Abstract
Transcriptomic data is accumulating rapidly; thus, scalable methods for extracting knowledge from this data are critical. Here, we assembled a top-down expression and regulation knowledge base for Escherichia coli. The expression component is a 1035-sample, high-quality RNA-seq compendium consisting of data generated in our lab using a single experimental protocol. The compendium contains diverse growth conditions, including: 9 media; 39 supplements, including antibiotics; 42 heterologous proteins; and 76 gene knockouts. Using this resource, we elucidated global expression patterns. We used machine learning to extract 201 modules that account for 86% of known regulatory interactions, creating the regulatory component. With these modules, we identified two novel regulons and quantified systems-level regulatory responses. We also integrated 1675 curated, publicly-available transcriptomes into the resource. We demonstrated workflows for analyzing new data against this knowledge base via deconstruction of regulation during aerobic transition. This resource illuminates the E. coli transcriptome at scale and provides a blueprint for top-down transcriptomic analysis of non-model organisms.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Over the past decade, RNA sequencing (RNA-seq) has emerged as an efficient, high-throughput method to determine the expression state of a cell population. Large RNA-seq datasets (1–5) have enabled the development and application of machine learning methods to advance our understanding of transcription and regulation (1,6–10). As datasets continue to grow, analytic methods must keep pace to convert this data to biological knowledge. A unified, large-scale resource integrating expression data, regulatory information, and analysis would address this need.
Large RNA-seq datasets compiled from multiple sources can be subject to batch effects that confound analysis. Mitigating these effects remains an important goal and an active area of research (11,12). Single-protocol, high-quality, curated RNA-seq datasets represent another strategy for batch effect limitation. However, generating such datasets is time- and cost-intensive.
A transcriptional regulatory network (TRN) is a key tool for analyzing regulation in an organism. A TRN is a directed graph with edges connecting regulators to the sets of genes they regulate (regulons). TRNs are also laborious to construct, as they depend on exhaustive bottom-up characterization of regulators binding to the promoter regions of their target genes and affecting transcription of those genes. Thus, top-down inference of regulatory signals directly from an RNA-seq dataset—without prior knowledge of the TRN—may provide a useful addition as the regulatory information component of a transcriptional resource.
Independent component analysis (ICA) (13) is a signal processing algorithm that outperforms other methods for the extraction of biologically meaningful regulatory modules from gene expression data (14). Application of this method to publicly-available prokaryotic expression data has consistently recovered TRN modules across organisms (1,15–19). ICA’s effectiveness results from its ability to identify independent groups of genes that vary consistently across samples, regardless of group size or overlapping membership. Thus, a dataset with sufficient scale and diversity in conditions to activate a broad range of regulatory signals is a key prerequisite for this method.
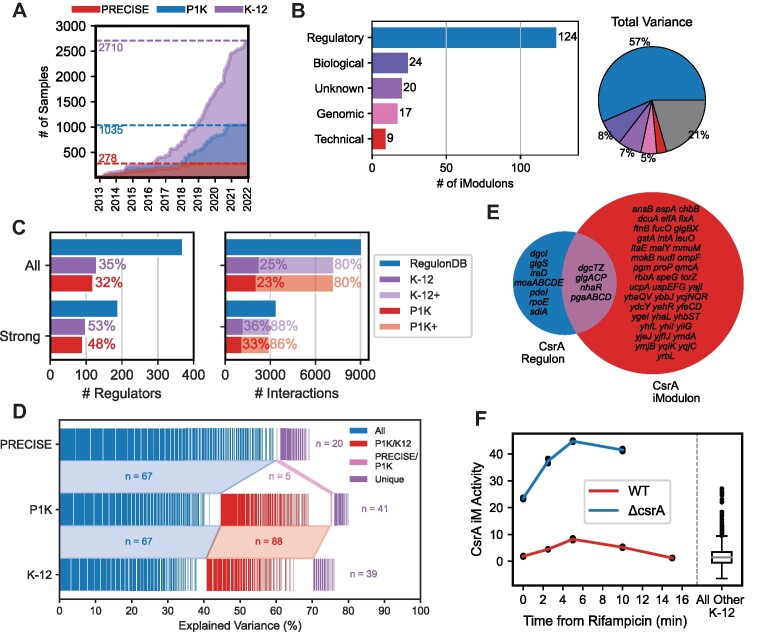
Here, we present an expression and regulation resource for the key model organism Escherichia coli K-12 MG1655. The expression component is PRECISE-1K, a 1035-sample, single-protocol RNA-seq dataset. This Precision RNA-seq Expression Compendium for Independent Signal Extraction contains 38% of all publicly-available high-quality RNA-seq data for E. coli K-12 and includes a broad range of growth conditions. These data were generated between 2013 and 2021 in our lab (Figure 1B). To create the resource's regulatory component, we use ICA to extract 201 independently modulated groups of genes (iModulons) that recover 86% of known regulatory interactions. Then, we demonstrate the use of this resource by: (i) describing genome-wide expression patterns; (ii) elucidating systems-level transcriptome properties and responses; (iii) proposing novel regulons for two putative transcription factors; (iv) identifying a promoter sequence basis for two regulatory modules; (v) adding 1675 high-quality publicly available K-12 samples and extracting similar regulatory modules and (vi) providing a workflow for systems-level transcriptome analysis of external data using our knowledge base. This example workflow, along with all analyses presented here, are available for use at our GitHub repositories, https://github.com/SBRG/precise1k-analyze and https://github.com/SBRG/precise1k. The PRECISE-1K and Public K-12 iModulons, along with those for the other organisms mentioned above, can also be explored at iModulonDB.org (20).
Figure 1.
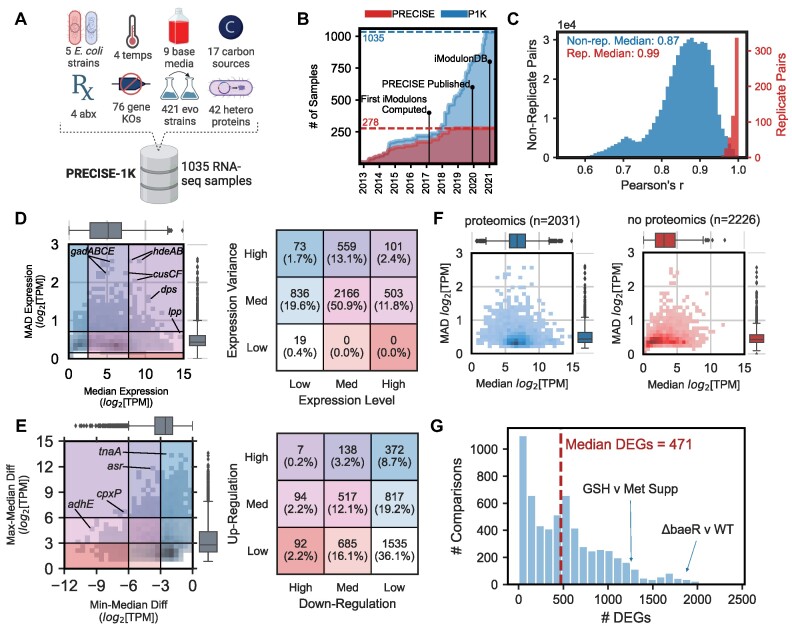
PRECISE-1K, a 1035-sample high-precision expression compendium, reveals expression trends in the E. coli transcriptome. (A) Overview of construction of PRECISE-1K compendium. Values indicate the number of unique categories for each condition (except evo strains). abx = antibiotics. (B) The growth in single-protocol transcriptomics samples contained in the PRECISE to PRECISE-1K databases. (C) Histogram of Pearson's r for both all replicate pairs and all non-replicate pairs (pairwise combinations of samples across projects that are not direct biological replicates). Samples included in PRECISE-1K are required to have replicate correlations of at least 0.95. (D) 2-D histogram of median expression level against median absolute deviation (MAD) of expression for all 4257 genes in PRECISE-1K. Table defines expression categories as per corresponding box color/location in histogram. For each axis, category splits are defined at median ± 1 standard deviation. (E) 2-D histogram of median-to-min expression difference against median-to-max expression difference for all 4257 genes in PRECISE-1K. Table defines regulatory categories as per corresponding box color/location in histogram. For each axis, low-to-medium split defined at 3 log2[TPM] units (8-fold change from median expression); medium-to-high split defined at 6 log2[TPM] units (32-fold change). (F) Median vs MAD expression 2D histogram, separated by availability of proteomics data in two large recent datasets (46,88). Blue = proteomics data available; red = no proteomics data available. (G) Histogram of the number of differentially expressed genes (DEGs) computed between condition pairs within the same project (n = 6103 pairs). GSH = glutathione, Met = methionine.
PRECISE-1K provides the expression component and iModulons provide the regulation component of a multi-scale transcriptomic knowledge base. This resource in turn empowers analyses that illuminate the transcriptomic responses of this critical model organism for cellular biology, pathogenicity, and systems biology. This resource may be used to inform novel experimental designs. Beyond its use in E. coli, this resource also provides a blueprint for regulatory information extraction in other organisms, especially those lacking exhaustive prior annotation.
MATERIALS AND METHODS
RNA sequencing
3 ml of cell broth (OD600 ∼ 0.5, unless otherwise specified in sample metadata file) was immediately added to two volumes Qiagen RNA-protect Bacteria Reagent (6 mL), vortexed for 5 s, incubated at room temperature for 5 min, and immediately centrifuged for 10 min at 11 000 × g. The supernatant was decanted, and the cell pellet was stored in the −80 °C. Cell pellets were thawed and incubated with Readylyse Lysozyme, SuperaseIn, Protease K and 20% SDS for 20 min at 37 °C. Total RNA was isolated and purified using the Qiagen RNeasy Mini Kit (Cat#74104) columns and following vendor procedures. An on-column DNase-treatment was performed for 30 min at room temperature. RNA was quantified using a Nanodrop and quality assessed by running an RNA-nano chip on a bioanalyzer. The rRNA was removed using Illumina Ribo-Zero rRNA removal kit (Cat#20037135) for Gram-negative bacteria. A KAPA stranded RNA-Seq Kit (Kapa Biosystems KK8401) was used following the manufacturer's protocol to create sequencing libraries with an average insert length of around ∼300 bp. Libraries were run on a HiSeq4000 or NextSeq (Illumina).
RNA-seq processing and quality control
Starting from 1055 candidate samples, data was processed using a Nextflow (21) pipeline designed for processing microbial RNA-seq datasets (22), and run on Amazon Web Services (AWS) Batch.
First, raw read trimming was performed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the default options, followed by FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) on the trimmed reads. Next, reads were aligned to the E. coli K-12 MG1655 reference genome (RefSeq accession number NC_000913.3) using Bowtie (23) with the following non-default options: −X 1000, −3 3, −n 2. The read direction was inferred using RSEQC (24) before generating read counts using featureCounts (25) with the following non-default options: -p -B -C -P -fracOverlap 0.5. Finally, all quality control metrics were compiled using MultiQC (26) and the final expression dataset was reported in units of log2-transformed Transcripts Per Million (log2[TPM]).
Samples were considered ‘high-quality’ if they met all of the following criteria:
‘Pass’ on the all of the following FastQC checks: per_base_sequence_quality, per_sequence_quality_scores, per_base_n_content, adapter_content
At least 500 000 reads mapped to coding sequences (CDS) from the reference genome (NC_000913.3)
Not an outlier in hierarchical clustering based on pairwise Pearson correlation between all samples (outlier defined as cluster with number of samples < 1% of the total number of samples)
Minimum Pearson correlation with biological replicates (if any) 0.95 (if more than two biological replicates, keep samples with high correlation in ‘greedy’ manner, dropping samples that have at least one sub-threshold correlation with all other replicates)
Short non-coding transcripts (<100 nucleotides) and extremely low-expression transcripts (FPKM < 10) were also removed to reduce noise.
Following this processing and QC workflow, 1035 high-quality RNA-seq samples (each with 4257 gene expression measurements) remained. These samples and their metadata define PRECISE-1K. log2[TPM], raw read count, QC data files, and sample metadata for all 1055 original samples may be found in the data directory of this project's GitHub repository.
Differentially expressed gene (DEG) computation
Differentially expressed genes (DEGs) were identified using the DESeq2 package (27) on the PRECISE-1K RNA-seq dataset. Genes with a log2 fold change greater than 1.5 and a false discovery rate (FDR) value less than 0.05 were considered to be differentially expressed genes. Genes with p-values assigned ‘NA’ based on extreme count outlier detection were not considered as potential DEGs. The number of DEGs was computed for each unique pair of conditions within each project in PRECISE-1K, for a total of 6104 pairwise computations.
iModulon computation
Log2[TPM] data (4257 gene rows by 1035 sample columns) was centered to the control condition (log-phase growth in M9 minimal media with glucose; sample IDs ‘p1k_00001’ and ‘p1k_00002’); the mean log2[TPM] of these two samples was computed, and the resultant 4257-gene log2[TPM] vector was subtracted from all 1035 samples (columns) of the log2[TPM] data table (including the control samples themselves, such that the mean of these samples was equal to 0).
No batch effect correction method (such as ComBat-Seq) was used—use of such methods significantly reduced regulatory signal discovery in testing. Many common types of batch variation—e.g. temperature, pH, growth phase—mediate expression changes through the TRN anyhow. Thus these minute perturbations - along with much larger variation across samples and projects - initiate the variant signals needed for ICA to identify regulatory activity.
The Scikit-learn (28) implementation of FastICA (29) was used to run ICA on the centered log2[TPM] data table. FastICA numerically solves the matrix decomposition equation X = MA; X is the input matrix; M is the ‘iModulon’ matrix, and A is the ‘Activity’ matrix; these terms will be used from here on in lieu of the traditional terminology X = SA (‘signal’ and ‘mixing’ matrices) to avoid confusion with the stoichiometric matrix S from metabolic modeling. In this context, M has dimensions of number of genes by number of components, and A has dimensions of number of components by number of samples. Thus, the M matrix contains weightings that specify how much each gene (row) belongs to each independent component (IC; column). The A matrix contains weightings that indicate how active each IC (row) is in each sample (column).
Unlike PCA, this method requires pre-specification of the number of components (parameter name n_components; also known as dimensionality) to use (the number of columns in M and number of rows in A). In order to choose an optimal dimensionality, the previously described OptICA method (30) was used.
For PRECISE-1K, the selected optimal dimensionality by this method was 290. The robust M and A matrices from this dimensionality run were selected, yielding 201 ICs. Thus, the final M matrix has dimensions of 4257 genes by 201 independent components, and the final activity matrix A has dimensions of 201 iModulons by 1035 samples.
The M matrix contains gene weightings, indicating how much each gene (row) ‘belongs’ to each component (column), with larger absolute values indicating more association of a particular gene with a particular IC. For a given IC, gene weightings are mostly normally distributed around 0, with a few outlier gene weightings deviating from 0. To define an iModulon, a cutoff must be defined that allows segmentation of the genes in an IC based on their gene weightings. These cutoffs were determined with a previously described method (1) using D’Agostino's K2 test for normality. In this way, the final 201 iModulons were computed from the 201 independent components. A binary matrix Mbinary was then constructed with the same dimensions as M; for a given gene (row)/iModulon (column) entry, a 1 indicates membership of the gene in the iModulon, and a 0 indicates that the gene is not a member of the iModulon.
The final matrices M, Mbinary and A (along with iModulon membership thresholds as defined above, regulatory annotation as described below, and all other iModulon metadata) are available in the supplementary data files and in this project's GitHub repository.
iModulon annotation and curation
Using the gold-standard TRN reference annotation downloaded from RegulonDB v10.5 (31), enrichment of the set of genes in each iModulon against known RegulonDB regulons was computed using Fisher's Exact Test, with false discovery rate controlled at 10−5 using the Benjamini–Hochberg correction. By default, iModulons were compared to all possible single regulons and all possible combinations of two regulons (intersection only). The regulons used by default consisted of only strong and confirmed evidence regulatory interactions, per RegulonDB. When multiple significant enrichments were available, the enrichment with the lowest adjusted P value was used for annotation. In the event of near equal P values (within an order of magnitude) across multiple enrichments, the priority was given to intersection regulons, followed by single regulons, followed by union regulons. If no significant enrichments were available, the following adjustments were used, in this order: relax evidence requirement to include weak evidence regulatory interactions; search only for single regulon enrichments; allow up to three regulons to be combined for enrichment; allow regulon unions as well as intersections (with priority given to intersections). If the iModulon consisted of genes with annotated co-regulation by four or more genes, a specific enrichment calculation was made to determine the enrichment statistics. If none of these adjustments yielded a significant enrichment, the iModulon was annotated as non-regulatory. All parameters and statistics related to calculation of TRN enrichments for regulatory iModulons are recorded in the iModulon metadata table, available in the GitHub repository. If any significant regulatory enrichments were found after applying this procedure, the iModulon was annotated as Regulatory and named according to the ruleset defined below in Case 1. Otherwise, the iModulon was assigned one of 4 additional categories (Genomic, Biological, Single-Gene Dominant, Uncharacterized), detailed in Cases 2–5 below, respectively.
iModulons were named and annotated according to the following ruleset:
General
Rule #1: iModulon names must be fewer than ∼15 characters.
Rule #2: iModulon names must be unique. If iModulons would otherwise have the same name, append ‘−1″, ‘−2″, etc., as needed to disambiguate. By default, order the suffixes by decreasing explained variance, unless another numbering is specifically preferred (e.g. aligning Crp-1 and Crp-2 with Crp binding site classes).
Case 1: Regulatory
The iModulon has a significant regulon enrichment chosen as described above:
Rule #1: Name the iModulon after the primary function of the enriched regulon(s) (e.g. the iModulon enriched for the CdaR regulon is named ‘Sugar Diacid’).
Rule #2: If no clear primary function is available for the iModulon, name the iModulon directly after the enriched regulon (e.g. the iModulon enriched for the CpxR regulon is named ‘CpxR’, as CpxR controls a diverse set of functions).
Exception #1: if the enriched regulon corresponds to a well-known global regulator (i.e. Fur, CRP, RpoS), name the iModulon after that regulator.
Exception #2: if the name per Rule #1 would violate General Rule #1, name the iModulon directly after the enriched regulon (e.g. the iModulon enriched for the union of the FucR and ExuR regulons is named ‘FucR/ExuR’ instead of ‘Fucose/Galacturonate/Glucuronate’).
Exception #3: if applying Rule #2, and the regulon enrichment involves an intersection between a global regulator and a local regulator (i.e. cooperative regulation), the global regulator may be dropped from the name (e.g. ‘NtrC-1″ instead of ‘RpoN + NtrC-1″, as RpoN is a larger-regulon sigma factor which co-regulates with the more-specific NtrC).
Case 2: Genomic
The iModulon activity profile has a clear correlation with a sample involving a specific genetic or genomic intervention:
Rule #1: if the iModulon captures intentional knockout of a gene (e.g. geneA is knocked out in sampleA, and the iModulon has a large positive gene weight for geneA and a large negative activity level for sampleA, accounting for the lack of geneA expression in sampleA), name the iModulon ‘[gene name] KO’ (e.g. baeR KO).
Rule #2: Similarly, if the iModulon captures intentional overexpression of a particular gene, name the iModulon ‘[gene name] OE’ (e.g. ‘malE OE’).
Rule #3: if the iModulon captures expression changes in relation to evolved samples (ALE), as determined by comparing the iModulon activities to known ALE samples, name the iModulon ‘[name of ALE project] Del’ (for deletions), ‘[name of ALE project] Amp’ (for amplifications), or ‘name of ALE project] Mut’ (for mixed effect mutations) (e.g. ROS TALE Del-1).
Rule #4: if the iModulon also has a significant regulon enrichment as described above, prioritize the specific genetic/genomic change.
Case 3: Biological
The iModulon does not have a significant regulon enrichment, does not relate to a specific genetic or genomic change, but the member genes share a clear biological function:
Rule #1: Name the iModulon after the shared biological function (e.g. the ‘LPS’ iModulon consists of many genes related to lipopolysaccharide biosynthesis and export, though no significant regulon enrichment was found for this iModulon's genes).
Case 4: Single-gene dominant
The iModulon contains one specific gene with a gene weight at least twice as large as the next closest gene, does not fall into Case 2—Genomic, and contains only the one highly-weighted genes, or at most 5 other genes with gene weights very close to the iModulon's threshold
Rule #1: Name the iModulon after the dominant gene (e.g. the ‘ymdG’ iModulon consists solely of the ymdG gene).
Case 5: Uncharacterized
The iModulon does not meet any of the previous criteria for naming
Rule #1: Name the iModulon ‘UC-#’ (short for ‘Uncharacterized’), with the number incrementing for each uncharacterized iModulon.
Differential iModulon activity computation
Differentially iModulon activities (DiMAs) were computed with a similar process as previously detailed (1). For each iModulon, the average activity of the iModulon between biological replicates, if available, was computed. Then, the absolute value of the difference in iModulon activities between the two conditions was compared to the fitted log-normal distribution of all differences in activity for the iModulon. iModulons that had an absolute value of activity >5, and an FDR <0.05 were considered to be significant. The number of DiMAs was computed for each unique pair of conditions within each project in the PRECISE-1K compendium, mirroring DEG computation.
Compiling the public K-12 dataset
Data was compiled from NCBI SRA as described previously (22). Initially, all data annotated as RNA-seq for E. coli was inspected. RNA-seq samples were discarded if the strain was not from a K-12 strain, if the strain was missing, or if the type of experiment was not actually RNA-seq. After initial curation, 3125 samples remained. Next, these data were processed and quality controlled as described previously. 74% of samples (2312) passed the RNA-seq quality control checks (FastQC, minimum reads mapped to coding sequences, non-outlier clustering). 58% of the original samples (1816) had sufficient metadata annotation to verify biological replicates. Only conditions with at least two biological replicates were kept at this step. Finally, the 0.95 minimum replicate correlation threshold was applied, yielding the final set of 1675 high-quality publicly-available samples (54% of the original set). Next, these 1675 samples were combined with the 1035 samples of PRECISE-1K to yield the ‘Public K-12’ dataset, comprising 2710 curated, high quality expression profiles for E. coli strain K-12. The log2[TPM], raw read count, QC data files and sample metadata for the high-quality public samples may be found in the data directory of this project's GitHub repository. After centering the Public K-12 dataset to the PRECISE-1K control condition, iModulons were computed and annotated in the same manner as described above.
RESULTS
PRECISE-1K is a 1035-sample, single-protocol, high-precision RNA-seq compendium
We constructed PRECISE-1K to enable a multi-scale analysis of transcription and regulation in E. coli K-12 MG1655 (Figure 1A; Supplemental Figure S1). PRECISE-1K is a large, high-fidelity expression compendium consisting of 1035 RNA-seq samples generated by a single research group using a standardized experimental and data processing protocol (see Methods). The samples come from 45 distinct projects. PRECISE-1K comprises a wide range of growth conditions, including: 5 strains, 4 temperatures, 5 pHs, 9 base media, 18 carbon sources, 38 supplements, 76 unique gene knockouts, 421 evolved samples and 87 fed-batch cultures (Supplemental Figure S2). PRECISE-1K features projects involving: adaptation to new growth conditions (32–36), expression of heterologous (37) and orthologous (38) genes, and a genome-reduced strain (39). PRECISE-1K constitutes a nearly 4-fold increase in size from the original 278-sample PRECISE(1) (Figure 1B). Replicates are tightly correlated, with a median Pearson's r of 0.99 (Figure 1C). PRECISE-1K thus represents a broad range of conditions under which changes in the composition of the E. coli transcriptome may be studied.
Principal component analysis (PCA) of PRECISE-1K reveals some expected batch effects. Separation between samples in principal component space largely stems from differences between project growth conditions (Supplemental Figure S3). In particular, projects that feature diverse growth media (e.g. the two-component system knockout (40) and antibiotic resistance project (41)) and projects that significantly alter the genome (e.g. a genome-reduced E. coli strain (39)) notably diverge from other projects. Clustering by library preparer is largely explained by project-based clustering, indicating - along with tight replicate correlations - that this commonly observed batch effect (11) is not prominent in PRECISE-1K.
PRECISE-1K segments genes by expression, variance and regulatory effect
Leveraging PRECISE-1K’s condition diversity and scale, we evaluated systems-level expression trends to compare data-driven observations to prior expectations. First, we compared genes’ median expression levels across PRECISE-1K to their median absolute deviations (MAD). This contrast enabled us to define expression-based categories for all genes (Figure 1D). For example, expression of gadABCE - four genes of the glutamate-dependent acid resistance system 2 - is medium in aggregate; however, these genes exhibit particularly high variation across conditions, likely due to the specificity of their response. The gene with the highest median expression is lipoprotein-encoding lpp, long known to be the most abundant protein in E. coli (42,43). Likely owing to its structural role in peptidoglycan, its cross-condition variation is medium. The plurality of genes have medium expression with medium variation, while only 101 genes—such as copper/silver export system component cusF—are both highly expressed and highly variable. Overall, most genes’ variation fell within one standard deviation of the overall median variation across all genes: 82% of genes (3505/4257). Only 19 genes have low variation and low overall expression, consisting mostly of insertion elements and prophage genes.
Next, we compared genes’ median expression levels to their minimum and maximum levels. In so doing, we identified the maximum extent to which regulation can influence the expression level of each gene in both the upwards and downwards directions (Figure 1E). Overall, 36.1% of genes are expressed in a tight range, exhibiting relatively low effects of up- or down-regulation. However, 45.6% of genes demonstrate medium or high upwards inducibility, and 36% have medium or high downwards inducibility. Thus, regulatory effects can influence expression level by an order of magnitude or more for a majority of genes. For example, cpxP - a protein responding to extracytoplasmic stresses as part of the CpxAR two-component system (44) - has a nearly unique tendency to be both highly up- and down-regulated from its median level. This characteristic may result from CpxP’s role as both a direct effector of various stress responses and a negative feedback regulator for the response pathway as a whole (45).
PRECISE-1K also highlights relationships between gene expression and other data types. Genes for which proteomics data is available in two large datasets (46,47) have significantly higher expression (P = 1.2E-150, Mann–Whitney U, m = 2031, n = 2226), consistent with a known bias towards higher-expressed genes amongst proteomics samples (Figure 1F). However, no significant difference in variability was found (P = 0.97). We also compared the expression of poorly-annotated genes (referred to as the ‘y-ome’ in E. coli (48)) to genes with more complete annotation. y-ome genes have significantly lower expression (P = 1.0E-75, Mann–Whitney U, m = 1473, n = 2784) than non-y-genes, highlighting the lack of transcription in standard laboratory conditions as a potential reason for these genes’ relative lack of annotation (Supplemental Figure S4). As expected, genes in the ‘Translation’ and ‘Cell Cycle’ functional categories are expressed most highly, while more specialized categories such as ‘Carbohydrate Metabolism’ have much lower median expression levels (Supplemental Figure S5).
We performed differential gene expression analysis within each member project for all projects in the PRECISE-1K compendium. A median of 471 differentially expressed genes (DEGs) were found across all pairwise within-project comparisons (Figure 1G). Many comparisons produced close to 0 DEGs—or example, comparison of a qseF deletion to a wild-type control after 6 h of batch culture yielded only six DEGs. Other in-project comparisons yielded far more DEGs. For example, the comparison between wild-type growth in minimal media and deletion of two-component system (TCS) response regulator baeR with ethanol supplementation yielded 1868 DEGs. In general, using DEGs alone to derive biological insight may require analysis of hundreds to thousands of genes.
Taken together, these results highlight PRECISE-1K’s capability to capture genome-wide expression patterns that both confirm existing expectations and reveal new knowledge. PRECISE-1K is thus an expression knowledge base, as it stores both expression data and informs knowledge-generating analyses. Quantifying the impact of regulation on gene expression at the systems level constitutes the next scale of knowledge extraction facilitated by this knowledge base.
Top-down extraction of independently-modulated groups of genes captures the transcriptome at the systems level
We used ICA to identify 201 iModulons from PRECISE-1K. iModulons are independently modulated groups of genes that vary in concert across the dataset. iModulons also have activity levels that quantify their response in each PRECISE-1K condition. iModulons account for 83% of the total variance in the dataset. 117 of these iModulons are classified as Regulatory, as they are significantly enriched in genes belonging to a known regulon (Figure 2A; see Materials and Methods for regulatory enrichment details). These regulatory iModulons explain 56% of the total variance in PRECISE-1K. iModulons capturing smaller regulons tend to align closely with the known regulon, while iModulons capturing larger regulons tend to recover smaller subsets of larger regulons’ genes, leading to lower precision and recall (Figure 2B). 36 genomic iModulons that capture known genetic alterations (e.g. gene knockouts) and 17 biological iModulons (composed of genes with shared function but lacking significant regulon enrichment) account for another 19% of the variance. 22 technical iModulons explaining just 2% of the variance are dominated by a single short, uncharacterized gene, including 12 consisting of only the one gene. These iModulons likely capture noise in the dataset. Nine uncharacterized iModulons account for just 6% of the variance in the dataset. Altogether, 88% of the variance captured by iModulons can be explained by either regulatory, genomic or biological phenomena.
Figure 2.
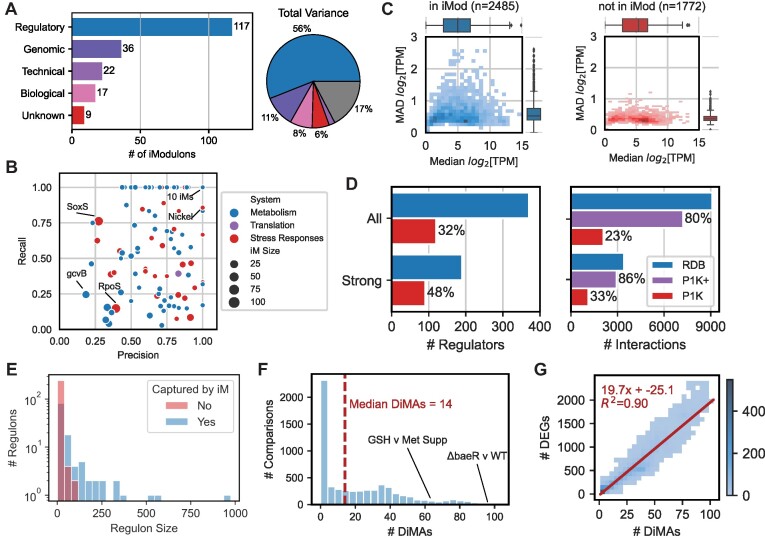
iModulons extracted from PRECISE-1K capture the transcriptional regulatory network. (A) A breakdown of PRECISE-1K iModulons by their annotation category: ‘Regulatory’ denotes significant enrichment of one or more known regulators; ‘Technical’ includes a single gene or technical artifact iModulon; ‘Genomic’ includes iModulons related to known genomic interventions (i.e. knockouts or segmental amplifications due to adaptive laboratory evolution); and ‘Biological’ includes iModulons containing genes of related function without significant regulator enrichment, or pointing to potential new regulons. Pie chart denotes iModulon annotation categories by percentage of variance explained. Gray wedge indicates variance unexplained by iModulons. (B) Summary of precision and recall for 117 regulatory iModulons. RegulonDB (http://regulondb.ccg.unam.mx) (31) regulons used as reference. (C) 2D histograms of median gene expression and median absolute deviation in gene expression by iModulon membership. (D) Comparison of regulators and regulatory interactions recovered by PRECISE-1K iModulons and available in RegulonDB. All = all evidence levels; Strong = only strong evidence interactions per RegulonDB; P1K+ = all interactions for which the corresponding regulator is captured by an iModulon. (E) Histogram of RegulonDB regulon sizes, colored depending on whether each RegulonDB regulon is or is not captured by at least one PRECISE-1K iModulon. (F) Histogram of the number of differential iModulon activities (DiMAs) computed between condition pairs within the same project (n = 6103; same as Figure 1G). (G) Comparison of number of DEGs and DiMAs for the same condition pairs. Linear best fit curve is shown in red, and indicates a ∼20-fold dimensionality reduction from DEGs to DiMAs. n = 4483 comparisons with non-zero DiMAs.
Fifty-eight percent of genes (2485/4257) are members of at least one iModulon. These genes have higher expression variation than genes not present in any iModulons (P = 1.03E-217, Mann–Whitney U test, m = 2485, n = 1772) (Figure 2C). Median expression itself does not differ significantly (P =0.33). Thus, iModulon membership is not restricted to higher-expressed genes. Indeed, 56% (823/1473) of y-ome genes—demonstrated above to be significantly less expressed—are members of at least one iModulon, highlighting the potential for iModulons to uncover putative functions for these uncharacterized genes (49). These observations highlight the need for genes to be differentially expressed under some conditions in order to be identified as a signal by ICA and incorporated into an iModulon.
The median iModulon consists of 10 genes, though many iModulons are much larger, such as global stress responses RpoS (122 genes) and SoxS (117) (Supplemental Figure S6A). Of the 189 multi-gene iModulons, 77% (145) consist of genes that are significantly intercorrelated compared to expectation (Supplemental Figure S7A). 88% of regulatory and 82% of biological iModulons have significantly intercorrelated genes, compared to just 47% of genomic and 13% of technical iModulons. Genomic iModulons capture genetic alterations present in small subsets of the total PRECISE-1K sample space—thus it is reasonable to expect that the genes perturbed in these limited samples need not be globally correlated across the compendium. Indeed, this observation indicates that iModulons can capture localized expression patterns beyond the reach of global correlation. Interestingly, eight out of nine uncharacterized iModulons contain significantly intercorrelated genes, highlighting an opportunity for further biologically-relevant discovery.
Thirty-five percent of genes in an iModulon (879/2485) are members of two or more iModulons, with two genes (ynfM and bhsA) appearing in seven each (Supplemental Figure S6B). Only 15% (131) of multi-iModulon genes are members of significantly correlated iModulons (Supplemental Figure SBLAH). However, within each of their iModulons, multi-iModulon genes rank in the 44th percentile in terms of intercorrelation with other iModulon genes (BLAH). These results suggest that multi-iModulon genes are influenced by distinct, recoverable signals, highlighting iModulons’ ability to capture overlapping regulatory modules of varying scale. iModulon-gene relationships are concentrated in a subset of large iModulons and genes present in multiple iModulons (Supplemental Figure S6C, D).
Eighty metabolism and 50 stress response iModulons account for 32% and 30% of the variance in PRECISE-1K, respectively (Supplemental Figure S8A). This breakdown emphasizes a ‘fear-greed’ tradeoff (50). Interestingly, the numbers of iModulons for these two functions differ considerably; the cell thus has a tendency towards more diversified regulation for metabolic capabilities and more centralized control for stress responses. Indeed, just two iModulons—RpoS and ppGpp, major stress response regulators—account for 6% of the variance in the dataset (Supplemental Figure S8B-C).
iModulons capturing the signals of global regulators (regulators with more than 25 regulatory targets) account for large proportions of the overall variance in the dataset. Flagella-related regulators FlhDC and FliA in combination explain over 5% of the expression variance, while anaerobic growth regulators FNR and ArcA combine to explain over 3% of the variance (Supplemental Figure S8C). These insights highlight the ability of global regulators to mobilize large-scale transcriptomic responses. Indeed, these regulators (along with iron regulator Fur) are responsible for variance between wild-type control samples run across projects, despite overall tight correlation between those samples (Supplemental Figure S9). Importantly, these batch variations are captured explicitly by these iModulon activities.
Regulatory modules represent the majority of the known transcriptional regulatory network
iModulons extracted from PRECISE-1K reconstruct a significant fraction of the total regulatory interactions available in RegulonDB (31), the premier database for curated and validated regulatory network information for E. coli. 32% of all known regulatory molecules (and 48% with strong evidence) are captured by regulatory iModulons (Figure 2D). Moreover, 23% of all specific regulatory interactions (33% of strong-evidence interactions) are reconstituted in iModulons. iModulons are known to capture regulatory signals by identifying the most strongly-regulated genes in a regulon based on promoter sequence (51). This sequence-based effect likely accounts for the relatively lower precision and recall enrichment statistics observed for larger iModulons that capture more global regulators. Thus, considering a regulatory iModulon as a biomarker for all of its regulator's interactions reveals that iModulons in fact reconstitute 80% of all known regulatory interactions (86% when considering only strong evidence). Importantly, iModulons preferentially capture the signals of larger regulons (Figure 2E), increasing their utility in describing transcriptome state across growth conditions.
Subsampling PRECISE-1K and recomputing iModulons demonstrates regulatory network coverages at different compendium sizes. On average across five trials, 20%-scale subsamples of PRECISE-1K (207 samples) yield 111 iModulons, of which 67% (75) are regulatory iModulons also captured from PRECISE-1K (Supplemental Figure S10A). As more samples are added, the total number of iModulons extracted also increases; however, the relative fraction of regulatory iModulons decreases. Nonetheless, regulatory recovery increases with scale: 33% of strong-evidence regulators are captured in iModulons from 20%-scale subsamples, compared with 48% from PRECISE-1K’s iModulons (Supplemental Figure S10B). Captured regulatory interactions follow a similar pattern. Critically, the step from 80%-scale subsamples (828 samples) to full PRECISE-1K elicits an increase in regulatory discovery following a plateau between the 60%- and 80%-scales, indicating that PRECISE-1K’s scale provides an advantage for regulatory recovery.
In all, iModulons provide the regulatory component of this transcriptome knowledge base. The subsequent sections demonstrate transcriptomic knowledge that can be derived from these regulatory modules.
Systems-level analysis of transcriptome states using regulatory modules
Because iModulons include an explicit representation of activity levels, they enable differential iModulon activity (DiMA) analysis. DiMA analysis allows for a systems-level comparison of transcriptome states by reducing hundreds or thousands of DEGs to a median of just 28 iModulons (Figure 2F). On average, a comparison between any two conditions in PRECISE-1K yields almost 20 times fewer differentially-activated iModulons than DEGs (Figure 2G), highlighting the particular usefulness of DiMA analysis for systems-level transcriptional analysis. On median, DiMAs directly explain 37% of variance between conditions. Because all iModulons explain a median of 80% of variance between conditions, DiMAs account for a median of 47% of variance explained by all iModulons (Supplemental Figure S11).
iModulon activities reflect the overall activity state of a transcriptional regulator across environmental conditions in PRECISE-1K. A stimulon is a higher-level regulatory structure composed of multiple regulons that respond to a particular stimulus (Supplemental Figure S1). While iModulons, by definition, include independently modulated groups of genes, in many instances these independent groups of genes are regulated in response to similar environmental stimuli, thus forming a stimulon. Two-component systems (TCS)—composed of a membrane-bound sensor and a cytoplasmic response regulator—enable the cell to sense and respond to important extracellular signals. iModulons derived from PRECISE-1K capture the response signal for 15 of 27 known TCS response regulators, providing insight into the cell's regulatory response to critical stimuli such as nitrogen, inorganic phosphate and alkali metals.
Additionally, iModulons can be clustered based on their activities to reveal higher order structures in the E. coli transcriptome. For example, one cluster captures the joint regulation of flagella formation by transcription factor complex FlhDC and sigma factor FliA (σ28) (Supplemental Figure S12). Six iron-related iModulons, five anaerobiosis-related iModulons and four amino acid-related iModulons also group together in this activity-based fashion. Thus, iModulons in combination can shed light on broad transcriptome patterns, providing a new definition of a stimulon.
Regulon discovery for putative transcription factors YgeV and YmfT
Functional annotation for putative transcription factors (TFs) remains challenging (52–54). However, iModulons are a powerful tool for the discovery and analysis of new regulons. PRECISE elucidated the regulons for three previously uncharacterized TFs (YieP, YiaJ/PlaR and YdhB/AdnB), and expanded the regulons of three known TFs (MetJ, CysB and KdgR) (1). Many of these regulatory interactions were confirmed through DNA-binding profiles (1,55,56). Furthermore, three novel regulons were predicted from iModulons derived from a microarray dataset (57). iModulons from PRECISE-1K recapitulate these previous results and reveal two new potential regulons.
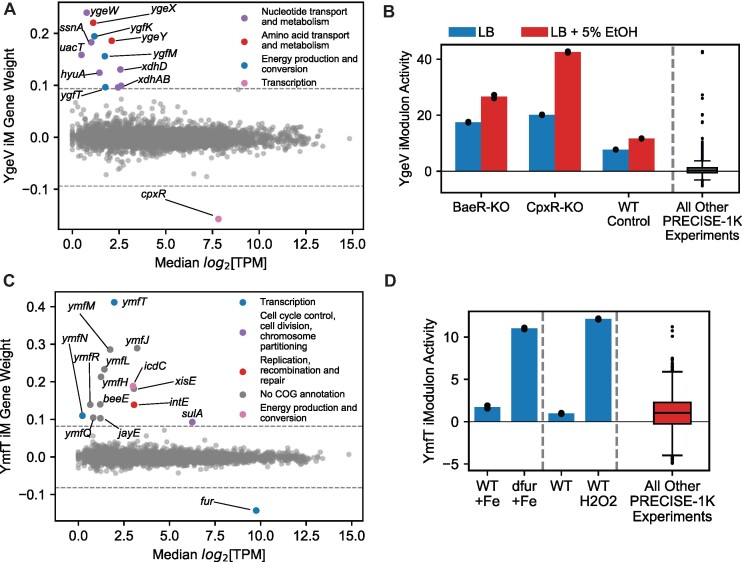
The putative YgeV regulon contains 13 genes, of which 7 are putatively involved in nucleotide transport and metabolism (Figure 3A). YgeV is predicted to be a Sigma54-dependent transcriptional regulator, and Sigma54-dependent promoters were previously identified upstream of the xdhABC and ygeWXY operons, which are in the YgeV iModulon (58). Although the iModulon does not contain the gene ygeV, ygeV is divergently transcribed from ygeWXY. A prior study (59) found that expression of ygfT was reduced in a YgeV mutant strain. Since ygfT is in the YgeV iModulon, this indicates that YgeV may serve as an activator for the genes in its iModulon. The activity of the YgeV iModulon rarely deviates from the reference condition; however, it is most active when knockouts of TCS response regulators BaeR or CpxR are exposed to ethanol (Figure 3B). Therefore, we hypothesize that the TF YgeV responds (either directly or indirectly) to ethanol to activate genes related to purine catabolism, and is repressed by TCS BaeRS and CpxAR.
Figure 3.
iModulons discover new regulons. (A) iModulon gene weights for the putative YgeV iModulon versus median log2[TPM]. (B) Activity of the YgeV iModulon in different media conditions. Each colored bar is the mean of two biological replicates (shown as individual black points). (C) iModulon gene weights for the putative YmfT iModulon vs. median log2[TPM]. (D) Activity of the YmfT iModulon in different media conditions. Each colored bar is the mean of two biological replicates (shown as individual black points).
The putative YmfT regulon contains 15 genes, including ymfT itself. It contains 12 of the 23 genes in the e14 prophage (60) (Figure 3C). The putative YmfT iModulon is most active in strains lacking the ferric uptake regulator Fur, or in strains challenged by oxidative stress through hydrogen peroxide (Figure 3D). Absence of Fur leads to overproduction of iron uptake proteins, oxidative damage, and, subsequently, mutagenesis (61). Therefore, we hypothesize that YmfT responds to oxidative stress to alter the expression of the e14 prophage.
These examples illustrate the potential for iModulons to predict new regulons and identify optimal conditions to study their activities.
Stratifying promoter-level mechanisms of crp regulation
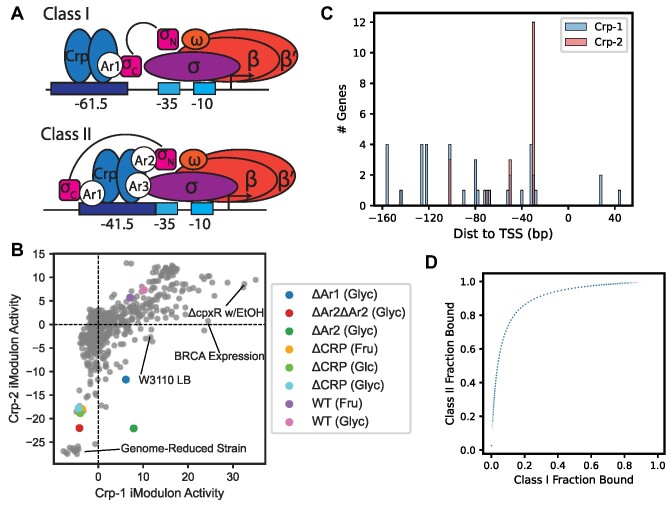
iModulons discover independent sub-groups of genes within global regulons that exhibit distinct regulatory dynamics. For example, the Fur-1 and Fur-2 iModulon activities (each capturing a subset of the Fur regulon) are non-linearly correlated based on both iron availability and aerobicity (41). In this section, we demonstrate that iModulons reflect biochemical mechanisms of TF binding by examining the relationship between two iModulons—Crp-1 and Crp-2—that stratify the CRP regulon. CRP contains multiple RNA polymerase-interacting domains (Ar1-3) (62) that facilitate its binding to Class I and Class II promoters. Class I promoters canonically involve binding centered 61.5 bp upstream of the transcription start site, and Class II are centered 41.5 bp upstream (63,64) (Figure 4A).
Figure 4.
iModulons stratify existing regulons by mode of binding. (A) Diagram of Class I and Class II CRP promoters. Arrow indicates transcription start site. σ = RNA polymerase (RNAP) sigma factor; σN and σC = sigma factor N- and C-terminal regions; β, β’, ω = RNAP core subunits; Ar1-3 = CRP activating regions (RNAP interaction sites). (B) iModulon phase plane between Crp-1 and Crp-2 iModulons. Colored points from samples involving partial and total CRP deletions. Ar regions correspond to panel A. Glyc = glycerol carbon source; fru = fructose; glc = glucose. (C) Histogram of CRP binding site locations for Crp-1 and Crp-2 iModulons. TSS = transcription start site of transcription unit for each gene. Data from RegulonDB. (D) Simulated binding curve for CRP Class I and Class II promoters. Each point indicates a particular CRP concentration. Binding modeled as 10× tighter at Class II versus Class I promoters.
The activities of the Crp-1 and Crp-2 iModulons across all PRECISE-1K conditions form a distinct nonlinear relationship (Figure 4B). As expected, low activities of both iModulons correspond with deletion of CRP, which is known to activate most of the genes in the two iModulons. Deletion of the Ar2 binding domain - implicated in Class II regulation - results in some Crp-1 activity but no Crp-2 activity (orange dot in Figure 4B). CRP binding sites for genes unique to Crp-1 are broadly distributed, while Crp-2-specific genes have CRP binding sites more consistently at the Class II location (Figure 4C). A steady-state biophysical model with 10-fold different binding affinities for Class I and Class II binding sites yields a similar binding site occupancy relationship as that between the Crp iModulon activities (Figure 4D). From this evidence, we propose that the Crp-1 and Crp-2 iModulons correspond to Crp regulatory activity at Class I and Class II promoter genes, respectively. This analysis highlights the capability of PRECISE-1K iModulons to capture multi-dimensional regulatory effects within a single regulon.
Incorporating 1675 high-quality publicly-available transcriptomes into the knowledgebase highlights method's scalability and robustness
To further expand our dataset, we sourced all publicly-available RNA-seq data for E. coli strain K-12 from NCBI’s Sequence Read Archive (SRA) (65). From 3230 K-12 samples, our processing and quality control pipeline yielded 1675 high-quality K-12 expression profiles. We combined these samples with PRECISE-1K to yield the ‘K-12 Dataset’, a high-quality transcriptomics dataset consisting of 2710 expression profiles (Figure 5A). These profiles come from 134 different projects, including 15 K-12 substrains and 9 distinct temperatures and pHs. ICA decomposition of the K-12 Dataset yields 194 iModulons.
Figure 5.
Adding public K-12 data to PRECISE-1K highlights PRECISE-1K’s stability. K-12 is a combined dataset composed of PRECISE-1K (1035 samples) plus all publicly-available high-quality RNA-seq data for E. coli K-12 (1675 samples). (A) The accumulation of high-quality RNA-seq data for K-12 over time. (B) K-12 iModulons by their annotation category (see Figure 2A legend). Pie chart denotes iModulon annotation categories by percentage of variance explained. The 194 annotated iModulons together explain 81% of the variance. Gray wedge indicates variance unexplained by iModulons. (C) Comparison of regulators and regulatory interactions recovered by K-12 and available in RegulonDB. All = all evidence levels; Strong = only strong evidence interactions per RegulonDB; K-12+ = all interactions for which the corresponding regulator is captured by the K-12 Dataset. P1K values from Figure 2D included for comparison. (D) Comparison of iModulons from three RNA-seq datasets: PRECISE(1); PRECISE-1K (this paper); and public K-12. Each small rectangle represents an iModulon for the corresponding dataset. Pairwise Pearson correlations were performed between PRECISE and P1K iModulons, and between P1K and K-12 iModulons; iModulons with correlations over 0.3 were considered to be the same iModulon (median correlation between PRECISE and P1K iModulons is 0.68; between P1K and K-12 is 0.70). Blue = iModulon exists in all three datasets; pink = iModulon only exists in PRECISE/PRECISE-1K; red = iModulon in PRECISE-1K/K-12 only; purple = iModulon unique to dataset. Explained variance is within each dataset (i.e. PRECISE iModulons explain ∼70% of variance in PRECISE, P1K iModulons explain ∼83% of variance in PRECISE-1K, etc.). iModulons are ordered by which dataset(s) they appear in, and sorted in decreasing order of explained variance within each dataset appearance category. (E) Overlap between the CsrA regulon per RegulonDB and the CsrA iModulon. (F) Activity of the CsrA iModulon after arrest of transcription initiation via addition of rifampicin (data from Potts et al (66)).
The distribution of iModulons by category – both in number and by explained variance—is broadly similar to that of PRECISE-1K. Regulatory iModulons account for 64% of the total number, and 57% of the total variance in the dataset (Figure 5B). Coverage of known regulatory network interactions increases only minutely as compared with PRECISE-1K alone, despite the more than doubling of the dataset's size (Figure 5C). Indeed, 89% of K-12′s explained variance comes from 155 iModulons highly correlated with iModulons extracted from either PRECISE or PRECISE-1K (Figure 5D). In contrast, 45% of explained variance from PRECISE-1K comes from 134 iModulons not present in PRECISE. Nonetheless, 67 iModulons captured in the original PRECISE are retained in both PRECISE-1K and K-12, accounting for sizable fractions of explained variance in each of the latter datasets. The iModulon structure remains largely consistent as dataset scale is increased; in general, higher-variance signals discovered by smaller-scale datasets are supplemented with new, more niche iModulons, rather than the entire iModulon structure shifting with scale. iModulons can also explain a slightly larger fraction of variance in PRECISE-1K than in the K-12 Dataset. iModulons extracted from just the 1675 publicly-available K-12 samples are similar to those extracted from the 2710-sample compendium, albeit with lower regulatory recovery (Supplemental Figure S13). Taken together, these results suggest that PRECISE-1K has sufficient scale and condition variety to represent the E. coli TRN, and additions of data beyond this scale may provide diminishing returns.
However, specific conditions in the K-12 dataset enable regulatory discovery. For example, 18 samples from a project exploring the post-transcriptional carbon storage regulator CsrA regulon (66) enabled recovery of a CsrA iModulon that is unique to the K-12 dataset. The CsrA iModulon is much larger than the known CsrA regulon: it contains 65 genes, of which 10 overlap with the 21-gene CsrA regulon (Figure 5E). Nonetheless, the enrichment of CsrA regulon genes in the iModulon is significant (adjusted P = 6.7E-9), and the genes in both the iModulon and regulon are particularly highly weighted in the iModulon. Moreover, the iModulon is much more highly active in a CsrA deletion strain after arrest of transcription initiation than the wild-type strain or other K-12 samples (Figure 5F), indicating relief of CsrA repression. Thus, the genes unique to the iModulon are candidates for expansion of the CsrA regulon.
Applying the knowledge base to new data: the anaerobic to aerobic transition
This knowledge base can be used to analyze new E. coli RNA-seq datasets. We demonstrate this capability for one project from the public K-12 Dataset. This project - called AAT for anaerobic-aerobic transition - captured six time-points in triplicate from 0 to 10 min after aeration of a previously anaerobic chemostat culture of E. coli K-12 W3110 (67). PRECISE-1K iModulon activities for the AAT project can be inferred, without necessitating full re-processing through the entire workflow. These inferred activities in turn enable analysis of AAT’s samples both within the project and within the context of all PRECISE-1K’s samples. The code used for this case study is available at https://github.com/SBRG/precise1k-analyze and can be used for analysis of any new data.
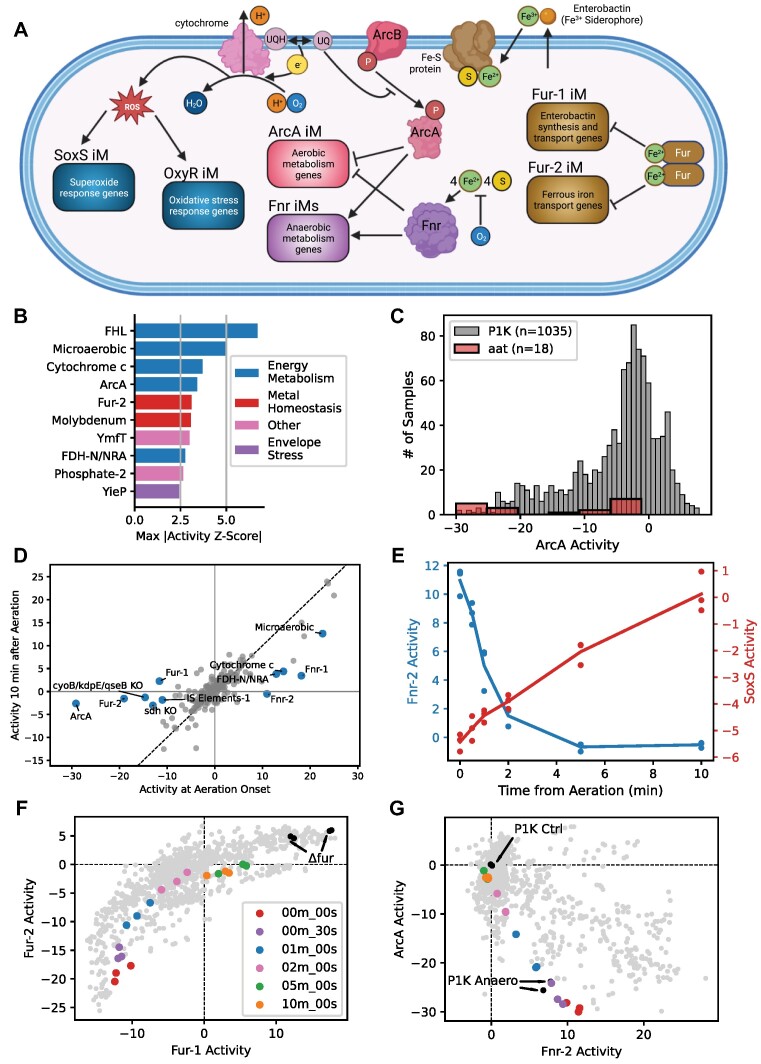
We hypothesized that certain iModulons would respond to the onset of aerobic growth (Figure 6A). For example, the regulators Fnr and ArcA are each influenced by oxygen availability. Fnr is activated by acquiring an iron-sulfur (4Fe–4S) cluster and dimerizing (68); oxygen directly inactivates Fnr by oxidizing the iron-sulfur cluster (69–71). While active, Fnr activates anaerobic metabolism genes and represses aerobic metabolism genes (72). ArcA is the TF component of a quinone-sensing two-component system. Under aerobic growth, quinols are oxidized to quinones as part of the electron transport chain; quinones in turn prevent sensor kinase ArcB from phosphorylating and activating ArcA (73,74). ArcA largely represses aerobic metabolism genes, while also activating a few fermentative genes (75–77). Many aerobic metabolism genes - especially oxidoreductases and electron transport chain components - require iron-sulfur clusters to function. Thus, the global iron regulator Fur - which represses iron acquisition genes when bound to iron (78) - is also implicated in this transition (79). Finally, oxidative phosphorylation under aerobic conditions generates reactive oxygen species (ROS), triggering the SoxS (80) and OxyR (81) responses.
Figure 6.
PRECISE-1K and iModulons provide key insight for assessing systems-level transcriptome changes for new data. For all graphs in this figure, the example new data comes from the public K-12 Dataset AAT (67) (anaerobic-aerobic transition) (not in PRECISE-1K, but in public K-12 metadata) which took 6 time-point samples of E. coli from 0 to 10 min after aeration of a previously anaerobic chemostat culture. (A) Schematic highlighting selected iModulons and systems involved in aerobic transition. (B) Top 10 regulatory iModulons by maximum activity difference between within-aat and PRECISE-1K activity (z-scored). For example, z-score of 5 for ‘Microaerobic’ iModulon indicates that the maximum activity of this iModulon amongst aat samples was 5 standard deviations from the mean activity of this iModulon in PRECISE-1K. (C) Histogram of iModulon activity across all PRECISE-1K samples and in new aat project (ArcA as example). (D) Differential iModulon activity (DiMA) plot comparing iModulon activities at aeration onset and 10 min after aeration. iModulons with significant activity differences between the two time points are in blue and labeled (see Methods for DiMA details). (E) iModulon activity by time from aeration for Fnr-2 and SoxS iModulons. (F) Phase plane comparing activities of Fur iModulons for all PRECISE-1K samples (gray) and aat samples (colored). Black dots indicate PRECISE-1K samples with fur knocked out. (G) Phase plane comparing activities of Fnr-2 and ArcA iModulons. aat color scheme same as (F).
We identified the iModulons with divergent activities in AAT compared to the rest of PRECISE-1K (Figure 6B). iModulons related to energy metabolism featured prominently; for example, the formate hydrogen lyase (FHL) iModulon had a maximum absolute activity in AAT six standard deviations off the PRECISE-1K median. FHL is known to be active under anaerobiosis during glucose fermentation (82). An activity histogram further contextualizes these observations: while ArcA iModulon activity is over three standard deviations away from the PRECISE-1K median at maximum in AAT, other AAT samples are closer to the PRECISE-1K median (Figure 6C).
To further characterize iModulon activity changes within AAT, DiMA analysis can identify iModulons that change significantly between any two sets of samples. Comparing aeration onset to 10 min post-aeration highlights the roles of key energy metabolism global regulators in facilitating this transition (Figure 6D). Fnr is more active at onset, while ArcA and Fur are significantly increased in activity 10 min after aeration. Fnr's activity decreases nonlinearly following aeration of the culture, reaching its aerobic growth reference level within 5 min (Figure 6E). In contrast, SoxS iModulon activity increases as aeration proceeds. Activity clustering highlights increased activity of the anaerobic stimulon at aeration onset, followed by increased activation of the iron stimulon 10 min post-aeration (Supplemental Figure S14).
Activity phase planes, which compare two iModulons’ activities across conditions, are another key tool for analyzing new data. The dynamic transcriptomic changes in the AAT project are notable in the Fur-1/Fur-2 (Figure 6F) and Fnr/ArcA (Figure 6G) phase planes. As aerobic metabolism takes over, iron-related genes repressed by Fur during anaerobiosis increase in activity as iron demand increases. Activity of anaerobic regulator Fnr decreases as aerobic regulator ArcA’s activity increases, with both arriving near the activity levels of PRECISE-1K’s aerobic growth control condition 10 min after aeration.
Taken together, these observations highlight the essential systems-level changes in the transcriptome composition during the anaerobic-aerobic transition while exemplifying PRECISE-1K’s function as an analysis resource. Further, they show the deep interpretation of TRN functions achieved through the use of iModulon activity phase planes.
DISCUSSION
This study establishes a multi-scale gene expression and regulation knowledge base for E. coli. The expression component is PRECISE-1K, a single protocol, high quality RNA-seq dataset containing 1035 samples covering a wide range of growth conditions. PRECISE-1K enables genome-wide categorization of genes based on expression level and expression variance across conditions. Using machine learning, we recover 117 regulatory modules (iModulons) from PRECISE-1K that reconstitute 86% of known regulatory interactions. iModulons—unlike principal components—explain variance in terms of knowledge of the TRN, not statistical magnitude. PRECISE-1K and its iModulons constitute the most complete top-down, computational transcription and regulation knowledge base yet generated for a microorganism. This resource enables regulon discovery and empowers novel experimental design. Most importantly, this resource empowers deep systems-level analysis of novel data.
We demonstrate that iModulons capture fundamental regulatory modes, not dataset-specific artifacts. iModulons from PRECISE-1K represent nearly all of the regulatory iModulons extracted from its predecessor PRECISE. Increasing the dataset size nearly four-fold does not hinder regulatory discovery; here we more than double the number of discovered regulatory iModulons. Conversely, decreasing the dataset's scale via subsampling yielded poorer regulatory recovery. This potential highlights the central role that top-down, data-driven methods must take in transcriptional regulatory discovery across organisms. Indeed, iModulons have already successfully generated top-down regulatory information for other organisms (1,15–19,83). Continued expansion of RNA-seq datasets for these and new organisms will likely drive further regulatory discovery.
Beyond their ability to systematically characterize a TRN, iModulons also provide a key tool: activity levels. This quantitative aspect of iModulons enables analysis of the functional transcriptome under specific environmental or genetic conditions. We demonstrate this capability by capturing two different functional regulatory modes of the Crp regulon based on binding site location. DiMA analysis also greatly simplifies differential expression analysis; with an average of nearly twenty times fewer significantly differential variables to analyze, DiMA analysis empowers systems-level analysis of transcriptomic changes, as demonstrated in the AAT case study.
Critically, PRECISE-1K and iModulon activities enable us to discover and partially characterize putative regulons for predicted transcription factors. We demonstrate this capability by assigning a putative function in ethanol stress tolerance related to nucleotide metabolism to the YgeV regulon, based on the YgeV iModulon activation pattern. In particular, this activation coincides with knockouts of two-component system response regulators BaeR and CpxR; thus, YgeV’s role in nucleotide metabolism upon ethanol stress response may arise as a compensatory mechanism following inactivation of these more prominent TCS regulators. The specificity of this activating condition may play a role in explaining why the functions of this regulator and the genes in its regulon remain unknown. Indeed, iModulons have already proven useful in studies to characterize regulators and their regulons (49,84,85). PRECISE-1K likely contains other instances of untapped insights and should continue to be mined for such discoveries.
However, we also highlight the need for judicious selection of growth conditions to maximize potential for regulatory elucidation. When we added all high-quality public K-12 data to PRECISE-1K, the iModulon structure remained quite similar, with the K-12 Dataset's 124 regulatory iModulons accounting for 88% of known TRN interactions. This result highlights two key points. Firstly, PRECISE-1K has sufficient scale and diversity to enable broad TRN discovery while avoiding noise introduced by combining data from multiple sources. Secondly, adding large numbers of RNA-seq samples beyond the scale of PRECISE-1K can yield diminishing returns. That said, certain specific new conditions from the K-12 Dataset were disproportionately useful - for example, a project perturbing the CsrA regulator enabled extraction of a corresponding regulatory iModulon. These observations likely highlight a limitation in the diversity of the available data, rather than of iModulons themselves. Thus, capturing additional unrecovered regulatory signals will likely rely on selection of growth conditions that activate niche transcriptional regulators with small regulons. Indeed, PRECISE-1K and the K-12 Dataset provide a blueprint for which conditions to prioritize for future discovery. Our knowledge base provides a centralized reference for assessment of gene expression and regulatory activity across conditions, empowering prudent study design. This capability is especially important for cost-, labor-, or time-intensive experiments, such as proteomics.
Our example analysis of the AAT project from the K-12 Dataset demonstrates perhaps the most exciting application of PRECISE-1K: analysis and contextualization of new RNA-seq datasets. PRECISE-1K’s iModulons clearly capture and summarize the regulatory dynamics at play during aerobic metabolism transition. We provide a variety of tools, both here and in our previously published code package (22) that will easily facilitate similar analyses for any other dataset. In this way, PRECISE-1K is not just useful in and of itself but as a backdrop for deriving regulatory insight from new data. Our example workflow for analyzing new data with PRECISE-1K is available at at https://github.com/SBRG/precise1k-analyze; all other analyses from this paper are available for use at https://github.com/SBRG/precise1k. These analyses have already enriched multi-omic studies of the aerobic respiration system (86), the adaptation of different E. coli strains (87), and the response of E. coli to antibiotics (41).
Overall, PRECISE-1K and iModulons represent a critical resource for studying expression and regulation in E. coli. We believe this resource should be a standard tool for systems-level analysis of E. coli RNA-seq data from all sources. As the number of publicly available datasets increases for other microorganisms, this study serves as a roadmap for interrogating similar datasets for less characterized organisms, with the potential to yield equally impactful insights into those organisms’ transcription and regulation characteristics. PRECISE-1K is disseminated through iModulonDB.org.
Limitations of the study
Although this resource presents many opportunities, some limitations also merit mention. First of all, assembling at least 200 high-quality, single-protocol RNA-seq profiles presents an up-front challenge for generating a PRECISE database for other organisms. While combining publicly available data can help, we have demonstrated that single-protocol datasets provide more regulatory elucidation on a per sample basis. Secondly, while PRECISE-1K does contain a broad range of growth conditions, this set is by no means exhaustive. Thus, minimal expression and regulatory knowledge can be provided for these missing conditions. Thirdly, iModulons are subject to limitations due to the ICA algorithm by which they are computed. For example, ICA assumes that each iModulon results from a single signal (regulator); therefore, genes with multiple regulators - or complex, multi-regulator regulons - can be more difficult to capture in iModulons. Also, ICA does not allow for hierarchy; thus, iModulons do not always capture the effects of regulators on other regulators, i.e. the activation of a set of local regulators by a global regulator. Additionally, while ICA does maximize statistical independence, its components are still based somewhat on variance. Thus, as dataset scale increases, signals that were captured from smaller datasets may not be captured from larger datasets because they account for relatively less variance in the larger dataset (hence the observation of some unique PRECISE iModulons). iModulons also cannot directly capture a true TRN—iModulons represent groupings of genes whose expression signals are intercorrelated while independent from other genes, not groupings of genes directly influenced by a regulator (as iModulons are computed without any prior knowledge of the TRN). Finally, DiMas—while indispensable for systems-level regulatory analysis - are not guaranteed to capture all individual gene-level changes in a given comparison. Indeed, the range of variance explained by DiMAs for any given condition comparison is wide; although a median of 47% of variance is explained by DiMAs, many comparisons are much more ‘lossy’ than this. Thus, it remains important to analyze gene expression data directly - for which PRECISE-1K itself may be used. These important caveats should be kept in mind when using this resource to analyze new data or analyzing this resource itself.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the National Energy Research Scientific Computing Center for support with high-performance computing. We acknowledge the UC San Diego Health Sciences Institute for Genomic Medicine for support with RNA-seq. We thank Hyun Gyu Lim for support with the anaerobic-aerobic transition analysis.
Author contributions: Conceptualization, B.O.P., C.R.L. and A.V.S.; Methodology, C.R.L., K.T.D., A.V.S. and K.R.; Software, C.R.L., K.T.D., A.V.S., K.R., J.L.M. and D.C.Z.; Validation, Y.G.; Investigation, C.R.L., K.T.D., A.V.S. and D.C.Z.; Writing - Original Draft, C.R.L., K.T.D. and A.V.S.; Writing - Review & Editing, C.R.L., K.R., D.C.Z. and B.O.P.; Visualization, C.R.L., K.T.D. and A.V.S.; Supervision, A.V.S. and B.O.P.; Funding Acquisition, B.O.P.
Contributor Information
Cameron R Lamoureux, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Katherine T Decker, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Anand V Sastry, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Kevin Rychel, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Ye Gao, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
John Luke McConn, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Daniel C Zielinski, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA.
Bernhard O Palsson, Department of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA; Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Kemitorvet, Building 220, 2800 Kgs. Lyngby, Denmark.
DATA AVAILABILITY
All data (aside from raw RNA-seq data) and code for analysis and figures are available on Zenodo at: https://doi.org/10.5281/zenodo.8284223. iModulons and related data are also available from iModulonDB.org under the dataset “E. coli PRECISE-1K” and “E. coli Modulome” (Public K-12). Raw RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the metadata file, located in the same Zenodo repository at the path: data/precise1k/metadata_qc.csv. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation [NNF20CC0035580]. Funding for open access charge: Novo Nordisk Fonden [NNF20CC0035580].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sastry A.V., Gao Y., Szubin R., Hefner Y., Xu S., Kim D., Choudhary K.S., Yang L., King Z.A., Palsson B.O.. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 2019; 10:5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ziemann M., Kaspi A., El-Osta A.. Digital expression explorer 2: a repository of uniformly processed RNA sequencing data. Gigascience. 2019; 8:giz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leader D.P., Krause S.A., Pandit A., Davies S.A., Dow J.A.T.. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-seq, miRNA-seq and sex-specific data. Nucleic Acids Res. 2018; 46:D809–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zrimec J., Börlin C.S., Buric F., Muhammad A.S., Chen R., Siewers V., Verendel V., Nielsen J., Töpel M., Zelezniak A.. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure. Nat. Commun. 2020; 11:6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Z., Pan Z., Ying Y., Xie Z., Adhikari S., Phillips J., Carstens R.P., Black D.L., Wu Y., Xing Y.. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods. 2019; 16:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwon M.S., Lee B.T., Lee S.Y., Kim H.U.. Modeling regulatory networks using machine learning for systems metabolic engineering. Curr. Opin. Biotechnol. 2020; 65:163–170. [DOI] [PubMed] [Google Scholar]
- 9. Kelley D.R., Reshef Y.A., Bileschi M., Belanger D., McLean C.Y., Snoek J.. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018; 28:739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Avsec Ž., Agarwal V., Visentin D., Ledsam J.R., Grabska-Barwinska A., Taylor K.R., Assael Y., Jumper J., Kohli P., Kelley D.R.. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods. 2021; 18:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Parmigiani G., Johnson W.E.. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020; 2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Q., Markatou M.. Evaluation of methods in removing batch effects on RNA-seq data. Infect. Dis. Transl. Med. 2016; 2:3–9. [Google Scholar]
- 13. Comon P. Independent component analysis, a new concept. Signal Process. 1994; 36:287–314. [Google Scholar]
- 14. Saelens W., Cannoodt R., Saeys Y.. A comprehensive evaluation of module detection methods for gene expression data. Nat. Commun. 2018; 9:1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rychel K., Sastry A.V., Palsson B.O.. Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome. Nat. Commun. 2020; 11:6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poudel S., Tsunemoto H., Seif Y., Sastry A.V., Szubin R., Xu S., Machado H., Olson C.A., Anand A., Pogliano J.et al.. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:17228–17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajput A., Tsunemoto H., Sastry A.V., Szubin R., Rychel K., Sugie J., Pogliano J., Palsson B.O.. Machine learning from Pseudomonas aeruginosa transcriptomes identifies independently modulated sets of genes associated with known transcriptional regulators. Nucleic Acids Res. 2022; 50:3658–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chauhan S.M., Poudel S., Rychel K., Lamoureux C., Yoo R., Al Bulushi T., Yuan Y., Palsson B.O., Sastry A.V. Machine learning uncovers a data-driven transcriptional regulatory network for the crenarchaeal thermoacidophile sulfolobus acidocaldarius. Front. Microbiol. 2021; 12:753521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoo R., Rychel K., Poudel S., Al-Bulushi T., Yuan Y., Chauhan S., Lamoureux C., Palsson B.O., Sastry A.. Machine learning of all Mycobacterium tuberculosis H37Rv RNA-seq data reveals a structured interplay between metabolism, stress response, and infection. mSphere. 2022; 7:e0003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rychel K., Decker K., Sastry A.V., Phaneuf P.V., Poudel S., Palsson B.O.. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res. 2021; 49:D112–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C.. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017; 35:316–319. [DOI] [PubMed] [Google Scholar]
- 22. Sastry A.V., Poudel S., Rychel K., Yoo R., Lamoureux C.R., Chauhan S., Haiman Z.B., Al Bulushi T., Seif Y., Palsson B.O. Mining all publicly available expression data to compute dynamic microbial transcriptional regulatory networks. 2021; bioRxiv doi:02 July 2021, preprint: not peer reviewed 10.1101/2021.07.01.450581. [DOI]
- 23. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L., Wang S., Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012; 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 25. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 26. Ewels P., Magnusson M., Lundin S., Käller M.. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016; 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V.et al.. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011; 12:2825–2830. [Google Scholar]
- 29. Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999; 10:626–634. [DOI] [PubMed] [Google Scholar]
- 30. McConn J.L., Lamoureux C.R., Poudel S., Palsson B.O., Sastry A.V.. Optimal dimensionality selection for independent component analysis of transcriptomic data. BMC Bioinf. 2021; 22:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santos-Zavaleta A., Salgado H., Gama-Castro S., Sánchez-Pérez M., Gómez-Romero L., Ledezma-Tejeida D., García-Sotelo J.S., Alquicira-Hernández K., Muñiz-Rascado L.J., Peña-Loredo P.et al.. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019; 47:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du B., Olson C.A., Sastry A.V., Fang X., Phaneuf P.V., Chen K., Wu M., Szubin R., Xu S., Gao Y.et al.. Adaptive laboratory evolution of Escherichia coli under acid stress. Microbiology. 2020; 166:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen K., Anand A., Olson C., Sandberg T.E., Gao Y., Mih N., Palsson B.O.. Bacterial fitness landscapes stratify based on proteome allocation associated with discrete aero-types. PLoS Comput. Biol. 2021; 17:e1008596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anand A., Chen K., Yang L., Sastry A.V., Olson C.A., Poudel S., Seif Y., Hefner Y., Phaneuf P.V., Xu S.et al.. Adaptive evolution reveals a tradeoff between growth rate and oxidative stress during naphthoquinone-based aerobic respiration. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:25287–25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anand A., Chen K., Catoiu E., Sastry A.V., Olson C.A., Sandberg T.E., Seif Y., Xu S., Szubin R., Yang L.et al.. OxyR is a convergent target for mutations acquired during adaptation to oxidative stress-prone metabolic states. Mol. Biol. Evol. 2020; 37:660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCloskey D., Xu S., Sandberg T.E., Brunk E., Hefner Y., Szubin R., Feist A.M., Palsson B.O.. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 2018; 9:3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan J., Sastry A.V., Fremming K.S., Bjørn S.P., Hoffmeyer A., Seo S., Voldborg B.G., Palsson B.O.. Independent component analysis of E. coli's transcriptome reveals the cellular processes that respond to heterologous gene expression. Metab. Eng. 2020; 61:360–368. [DOI] [PubMed] [Google Scholar]
- 38. Sandberg T.E., Szubin R., Phaneuf P.V., Palsson B.O.. Synthetic cross-phyla gene replacement and evolutionary assimilation of major enzymes. Nat. Ecol. Evol. 2020; 4:1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hirokawa Y., Kawano H., Tanaka-Masuda K., Nakamura N., Nakagawa A., Ito M., Mori H., Oshima T., Ogasawara N.. Genetic manipulations restored the growth fitness of reduced-genome Escherichia coli. J. Biosci. Bioeng. 2013; 116:52–58. [DOI] [PubMed] [Google Scholar]
- 40. Choudhary K.S., Kleinmanns J.A., Decker K., Sastry A.V., Gao Y., Szubin R., Seif Y., Palsson B.O.. Elucidation of regulatory modes for five two-component systems in Escherichia coli reveals novel relationships. Msystems. 2020; 5:e00980-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sastry A., Dillon N., Poudel S., Hefner Y., Xu S., Szubin R., Feist A., Nizet V., Palsson B.. Decomposition of transcriptional responses provides insights into differential antibiotic susceptibility. 2020; bioRxiv doi:04 May 2020, preprint: not peer reviewed 10.1101/2020.05.04.077271. [DOI]
- 42. Braun V., Rehn K.. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 1969; 10:426–438. [DOI] [PubMed] [Google Scholar]
- 43. Li G.-W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fleischer R., Heermann R., Jung K., Hunke S.. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J. Biol. Chem. 2007; 282:8583–8593. [DOI] [PubMed] [Google Scholar]
- 45. Tschauner K., Hörnschemeyer P., Müller V.S., Hunke S.. Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS One. 2014; 9:e107383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt A., Kochanowski K., Vedelaar S., Ahrné E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M.. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016; 34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heckmann D., Campeau A., Lloyd C.J., Phaneuf P.V., Hefner Y., Carrillo-Terrazas M., Feist A.M., Gonzalez D.J., Palsson B.O.. Kinetic profiling of metabolic specialists demonstrates stability and consistency of in vivo enzyme turnover numbers. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:23182–23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghatak S., King Z.A., Sastry A., Palsson B.O.. The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acids Res. 2019; 47:2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodionova I.A., Gao Y., Sastry A., Hefner Y., Lim H.G., Rodionov D.A., Saier M.H. Jr, Palsson B.O. Identification of a transcription factor, PunR, that regulates the purine and purine nucleoside transporter punC in E. coli. Commun. Biol. 2021; 4:991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Utrilla J., O’Brien E.J., Chen K., McCloskey D., Cheung J., Wang H., Armenta-Medina D., Feist A.M., Palsson B.O.. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst. 2016; 2:260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu S., Lamoureux C., Akbari A., Palsson B.O., Zielinski D.C.. Quantitative sequence basis for the E. coli transcriptional regulatory network. 2022; bioRxiv doi:20 February 2022, preprint: not peer reviewed 10.1101/2022.02.20.481200. [DOI]
- 52. Gao Y., Yurkovich J.T., Seo S.W., Kabimoldayev I., Dräger A., Chen K., Sastry A.V., Fang X., Mih N., Yang L.et al.. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2018; 46:10682–10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao Y., Lim H.G., Verkler H., Szubin R., Quach D., Rodionova I., Chen K., Yurkovich J.T., Cho B.-K., Palsson B.O.. Unraveling the functions of uncharacterized transcription factors in Escherichia coli using ChIP-exo. Nucleic Acids Res. 2021; 49:9696–9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim G.B., Gao Y., Palsson B.O., Lee S.Y.. DeepTFactor: a deep learning-based tool for the prediction of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2021171118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodionova I.A., Gao Y., Sastry A., Yoo R., Rodionov D.A., Saier M.H., Palsson B.Ø.. Synthesis of the novel transporter YdhC, is regulated by the YdhB transcription factor controlling adenosine and adenine uptake. 2020; bioRxiv doi:03 May 2020, preprint: not peer reviewed 10.1101/2020.05.03.074617. [DOI]
- 56. Rodionova I.A., Gao Y., Sastry A.V., Monk J., Szubin R.. PtrR (YneJ) is a novel E. coli transcription factor regulating the putrescine stress response and glutamate utilization. 2020; bioRxiv doi:29 April 2020, preprint: not peer reviewed 10.1101/2020.04.27.065417. [DOI]
- 57. Sastry A.V., Hu A., Heckmann D., Poudel S., Kavvas E., Palsson B.O.. Independent component analysis recovers consistent regulatory signals from disparate datasets. PLoS Comput. Biol. 2021; 17:e1008647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reitzer L., Schneider B.L.. Metabolic context and possible physiological themes of ς54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 2001; 65:422–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. DeLisa M.P., Wu C.F., Wang L., Valdes J.J., Bentley W.E.. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 2001; 183:5239–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mehta P., Casjens S., Krishnaswamy S.. Analysis of the lambdoid prophage element e14 in the E. coli K-12 genome. BMC Microbiol. 2004; 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Touati D., Jacques M., Tardat B., Bouchard L., Despied S.. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 1995; 177:2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lawson C.L., Swigon D., Murakami K.S., Darst S.A., Berman H.M., Ebright R.H.. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004; 14:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Busby S., Ebright R.H.. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 1999; 293:199–213. [DOI] [PubMed] [Google Scholar]
- 64. Latif H., Federowicz S., Ebrahim A., Tarasova J., Szubin R., Utrilla J., Zengler K., Palsson B.O.. ChIP-exo interrogation of crp, DNA, and RNAP holoenzyme interactions. PLoS One. 2018; 13:e0197272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. International Nucleotide Sequence Database Collaboration Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Potts A.H., Vakulskas C.A., Pannuri A., Yakhnin H., Babitzke P., Romeo T.. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 2017; 8:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bui T.T., Selvarajoo K.. Attractor concepts to evaluate the transcriptome-wide dynamics guiding anaerobic to aerobic state transition in Escherichia coli. Sci. Rep. 2020; 10:5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moore L.J., Kiley P.J.. Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 2001; 276:45744–45750. [DOI] [PubMed] [Google Scholar]
- 69. Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P.J.. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:6087–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sutton V.R., Mettert E.L., Beinert H., Kiley P.J.. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J. Bacteriol. 2004; 186:8018–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jervis A.J., Crack J.C., White G., Artymiuk P.J., Cheesman M.R., Thomson A.J., Le Brun N.E., Green J.. The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4659–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Salmon K., Hung S.-P., Mekjian K., Baldi P., Hatfield G.W., Gunsalus R.P.. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003; 278:29837–29855. [DOI] [PubMed] [Google Scholar]
- 73. Bekker M., Alexeeva S., Laan W., Sawers G., Teixeira de Mattos J., Hellingwerf K.. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 2010; 192:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Beilen J.W.A., Hellingwerf K.J.. All three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/Anaerobic response regulator ArcA. Front. Microbiol. 2016; 7:1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iuchi S., Lin E.C.. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:1888–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iuchi S., Lin E.C.C.. Adaptation of Escherichia coli to respiratory conditions: regulation of gene expression. Cell. 1991; 66:5–7. [DOI] [PubMed] [Google Scholar]
- 77. Gunsalus R.P., Park S.J.. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 1994; 145:437–450. [DOI] [PubMed] [Google Scholar]
- 78. Mills S.A., Marletta M.A.. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry. 2005; 44:13553–13559. [DOI] [PubMed] [Google Scholar]
- 79. Beauchene N.A., Myers K.S., Chung D., Park D.M., Weisnicht A.M., Keleş S., Kiley P.J.. Impact of anaerobiosis on expression of the iron-responsive fur and RyhB regulons. MBio. 2015; 6:e01947-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nunoshiba T., Hidalgo E., Amábile Cuevas C.F., Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1992; 174:6054–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zheng M., Wang X., Templeton L.J., Smulski D.R., LaRossa R.A., Storz G.. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 2001; 183:4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stephenson M., Stickland L.H.. Hydrogenlyases: bacterial enzymes liberating molecular hydrogen. Biochem. J. 1932; 26:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lim H.G., Rychel K., Sastry A.V., Bentley G.J., Mueller J., Schindel H.S., Larsen P.E., Laible P.D., Guss A.M., Niu W.et al.. Machine-learning from Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network. Metab. Eng. 2022; 72:297–310. [DOI] [PubMed] [Google Scholar]
- 84. Rodionova I.A., Hosseinnia A., Kim S., Goodacre N., Zhang L., Zhang Z., Palsson B., Uetz P., Babu M., Saier M.H. Jr. E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding. Sci. Rep. 2023; 13:7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rodionova I.A., Gao Y., Monk J., Hefner Y., Wong N., Szubin R., Lim H.G., Rodionov D.A., Zhang Z., Saier M.H. Jret al.. A systems approach discovers the role and characteristics of seven LysR type transcription factors in Escherichia coli. Sci. Rep. 2022; 12:7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Anand A., Patel A., Chen K., Olson C.A., Phaneuf P.V., Lamoureux C., Hefner Y., Szubin R., Feist A.M., Palsson B.O.. Laboratory evolution of synthetic electron transport system variants reveals a larger metabolic respiratory system and its plasticity. Nat. Commun. 2022; 13:3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kavvas E.S., Long C.P., Sastry A., Poudel S., Antoniewicz M.R., Ding Y., Mohamed E.T., Szubin R., Monk J.M., Feist A.M.et al.. Experimental evolution reveals unifying systems-level adaptations but diversity in driving genotypes. mSystems. 2022; 7:e0016522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Heckmann D., Lloyd C.J., Mih N., Ha Y., Zielinski D.C., Haiman Z.B., Desouki A.A., Lercher M.J., Palsson B.O.. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models. Nat. Commun. 2018; 9:5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data (aside from raw RNA-seq data) and code for analysis and figures are available on Zenodo at: https://doi.org/10.5281/zenodo.8284223. iModulons and related data are also available from iModulonDB.org under the dataset “E. coli PRECISE-1K” and “E. coli Modulome” (Public K-12). Raw RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the metadata file, located in the same Zenodo repository at the path: data/precise1k/metadata_qc.csv. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.