from new ΔmENH+/– crosses were collected for further histological analyses.
All procedures involving animals were performed in compliance with the Animals for Research Act of Ontario and the Guidelines of the Canadian Council on Animal Care. The TCP Animal Care Committee reviewed and approved all procedures conducted on animals at the facility. Sperm from male ΔmENH+/– mice has been cryopreserved at the Canadian Mouse Mutant Repository (CMMR) and is available upon request.
Histological analyses
A total of 46 embryos were collected at E18.5 and fixed in 4% paraformaldehyde. Each of these embryos was genotyped. A total of 15 embryos (Supplementary Table S10), five of each genotype (ΔmENH+/+, ΔmENH+/–, ΔmENH–/–), were randomly selected, processed and embedded in paraffin for sectioning and further analysis. Tissue sections were collected at 4 μm thickness roughly at the start of the thymus. Sections were prepared by the Pathology Core at TCP.
Tissue sections were stained with hematoxylin and eosin (H&E) using an auto-stainer to ensure batch consistency. Slides were scanned using a Hamamatsu Nanozoomer slide scanner at ×20 magnification. For immunohistochemistry staining, E18.5 embryo cross-sections were submitted to heat-induced epitope retrieval with Tris-EDTA (pH 9.0) for 10 min, followed by quenching of endogenous peroxidase with Bloxall reagent (Vector). Non-specific antibody binding was blocked with 2.5% normal horse serum (Vector), followed by incubation for 1 hour in rabbit anti-SOX2 (Abcam, ab92494, 1:500). After washes, sections were incubated for 30 min with ImmPRESS anti-rabbit horseradish peroxidase (HRP; Vector), followed by 3,3′-diaminobenzidine (DAB) reagent and counterstained in Mayer's hematoxylin.
For immunofluorescence staining, E18.5 embryo cross-sections were collected onto charged slides and then baked at 60°C for 30 min. Tissue sections were submitted to heat-induced epitope retrieval with citrate buffer pH 6.0 for 10 min. Non-specific antibody binding was blocked with Protein Block Serum-Free (Dako) for 10 min, followed by overnight incubation at 4°C in a primary antibody cocktail (rabbit anti-NKX2.1, Abcam ab76013 at 1:200; rat anti-SOX2, Thermo Fisher Scientific 14–9811-80 at 1:100). After washes with TBS-T, sections were incubated for 1 hour with a cocktail of Alexa Fluor-conjugated secondary antibodies at 1:200 (goat anti-rabbit IgG AF488, Thermo Fisher Scientific A32731; goat anti-rat IgG AF647, Thermo Fisher Scientific A21247), followed by counterstaining with 4′,6-diamidino-2-phenylindole (DAPI). Scanning was performed using an Olympus VS-120 slide scanner and imaged using a Hamamatsu ORCA-R2 C10600 digital camera for all dark-field and fluorescent images.
RESULTS
Two regions downstream of SOX2 gain enhancer features in cancer cells
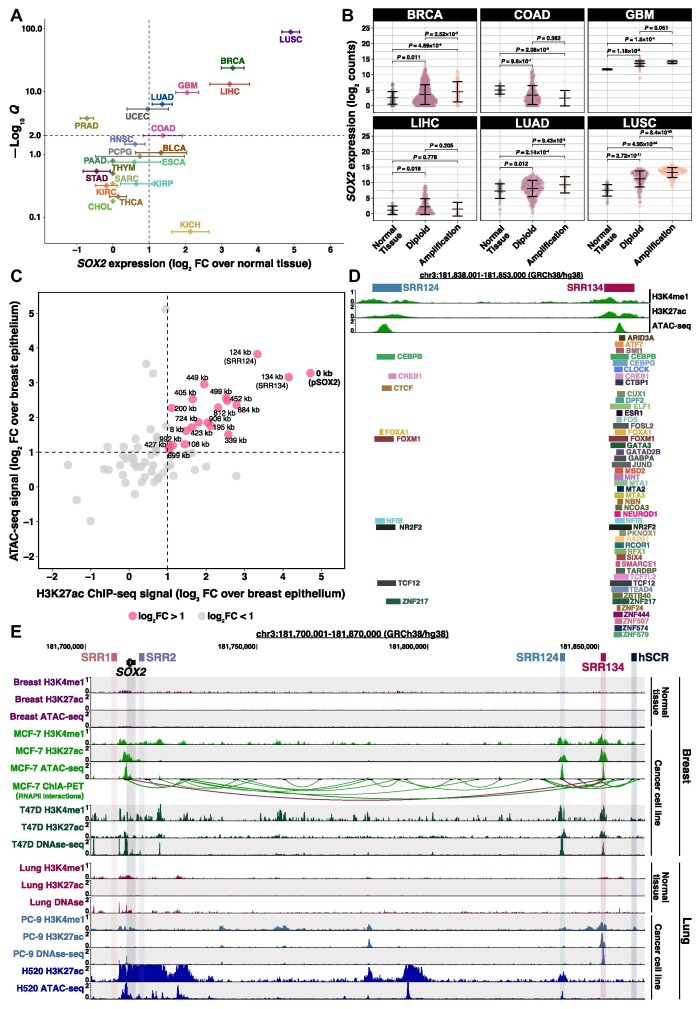
SOX2 overexpression occurs in multiple types of cancer (reviewed in 21,22). To examine which cancer types have the highest levels of SOX2 up-regulation, we performed differential expression analysis by calculating the log2 FC of SOX2 transcription from 21 TCGA primary solid tumors (see Supplementary Table S11 for cancer type abbreviations) compared with normal tissue samples (90). We found that BRCA (log2 FC = 3.31), COAD (log2 FC = 1.38), GBM (log2 FC = 2.05), LIHC (log2 FC = 3.22), LUAD (log2 FC = 1.36) and LUSC (log2 FC = 4.91) tumors had the greatest SOX2 up-regulation (log2 FC > 1; FDR-adjusted Q < 0.01; Figure 1A; Supplementary Table S12). As a negative control, we ran this same analysis using the housekeeping gene PUM1 (81) and found no cancer types with significant up-regulation of this gene (Supplementary Figure S1A; Supplementary Table S13).
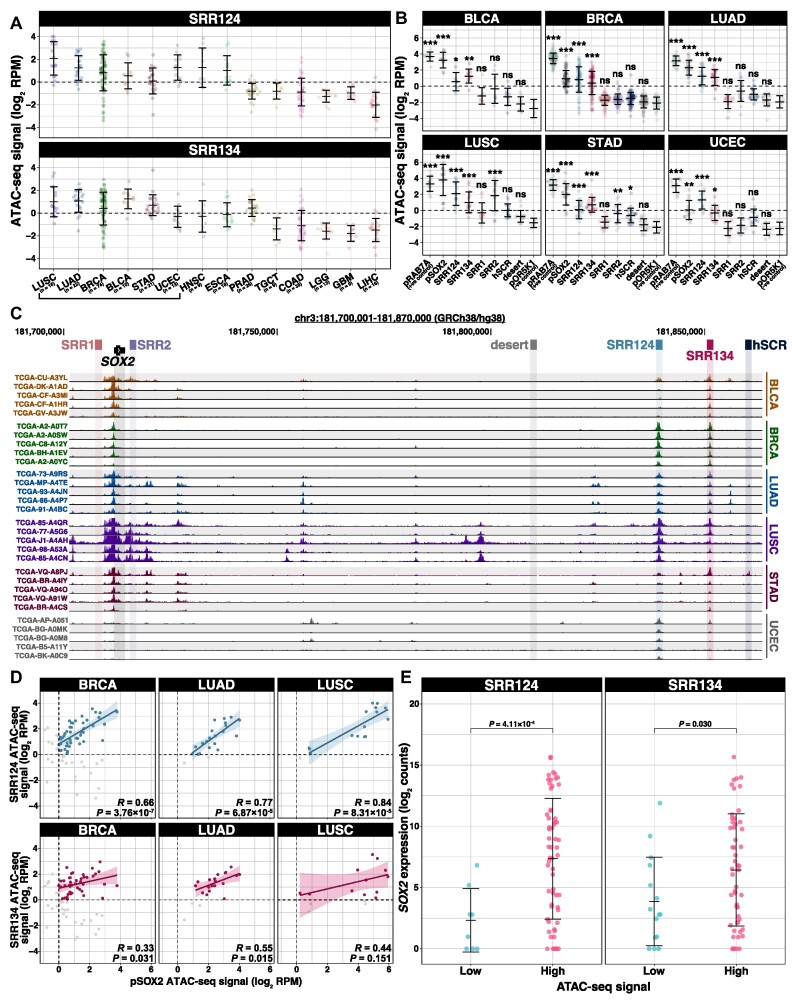
Figure 1.
A cluster 124–134 kb downstream of SOX2 gains enhancer features in cancer cells. (A) Super-logarithmic RNA-seq volcano plot of SOX2 expression from 21 cancer types compared with normal tissue (90). Cancer types with log2 FC > 1 and FDR-adjusted Q< 0.01 were considered to significantly overexpress SOX2. Error bars: standard deviation (SD). (B) SOX2 log2-normalized expression (log2 counts) associated with the SOX2 copy number from BRCA (n = 1174), COAD (n = 483), GBM (n = 155), LIHC (n = 414), LUAD (n = 552) and LUSC (n = 546) patient tumors (90). RNA-seq reads were normalized to library size using DESeq2 (88). Error bars: SD. Significance analysis by Dunn's test (180) with Holm correction (181). (C) 1500 bp genomic regions within ± 1 Mb from the SOX2 transcription start site (TSS) that gained enhancer features in MCF-7 cells (85) compared with normal breast epithelium (86). Regions that gained both ATAC-seq and H3K27ac ChIP-seq signal above our threshold (log2 FC > 1, dashed line) are highlighted in pink. Each region was labeled according to their distance in kilobases to the SOX2 promoter (pSOX2, bold). (D) ChIP-seq signal for H3K4me1 and H3K27ac, ATAC-seq signal and transcription factor ChIP-seq peaks at the SRR124–134 cluster in MCF-7 cells. Datasets are from ENCODE (85). (E) UCSC Genome Browser (102) display of H3K4me1 and H3K27ac ChIP-seq signal, DNase-seq and ATAC-seq chromatin accessibility signal, and ChIA-PET RNA polymerase II (RNAPII) interactions around the SOX2 gene within breast (normal tissue and 2 BRCA cancer cell lines) and lung (normal tissue, one LUAD and one LUSC cancer cell line) samples (85,106,108,127). Relevant RNAPII interactions (between SRR124 and SRR134, and between SRR134 and pSOX2) are highlighted in maroon.
Next, we divided BRCA, COAD, GBM, LIHC, LUAD and LUSC patients (n = 3064) into four groups according to their SOX2 expression. Gene expression levels were measured by RNA-seq counts normalized to library size and transformed to a log2 scale, hereinafter referred to as log2 counts. Cancer patients within the top group (25% highest SOX2 expression; log2 counts > 10.06) have a significantly (P = 1.27 × 10−23, log-rank test) lower overall probability of survival compared with cancer patients within the bottom group (25% lowest SOX2 expression; log2 counts < 1.68) (Supplementary Figure S1B; Supplementary Table S14). We also examined the relationship between SOX2 copy number and SOX2 overexpression within these six tumor types. Although previous studies have shown that SOX2 is frequently amplified in squamous cell carcinoma (58,59,111,112), we found that most BRCA (88%), COAD (98%), GBM (91%), LIHC (94%) and LUAD (92%) tumors were diploid for SOX2. In addition, BRCA (P = 0.011, Holm-adjusted Dunn's test), GBM (P = 1.18 × 10−3), LIHC (P = 0.016), LUAD (P = 0.012) and LUSC (P = 2.72 × 10−11) diploid tumors significantly overexpressed SOX2 compared with normal tissue (Figure 1B; Supplementary Table S15). This indicates that gene amplification is dispensable for driving SOX2 overexpression in most cancer types.
We investigated whether the SOX2 locus gains epigenetic features associated with active enhancers in cancer cells. Enhancer features commonly include accessible chromatin determined by either Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq) (113) or DNase I-hypersensitive sites sequencing (DNase-seq) (114), and histone modifications including histone H3 lysine 4 monomethylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac) (115,116). To study gains in enhancer features within the SOX2 locus, we initially focused our analyses on luminal A breast cancer, the most common subtype of BRCA to significantly (P = 0.021, Tukey's test) overexpress SOX2 (Supplementary Figure S1C) (90,117). MCF-7 cells are a widely used ER+/PR+/HER2− luminal A breast adenocarcinoma model (118), which have been previously described to overexpress SOX2 (41,69,119,120). After confirming that SOX2 is one of the most up-regulated genes in MCF-7 cells (log2 FC = 10.75; FDR-adjusted Q = 2.20 × 10−36; Supplementary Figure S1D; Supplementary Table S16) compared with normal breast epithelium (86), we contrasted their chromatin accessibility and histone modifications (85). By intersecting 1500 bp regions that contain at least a 500 bp overlap between H3K27ac and ATAC-seq peaks, we found that 19 putative enhancers gained (log2 FC > 1) both these features within ± 1 Mb from the SOX2 transcription start site (TSS) in MCF-7 cells (Figure 1C; Supplementary Table S17). Besides the SOX2 promoter (pSOX2), we identified a downstream cluster containing two regions that have gained the highest ATAC-seq and H3K27ac signal in MCF-7 cells: SRR124 (124 kb downstream of pSOX2) and SRR134 (134 kb downstream of pSOX2). The previously described SRR1, SRR2 (23,70,71) and hSCR (72,73), however, lacked substantial gains in enhancer features within MCF-7 cells.
Alongside gains in chromatin features, another characteristic of active enhancers is the binding of numerous (> 10) transcription factors (121–123). Chromatin immunoprecipitation sequencing (ChIP-seq) data from ENCODE (85) on 117 transcription factors revealed 48 different factors present at the SRR124–134 cluster in MCF-7 cells, with the majority (47) of these factors present at SRR134 (Figure 1D). Transcription factors bound at both SRR124 and SRR134 include CEBPB, CREB1, FOXA1, FOXM1, NFIB, NR2F2, TCF12 and ZNF217. An additional feature of distal enhancers is that they contact their target genes through long-range chromatin interactions (124,125). We analyzed Chromatin Interaction Analysis by Paired-End-Tag sequencing (ChIA-PET) data from MCF-7 cells (126) and found two interesting RNA polymerase II (RNAPII)-mediated chromatin interactions: one between the SOX2 gene and SRR134, and one between SRR124 and SRR134 (Figure 1E). Beyond the interactions with SOX2, we also identified long-range interactions between SRR124 and the upstream long non-coding RNA (lncRNA) SOX2-OT (∼665 kb away), between SRR134 and the downstream lncRNA LINC01206 (∼150 kb away), and between SRR134 and the upstream RSRC1 gene (∼23 Mb away) (Supplementary Table S18). In addition to MCF-7 cells, we found that H520 (LUSC), PC-9 (LUAD) and T47D (luminal A BRCA) cancer cell lines, which display varying levels of SOX2 expression (Supplementary Figure S1E), also gained substantial enhancer features at SRR124 and SRR134 when compared with normal tissue (Figure 1E) (85,106,108,127). Together, these data suggest that SRR124 and SRR134 could be active enhancers driving SOX2 transcription in BRCA, LUAD and LUSC.
The SRR124–134 cluster is essential for SOX2 expression in BRCA and LUAD cells
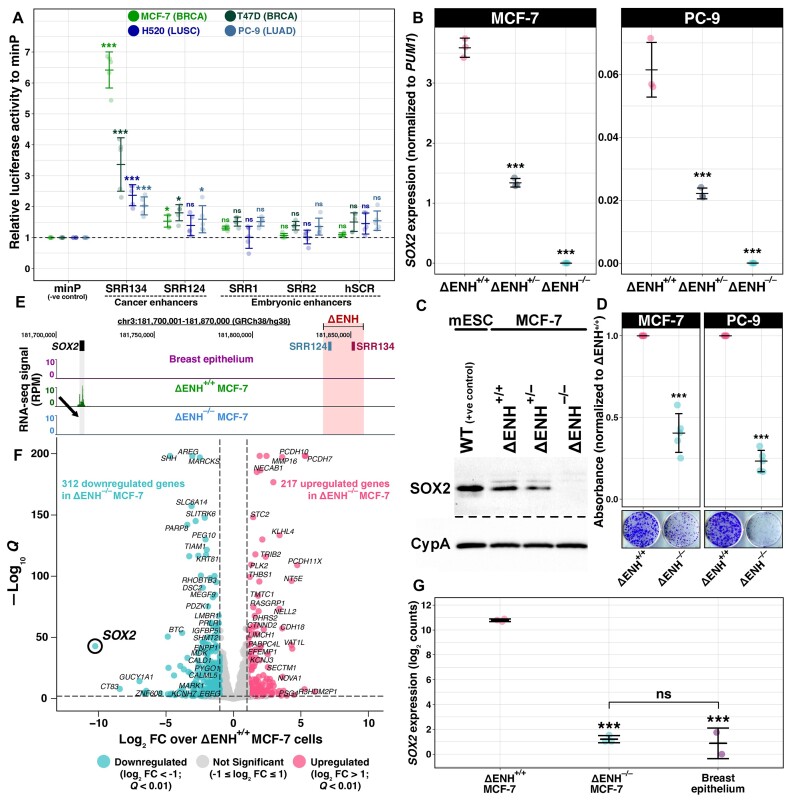
To assess SRR124 and SRR134 enhancer activity alongside the embryonic-associated SRR1, SRR2 and hSCR regions, we used a reporter vector containing the firefly luciferase gene under the control of a minimal promoter (minP, pGL4.23). We transfected each enhancer construct into the BRCA (MCF-7, T47D), LUAD (PC-9) and LUSC (H520) cell lines and measured luciferase activity as a relative FC compared with the empty minP vector. SRR134 demonstrated the strongest enhancer activity, with the MCF-7 (FC = 6.42; P < 2 × 10−16, Dunnett's test), T47D (FC = 3.36; P = 9.34 × 10−10), H520 (FC = 2.37; P = 1.22 × 10−6) and PC-9 (FC = 2.03; P = 9.79 × 10−5) cell lines displaying a significant increase in luciferase activity compared with minP (Figure 2A). SRR124 also showed a modest, significant increase in luciferase activity compared with minP in the MCF-7 (FC = 1.53; P = 4.27 × 10−2), T47D (FC = 1.80; P = 4.57 × 10−2) and PC-9 (FC = 1.60; P = 4.27 × 10−2) cell lines. The SRR1, SRR2 and hSCR enhancers, however, showed no significant enhancer activity (P > 0.05) in any of the four cell lines.
Figure 2.
The SRR124–134 cluster drives SOX2 overexpression in BRCA and LUAD cells. (A) Enhancer reporter assay comparing luciferase activity driven by the SRR1, SRR2, SRR124, SRR134 and hSCR regions with an empty vector containing only a minimal promoter (minP). Enhancer constructs were assayed in the BRCA (MCF-7, T47D), LUAD (PC-9) and LUSC (H520) cell lines. Dashed line: average activity of minP. Error bars: SD. Significance analysis by Dunnett's test (n = 5; *P < 0.05, ***P < 0.001, ns: not significant) (182). (B) RT–qPCR analysis of SOX2 transcript levels in SRR124–134 heterozygous- (ΔENH+/–) and homozygous- (ΔENH–/–) deleted MCF-7 (BRCA) and PC-9 (LUAD) clones compared with WT (ΔENH+/+) cells. Error bars: SD. Significance analysis by Dunnett's test (n = 3; ***P < 0.001). (C) SOX2 protein levels in mouse embryonic stem cells (mESCs, positive control), ΔENH+/+, ΔENH+/– and ΔENH–/– MCF-7 clones. Cyclophilin A (CypA) was used as a loading control across all samples. (D) Colony formation assay with ΔENH+/+ and ΔENH–/– MCF-7 and PC-9 cells. Total crystal violet absorbance was normalized relative to the average absorbance from ΔENH+/+ cells for each respective cell line. Significance analysis by t-test with Holm correction (n = 5; ***P < 0.001). (E) UCSC Genome Browser (102) view of the SRR124–134 cluster deletion in ΔENH–/– MCF-7 cells with RNA-seq tracks from normal breast epithelium (86), ΔENH+/+ and ΔENH–/– MCF-7 cells. Arrow: reduction in RNA-seq signal at the SOX2 gene in ΔENH–/– MCF-7 cells. (F) Volcano plot with DESeq2 (88) differential expression analysis between ΔENH–/– and ΔENH+/+ MCF-7 cells. Blue: 312 genes that significantly lost expression (log2 FC < –1; FDR-adjusted Q < 0.01) in ΔENH–/– MCF-7 cells. Pink: 217 genes that significantly gained expression (log2 FC > 1; Q < 0.01) in ΔENH–/– MCF-7 cells. Gray: 35 891 genes that maintained similar (–1 ≤ log2 FC ≤ 1) expression between ΔENH–/– and ΔENH+/+ MCF-7 cells. (G) Comparison of SOX2 transcript levels between ΔENH+/+ and either ΔENH–/– MCF-7 or normal breast epithelium cells (86), and between ΔENH–/– MCF-7 and normal breast epithelium cells. RNA-seq reads were normalized to library size using DESeq2 (88). Error bars: SD. Significance analysis by Tukey's test (***P < 0.001, ns: not significant) (183).
Although reporter assays can be used to assess enhancer activity, enhancer knockout approaches remain the current gold standard method for enhancer validation (128,129). To investigate whether the SRR124–134 cluster drives SOX2 expression in cancer cells, we used CRISPR/Cas9 to delete this cluster from breast (MCF-7, T47D) and lung (H520, PC-9) cancer cell lines (Supplementary Figure S2A). Reverse transcription–qPCR (RT–qPCR) showed that homozygous SRR124–134 deletion (ΔENH–/–) causes a profound (> 99.5%) and significant (P < 0.001, Dunnett's test) loss of SOX2 expression compared with non-deleted cells (ΔENH+/+) in both the MCF-7 and PC-9 cell lines (Figure 2B). Heterozygous SRR124–134 deletion (ΔENH+/–) also significantly (P < 0.001) reduced SOX2 expression by ∼60% in both MCF-7 and PC-9 cells (Figure 2B). Immunoblot analysis confirmed the depletion of the SOX2 protein in ΔENH–/– MCF-7 cells (Figure 2C). Although we were unable to isolate a homozygous deletion clone from T47D cells, multiple independent heterozygous ΔENH+/– T47D clonal isolates also showed a significant down-regulation (>50%; P < 0.001) in SOX2 expression (Supplementary Figure S2B). H520 cells, on the other hand, showed no significant (P > 0.05) impact on SOX2 expression following either heterozygous or homozygous deletions (Supplementary Figure S2C), which indicates that SOX2 transcription is sustained by a different mechanism in these cells. To assess the impact of the loss of SOX2 expression in the tumor initiation capacity of enhancer-deleted cells, we performed a colony formation assay with MCF-7 and PC-9 ΔENH–/– cells. We found that both MCF-7 (P = 3.53 × 10−4, t-test) and PC-9 (P = 1.26 × 10−5) ΔENH–/– cells showed a significant decrease (> 50%) in their ability to form colonies compared with ΔENH+/+ cells (Figure 2D), further underscoring the crucial role of SRR124–134-driven SOX2 overexpression in sustaining the elevated tumor initiation potential in both BRCA and LUAD.
Next, we performed total RNA sequencing (RNA-seq) to measure changes in the transcriptome of ΔENH–/– MCF-7 cells compared with ΔENH+/+ MCF-7 cells. Although RNA-seq mainly measures the steady-state level of RNA molecules in the cell, we opted for this approach to provide a broad perspective on the transcriptional changes resulting from the SRR124–134 deletion and to detect any SOX2 transcripts if they were present. As expected, all three replicates of each genotype clustered together (Supplementary Figure S2D). In addition to SOX2 down-regulation (Figure 2E), differential expression analysis showed a total of 529 genes differentially (|log2 FC| > 1; FDR-adjusted Q < 0.01) expressed in ΔENH–/– MCF-7 cells (Figure 2F; Supplementary Table S19). From these, 312 genes significantly lost expression (59%), whereas 217 (41%) genes significantly gained expression in ΔENH–/– compared with ΔENH+/+ MCF-7 cells (Supplementary Figure S2E). SOX2 was the gene with the greatest loss in expression (log2 FC = –10.24; Q = 1.23 × 10−43) in ΔENH–/– MCF-7 cells, followed by CT83 (log2 FC = –8.43; Q = 1.07 × 10−8) and GUCY1A1 (log2 FC = –6.96; Q = 5.09 × 10−15). Interestingly, the expression of the lncRNA SOX2-OT was also significantly down-regulated (log2 FC = –2.23; Q = 4.64 × 10−4) in ΔENH–/– MCF-7 cells (Supplementary Table S19). However, since this transcript overlaps the SOX2 coding region, it is unclear if this reduction is a direct result of the SRR124–134 deletion or secondary to SOX2 down-regulation. Despite showing chromatin interactions with the SRR124–134 cluster, transcription of the RSRC1 gene and the lncRNA LINC01206 remained unchanged (Q > 0.05) in ΔENH–/– MCF-7 cells. Genes with the most substantial gains in expression within ΔENH–/– MCF-7 cells included the protocadherins PCDH7 (log2 FC = 5.34; Q < 1 × 10−200), PCDH10 (log2 FC = 5.29; Q < 1 × 10−200) and PCDH11X (log2 FC = 4.73; Q = 9.29 × 10−110). Finally, deletion of the SRR124–134 cluster reduced SOX2 expression back to the levels found in normal breast epithelium (P = 0.48, Tukey's test) (85,86) (Figure 2G). Together, these data confirm that the SRR124–134 cluster drives SOX2 overexpression in BRCA and LUAD.
SOX2 regulates pathways associated with epithelium development in luminal A BRCA
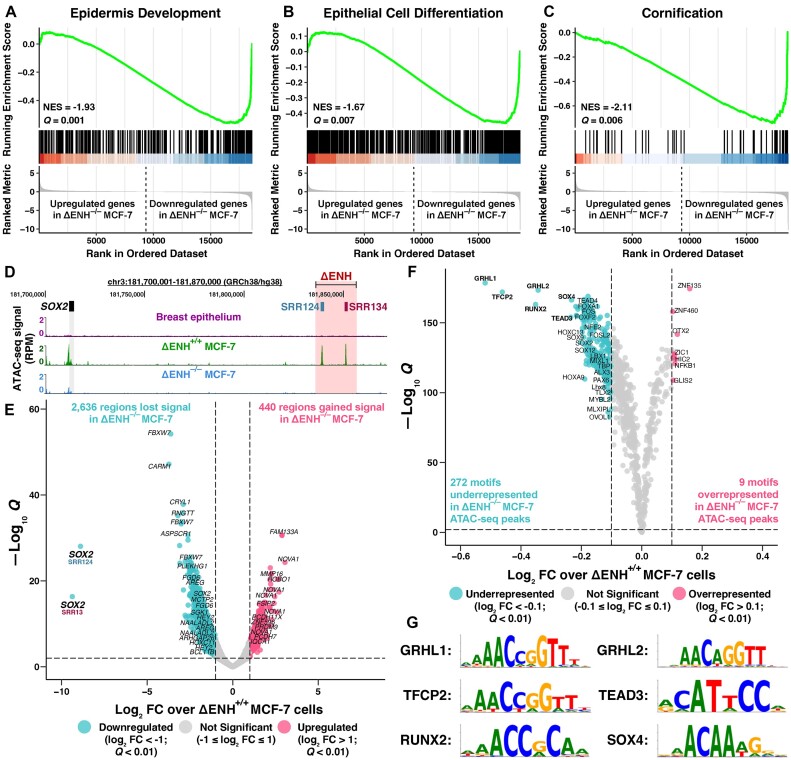
Given the established role of SOX2 in regulating proliferation and differentiation pathways in other epithelial cells (40,130), we decided to further investigate the molecular function of SOX2 in luminal A BRCA cells by leveraging our SOX2-depleted ΔENH–/– MCF-7 cell model. GSEA showed a significant (FDR-adjusted Q < 0.05) depletion of multiple epithelium-associated processes within the transcriptome of ΔENH–/– MCF-7 cells, as indicated by the normalized enrichment score (NES) < 1 (Supplementary Table S20). These processes included epidermis development (NES = –1.93; Q = 0.001; Figure 3A), epithelial cell differentiation (NES = –1.67; Q = 0.007; Figure 3B) and cornification (NES = –2.11; Q = 0.006; Figure 3C). Cornification is the process of terminal differentiation of epidermal cells, wherein these cells undergo a specialized form of programmed cell death to produce a layer of flattened, dead cells with a high keratin content (reviewed in 131). This suggests that SOX2 has a pivotal role in regulating epithelial development and differentiation pathways in luminal A BRCA cells.
Figure 3.
SOX2 down-regulation impacts chromatin accessibility in luminal A BRCA. (A–C) GSEA in the transcriptome of ΔENH–/– compared with ΔENH+/+ MCF-7 cells. Genes were ranked according to their change in expression (log2 FC). A subset of Gene Ontology (GO) terms significantly enriched among down-regulated genes in ΔENH–/– MCF-7 cells are displayed, indicated by the NES < 1: (A) epidermis development, (B) epithelial cell differentiation and (C) cornification. GSEA was performed using clusterProfiler (94) with an FDR-adjusted Q < 0.05 threshold. Green line: running enrichment score. (D) UCSC Genome Browser (102) view of the SRR124–134 deletion in ΔENH–/– MCF-7 cells with ATAC-seq tracks from breast epithelium (86), ΔENH+/+ and ΔENH–/– MCF-7 cells. (E) Volcano plot with differential ATAC-seq analysis between ΔENH–/– and ΔENH+/+ MCF-7 cells. Blue: 2638 regions that lost (log2 FC < –1; FDR-adjusted Q < 0.01) chromatin accessibility in ΔENH–/– MCF-7 cells. Pink: 440 regions that gained (log2 FC > 1; Q < 0.01) chromatin accessibility in ΔENH–/– MCF-7 cells. Gray: 132 726 regions that retained chromatin accessibility in ΔENH–/– MCF-7 cells (–1 ≤ log2 FC ≤ 1). Regions were labeled with their closest gene within a ± 1 Mb distance threshold. Differential chromatin accessibility analysis was performed using diffBind (96). (F) Volcano plot with ATAC-seq footprint analysis of differential transcription factor binding in ΔENH–/– compared with ΔENH+/+ MCF-7 cells. Blue: 272 under-represented (log2 FC < –0.1; FDR-adjusted Q < 0.01) motifs in ATAC-seq peaks from ΔENH–/– MCF-7 cells. Pink: nine over-represented (log2 FC > 0.1; Q < 0.01) motifs in ATAC-seq peaks from ΔENH–/– MCF-7 cells. Gray: 560 motifs with no representative change (–0.1 ≤ log2 FC ≤ 0.1) within ATAC-seq peaks from ΔENH–/– MCF-7 cells. (G) Sequence motifs of the top six transcription factors with the lowest binding score in ΔENH–/– compared with ΔENH+/+ MCF-7 cells: GRHL1, TFCP2, RUNX2, GRHL2, TEAD3 and SOX4. Footprint analysis was performed using TOBIAS (97) utilizing the JASPAR 2022 motif database (79).
SOX2 is a pioneer transcription factor that associates with its motif in heterochromatin (132) and recruits chromatin-modifying complexes (133) in embryonic and reprogrammed stem cells. We performed ATAC-seq in ΔENH–/– MCF-7 cells and compared chromatin accessibility with ΔENH+/+ MCF-7 cells to identify genome-wide loci that are dependent on SOX2 to remain accessible in luminal A BRCA. As expected, the ATAC-seq signal from all replicates was highly enriched around the gene TSS (Supplementary Figure S3A), with both ΔENH+/+ (Supplementary Figure S3B) and ΔENH–/– (Supplementary Figure S3C) MCF-7 cells having higher chromatin accessibility at the TSS of highly expressed genes. Correlation analysis also confirmed the clustering of all three replicates from each genotype (Supplementary Figure S3D). Including the SRR124–134 cluster and pSOX2 (Figure 3D), a total of 3076 regions of 500 bp had significant (|log2 FC| > 1; FDR-adjusted Q < 0.01) changes in chromatin accessibility in ΔENH–/– compared with ΔENH+/+ MCF-7 cells (Figure 3E; Supplementary Table S21). Most regions (86%, 2636 regions) significantly lost chromatin accessibility in ΔENH–/– MCF-7 cells and 76% (2024 regions) of these regions also gained chromatin accessibility in ΔENH+/+ MCF-7 cells compared with normal breast epithelium (86) (Supplementary Table S22). Together, this supports the important role that SOX2 plays in modulating the chromatin accessibility changes acquired in luminal A BRCA.
We used TOBIAS (97) to further analyze changes in transcription factor footprints within differential ATAC-seq peaks between ΔENH–/– and ΔENH+/+ MCF-7 cells. From 841 vertebrate motifs (79), we found a total of 281 motifs with a significant (|log2 FC| > 0.1; FDR-adjusted Q < 0.01) differential binding score (Figure 3F; Supplementary Table S23). Most of these motifs (97%, 272 motifs) were under-represented within ATAC-seq peaks in ΔENH–/– compared with ΔENH+/+ MCF-7 cells, indicating that reduced SOX2 expression affects the binding of multiple other transcription factors. Among them, the GRHL1 (log2 FC = –0.519; Q = 3 × 10−179), TFCP2 (log2 FC = –0.462; Q = 1.03 × 10−172), RUNX2 (log2 FC = –0.352; Q = 8.02 × 10−164), GRHL2 (log2 FC = –0.343; Q = 4.43 × 10−174), TEAD3 (log2 FC = –0.235; Q = 9.74 × 10−155) and SOX4 (log2 FC = –0.232; Q = 5.33 × 10−167) motifs (Figure 3G) had the most reduced binding score in ΔENH–/– MCF-7 cells compared with ΔENH+/+ MCF-7 cells. These factors belong to three main JASPAR (79) motif clusters: GRHL/TFCP (cluster 33; aaAACAGGTTtcAgtt), RUNX (cluster 60; ttctTGtGGTTttt), TEAD (cluster 2; tccAcATTCCAggcCTTta) and SOX (cluster 8; acggaACAATGgaagTGTT). The SOX cluster also included the SOX2 (log2 FC = –0.175; Q = 6.61 × 10−139) motif.
Next, we aimed to analyze ChIP-seq data from transcription factors within these motif clusters in MCF-7 cells. We utilized two published datasets: GRHL2 (107) and RUNX2 (134). Regions that lost (log2 FC < –1; Q < 0.01) chromatin accessibility in ΔENH–/– compared with ΔENH+/+ MCF-7 cells significantly (P < 2 × 10−16, hypergeometric test) overlapped regions with binding of either of these transcription factors. Among the 2636 regions that lost chromatin accessibility, 40% (750 regions) also show GRHL2 binding (Supplementary Figure S3E), whereas 21% (552 regions) share RUNX2 binding (Supplementary Figure S3F). In addition, we found multiple SOX motifs significantly (FDR-adjusted Q < 0.001) enriched within peaks from both GRHL2 (Supplementary Table S24) and RUNX2 (Supplementary Table S25) ChIP-seq datasets, further suggesting that SOX2 collaborates with GRHL2 and RUNX2 to maintain chromatin accessibility in luminal A BRCA. Expression levels of either GRHL2 or RUNX2, however, were not significantly affected by SOX2 down-regulation in ΔENH–/– MCF-7 cells (–1 ≤ log2 FC ≤ 1; Supplementary Table S19), indicating that they are not directly regulated by SOX2 at the transcriptional level but may interact at the protein level.
The SRR124–134 cluster is associated with SOX2 overexpression in primary tumors
With the confirmation that the SRR124–134 cluster drives SOX2 overexpression in the BRCA and LUAD cell lines, we investigated chromatin accessibility at this enhancer cluster within primary tumors isolated from cancer patients. By analyzing the pan-cancer ATAC-seq dataset from TCGA (100), we found that SRR124 and SRR134 are most accessible within LUSC, LUAD, BRCA, bladder carcinoma (BLCA), stomach adenocarcinoma (STAD) and uterine endometrial carcinoma (UCEC) patient tumors (Figure 4A). We also quantified the ATAC-seq signal at six other regions: the SOX2 embryonic-associated enhancers (SRR1, SRR2 and hSCR), the SOX2 promoter (pSOX2), a gene regulatory desert with no enhancer features located between the SOX2 gene and the SRR124–134 cluster (desert), and the promoter of the housekeeping gene RAB7A (pRAB7A, positive control). We then compared the chromatin accessibility levels at each of these regions with the promoter of the repressed olfactory gene OR5K1 (pOR5K1, negative control). Both SRR124 and SRR134 showed significantly increased (P < 0.05, Holm-adjusted Dunn's test) chromatin accessibility when compared with pOR5K1 in BLCA (SRR124 P = 0.014; SRR134 P = 1.52 × 10−3; Holm-adjusted Dunn's test), BRCA (SRR124 P = 1.70 × 10−20; SRR134 P = 1.03 × 10−16), LUAD (SRR124 P = 6.76 × 10−7; SRR134 P = 3.26 × 10−6), LUSC (SRR124 P = 1.62 × 10−6; SRR134 P = 7.08 × 10−4), STAD (SRR124 P = 1.15 × 10−4; SRR134 P = 1.96 × 10−7) and UCEC (SRR124 P = 3.15 × 10−5; SRR134 P = 0.025) patient tumors (Figure 4B).
Figure 4.
The SRR124–134 cluster is associated with SOX2 overexpression in cancer patient tumors. (A) ATAC-seq signal (log2 RPM) at SRR124 and SRR134 for 294 patient tumors from 14 cancer types (100). Cancer types are sorted in descending order by the median signal between all three regions. Dashed line: regions with a sum of reads above our threshold (log2 RPM > 0) were considered ‘accessible’. Error bars: SD. Underscore: top six cancer types with the highest ATAC-seq median signal. (B) ATAC-seq signal (log2 RPM) at the RAB7A promoter (pRAB7A), SOX2 promoter (pSOX2), SRR1, SRR2, SRR124, SRR134, hSCR and a desert region within the SOX2 locus (desert) compared with the background signal at the repressed OR5K1 promoter (pOR5K1) in BLCA (n = 10), BRCA (n = 74), LUAD (n = 22), LUSC (n = 16), STAD (n = 21) and UCEC (n = 13) patient tumors. Dashed line: regions with a sum of reads above our threshold (log2 RPM > 0) were considered ‘accessible’. Error bars: SD. Significance analysis by Dunn's test with Holm correction (*P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant). (C) UCSC Genome Browser (102) visualization of the SOX2 region with ATAC-seq data from BLCA, BRCA, LUAD, LUSC, STAD and UCEC patient tumors (n = 5 in each cancer type) (100). ATAC-seq reads were normalized by library size (RPM). Scale: 0–250 RPM. (D) ATAC-seq signal at SRR124 and SRR134 regions against ATAC-seq signal for the SOX2 promoter (pSOX2) from 74 BRCA, 22 LUAD and 16 LUSC patient tumors. Correlation is shown for accessible chromatin (log2 RPM > 0). Gray: tumors with closed chromatin (log2 RPM < 0) at either region, not included in the correlation analysis. Significance analysis by Pearson correlation. Bold line: fitted linear regression model. Shaded area: 95% confidence region for the regression fit. (E) Comparison of log2-normalized SOX2 transcript levels (log2 counts) between BRCA, LUAD and LUSC patient tumors according to the chromatin accessibility at SRR124 and SRR134 regions. Chromatin accessibility at each region was considered ‘low’ if log2 RPM < –1, or ‘high’ if log2 RPM > 1. RNA-seq reads were normalized to library size using DESeq2 (88). Error bars: SD. Significance analysis by a two-sided t-test with Holm correction.
One potential explanation for increased chromatin accessibility could be locus amplification. While LUSC had high levels of chromatin accessibility probably related to previously described SOX2 amplifications (58,59,111,112), most patient tumors showed no evidence of locus amplifications extending to the SRR124–134 cluster, as evidenced by the lack of significant (P > 0.05) accessibility at the intermediate desert region. In contrast, the SRR124–134 cluster displayed a consistent pattern of accessible chromatin across multiple cancer types: BLCA, BRCA, LUAD, LUSC, STAD and UCEC (Figure 4C). GBM and LGG tumors lacked accessible chromatin at this cluster but displayed increased chromatin accessibility at the SRR1 and SRR2 enhancers (Supplementary Figure S4A; Supplementary Table S26), which is consistent with the evidence that SRR1 and SRR2 drive SOX2 expression in the neural lineage (23,71,135).
Next, we reasoned that an accessible SRR124–134 cluster drives subsequent SOX2 transcription within patient tumors. If this was the case, we anticipated finding positive and significantly correlated chromatin accessibility between this enhancer cluster and pSOX2. Indeed, we found that the majority of BRCA (58%), LUAD (82%) and LUSC (69%) tumors have concurrent accessibility (log2 RPM > 0) at pSOX2, SRR124 and SRR134. Patient tumors also showed a significant (P < 0.05) correlation (Pearson R) between accessible chromatin signal at pSOX2 and at both SRR124 and SRR134 in BRCA and LUAD (Figure 4D). LUSC tumors showed a significant correlation between accessible chromatin at pSOX2 and SRR124, but not at SRR134 (Figure 4D). As a negative control, we measured the correlation between chromatin accessibility at pSOX2 and at the SOX2 desert region and found no significant (P > 0.05) correlation in any of these cancer types (Supplementary Figure S4B). We also conducted a similar analysis after segregating BRCA tumors into luminal A, luminal B, HER2+ and basal-like subtypes (100,117). Interestingly, we found that both luminal A and luminal B tumors possess a significant (P < 0.05) correlation between enhancer accessibility and pSOX2 accessibility, whereas for HER2+ tumors the correlation was weaker (Supplementary Figure S4C). Basal-like tumors, on the other hand, display no accessible chromatin at either SRR124 or SRR134. This supports that luminal BRCA and LUAD subtypes are strongly associated with increased accessibility at the SRR124–134 cluster.
Finally, by separating BRCA, LUAD and LUSC patient tumors according to their chromatin accessibility at SRR124 and SRR134, we found that tumors with the most accessible chromatin at each of these regions also significantly (P < 0.05, t-test) overexpress SOX2 compared with tumors with low chromatin accessibility at these regions (Figure 4E; Supplementary Table S27). Together, these data are consistent with a model in which increased chromatin accessibility at the SRR124–134 cluster drives SOX2 overexpression in breast and lung patient tumors.
FOXA1 and NFIB are upstream regulators of the SRR124–134 cluster
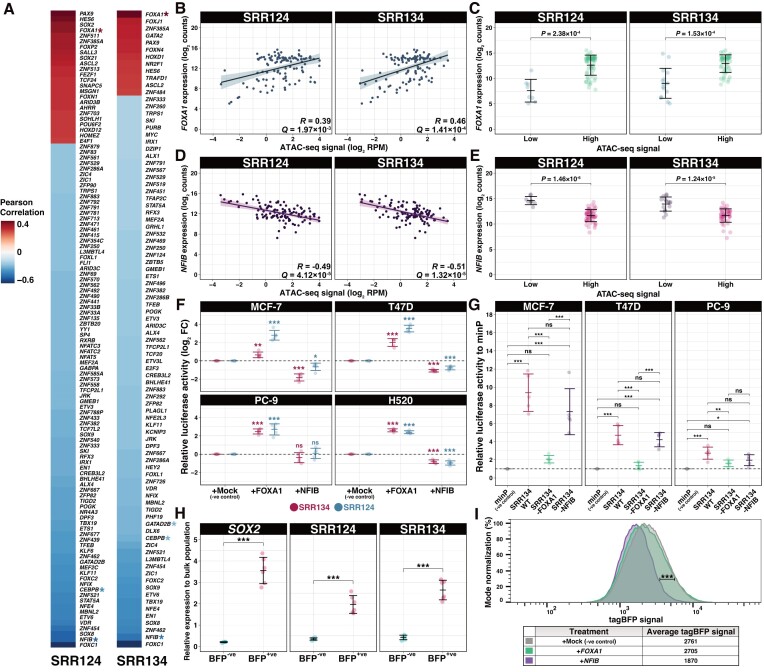
Given the evidence that the SRR124–134 cluster is driving SOX2 overexpression in cancer patient tumors, we investigated which transcription factors regulate this cluster in BRCA, LUAD and LUSC tumors from TCGA (90,100). From a comprehensive list of 1622 human transcription factors (136), we found 115 transcription factors whose expression significantly correlated (FDR-adjusted Q < 0.05) with chromatin accessibility at SRR124 and 90 transcription factors whose expression correlated with accessibility at SRR134 (Figure 5A; Supplementary Table S28). From this list, we focused our investigation on FOXA1 and NFIB, which show binding at both SRR124 and SRR134 in ChIP-seq data from MCF-7 cells (85).
Figure 5.
FOXA1 and NFIB are upstream regulators of SRR124 and SRR134. (A) Heatmap of the Pearson correlation between transcription factor expression (90) and chromatin accessibility (100) at SRR124 and SRR134 in BRCA, LUAD and LUSC patient tumors (n = 111). Transcription factors are ordered according to their correlation to chromatin accessibility at each region. Red: transcription factors with a positive correlation (R > 0; FDR-adjusted Q < 0.05) to chromatin accessibility. Blue: transcription factors with a negative correlation (R < 0; Q < 0.05) to chromatin accessibility. Asterisk: transcription factors that show binding at SRR124 or SRR134 by ChIP-seq (85). (B) Correlation analysis between FOXA1 expression (log2 counts) and chromatin accessibility (log2 RPM) at SRR124 and SRR134 regions in BRCA (n = 74), LUAD (n = 21) and LUSC (n = 16) tumors. RNA-seq reads were normalized to library size using DESeq2 (88). Significance analysis by Pearson correlation (n = 111). Bold line: fitted linear regression model. Shaded area: 95% confidence region for the regression fit. (C) Comparison of FOXA1 expression (log2 counts) from BRCA, LUAD and LUSC patient tumors according to their chromatin accessibility at the SRR124 and SRR134 regions. Chromatin accessibility at each region was considered ‘low’ if log2 RPM < 1, or ‘high’ if log2 RPM > 1. RNA-seq reads were normalized to library size using DESeq2 (88). Error bars: SD. Significance analysis by a two-sided t-test with Holm correction. (D) Correlation analysis between NFIB expression (log2 counts) and chromatin accessibility (log2 RPM) at SRR124 and SRR134 regions in BRCA (n = 74), LUAD (n = 21) and LUSC (n = 16) tumors. RNA-seq reads were normalized to library size using DESeq2 (88). Significance analysis by Pearson correlation (n = 111). Boldline: fitted linear regression model. Shaded area: 95% confidence region for the regression fit. (E) Comparison of NFIB expression (log2 counts) from BRCA, LUAD and LUSC patient tumors according to their chromatin accessibility at the SRR124 and SRR134 regions. Chromatin accessibility at each region was considered ‘low’ if log2 RPM < 1, or ‘high’ if log2 RPM > 1. RNA-seq reads were normalized to library size using DESeq2 (88). Error bars: SD. Significance analysis by a two-sided t-test with Holm correction. (F) Relative fold change (log2 FC) in luciferase activity driven by SRR124 and SRR134 after overexpression of either FOXA1 or NFIB compared with an empty vector (mock negative control, miRFP670). Dashed line: average activity of the mock control. Error bars: SD. Significance analysis by Tukey's test (n = 5; *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant). (G) Relative luciferase activity driven by WT, FOXA1-mutated and NFIB-mutated SRR134 constructs compared with a minimal promoter (minP) vector in the MCF-7, PC-9 and T47D cell lines. Dashed line: average activity of minP. Error bars: SD. Significance analysis by Tukey's test (n = 5; *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant). (H) RT–qPCR comparison of transcripts at SOX2, SRR124 and SRR134 between sorted BFP−ve and BFP+ve MCF-7 cells relative to the unsorted population. Error bars: SD. Significance analysis by paired t-test with Holm correction (n = 6; ***P < 0.001). (I) FACS density plot comparing tagBFP signal between SOX2-P2A-tagBFP MCF-7 cells transfected with an empty vector (mock negative control, miRFP670), FOXA1-T2A-miRFP670 or NFIB-T2A-miRFP670. tagBFP signal was acquired from successfully transfected live cells (miRFP+/PI–) after 5 days post-transfection. Significance analysis by FlowJo's chi-squared T(x) test. T(x) scores >1000 were considered ‘strongly significant’ (***P < 0.001), whereas T(x) scores <100 were considered ‘non-significant’.
The expression of FOXA1 is positively (Pearson correlation R > 0) and significantly correlated to chromatin accessibility at both SRR124 (R = 0.39; FDR-adjusted Q = 1.97 × 10−3) and SRR134 (R = 0.46; Q = 1.41 × 10−4) (Figure 5B). By separating BRCA, LUAD and LUSC patient tumors according to the chromatin accessibility levels at each region, we found that tumors with the most accessible chromatin within SRR124 (P = 2.38 × 10−4, t-test) and SRR134 (P = 1.53 × 10−4) also significantly overexpress FOXA1 compared with tumors with low accessibility at these regions (Figure 5C; Supplementary Table S29). On the other hand, we found the expression of NFIB to be negatively (R < 0) and significantly correlated with chromatin accessibility at both SRR124 (R = –0.49; Q = 4.12 × 10−5) and SRR134 (R = –0.51; Q = 1.32 × 10−5) (Figure 5D). Patient tumors with highly accessible chromatin within SRR124 (P = 1.46 × 10−6) and SRR134 (P = 1.24 × 10−5) also display significantly down-regulated NFIB expression (Figure 5E; Supplementary Table S30). These data suggest that whereas FOXA1 could be inducing increased accessibility at the SRR124–134 cluster, NFIB expression could counteract FOXA1 by acting as a repressor.
To assess the influence of these transcription factors on enhancer activity, we overexpressed either FOXA1 or NFIB in H520, MCF-7, PC-9 and T47D cells and compared SRR124 and SRR134 enhancer activity measured by luciferase reporter assay with cells transfected with an empty vector (mock). Despite the high endogenous expression of FOXA1 and NFIB in MCF-7 and T47D cells, but not in H520 and PC-9 cells (Supplementary Figure S5A), we found that overexpression of FOXA1 significantly increased (log2 FC > 1; P < 0.05, Tukey's test) the enhancer activity of both SRR124 and SRR134 in all four cell lines, whereas NFIB overexpression led to a significant decrease (log2 FC < 1; P < 0.05) in SRR124 and SRR134 enhancer activity in the H520, MCF-7 and T47D cell lines (Figure 5F). This further indicates that FOXA1 overexpression increases SRR124–134 activity, whereas NFIB represses the enhancer activity of this cluster.
To assess the importance of FOXA1 and NFIB motifs in modulating enhancer activity, we analyzed the SRR134 sequence using the JASPAR2022 motif database (79) and mutated FOXA1 (GTAAACA) or NFIB (TGGCAnnnnGCCAA) motifs to eliminate their binding. We found that mutation of the FOXA1 motif abolished SRR134 enhancer activity measured by luciferase reporter assay compared with the WT SRR134 sequence within MCF-7 (P = 1.53 × 10−5, Tukey's test), PC-9 (P = 1 × 10−2) and T47D (P = 4.48 × 10−6) cells, whereas no significant change (P > 0.05) in enhancer activity was found for the NFIB-mutated construct (Figure 5G). These findings underscore the pivotal role of the FOXA1 motif in maintaining SRR134 activity, whereas the NFIB motif is dispensable in this context, consistent with the behavior of a negative regulator when the target activity is elevated.
With the evidence that these two transcription factors are modulating SRR124–134 activity, we investigated their transcriptional effects on SOX2 expression. We used CRISPR HDR to create an MCF-7 cell line in which the SOX2 gene is tagged with a 2A self-cleaving peptide (P2A) followed by a blue fluorescent protein (tagBFP). This cell line, MCF-7 SOX2-P2A-tagBFP, allows rapid visualization of SOX2 transcriptional changes by measuring tagBFP signal through FACS. To validate this model, we sorted cells within the top 10% (BFP+ve) and bottom 10% (BFP−ve) tagBFP signal (Supplementary Figure S5B). We found that BFP+ve cells showed a significant (P = 4.25 × 10−5, paired t-test) increase in SOX2 expression, and displayed significantly up-regulated transcription of enhancer RNA (eRNA) at SRR124 (P = 1.54 × 10−4) and SRR134 (P = 5.13 × 10−5) compared with BFP−ve cells (Figure 5H). This confirms that the tagBFP signal is directly correlated to SOX2 transcription levels and enhancer output in MCF-7 SOX2-P2A-tagBFP cells.
Finally, we overexpressed FOXA1 or NFIB in MCF-7 SOX2-P2A-tagBFP to assess changes in SOX2 transcription. Although overexpression of FOXA1 did not significantly [chi-squared T(x) = 63.70] change the tagBFP signal, we found that overexpression of NFIB significantly [chi-squared T(x) = 1168.88] reduced the tagBFP signal compared with transfection of an empty vector (mock) (Figure 5I). This confirms the repressive effect of NFIB over SOX2 expression and illustrates a potential mechanism upstream of SOX2 that modulates chromatin accessibility at the SRR124–134 cluster and subsequent control of SOX2 transcription in cancer cells.
SRR124 and SRR134 are conserved enhancers across mammals and are required for the separation of the anterior foregut
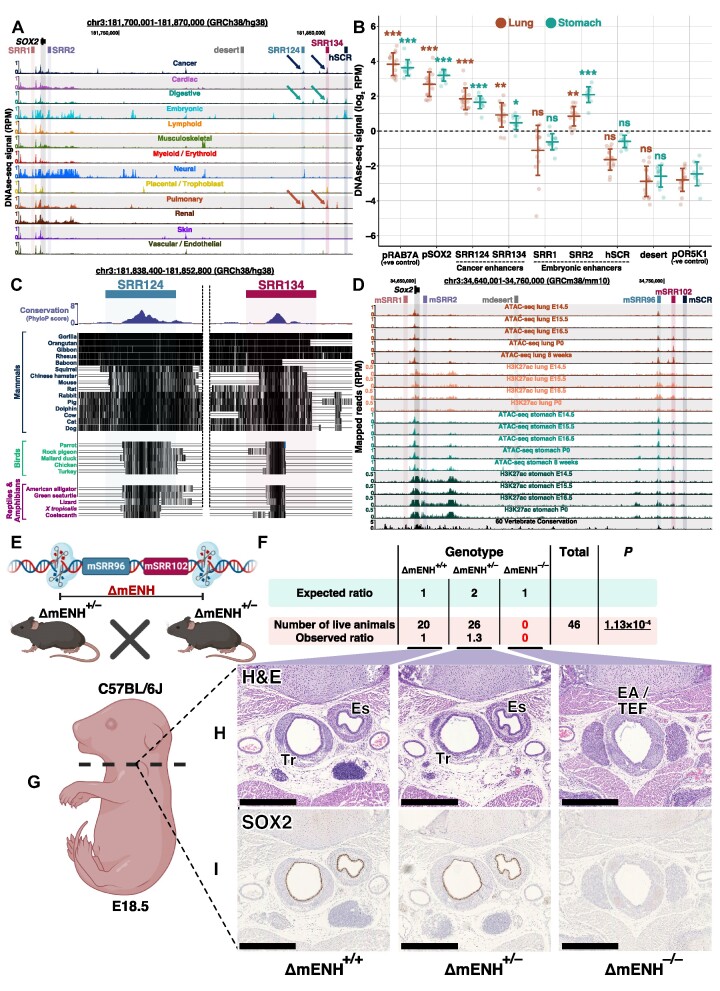
SOX2 is required for the proper development of multiple tissues (39), including the digestive and respiratory systems in the mouse (25,27,29,31,32,40) and in humans (34–36). Therefore, we questioned whether the SRR124–134 cluster drives SOX2 expression in additional contexts other than cancer. An analysis of chromatin accessibility data spanning a range of tissue types—cardiac, digestive, embryonic, lymphoid, musculoskeletal, myeloid, neural, placental, pulmonary, renal, skin and vascular tissues (85,86,137)—showed that both SRR124 and SRR134 display increased chromatin accessibility in digestive and respiratory tissues alongside cancer samples (Figure 6A). By comparing DNase-seq signal from fetal lung and stomach tissues (85), we found that both SRR124 (lung P = 1.25 × 10−6; stomach P = 9.64 × 10−4; Holm-adjusted Dunn's test) and SRR134 (lung P = 1.14 × 10−3; stomach P = 0.045), together with SRR2 (lung P = 1.55 × 10−3; stomach P = 5.74 × 10−5), are significantly more accessible than pOR5K1 (Figure 6B; Supplementary Table S31). This suggests that SRR124 and SRR134 are contributing to SOX2 expression during the development of the digestive and respiratory systems.
Figure 6.
The SRR124 and SRR134 enhancers are conserved across species and are required for the separation of the esophagus and trachea in the mouse. (A) UCSC Genome Browser (102) view of the SOX2 region containing a compilation of chromatin accessibility tracks of multiple human tissues (85,86,137). Arrow: increased chromatin accessibility at the SRR124–134 cluster in cancer and in digestive and respiratory tissues. (B) DNase-seq quantification (log2 RPM) at the RAB7A promoter (pRAB7A), SOX2 promoter (pSOX2), SRR1, SRR2, SRR124, SRR134, hSCR and a desert region within the SOX2 locus (desert) compared with the background signal at the repressed OR5K1 promoter (pOR5K1) in lung and stomach embryonic tissues (85). Dashed line: regions with a sum of reads above our threshold (log2 RPM > 0) were considered ‘accessible’. Error bars: SD. Significance analysis by Dunn's test with Holm correction (*P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant). (C) UCSC Genome Browser (102) with PhyloP conservation scores (103) at the SRR124 and SRR134 enhancers across mammals, birds, reptiles and amphibians. Black lines: highly conserved sequences. Empty lines: variant sequences. (D) UCSC Genome Browser (102) view of the Sox2 region in the mouse. ATAC-seq and H3K27ac ChIP-seq data from lung and stomach tissues throughout developmental days E14.5 to the eighth post-natal week (85,101). mSRR96: homologous to SRR124. mSRR102: homologous to SRR134. Reads were normalized to library size (RPM). (E) Illustration demonstrating the mSRR96–102 enhancer cluster CRISPR deletion (ΔmENH) in C57BL/6J mouse embryos. (F) Quantification and genotype of the C57BL/6J progeny from mSRR96–102-deleted crossings (ΔmENH+/–). Pups were counted and genotyped at weaning (P21). Significance analysis by chi-squared test to measure the deviation in the number of obtained pups from the expected Mendelian ratio of 1:2:1 (ΔmENH+/+:ΔmENH+/–:ΔmENH–/–). (G) Transverse cross-section of fixed E18.5 embryos at the start of the thymus. (H) Embryo sections stained with H&E. Scale bar: 500 μm. Es, esophagus; Tr, trachea; EA/TEF, esophageal atresia with distal tracheoesophageal fistula. (I) Embryo cross-sections stained for SOX2. Scale bar: 500 μm. Es, esophagus; Tr, trachea; EA/TEF, esophageal atresia with distal tracheoesophageal fistula.
Since critical developmental genes are often controlled by highly conserved enhancers across species (138,139), we hypothesized that the SRR124–134 cluster might regulate SOX2 expression during the development of other species. By analyzing PhyloP conservation scores (102,103), we discovered that both SRR124 and SRR134 contain a highly conserved core sequence that is preserved across mammals, birds, reptiles and amphibians (Figure 6C). After aligning and comparing enhancer sequences between humans and mice, we found that the core sequences at both SRR124 and SRR134 are highly conserved (> 80%) in the mouse genome (Supplementary Figure S6A). We termed these homologous regions mSRR96 (96 kb downstream of the mouse Sox2 promoter; homologous to the human SRR124) and mSRR102 (102 kb downstream of the mouse Sox2 promoter; homologous to the human SRR134). Enhancer feature analysis in the developing lung and stomach tissues in the mouse (85,101) showed that both mSRR96 and mSRR102 display increased chromatin accessibility and H3K27ac signal throughout developmental days E14.5 to the eighth post-natal week (Figure 6D). Interestingly, mSRR96 and mSRR102 display higher ATAC-seq and H3K27ac signal towards the later stages of development in the lungs, but at early stages of development in the stomach. This suggests a distinct spatiotemporal contribution of this homologous cluster to Sox2 expression during the development of these tissues in the mouse. Furthermore, ATAC-seq quantification showed that both mSRR96 (lung P = 5.54 × 10−5; stomach P = 2.37 × 10−4; Holm-adjusted Dunn's test) and mSRR102 (lung P = 1.27 × 10−3; stomach P = 0.046) are significantly more accessible than the repressed promoter of the olfactory gene Olfr266 (pOlfr266, negative control) during the development of the lungs and stomach in the mouse (Supplementary Figure S6B; Supplementary Table S32). Together, these results suggest a conserved SOX2 regulatory mechanism across multiple species and support a model in which the SRR124 and SRR134 enhancers and their homologs regulate SOX2 expression during the development of the digestive and respiratory systems.
To assess the contribution of the mSRR96 and mSRR102 regions to the development of the mouse, we generated a C57BL/6J knockout containing a deletion spanning the mSRR96–102 enhancer cluster (ΔmENH) (Figure 6E). We crossed animals carrying a heterozygous mSRR96–102 deletion (ΔmENH+/–) and determined the number of pups alive at weaning (P21) from each genotype. We found a significant (P = 1.13 × 10−4, Chi-squared test) deviation from the expected Mendelian ratio, with no homozygous mice (ΔmENH–/–) alive at weaning (Figure 6F), demonstrating that the mSRR96–102 enhancer cluster is crucial for survival in the mouse. To investigate the resulting phenotype in a homozygous mSRR96–102 enhancer deletion, we collected E18.5 littermate embryos and prepared cross-sections at the thymus level from five animals of each genotype (ΔmENH+/+, ΔmENH+/– and ΔmENH–/–) (Figure 6G). Similar to other studies that interfered with Sox2 expression during development (25,32,33), we found that all five ΔmENH–/– embryos developed EA/TEF, where the esophagus and trachea fail to separate during embryonic development (Figure 6H; Supplementary Figure S6C). In contrast, ΔmENH+/+ and ΔmENH+/– embryos displayed normal development of the esophageal and tracheal tissues. Immunohistochemistry revealed the complete absence of the SOX2 protein within the EA/TEF tissue in ΔmENH–/– embryos, whereas ΔmENH+/+ and ΔmENH+/– embryos showed high levels of SOX2 protein within both the esophagus and tracheal tubes (Figure 6I). Finally, immunofluorescence staining for NKX2.1, a transcription factor associated with the inner epithelium of the respiratory tract (140), showed high protein levels within the inner layer of the EA/TEF tissue in ΔmENH–/– embryos, indicating that this aberrant tissue resembles a tracheal-like structure lacking SOX2 (Supplementary Figure S7A). Together, these results demonstrate that mSRR96 and mSRR102 are required to drive Sox2 expression during the development and separation of the esophagus and trachea.
DISCUSSION
Our findings reveal that the SRR124–134 enhancer cluster is essential for Sox2 expression in the developing digestive and respiratory systems as it is required for the separation of the esophagus and trachea during mouse development. When embryogenesis is complete, Sox2 expression is down-regulated in most differentiated cell types as its developmental enhancers are decommissioned. We propose that aberrant up-regulation of the pioneer factor FOXA1 recommissions both SRR124 and SRR134 in tumor cells, driving SOX2 overexpression in breast and lung adenocarcinoma. Given that SOX2 itself acts as a pioneer transcription factor throughout development, we determined that increased levels of this protein further reprogram the chromatin landscape of cancer cells, binding at multiple regulatory regions, increasing chromatin accessibility, and driving subsequent up-regulation of genes associated with epithelium development. Previous studies have already underscored the indispensable role of SOX2 in both preserving gene expression patterns and orchestrating long-range chromatin interactions in neural stem cells (141), where SOX2 acts as a master regulator (23,142). Considering our observation that the loss of SOX2 expression leads to a genome-wide reduction in chromatin accessibility and transcription, our results position SOX2 as a central agent in the aberrant activation of gene regulatory pathways that ultimately support a tumor-initiating phenotype in breast and lung adenocarcinomas.
Our discovery that enhancers involved in the development of the digestive and respiratory systems are reprogrammed to support SOX2 up-regulation during tumorigenesis is in line with previous observations that tumor-initiating cells acquire a less differentiated phenotype (143–146). It is more surprising, however, that the SOX2 gene is regulated by common enhancers in both breast and lung adenocarcinoma cells as enhancers are usually highly tissue specific (6,138,139,147). Our observation that FOXA1 expression is significantly correlated to chromatin accessibility at the SRR124–134 cluster and increases the transcriptional output of the SRR124 and SRR134 enhancers provides a mechanistic link between breast and lung developmental programs and cancer progression. FOXA1 is directly involved in the branching morphogenesis of the epithelium in breast (148,149) and lung (150,151) tissues, where SOX2 also plays an important role (27,60). Overexpression of both FOXA1 (6,9,10,13,152–154) and SOX2 (55,66,155) have been individually linked to the activation of transcriptional programs associated with multiple types of cancer. Therefore, we propose that FOXA1 is one of the key players responsible for the reprogramming of the SRR124–134 cluster in cancer, which then drives SOX2 overexpression in breast and lung tumors. It remains intriguing, however, that we were unable to detect a further increase in SOX2 expression in MCF-7 cells overexpressing FOXA1 despite observing an up-regulation in SRR124 and SRR134 activity measured by luciferase assay. Since FOXA1 is already highly expressed in MCF-7 cells, we reason that exogenous overexpression of FOXA1 may be incapable of further increasing SOX2 expression if transcriptional levels are already high, such as in the case of MCF-7 cells. Furthermore, our approach to detect changes in SOX2 transcription using BFP as a fluorescent reporter may have limited our ability to detect small changes in gene expression compared with the higher sensitivity obtained from the luciferase reporter. As mutation of the FOXA1 motif disrupted SRR134 enhancer activity, and this motif is shared among other members of the forkhead box (FOX) transcription factor family (156), it also remains possible that other FOX proteins are involved in activating the SRR124–134 cluster. For example, FOXM1 overexpression, which also showed binding at both SRR124 and SRR134 in MCF-7 cells, has similarly been associated with poor patient outcomes in multiple types of cancer (157).
In addition to the activating role of FOXA1, we identified NFIB as a negative regulator of SOX2 expression through inhibition of SRR124–134 activity. NFIB is normally required for the development of multiple tissues (reviewed in 158), including the brain and lungs (159–161), tissues in which SOX2 expression is also tightly regulated (27,142). In the lungs, NFIB is essential for promoting the maturation and differentiation of progenitor cells (159,160). This is in stark contrast to SOX2, which inhibits the differentiation of lung cells (27). Interestingly, NFIB seems to have paradoxical roles in cancer, acting both as a tumor suppressor and as an oncogene in different tissues (162). Among its tumor suppressor activities, NFIB acts as a barrier to skin carcinoma progression (163), and its down-regulation is associated with dedifferentiation and aggressiveness in LUAD (164). On the other hand, SOX2 promotes skin (66) and lung (165) cancer progression. As an oncogene, NFIB promotes cell proliferation and metastasis in STAD (166), where SOX2 down-regulation is associated with poor patient outcomes (167–169). With this contrasting relationship between SOX2 and NFIB across multiple tissues, we propose that NFIB normally acts as a suppressor of SRR124–134 activity and SOX2 expression during the differentiation of progenitor cells; down-regulation of NFIB expression then results in SOX2 overexpression during breast and lung tumorigenesis.
We initially hypothesized that SRR1 and SRR2 (70,71,170), and/or the SCR (72,73), might be recommissioned during cancer progression, as stem cell-related enhancers have been shown to acquire enhancer features in tumorigenic cells (171). Although other studies have also proposed the activation of either SRR1 (42,69) or SRR2 (172,173) as the main drivers of SOX2 overexpression in BRCA, we found no evidence of this mechanism and instead identified the SRR124–134 cluster as the main driver of SOX2 expression in BRCA and LUAD. Our patient tumor analysis did show that GBM and LGG were the only cancer types that display a unique and consistent pattern of accessible chromatin at SRR1 and SRR2, which is probably related to glioma cells assuming a neural stem cell-like identity to sustain high levels of cell proliferation in the brain (62). In fact, SRR2 deletion was shown to down-regulate SOX2 and reduce cell proliferation in GBM cells (174), highlighting enhancer specificity to different tumor types. In line with these findings, our observation that PC-9 LUAD cells are dependent on SRR124–134 for SOX2 transcription, whereas in H520 LUSC cells SRR124–134 is dispensable, again underscores these tumor type-specific regulatory mechanisms. LUSC tumors frequently amplify the SOX2 locus (58,59,111,112), whereas LUAD tumors do not (175), indicating that different mechanisms are involved in genome dysregulation in these two subtypes of lung cancer. Indeed, we found FOXA1 expression to be the lowest in H520 cells, which may explain the diminished transcriptional activity of the SRR124–134 cluster in this cell line. Interestingly, a further downstream enhancer cluster located ∼55 kb away from SRR124–134 exhibits high H3K27ac signal and is co-amplified with SOX2 in H520 cells and other LUSC cell lines (112), revealing an alternative mechanism that could sustain SOX2 overexpression in the absence of the SRR124–134 cluster in certain types of LUSC but not in LUAD.
Enhancer clusters often contain individual enhancers with partially redundant functions (128,176,177). Our analyses positioned SRR134 as the most potent enhancer within the SRR124–134 cluster. This is not surprising since SRR134 also shows a higher amount of transcription factor binding in MCF-7 cells, a key feature associated with enhancer activity (123). However, while both SRR124 and SRR134 display similar chromatin accessibility in MCF-7 cells, PC-9 cells showed much greater accessibility at the SRR134 enhancer, whereas T47D and H520 cells showed a more accessible SRR124 region. Given that SOX2 expression is more elevated in MCF-7, T47D and H520 compared with PC-9 cells, we postulate that simultaneous activation of both SRR124 and SRR134 enhancers may be crucial for optimal SOX2 transcription. Another distinguishing feature between these enhancers is the exclusive binding of CTCF at SRR124. CTCF is a transcription factor involved in chromatin structure and distal enhancer–promoter loop formation at some loci (178,179). Based on these findings, we propose that SRR124 acts as a tether between pSOX2 and SRR134, the latter functioning as a docking region for the binding of multiple transcription factors that ultimately drive SOX2 overexpression. Therefore, in a scenario where both enhancers are accessible, we believe the chromatin dynamics facilitate enhanced interactions between pSOX2 and the entire SRR124–134 cluster, ultimately elevating the transcription of SOX2.
Deletion of mSRR96–102, a homolog of the human SRR124–134 cluster, resulted in EA/TEF, which is also observed in human cases with SOX2 heterozygous mutations (34–36). A recent study showed that insertion of a CTCF insulation cluster downstream of the Sox2 gene, but upstream of mSRR96–102, disrupts Sox2 expression, impairs separation of the esophagus and trachea, and results in perinatal lethality due to EA/TEF in the mouse (33). This was of particular interest for understanding enhancer functional nuances during development since the SCR, which is required for Sox2 transcription at implantation, can partially overcome the insulator effect of this insertion. The authors proposed that enhancer density might explain the EA/TEF phenotype, as chromatin features suggested that enhancers in the developing lung and stomach tissues might be spread over a 400 kb domain (33). However, the 6 kb deletion that removes the mSRR96–102 cluster causing EA/TEF suggests that this is not the case. Instead, we propose that the sensitivity of each cell type to gene dosage is behind the differing ability of CTCF to block distal enhancers. This is based on two observations: in humans, heterozygous SOX2 mutations are linked with the anophthalmia–esophageal–genital syndrome (34–36); in mice, hypomorphic Sox2 alleles display similar phenotypes in the eye (24) and EA/TEF (25,32). This suggests that cells from the peri-implantation phase are less sensitive to lower Sox2 dosages compared with cells from the developing airways and digestive systems in both species, and explains the aberrant phenotypes observed at term.
Overall, our findings illustrate how cis-regulatory regions can similarly drive gene expression in both normal and diseased contexts and serve as a prime example of how decommissioned developmental enhancers may be reprogrammed during tumorigenesis. The fact that we have found a digestive/respiratory-associated enhancer cluster driving gene expression in a non-native context such as BRCA remains intriguing and reinforces a model in which tumorigenic cells often revert to a progenitor-like state that combines cis-regulatory features of progenitor cells from multiple developing lineages (6). This ‘dys-differentiation’ mechanism seems to be centered around the overexpression of a few key development-associated pioneer transcription factors such as FOXA1 and SOX2. Identifying additional mechanisms that regulate the reprogramming of these enhancers could lead to new approaches to target tumor-initiating cells that depend on SOX2 overexpression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of the Mitchell laboratory for helpful discussions, and Mathieu Lupien for manuscript review. We also thank the ENCODE Consortium and the TCGA project for generating and releasing data to the scientific community. Finally, we thank the contribution of the staff at TCP (The Centre for Phenogenomics), including Kyle Roberton, who handled the embedding, cutting and H&E staining of E18.5 mouse embryos, and Vivian Bradaschia, responsible for the IHC staining. BioRender.com was used to create parts of Figure 6E and G and the graphical abstract.
Author contributions: L.E.A. designed and performed bioinformatic analyses, cell culture work, CRISPR deletions, data curation and gene expression quantification, and led the conceptualization and writing of the manuscript; P.L.F. assessed cellular phenotypes, including the colony formation assay; L.H. acquired and processed TCGA ATAC-seq data, and assisted in writing review & editing; M.C. assisted in the writing review & editing; M.M.H. provided TCGA data access and assisted in writing review & editing; J.A.M. was involved in supervision, funding acquisition, data interpretation, experimental design and writing review & editing. All authors have participated in the editing and approval of the manuscript.
Notes
Present address: Jennifer A. Mitchel, Department of Cell and Systems Biology, University of Toronto, Toronto, Canada.
Contributor Information
Luis E Abatti, Department of Cell and Systems Biology, University of Toronto, Toronto, Ontario, Canada.
Patricia Lado-Fernández, Laboratory of Cell Senescence, Cancer and Aging, Health Research Institute of Santiago de Compostela (IDIS), Xerencia de Xestión Integrada de Santiago (XXIS/SERGAS), Santiago de Compostela, Spain; Department of Physiology and Center for Research in Molecular Medicine and Chronic Diseases (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela, Spain.
Linh Huynh, Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada.
Manuel Collado, Laboratory of Cell Senescence, Cancer and Aging, Health Research Institute of Santiago de Compostela (IDIS), Xerencia de Xestión Integrada de Santiago (XXIS/SERGAS), Santiago de Compostela, Spain.
Michael M Hoffman, Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada; Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada; Department of Computer Science, University of Toronto, Toronto, Ontario, Canada; Vector Institute for Artificial Intelligence, Toronto, Ontario, Canada.
Jennifer A Mitchell, Department of Cell and Systems Biology, University of Toronto, Toronto, Ontario, Canada; Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada.
DATA AVAILABILITY
Sequencing and processed data files were submitted to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) repository (GSE132344).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Canadian Institutes of Health Research [FRN PJT153186 and PJT180312]; the Canada Foundation for Innovation; and the Ontario Ministry of Research and Innovation [operating and infrastructure grants held by J.A.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L.et al.. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013; 152:642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hawkins R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S.et al.. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010; 6:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A.et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rubin A.J., Barajas B.C., Furlan-Magaril M., Lopez-Pajares V., Mumbach M.R., Howard I., Kim D.S., Boxer L.D., Cairns J., Spivakov M.et al.. Lineage-specific dynamic and pre-established enhancer–promoter contacts cooperate in terminal differentiation. Nat. Genet. 2017; 49:1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stergachis A.B., Neph S., Reynolds A., Humbert R., Miller B., Paige S.L., Vernot B., Cheng J.B., Thurman R.E., Sandstrom R.et al.. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013; 154:888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu X., Pereira R., De Angelis C., Veeraraghavan J., Nanda S., Qin L., Cataldo M.L., Sethunath V., Mehravaran S., Gutierrez C.et al.. FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc. Natl Acad. Sci. USA. 2019; 116:26823–26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roe J.-S., Hwang C.-I., Somerville T.D.D., Milazzo J.P., Lee E.J., Da Silva B., Maiorino L., Tiriac H., Young C.M., Miyabayashi K.et al.. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017; 170:875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bi M., Zhang Z., Jiang Y.-Z., Xue P., Wang H., Lai Z., Fu X., De Angelis C., Gong Y., Gao Z.et al.. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat. Cell Biol. 2020; 22:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pomerantz M.M., Li F., Takeda D.Y., Lenci R., Chonkar A., Chabot M., Cejas P., Vazquez F., Cook J., Shivdasani R.A.et al.. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015; 47:1346–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M.. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richart L., Bidard F.-C., Margueron R.. Enhancer rewiring in tumors: an opportunity for therapeutic intervention. Oncogene. 2021; 40:3475–3491. [DOI] [PubMed] [Google Scholar]
- 15. Okabe A., Kaneda A.. Transcriptional dysregulation by aberrant enhancer activation and rewiring in cancer. Cancer Sci. 2021; 112:2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Avilion A.A. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003; 17:126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chew J.-L., Loh Y.-H., Zhang W., Chen X., Tam W.-L., Yeap L.-S., Li P., Ang Y.-S., Lim B., Robson P.et al.. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005; 25:6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 19. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S.. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. [DOI] [PubMed] [Google Scholar]
- 20. Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R.et al.. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007; 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 21. Wuebben E.L., Rizzino A.. The dark side of SOX2: cancer—a comprehensive overview. Oncotarget. 2017; 8:44917–44943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Novak D., Hüser L., Elton J.J., Umansky V., Altevogt P., Utikal J.. SOX2 in development and cancer biology. Semin. Cancer Biol. 2019; 67:74–82. [DOI] [PubMed] [Google Scholar]
- 23. Ferri A.L.M., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P.P., Sala M., DeBiasi S.et al.. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004; 131:3805–3819. [DOI] [PubMed] [Google Scholar]
- 24. Taranova O.V., Magness S.T., Fagan B.M., Wu Y., Surzenko N., Hutton S.R., Pevny L.H.. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006; 20:1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Que J., Okubo T., Goldenring J.R., Nam K.-T., Kurotani R., Morrisey E.E., Taranova O., Pevny L.H., Hogan B.L.M.. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007; 134:2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiernan A.E., Pelling A.L., Leung K.K.H., Tang A.S.P., Bell D.M., Tease C., Lovell-Badge R., Steel K.P., Cheah K.S.E.. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005; 434:1031–1035. [DOI] [PubMed] [Google Scholar]
- 27. Gontan C., de Munck A., Vermeij M., Grosveld F., Tibboel D., Rottier R.. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev. Biol. 2008; 317:296–309. [DOI] [PubMed] [Google Scholar]
- 28. Driskell R.R., Giangreco A., Jensen K.B., Mulder K.W., Watt F.M.. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development. 2009; 136:2815–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Francis R., Guo H., Streutker C., Ahmed M., Yung T., Dirks P.B., He H.H., Kim T.-H.. Gastrointestinal transcription factors drive lineage-specific developmental programs in organ specification and cancer. Sci. Adv. 2019; 5:eaax8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okubo T., Pevny L.H., Hogan B.L.M.. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006; 20:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Que J., Luo X., Schwartz R.J., Hogan B.L.M.. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009; 136:1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teramoto M., Sugawara R., Minegishi K., Uchikawa M., Takemoto T., Kuroiwa A., Ishii Y., Kondoh H.. The absence of SOX2 in the anterior foregut alters the esophagus into trachea and bronchi in both epithelial and mesenchymal components. Biology Open. 2020; 9:bio048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chakraborty S., Kopitchinski N., Zuo Z., Eraso A., Awasthi P., Chari R., Mitra A., Tobias I.C., Moorthy S.D., Dale R.K.et al.. Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat. Genet. 2023; 55:280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zenteno J.C., Perez-Cano H.J., Aguinaga M.. Anophthalmia–esophageal atresia syndrome caused by an SOX2 gene deletion in monozygotic twin brothers with markedly discordant phenotypes. Am. J. Med. Genet. A. 2006; 140:1899–1903. [DOI] [PubMed] [Google Scholar]
- 35. Williamson K.A., Hever A.M., Rainger J., Rogers R.C., Magee A., Fiedler Z., Keng W.T., Sharkey F.H., McGill N., Hill C.J.et al.. Mutations in SOX2 cause anophthalmia–esophageal–genital (AEG) syndrome. Hum. Mol. Genet. 2006; 15:1413–1422. [DOI] [PubMed] [Google Scholar]
- 36. Chassaing N., Gilbert-Dussardier B., Nicot F., Fermeaux V., Encha-Razavi F., Fiorenza M., Toutain A., Calvas P.. Germinal mosaicism and familial recurrence of a SOX2 mutation with highly variable phenotypic expression extending from AEG syndrome to absence of ocular involvement. Am. J. Med. Genet. A. 2007; 143:289–291. [DOI] [PubMed] [Google Scholar]
- 37. Brunner H.G., van Bokhoven H.. Genetic players in esophageal atresia and tracheoesophageal fistula. Curr. Opin. Genet. Dev. 2005; 15:341–347. [DOI] [PubMed] [Google Scholar]
- 38. Que J., Choi M., Ziel J.W., Klingensmith J., Hogan B.L.M.. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and bmps. Differentiation. 2006; 74:422–437. [DOI] [PubMed] [Google Scholar]
- 39. Arnold K., Sarkar A., Yram M.A., Polo J.M., Bronson R., Sengupta S., Seandel M., Geijsen N., Hochedlinger K.. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011; 9:317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tompkins D.H., Besnard V., Lange A.W., Wert S.E., Keiser A.R., Smith A.N., Lang R., Whitsett J.A.. Sox2 Is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS One. 2009; 4:e8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y., Shi L., Zhang L., Li R., Liang J., Yu W., Sun L., Yang X., Wang Y., Zhang Y.et al.. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J. Biol. Chem. 2008; 283:17969–17978. [DOI] [PubMed] [Google Scholar]
- 42. Leis O., Eguiara A., Lopez-Arribillaga E., Alberdi M.J., Hernandez-Garcia S., Elorriaga K., Pandiella A., Rezola R., Martin A.G.. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012; 31:1354–1365. [DOI] [PubMed] [Google Scholar]
- 43. Liu P., Tang H., Song C., Wang J., Chen B., Huang X., Pei X., Liu L.. SOX2 promotes cell proliferation and metastasis in triple negative breast cancer. Front. Pharmacol. 2018; 9:942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meng Y., Xu Q., Chen L., Wang L., Hu X.. The function of SOX2 in breast cancer and relevant signaling pathway. Pathol. Res. Pract. 2020; 216:153023. [DOI] [PubMed] [Google Scholar]
- 45. Piva M., Domenici G., Iriondo O., Rábano M., Simões B.M., Comaills V., Barredo I., López-Ruiz J.A., Zabalza I., Kypta R.et al.. Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol. Med. 2014; 6:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takeda K., Mizushima T., Yokoyama Y., Hirose H., Wu X., Qian Y., Ikehata K., Miyoshi N., Takahashi H., Haraguchi N.et al.. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018; 8:17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Talebi A., Kianersi K., Beiraghdar M.. Comparison of gene expression of SOX2 and OCT4 in normal tissue, polyps, and colon adenocarcinoma using immunohistochemical staining. Adv. Biomed. Res. 2015; 4:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X.-H., Wang W., Wang Y.-Q., Zhu L., Ma L.. The association of SOX2 with clinical features and prognosis in colorectal cancer: a meta-analysis. Pathol. Res. Pract. 2020; 216:152769. [DOI] [PubMed] [Google Scholar]
- 49. Zhu Y., Huang S., Chen S., Chen J., Wang Z., Wang Y., Zheng H.. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial–mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021; 12:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alonso M.M., Diez-Valle R., Manterola L., Rubio A., Liu D., Cortes-Santiago N., Urquiza L., Jauregi P., de Munain A.L., Sampron N.et al.. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011; 6:e26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cox J.L., Wilder P.J., Desler M., Rizzino A.. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PLoS One. 2012; 7:e44087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gangemi R.M.R., Griffero F., Marubbi D., Perera M., Capra M.C., Malatesta P., Ravetti G.L., Zona G.L., Daga A., Corte G.. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009; 27:40–48. [DOI] [PubMed] [Google Scholar]
- 53. Hägerstrand D., He X., Bradic Lindh M., Hoefs S., Hesselager G., Ostman A., Nistér M.. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011; 13:1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun C., Sun L., Li Y., Kang X., Zhang S., Liu Y.. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med. Oncol. 2013; 30:503. [DOI] [PubMed] [Google Scholar]
- 55. Chou Y.-T., Lee C.-C., Hsiao S.-H., Lin S.-E., Lin S.-C., Chung C.-H., Chung C.-H., Kao Y.-R., Wang Y.-H., Chen C.-T.et al.. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells. 2013; 31:2607–2619. [DOI] [PubMed] [Google Scholar]
- 56. Sholl L.M., Barletta J.A., Yeap B.Y., Chirieac L.R., Hornick J.L.. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am. J. Surg. Pathol. 2010; 34:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakatsugawa M., Takahashi A., Hirohashi Y., Torigoe T., Inoda S., Murase M., Asanuma H., Tamura Y., Morita R., Michifuri Y.et al.. SOX2 is overexpressed in stem-like cells of human lung adenocarcinoma and augments the tumorigenicity. Lab. Invest. 2011; 91:1796–1804. [DOI] [PubMed] [Google Scholar]
- 58. Bass A.J., Watanabe H., Mermel C.H., Yu S., Perner S., Verhaak R.G., Kim S.Y., Wardwell L., Tamayo P., Gat-Viks I.et al.. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009; 41:1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hussenet T., Dali S., Exinger J., Monga B., Jost B., Dembelé D., Martinet N., Thibault C., Huelsken J., Brambilla E.et al.. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010; 5:e8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Domenici G., Aurrekoetxea-Rodríguez I., Simões B.M., Rábano M., Lee S.Y., Millán J.S., Comaills V., Oliemuller E., López-Ruiz J.A., Zabalza I.et al.. A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene. 2019; 38:3151–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simões B.M., Piva M., Iriondo O., Comaills V., López-Ruiz J.A., Zabalza I., Mieza J.A., Acinas O., Vivanco M.d.M.. Effects of estrogen on the proportion of stem cells in the breast. Breast Cancer Res. Treat. 2011; 129:23–35. [DOI] [PubMed] [Google Scholar]
- 62. Bulstrode H., Johnstone E., Marques-Torrejon M.A., Ferguson K.M., Bressan R.B., Blin C., Grant V., Gogolok S., Gangoso E., Gagrica S.et al.. Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev. 2017; 31:757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jeon H.-M., Sohn Y.-W., Oh S.-Y., Oh S.-Y., Kim S.-H., Beck S., Kim S., Kim H.. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011; 71:3410–3421. [DOI] [PubMed] [Google Scholar]
- 64. Zhang L.-H., Yin Y.-H., Chen H.-Z., Feng S.-Y., Liu J.-L., Chen L., Fu W.-L., Sun G.-C., Yu X.-G., Xu D.-G.. TRIM24 promotes stemness and invasiveness of glioblastoma cells via activating Sox2 expression. Neuro Oncol. 2020; 22:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Singh S., Trevino J., Bora-Singhal N., Coppola D., Haura E., Altiok S., Chellappan S.P.. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol. Cancer. 2012; 11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E.et al.. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014; 511:246–250. [DOI] [PubMed] [Google Scholar]
- 67. Berezovsky A.D., Poisson L.M., Cherba D., Webb C.P., Transou A.D., Lemke N.W., Hong X., Hasselbach L.A., Irtenkauf S.M., Mikkelsen T.et al.. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014; 16:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fang X., Yoon J.-G., Li L., Yu W., Shao J., Hua D., Zheng S., Hood L., Goodlett D.R., Foltz G.et al.. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics [Electronic Resource]. 2011; 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stolzenburg S., Rots M.G., Beltran A.S., Rivenbark A.G., Yuan X., Qian H., Strahl B.D., Blancafort P.. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012; 40:6725–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A.. Identification of Sox-2 regulatory region which is under the control of Oct-3/4–Sox-2 complex. Nucleic Acids Res. 2002; 30:3202–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zappone M.V., Galli R., Catena R., Meani N., De Biasi S., Mattei E., Tiveron C., Vescovi A.L., Lovell-Badge R., Ottolenghi S.et al.. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000; 127:2367–2382. [DOI] [PubMed] [Google Scholar]
- 72. Zhou H.Y., Katsman Y., Dhaliwal N.K., Davidson S., Macpherson N.N., Sakthidevi M., Collura F., Mitchell J.A.. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014; 28:2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li Y., Rivera C.M., Ishii H., Jin F., Selvaraj S., Lee A.Y., Dixon J.R., Ren B.. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014; 9:e114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ding Q., Regan S.N., Xia Y., Oostrom L.A., Cowan C.A., Musunuru K.. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013; 12:393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ahier A., Jarriault S.. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics. 2014; 196:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang J.-P., Li X.-L., Li G.-H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.-W.et al.. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Berhanu Lemma R., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N.et al.. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022; 50:D165–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kılıç Y., Çelebiler A.Ç., Sakızlı M.. Selecting housekeeping genes as references for the normalization of quantitative PCR data in breast cancer. Clin. Transl. Oncol. 2014; 16:184–190. [DOI] [PubMed] [Google Scholar]
- 81. Krasnov G.S., Kudryavtseva A.V., Snezhkina A.V., Lakunina V.A., Beniaminov A.D., Melnikova N.V., Dmitriev A.A.. Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front. Genet. 2019; 10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lyng M.B., Lænkholm A.-V., Pallisgaard N., Ditzel H.J.. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer. 2008; 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Consortium R.E., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 88. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shen L., Shao N., Liu X., Nestler E.. ngs.Plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMCGenomics [Electronic Resource]. 2014; 15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hoadley K.A., Yau C., Wolf D.M., Cherniack A.D., Tamborero D., Ng S., Leiserson M.D.M., Niu B., McLellan M.D., Uzunangelov V.et al.. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014; 158:929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I.et al.. TCGAbiolinks: an R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016; 44:e71–e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kaplan E.L., Meier P.. Nonparametric estimation from incomplete observations. J. Am. Statist. Assoc. 1958; 53:457–481. [Google Scholar]
- 93. Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V.et al.. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yu G., Wang L.-G., Han Y., He Q.-Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B.et al.. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017; 14:959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A, Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R.et al.. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T.et al.. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 2020; 11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W.et al.. The chromatin accessibility landscape of primary human cancers. Science. 2018; 362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu C., Wang M., Wei X., Wu L., Xu J., Dai X., Xia J., Cheng M., Yuan Y., Zhang P.et al.. An ATAC-seq atlas of chromatin accessibility in mouse tissues. Sci Data. 2019; 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J.et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Seibt K.M., Schmidt T., Heitkam T.. FlexiDot: highly customizable, ambiguity-aware dotplots for visual sequence analyses. Bioinformatics. 2018; 34:3575–3577. [DOI] [PubMed] [Google Scholar]
- 106. Chan H.L., Beckedorff F., Zhang Y., Garcia-Huidobro J., Jiang H., Colaprico A., Bilbao D., Figueroa M.E., LaCava J., Shiekhattar R.et al.. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018; 9:3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cocce K.J., Jasper J.S., Desautels T.K., Everett L., Wardell S., Westerling T., Baldi R., Wright T.M., Tavares K., Yllanes A.et al.. The lineage determining factor GRHL2 collaborates with FOXA1 to establish a targetable pathway in endocrine therapy-resistant breast cancer. Cell Rep. 2019; 29:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sato T., Yoo S., Kong R., Sinha A., Chandramani-Shivalingappa P., Patel A., Fridrikh M., Nagano O., Masuko T., Beasley M.B.et al.. Epigenomic profiling discovers trans-lineage SOX2 partnerships driving tumor heterogeneity in lung squamous cell carcinoma. Cancer Res. 2019; 79:6084–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gertsenstein M., Nutter L.M.J.. Production of knockout mouse lines with Cas9. Methods. 2021; 191:32–43. [DOI] [PubMed] [Google Scholar]
- 110. Peterson K.A., Khalouei S., Hanafi N., Wood J.A., Lanza D.G., Lintott L.G., Willis B.J., Seavitt J.R., Braun R.E., Dickinson M.E.et al.. Whole genome analysis for 163 gRNAs in Cas9-edited mice reveals minimal off-target activity. Commun. Biol. 2023; 6:626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Maier S., Wilbertz T., Braun M., Scheble V., Reischl M., Mikut R., Menon R., Nikolov P., Petersen K., Beschorner C.et al.. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum. Pathol. 2011; 42:1078–1088. [DOI] [PubMed] [Google Scholar]
- 112. Liu Y., Wu Z., Zhou J., Ramadurai D.K.A., Mortenson K.L., Aguilera-Jimenez E., Yan Y., Yang X., Taylor A.M., Varley K.E.et al.. A predominant enhancer co-amplified with the SOX2 oncogene is necessary and sufficient for its expression in squamous cancer. Nat. Commun. 2021; 12:7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for multimodal regulatory analysis and personal epigenomics. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E.. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008; 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A.et al.. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 116. Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W.et al.. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Berger A.C., Korkut A., Kanchi R.S., Hegde A.M., Lenoir W., Liu W., Liu Y., Fan H., Shen H., Ravikumar V.et al.. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018; 33:690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Soule H.D., Vazquez J., Long A., Albert S., Brennan M.. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl Cancer Inst. 1973; 51:1409–1416. [DOI] [PubMed] [Google Scholar]
- 119. Liang S., Furuhashi M., Nakane R., Nakazawa S., Goudarzi H., Hamada J., Iizasa H.. Isolation and characterization of human breast cancer cells with SOX2 promoter activity. Biochem. Biophys. Res. Commun. 2013; 437:205–211. [DOI] [PubMed] [Google Scholar]
- 120. Ling G.-Q., Chen D.-b., Wang B.-Q., Zhang L.-S.. Expression of the pluripotency markers Oct3/4, Nanog and Sox2 in human breast cancer cell lines. Oncol. Lett. 2012; 4:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chen C., Morris Q., Mitchell J.A.. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics [Electronic Resource]. 2012; 13:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mitchell J.A., Clay I., Umlauf D., Chen C.-Y., Moir C.A., Eskiw C.H., Schoenfelder S., Chakalova L., Nagano T., Fraser P.. Nuclear RNA sequencing of the mouse erythroid cell transcriptome. PLoS One. 2012; 7:e49274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Singh G., Mullany S., Moorthy S.D., Zhang R., Mehdi T., Tian R., Duncan A.G., Moses A.M., Mitchell J.A.. A flexible repertoire of transcription factor binding sites and a diversity threshold determines enhancer activity in embryonic stem cells. Genome Res. 2021; 31:564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tolhuis B., Palstra R.-J., Splinter E., Grosveld F., de Laat W.. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002; 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 125. Carter D., Chakalova L., Osborne C.S., Dai Y., Fraser P.. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002; 32:623. [DOI] [PubMed] [Google Scholar]
- 126. Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H.et al.. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009; 462:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gopi L.K., Kidder B.L.. Integrative pan cancer analysis reveals epigenomic variation in cancer type and cell specific chromatin domains. Nat. Commun. 2021; 12:1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Moorthy S.D., Davidson S., Shchuka V.M., Singh G., Malek-Gilani N., Langroudi L., Martchenko A., So V., Macpherson N.N., Mitchell J.A.. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017; 27:246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tobias I.C., Abatti L.E., Moorthy S.D., Mullany S., Taylor T., Khader N., Filice M.A., Mitchell J.A.. Transcriptional enhancers: from prediction to functional assessment on a genome-wide scale. Genome. 2021; 64:426–448. [DOI] [PubMed] [Google Scholar]
- 130. Tompkins D.H., Besnard V., Lange A.W., Keiser A.R., Wert S.E., Bruno M.D., Whitsett J.A.. Sox2 activates cell proliferation and differentiation in the respiratory epithelium. Am. J. Respir. Cell Mol. Biol. 2011; 45:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Eckhart L., Lippens S., Tschachler E., Declercq W.. Cell death by cornification. Biochim. Biophys. Acta. 2013; 1833:3471–3480. [DOI] [PubMed] [Google Scholar]
- 132. Soufi A., Donahue G., Zaret K.S.. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012; 151:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S.. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015; 161:555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jeselsohn R., Cornwell M., Pun M., Buchwalter G., Nguyen M., Bango C., Huang Y., Kuang Y., Paweletz C., Fu X.et al.. Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc. Natl Acad. Sci. USA. 2017; 114:E4482–E4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Miyagi S., Nishimoto M., Saito T., Ninomiya M., Sawamoto K., Okano H., Muramatsu M., Oguro H., Iwama A., Okuda A.. The Sox2 regulatory region 2 functions as a neural stem cell-specific enhancer in the telencephalon. J. Biol. Chem. 2006; 281:13374–13381. [DOI] [PubMed] [Google Scholar]
- 136. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 137. Meuleman W., Muratov A., Rynes E., Halow J., Lee K., Bates D., Diegel M., Dunn D., Neri F., Teodosiadis A.et al.. Index and biological spectrum of human DNase I hypersensitive sites. Nature. 2020; 584:244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D.et al.. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006; 444:499–502. [DOI] [PubMed] [Google Scholar]
- 139. Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K.et al.. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005; 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Minoo P., Su G., Drum H., Bringas P., Kimura S.. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(-/-) mouse embryos. Dev. Biol. 1999; 209:60–71. [DOI] [PubMed] [Google Scholar]
- 141. Bertolini J.A., Favaro R., Zhu Y., Pagin M., Ngan C.Y., Wong C.H., Tjong H., Vermunt M.W., Martynoga B., Barone C.et al.. Mapping the global chromatin connectivity network for Sox2 function in neural stem cell maintenance. Cell Stem Cell. 2019; 24:462–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Favaro R., Valotta M., Ferri A.L.M., Latorre E., Mariani J., Giachino C., Lancini C., Tosetti V., Ottolenghi S., Taylor V.et al.. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 2009; 12:1248–1256. [DOI] [PubMed] [Google Scholar]
- 143. Bonnet D., Dick J.E.. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997; 3:730–737. [DOI] [PubMed] [Google Scholar]
- 144. Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O., Brooks M., Reinhardt F., Su Y., Polyak K.et al.. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl Acad. Sci. USA. 2011; 108:7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E.. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994; 367:645–648. [DOI] [PubMed] [Google Scholar]
- 146. Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S.. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011; 146:633–644. [DOI] [PubMed] [Google Scholar]
- 147. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B.et al.. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Bernardo G.M., Lozada K.L., Miedler J.D., Harburg G., Hewitt S.C., Mosley J.D., Godwin A.K., Korach K.S., Visvader J.E., Kaestner K.H.et al.. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010; 137:2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Liu Y., Zhao Y., Skerry B., Wang X., Colin-Cassin C., Radisky D.C., Kaestner K.H., Li Z.. Foxa1 is essential for mammary duct formation. Genesis. 2016; 54:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Besnard V., Wert S.E., Kaestner K.H., Whitsett J.A.. Stage-specific regulation of respiratory epithelial cell differentiation by Foxa1. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005; 289:L750–L759. [DOI] [PubMed] [Google Scholar]
- 151. Paranjapye A., Mutolo M.J., Ebron J.S., Leir S.-H., Harris A.. The FOXA1 transcriptional network coordinates key functions of primary human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020; 319:L126–L136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Camolotto S.A., Pattabiraman S., Mosbruger T.L., Jones A., Belova V.K., Orstad G., Streiff M., Salmond L., Stubben C., Kaestner K.H.et al.. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. eLife. 2018; 7:e38579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fu X., Jeselsohn R., Pereira R., Hollingsworth E.F., Creighton C.J., Li F., Shea M., Nardone A., Angelis C.D., Heiser L.M.et al.. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc. Natl Acad. Sci. USA. 2016; 113:E6600–E6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Orstad G., Fort G., Parnell T.J., Jones A., Stubben C., Lohman B., Gillis K.L., Orellana W., Tariq R., Klingbeil O.et al.. FoxA1 and FoxA2 control growth and cellular identity in NKX2-1-positive lung adenocarcinoma. Dev. Cell. 2022; 57:1866–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Liu K.-C., Lin B.-S., Zhao M., Yang X., Chen M., Gao A., Que J., Lan X.-P.. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013; 25:1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pierrou S., Hellqvist M., Samuelsson L., Enerbäck S., Carlsson P.. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994; 13:5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Li L., Wu D., Yu Q., Li L., Wu P. Prognostic value of FOXM1 in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017; 8:32298–32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Harris L., Genovesi L.A., Gronostajski R.M., Wainwright B.J., Piper M.. Nuclear factor one transcription factors: divergent functions in developmental versus adult stem cell populations. Dev. Dyn. 2015; 244:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Steele-Perkins G., Plachez C., Butz K.G., Yang G., Bachurski C.J., Kinsman S.L., Litwack E.D., Richards L.J., Gronostajski R.M.. The transcription factor gene nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 2005; 25:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gründer A., Ebel T.T., Mallo M., Schwarzkopf G., Shimizu T., Sippel A.E., Schrewe H.. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech. Dev. 2002; 112:69–77. [DOI] [PubMed] [Google Scholar]
- 161. Hsu Y.-C., Osinski J., Campbell C.E., Litwack E.D., Wang D., Liu S., Bachurski C.J., Gronostajski R.M.. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 2011; 354:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Becker-Santos D.D., Lonergan K.M., Gronostajski R.M., Lam W.L.. Nuclear factor I/B: a master regulator of cell differentiation with paradoxical roles in cancer. EBioMedicine. 2017; 22:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Zhou M., Zhou L., Zheng L., Guo L., Wang Y., Liu H., Ou C., Ding Z.. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB). PLoS One. 2014; 9:e100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Becker-Santos D.D., Thu K.L., English J.C., Pikor L.A., Martinez V.D., Zhang M., Vucic E.A., Luk M.T., Carraro A., Korbelik J.et al.. Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma. J. Pathol. 2016; 240:161–172. [DOI] [PubMed] [Google Scholar]
- 165. Ferone G., Song J.-Y., Sutherland K.D., Bhaskaran R., Monkhorst K., Lambooij J.-P., Proost N., Gargiulo G., Berns A.. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell. 2016; 30:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wu C., Zhu X., Liu W., Ruan T., Wan W., Tao K.. NFIB promotes cell growth, aggressiveness, metastasis and EMT of gastric cancer through the Akt/Stat3 signaling pathway. Oncol. Rep. 2018; 40:1565–1573. [DOI] [PubMed] [Google Scholar]
- 167. Otsubo T., Akiyama Y., Yanagihara K., Yuasa Y.. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br. J. Cancer. 2008; 98:824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Wang S., Tie J., Wang R., Hu F., Gao L., Wang W., Wang L., Li Z., Hu S., Tang S.et al.. SOX2, a predictor of survival in gastric cancer, inhibits cell proliferation and metastasis by regulating PTEN. Cancer Lett. 2015; 358:210–219. [DOI] [PubMed] [Google Scholar]
- 169. Zhang X., Yu H., Yang Y., Zhu R., Bai J., Peng Z., He Y., Chen L., Chen W., Fang D.et al.. SOX2 in gastric carcinoma, but not Hath1, is related to patients’ clinicopathological features and prognosis. J. Gastrointest. Surg. 2010; 14:1220–1226. [DOI] [PubMed] [Google Scholar]
- 170. Miyagi S., Saito T., Mizutani K., Masuyama N., Gotoh Y., Iwama A., Nakauchi H., Masui S., Niwa H., Nishimoto M.et al.. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol. Cell. Biol. 2004; 24:4207–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Aran D., Abu-Remaileh M., Levy R., Meron N., Toperoff G., Edrei Y., Bergman Y., Hellman A.. Embryonic stem cell (ES)-specific enhancers specify the expression potential of ES genes in cancer. PLoS Genet. 2016; 12:e1005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Iglesias J.M., Leis O., Pérez Ruiz E., Gumuzio Barrie J., Garcia-Garcia F., Aduriz A., Beloqui I., Hernandez-Garcia S., Lopez-Mato M.P., Dopazo J.et al.. The activation of the Sox2 RR2 pluripotency transcriptional reporter in human breast cancer cell lines is dynamic and labels cells with higher tumorigenic potential. Front. Oncol. 2014; 4:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Jung K., Gupta N., Wang P., Lewis J.T., Gopal K., Wu F., Ye X., Alshareef A., Abdulkarim B.S., Douglas D.N.et al.. Triple negative breast cancers comprise a highly tumorigenic cell subpopulation detectable by its high responsiveness to a Sox2 regulatory region 2 (SRR2) reporter. Oncotarget. 2015; 6:10366–10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Saenz-Antoñanzas A., Moncho-Amor V., Auzmendi-Iriarte J., Elua-Pinin A., Rizzoti K., Lovell-Badge R., Matheu A.. CRISPR/Cas9 deletion of SOX2 regulatory region 2 (SRR2) decreases SOX2 malignant activity in glioblastoma. Cancers (Basel). 2021; 13:1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Björkqvist A.M., Husgafvel-Pursiainen K., Anttila S., Karjalainen A., Tammilehto L., Mattson K., Vainio H., Knuutila S.. DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes Cancer. 1998; 22:79–82. [PubMed] [Google Scholar]
- 176. Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M.. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007; 5:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S.et al.. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018; 554:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., Hocker J.D., Conte M., Gorkin D., Yu M.et al.. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021; 28:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Splinter E., Heath H., Kooren J., Palstra R.-J., Klous P., Grosveld F., Galjart N., de Laat W.. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006; 20:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dunn O.J. Multiple comparisons using rank sums. Technometrics. 1964; 6:241–252. [Google Scholar]
- 181. Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979; 6:65–70. [Google Scholar]
- 182. Dunnett C.W. A multiple comparison procedure for comparing several treatments with a control. J. Am. Statist. Assoc. 1955; 50:1096–1121. [Google Scholar]
- 183. Tukey J.W. Comparing individual means in the analysis of variance. Biometrics. 1949; 5:99. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing and processed data files were submitted to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) repository (GSE132344).