Abstract
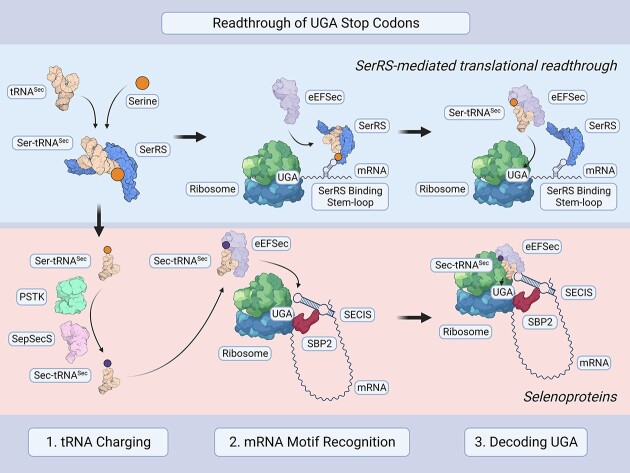
Translational readthrough of UGA stop codons by selenocysteine-specific tRNA (tRNASec) enables the synthesis of selenoproteins. Seryl-tRNA synthetase (SerRS) charges tRNASec with serine, which is modified into selenocysteine and delivered to the ribosome by a designated elongation factor (eEFSec in eukaryotes). Here we found that components of the human selenocysteine incorporation machinery (SerRS, tRNASec, and eEFSec) also increased translational readthrough of non-selenocysteine genes, including VEGFA, to create C-terminally extended isoforms. SerRS recognizes target mRNAs through a stem-loop structure that resembles the variable loop of its cognate tRNAs. This function of SerRS depends on both its enzymatic activity and a vertebrate-specific domain. Through eCLIP-seq, we identified additional SerRS-interacting mRNAs as potential readthrough genes. Moreover, SerRS overexpression was sufficient to reverse premature termination caused by a pathogenic nonsense mutation. Our findings expand the repertoire of selenoprotein biosynthesis machinery and suggest an avenue for therapeutic targeting of nonsense mutations using endogenous factors.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
mRNA translation is a finely tuned process by which proteins are generated. tRNAs are charged by their cognate aminoacyl-tRNA synthetase (aaRS) with the specific amino acid their anticodon deciphers (1). These charged tRNAs are then used by the ribosome to produce a polypeptide chain following instructions encoded by mRNA. However, in order for the ribosome to know where to begin and end, specific translation initiation, elongation, and termination factors are needed to facilitate the recognition of signals embedded in the mRNA (2,3). In fact, the same mRNA can produce different protein isoforms with vastly different functions (4). Regulation by the flanking untranslated regions (UTRs), especially the 3′UTR, dictates the proportions of different protein variants that are synthesized (5). This allows for an additional level of gene product regulation unrestricted by genetically encoded information.
Translational readthrough (TR) is one mode of translational regulation involving the 3′UTR. Suppression of a stop codon leads to TR, upon which additional amino acids are attached to the C-terminus of a nascent peptide chain (6). Stop codon suppression is diversely utilized in cells – apart from the generation of protein isoforms by functional TR, it is also key to introducing the 21st amino acid of the genetic code, selenocysteine (7).
The incorporation of selenocysteine via TR occurs in all three domains of life. The first step, strictly conserved across kingdoms, is the aminoacylation of tRNASec by seryl-tRNA synthetase (SerRS, gene name SARS1) with a serine (8,9). The serine residue is further converted to selenocysteine in the second step, where eukaryotes use a divergent system from that of bacteria (10,11). In eukaryotes, this step is catalyzed by two consecutive enzymes: O-phosphoseryl-tRNASec kinase (PSTK) and O-phosphoseryl-tRNASec selenium transferase (SepSecS). The resulting Sec-tRNASec is delivered to the ribosome by a designated alternative translational elongation factor, eEFSec, to decode the UGA stop codon (12). In all cases, whether to synthesize selenoproteins or to extend specific protein forms, TR must be strictly regulated as random disregard of stop codons would be highly detrimental. Thus, complex RNA structures, which dictate translational speed to allow for the recruitment of nearby factors, regulate the readthrough of the stop codon for selenocysteine incorporation (13).
Numerous diseases are caused by nonsense mutations leading to in-frame premature termination codons (PTCs); in fact, about 11% of all genetic lesions that cause human disease can be traced to PTCs (14). Nonsense mutations can lead to a breadth of symptoms, ranging from developmental disorders early in life to increased susceptibility towards cancer during adulthood (15,16). Methods to utilize TR as a therapeutic paradigm in these diseases are currently being evaluated (17), such as using aminoglycoside drugs (18), nonsense suppressor tRNAs (19,20), and nonsense-mediated mRNA decay (NMD) inhibitors.
A major concern with these techniques is side effects resulting from possible readthrough of non-PTC genes (21,22), necessitating the development of targeted therapeutics. Deeper knowledge into the mechanisms of naturally occurring readthrough would provide clues on how to rescue nonsense mutations without the need to introduce foreign components.
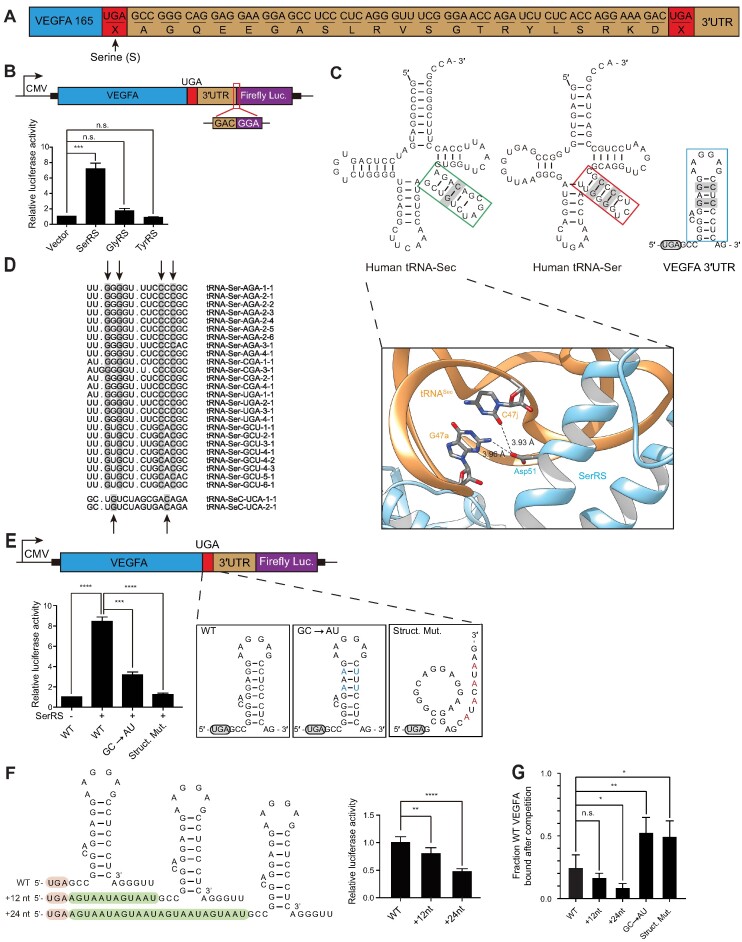
TR has been described for the master angiogenesis regulator VEGFA, a pivotal protein in cancer and development (23,24). Attachment of 22 additional amino acids (Figure 1A) leads to novel VEGFA functions: instead of eliciting a pro-angiogenic response as found for the prototypical VEGFA variant, the so-termed VEGF-Ax readthrough product no longer strongly induces angiogenesis, although its exact function is debated (25–27). VEGFA TR is dependent on heterogeneous nuclear ribonucleoprotein HNRNPA2/B1 binding to a regulatory element following the stop codon. However, this mechanism does not directly explain how the stop codon is suppressed nor why serine is exclusively incorporated at the stop codon position (25). We further investigated the mechanism of VEGFA TR and found that it involved SerRS and select components of the selenocysteine incorporation machinery. This function was dependent on the conserved catalytic function of SerRS and a unique appended domain that appeared in vertebrates. Moreover, we performed enhanced cross-linking immunoprecipitation (eCLIP) in human cells and identified other candidate genes beyond VEGFA whose TR may be regulated by SerRS. We further showed that overexpression of human SerRS alone was sufficient to alleviate protein truncation in a cell-based model of a familial nonsense mutation in the tumor suppressor MSH2 that causes high risk for colorectal cancer.
Figure 1.
SerRS binds to the 3′UTR of VEGFA to facilitate translational readthrough. (A) Scheme of VEGFA downstream of the first stop codon. VEGFA mRNA can be read through its first stop codon, resulting in VEGF-Ax with 22 appended amino acids. (B) Luciferase assay to quantify VEGFA translational readthrough. SerRS overexpression increased VEGFA TR while expression of two other aminoacyl-tRNA synthetases, GlyRS and TyrRS, did not impact VEGFA TR. (C) Upper panel: Secondary structure analysis of VEGFA predicts a stem-loop in the 3′UTR. Comparison between the VEGFA stem-loop motif and variable loops of tRNASer and tRNASec. Both variable loops and the VEGFA stem-loop contain G–C base pairs, which are recognized by SerRS (PDB-ID: 4RQF, lower panel). (D) Alignment of human tRNASec and tRNASer isoacceptor variable loops. The conserved G–C base pairs in each variable loop are highlighted. (E) Disruption of the VEGFA stem-loop motif abrogated SerRS-mediated TR, quantified by luciferase reporter assay. Exchanging G–C base pairs to A–U reduced TR, as did disrupting the stem-loop structure entirely. (F) Addition of 12 or 24 nucleotide spacers between the VEGFA UGA stop codon and stem-loop reduced TR as quantified by luciferase reporter assay. (G) Quantification of competitive EMSA results showing how different VEGFA constructs compete WT VEGFA mRNA off SerRS. (A–G) (n.s., not significant; * P< 0.05, ** P< 0.01, *** P< 0.001; **** P< 0.0001).
MATERIALS AND METHODS
Cell culture and constructs
All cells were cultured in a humidified incubator at 37°C with 5% CO2. Human HEK293 and MDA-MB-231 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and the human 293AD cell line was purchased from Agilent (Agilent, Santa Clara, CA, USA). These were cultured in Dulbecco's Modified Eagle Medium (ThermoFisher Scientific, Grand Island, NY, USA) supplemented with heat-inactivated fetal bovine serum (Omega Scientific, Tarzana, CA, USA) to a final concentration of 10% and 1% Penicillin-Streptomycin (ThermoFisher Scientific). Transient transfections were performed using Lipofectamine 2000 (ThermoFisher Scientific). Human SerRS variants, GlyRS, and TyrRS were cloned into pCDNA6c-V5/His6 vector (ThermoFisher Scientific), and human full-length SerRS was cloned into pBabe-puro vector (Addgene, pBABE-puro was a gift from Hartmut Land & Jay Morgenstern & Bob Weinberg) (Addgene plasmid # 1764; http://n2t.net/addgene:1764; RRID:Addgene_1764) (28). For mutations in SerRS, we performed site-directed mutagenesis PCR to obtain the SerRST429A construct. We established MDA-MB-231 cell lines stably expressing human SerRS mutants by using pBabe-puro (28) vector-based retroviral infections. Stable polyclonal cell lines with either SerRS overexpression or the corresponding control were generated through selection with puromycin.
Protein purification
For protein purification, human SerRS was subcloned into pET-20b(+) plasmid (Novagen, Darmstadt, Germany) and expressed in E. coli. The recombinant C-terminal His6-tagged proteins were purified using Ni-NTA beads (Qiagen, Valencia, CA, USA) followed by a heparin column and size exclusion chromatography. The purities of the recombinant proteins were assessed by Coomassie blue staining and electrophoresis with 4–12% Bis-Tris Mini gels (ThermoFisher Scientific). Protein concentrations were determined by measuring absorbance at 280 nm.
Cell fractionation analysis
Cell fractionation was performed according to a previously described protocol (29). Briefly, the cells were harvested with 0.25% Trypsin-EDTA, and cells were lysed with swelling buffer (10 mM Tris–HCl pH 7.4, 2 mM EDTA, proteinase inhibitor cocktail (Roche, Basel, Switzerland)). The cytoplasmic fraction was separated by centrifugation at 800 × g for 5 min (supernatant) and the pellet containing nuclear proteins was washed. The nuclear fractions were extracted using nuclear extraction buffer (20 mM HEPES pH 7.6, 300 mM NaCl, 2 mM EDTA, 1 mM 1,4-dithiothreitol, 10% glycerol, 1% Triton X-100, protease inhibitor cocktail). SerRS was detected with an anti-SerRS antibody (made in-house by immunizing rabbits with recombinant human SerRS protein). Lamin A/C and α-tubulin, nuclear and cytoplasmic markers, respectively, were detected by western blot to test the purity of cellular fractions.
Western blot
Cells were washed with phosphate-buffered saline (PBS) and lysed with cell lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM of EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4 and protease inhibitor cocktail). Protein concentration of each sample was quantified with a BCA Protein Assay Kit (ThermoFisher Scientific). Equal amounts of protein extract were mixed with SDS loading buffer and boiled at 95°C for 5 minutes. Samples were loaded on 4–12% Tris-acrylamide gels, then transferred onto polyvinylidene difluoride membranes. Membranes were first blocked with 5% milk, then incubated with the indicated antibodies, followed by secondary antibody conjugated to horseradish peroxidase. Blots were visualized with an ECL chemiluminescence kit (ThermoFisher Scientific, USA) in a FluorChem M (Proteinsimple). Custom-made rabbit anti-human SerRS antibody was made by the Scripps Research antibody core. Monoclonal anti-His6-Tag antibody (#HRP-66005) was purchased from Proteintech. Monoclonal anti-V5 antibody (#R960CUS) and polyclonal anti-eEFSec antibody (#PA5-31764) were purchased from ThermoFisher Scientific. The anti-MSH2 (#2017), anti-Lamin A/C (#2032), anti-β-actin (#3700), and anti-α-tubulin (#3873) antibodies were purchased from Cell Signaling Technology. Monoclonal anti-SBP2 antibody (#sc-130639) was purchased from Santa Cruz (Santa Cruz, CA, USA).
Luciferase reporter system
Firefly luciferase (FLuc) reporters downstream of sequences of interest (VEGFA, MSH2, BAK1, TIMP1, CFL1) were transfected (500 ng/well) into HEK293 cells in 24-well plates using Lipofectamine 2000. Renilla luciferase (RLuc) (50 ng/well) was co-transfected as a control. After 48 h, cells were lysed and FLuc and RLuc activity were measured using Dual-Luciferase Reporter Assay System (Promega) in a Victor3 1420 Multilabel Plate Counter (PerkinElmer).
Cross-linking immunoprecipitation (CLIP)
CLIP was performed as previously described (30). Briefly, 2 × 108 SerRS-overexpressing MDA-MB-231 cells were crosslinked by exposure to short-wave UV light at 300 mJ per cm2. Cells were harvested by scraping and solubilized in RIPA buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and frozen at −80°C. Immediately preceding immunoprecipitation, lysates were thawed on ice, vortexed until homogeneous, and clarified by centrifugation at 16 000 × g for 5 min in a 4°C refrigerated microcentrifuge. Clarified lysate was transferred to a fresh microcentrifuge tube and pre-depleted with protein G Sepharose beads for 30 min at 4°C. The resin was removed from the clarified lysate by centrifugation at 5000 × g for 5 min, and the depleted lysate was transferred to a fresh microcentrifuge tube, followed by the addition of rabbit polyclonal SerRS antibody or rabbit IgG control. The extracts were incubated for 2 h at 4°C under constant agitation. After antibody binding, the immunocomplexes were captured from the lysate by the addition of protein G Sepharose resin and incubated for 2 h at 4°C under agitation. After binding, RNA-protein complexes were released by proteinase K digestion at 37°C in the presence of 1% SDS. RNA fragments were TRIzol extracted.
eCLIP-seq
eCLIP-seq was performed using the RBP-eCLIP Kit (#ECK001, Eclipse Bioinnovations, San Diego, CA). 2 × 107 MDA-MB-231 cells were used for each of the three replicates at different cell passages. Cell culture medium was replaced with ice-cold PBS and cross-linking was performed at 254 nM UV with a setting of 400 mJ/cm2. After cross-linking, cells were scraped, washed, counted, centrifuged and snap-frozen. eCLIP was performed by following the manufacturer's instructions. Briefly, cells were lysed, sonicated using a Bioruptor Pico (Diagenode, Denville, NJ), and digested with DNase and Ambion RNase I, cloned (#AM2294, ThermoFisher Scientific). Immunoprecipitation was performed with mouse monoclonal anti-SerRS antibody (#sc-271032, Santa Cruz Biotechnology) bound to goat anti-mouse IgG magnetic beads (#S1430, New England Biolabs). Size-matched input (SMInput) samples were taken before antibody enrichment. Samples were washed, adapters were added, protein-RNA complexes were separated by SDS-PAGE electrophoresis and transferred to nitrocellulose membranes by wet transfer. The membrane section containing the complexes was cut out and digested with Proteinase mix supplied by the manufacturer. Resulting RNAs were purified and reverse transcribed. Resulting cDNA was purified and adapters were ligated on the ends. cDNA was quantified by qRT-PCR, and libraries were amplified by PCR. Libraries were gel-purified from a 3% low-melting temperature agarose gel, pooled, analyzed by Agilent TapeStation, and sequenced on a NextSeq 2000 (Illumina).
eCLIP-seq analysis
Reads were processed using the Skipper processing pipeline, available at https://github.com/YeoLab/skipper (31). In short, reads were trimmed of adapters with skewer (32), mapped with STAR (2.7.10a_alpha_220314) (33) and PCR-deduped with UMIcollapse (34). Binding candidates were identified using a tiled window approach, where the 5′ read ends (representing the crosslinking site) were counted across evenly sized windows for each genic region. Windows were then binned according to GC content to estimate and adjust for GC biases, and the comparison of IP reads to corresponding size-matched input (SMinput) reads were used to determine enrichment of signal above background. Browser tracks were created using IGV (35). Pathway analysis was performed using Metascape (36).
Ribosome profiling library construction
Ribosome profiling libraries were generated as previously described (37,38) with some minor modifications. Briefly, two 10 cm dishes of cells were used for each biological replicate, and three biological replicates were prepared for each cell line (MDA-MB-231-empty vector, MDA-MB-231-SARS). Cell homogenization was performed in 1 ml lysis buffer (20 mM Tris–Cl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml CHX, 1% (v/v) TritonX-100, 50 units/ml Turbo DNaseI). RNase I-treated lysates were overlaid on top of a sucrose cushion in 5 ml Beckman Ultraclear tubes and centrifuged in an SW55Ti rotor for 4 hours at 4°C at 46 700 rpm to isolate monosomes. Pellets were resuspended and RNA was extracted using the miRNeasy kit (Qiagen) according to manufacturer's instructions. 26–34 nucleotide RNA fragments were purified by electrophoresis on a 15% denaturing gel. Linker addition, cDNA generation (first-strand synthesis was performed at 50°C for 1 h), circularization, rRNA depletion, and amplification of cDNAs with indexing primers were performed. Library quality and concentration were assessed using high sensitivity D1000 screen tape on the Agilent tape station, Qubit 2.0 Fluorometer, and qPCR. All libraries were pooled and run on HiSeq4000 (SR75).
Ribosome profiling analysis
Ribosomal footprints were analyzed as described by Ingolia et al. (37) with these modifications: Trimgalore was used to trim off adapters and clip the first nucleotide off the 5′ end. Reads were then mapped to ribosomal RNA using bowtie2 (39) and unmapped reads were further mapped to the human transcriptome (v19) with STAR aligner (33). Expected read length distribution was tested with the R package RiboProfiling. To center ribosomes and obtain a list of genes with P-sites in their 3′ UTR, we used functionalities within Ribowaltz (40) and a custom python script by Scott Adamson, UConn, and Jax Laboratories.
Quantitative real-time PCR assay (qRT-PCR)
Total RNA was isolated from cells with TRIzol Reagent (ThermoFisher Scientific) as a control. For assaying mRNA stability, cells were treated with 5 μg/ml actinomycin D for up to 4 h before isolating total RNA. One microgram (μg) of total RNA from each CLIP experiment was reverse transcribed to cDNA with M-MLV reverse transcriptase (Promega, Madison, WI, USA) or SuperScript III (Invitrogen). All real-time PCR reactions were performed using the StepOnePlus Real-Time PCR system (ThermoFisher Scientific) with SYBR Select Master Mix (Applied Biosystems) or Power Sybr Green Master Mix (Thermo Fisher). Primers used for the PCR reactions are listed in Supplementary Table S1. The PCR reaction started at 95°C for 10 min, followed by 45 cycles of 95°C for 20 s and 60°C for 1 min.
RNAi
DNA oligos encoding short-hairpin RNAs (shRNA) designed against human eEFSec (5′-GAT CCG CTA GAT GCG GAC ATT CAC ACC TCG AGG TGT GAA TGT CCG CAT CTA GCT TTT TTC -3′), SECISBP2 (5′- GAT CCG CCA GTC CTT TCC AAA GAA TGC TCG AGC ATT CTT TGG AAA GGA CTG GCT TTT TTC -3′), and tRNASec (5′-GAT CCG TGC AGG CTT CAA ACC TGT AGC TCG AGC TAC AGG TTT GAA GCC TGC ACT TTT TTC-3′) were inserted into pLentiLox-hH1 plasmid, modified from the pLentiLox 3.7 plasmid to contain a H1 promoter (between Xba I and Xho I sites) to drive the shRNA expression. For non-targeting control shRNA, we used the sequence 5′-TAA GGC TAT GAA GAG ATA C-3′. Cells were transfected with shRNA plasmids by using Lipofectamine 2000 reagent (ThermoFisher Scientific). 48 h post-transfection, cells were subjected to analysis.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as previously described (41). Briefly, the 69 bp RNA oligonucleotides corresponding to the SerRS binding site on the VEGFA 3′UTR (5′-UGA GCC GGG CAG GAG GAA GGA GCC UCC CUC AGG GUU UCG GGA ACC AGA UCU CUC ACC AGG AAA GAC UGA-3′) and its variants were synthesized. For MSH2, a 30 bp RNA oligonucleotide (5′-UCA AAU GGA GCA CCU GUU CCA UAU GUA CGA-3′) was synthesized. The RNA products were annealed and [32P]-labeled at the 5′ end by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) before desalting using a Sephadex G-25 spin column (GE Healthcare, Pittsburgh, PA, USA). The labeled oligonucleotides (5 nM final concentration) were incubated with recombinant SerRS at the indicated concentrations (and cold RNA competitors in competitive EMSAs) in binding buffer [20 mM Tris–HCl pH 8.0, 60 mM KCl, 5 mM MgCl2, 0.1 mg/mL BSA, 10 ng/μl poly(I:C), 1 mM DTT, 5% glycerol] for 1 h at room temperature. The samples were loaded on a 5% native polyacrylamide gel and underwent electrophoresis at 250 V in running buffer (25 mM Tris, pH 8.3, 190 mM glycine). Afterwards, the gel was dried and examined by autoradiography.
Northern blot
Total RNAs of HEK293 cells transfected with shRNA-encoding plasmids were extracted by using TRIzol (Invitrogen) according to manufacturer's instructions with minor modifications. RNAs were precipitated by adding 2.5 volumes of ethanol and incubating at –20°C overnight. The RNA samples were subjected to electrophoresis on 10% TBE-Urea gels (Invitrogen), followed by electroblotting to Hybond-N + nylon membranes (GE Healthcare). The membranes were blocked and incubated with [32P]-labeled DNA probes at 50°C. The probes used for detecting human tRNASec and U6 snRNA are as follows: 5′- GAA AGG TGG AAT TGA ACC ACT CTG TCG CTA GAC AGC TAC AGG TTT GAA GCC TGC ACC CCA GAC CAC TGA GGA TCA TCC G -3′ and 5′-GCA GGG GCC ATG CTA ATC TTC TCT GTA TCG-3′.
Patient-derived B cell isolation and immortalization
Blood samples were collected from the mother (P) and her three children (F1, M1, M2) (Figure 3A) at the National Cheng Kung University Hospital following institutional guidelines for research with human subjects (IRB Protocol Number: B-ER-106–186). B cells were isolated by positive selection using CD19 Dynabeads PanB magnetic beads (Invitrogen). Purified primary B cells were immortalized by incubation with supernatant from sodium butyrate (3 mM) and TPA (tetradecanoyl phorbol acetate, 40 ng/ml) treated B95.8 cell for 1.5 h at 37°C to infect them with EBV. Aliquots of infected cells were collected at different time points for analysis: day 16, week 3 and week 6 (to ensure that EBV immortalization had occurred).
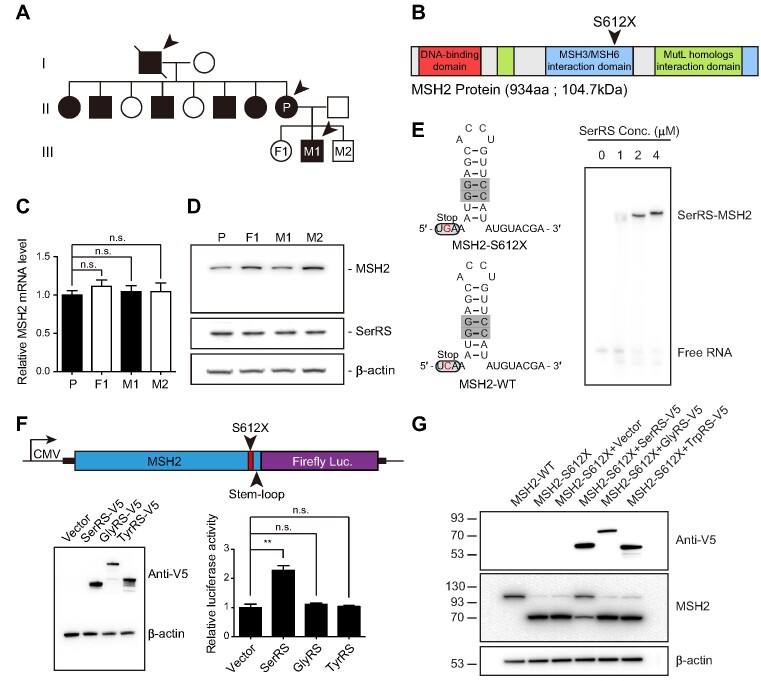
Figure 3.
SerRS-mediated translational readthrough rescues protein levels reduced by a cancer-causing nonsense mutation found in a Taiwanese family. (A) Pedigree of the Taiwanese family with a hereditary MSH2 nonsense mutation (p.S612X, annotated as S611X in a previous study (16)). Female carriers are indicated by filled black circles, male carriers by filled black squares, and affected individuals (diagnosed with gastrointestinal cancers) by arrows. Open circles and squares indicate family members carrying the WT MSH2 gene. (B) Domain structure of MSH2. The monoallelic p.S612X nonsense mutation is located in the domain that interacts with MSH3 and MSH6. (C) qRT-PCR of MSH2 mRNA levels in patient-derived B cells (n = 3) showed no difference between mutant carriers and wild type individuals. (D) Western blot result of MSH2 protein levels in patient-derived B cells showed reduced full-length MSH2 protein in mutant carriers. (E) RNA structure prediction of the stem-loop motif following the p.S612X mutation on MSH2 that is recognized by SerRS. An EMSA showed SerRS binding to the stem-loop structure of MSH2. (F) Luciferase assay to quantify MSH2-S612X translational readthrough. Signal amplification was obtained from a reporter containing the N-terminal MSH2-S612X sequence including the 30 nucleotides downstream of the premature stop codon, followed by the coding sequence for luciferase. SerRS overexpression increased MSH2-S612X translational readthrough while expression of two other aminoacyl-tRNA synthetases, GlyRS and TyrRS, did not impact MSH2-S612X translational readthrough (n = 3). (G) Western blot results showing SerRS-mediated rescue of full-length MSH2-S612X protein levels after overexpression of SerRS, GlyRS and TrpRS with MSH2-S612X in HEK293 cells. (A–G) (n.s., not significant; ** P< 0.01).
tRNA alignment
Statistical analyses
Statistical significance was tested with Student's t-test using GraphPad Prism (Graphpad Software, Inc.). n.s., not significant; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
RESULTS
SerRS overexpression promotes translational readthrough of VEGFA in a reporter system
Eswarappa et al. reported that the amino acid serine was incorporated in the position of the UGA stop codon to generate VEGF-Ax (25) (Figure 1A). As SerRS charges serine onto tRNASec, which can suppress the UGA stop codon, during the initial step of selenocysteine incorporation, we were interested in investigating whether these processes are related. The efficiency of stop codon suppression is increased if local concentrations of the charged tRNA are high (44), so we tested whether increasing the levels of SerRS would affect VEGFA TR. However, detection of VEGF-Ax on the protein level is challenging as the TR event likely makes up only a small percentage of the total VEGFA protein produced and no commercial antibodies are available against the appended amino acid sequence to specifically detect the VEGF-Ax isoform. To facilitate VEGF-Ax detection, we used a luciferase reporter assay to allow sensitive detection and quantification of TR events while controlling for other regulatory effects that might affect general protein synthesis (Figure 1B). Expression of VEGFA from a plasmid under a CMV promoter additionally enabled us to easily exchange regulatory elements in the mRNA and avoided interference with potential SerRS-dependent transcriptional regulation of VEGFA (45). Overexpression of SerRS in 293AD cells increased TR by ∼7-fold, while overexpression of other aaRSs (glycyl-tRNA synthetase, GlyRS, GARS1, and tyrosyl-tRNA synthetase, TyrRS, YARS1) did not alter TR significantly (Figure 1B, Supplementary Figure S1A), demonstrating that SerRS specifically induced VEGFA TR.
SerRS recognizes stem–loop structures resembling its cognate tRNAs
As unregulated readthrough of genes caused by the expression of a constitutively expressed protein would be highly undesirable, we investigated whether SerRS-dependent readthrough would be specific to VEGFA transcripts due to the recognition of internal RNA sequences or structures. SerRS does not use the tRNA anticodon as an identity element for recognition (46). Instead, a long variable loop unique to both tRNASer and tRNASec is recognized by SerRS (9,46,47) (Figure 1C). Recognition of tRNASec by SerRS is mediated by contacts with a G-C base pair in the middle of the variable loop (9) (Figure 1C). Conserved G-C base pairs are also found in the tRNASer variable loop (43) (Figure 1C and D). We found a stem-loop structure following the regular UGA stop codon in the 3′UTR of VEGFA mRNA that shared similarities with the variable loop in tRNASer and tRNASec (Figure 1C and Supplementary Figure S1B). Specifically, two G–C base pairs are similarly located in the stem-loop compared with the conserved G-C pairs in the variable loops of tRNASer and tRNASec (Figure 1C). To test the importance of the stem-loop structure and the G–C base pairs for TR, we introduced mutations in the VEGFA mRNA stem-loop which substituted these two G–C base pairs for A–U pairs or with mutations that were predicted to disrupt stem-loop formation entirely (Figure 1E and Supplementary Figure S1B). Substitution of the two G–C base pairs with A-U or disruption of the stem-loop structure both significantly reduced the SerRS-mediated increase of VEGFA TR using our luciferase TR assay (Figure 1E). Through competitive EMSA analysis, we confirmed that SerRS binds to the VEGFA mRNA and that the A–U and structural mutants displayed weaker ability to compete off WT VEGFA mRNA from SerRS (Figure 1G and Supplementary Figure S1C), demonstrating that binding by SerRS was dependent on stem-loop G–C base pairs in its target mRNA. Additionally, we introduced either a 12 or 24 nucleotide spacer between the UGA stop codon and the stem-loop, both of which also decreased the efficiency of TR (Figure 1F). In contrast to the A-U and structural mutants, extending the distance between stop codon and the stem-loop either did not affect or improved WT VEGFA mRNA binding to SerRS (Figure 1G and Supplementary Figure S1C), despite the observed reduction of TR in cells (Figure 1F). This strongly suggests that the close proximity between the SerRS binding site and the stop codon is also important for TR. To further rule out the possibility that the observed TR increase was due to a mRNA stabilization effect of SerRS binding, we performed qPCR of the VEGFA reporter mRNA with and without SerRS overexpression (Supplementary Figure S1D). No significant difference in the levels and the stability of VEGFA reporter mRNA between the two conditions was observed, suggesting SerRS does not increase TR by mRNA stabilization.
Translational readthrough relies on SerRS aminoacylation function and a vertebrate-specific domain
Using the luciferase assay, we further mapped the involvement of different domains of SerRS in TR. SerRS consists of a N-terminal tRNA-binding domain (TBD) that binds to the long variable loop of its cognate tRNAs, a catalytic domain (CD) containing the active site, and a C-terminal UNE-S domain (Figure 2A), which is an evolutionarily new addition found in vertebrate SerRS from fish to humans (45). We found both the tRNA-binding and UNE-S domains are necessary for mediating TR, as expression of truncated SerRS lacking either domain did not stimulate TR (Figure 2B). Unlike the conserved tRNA-binding domain and catalytic domain, the UNE-S domain does not contribute to tRNA binding and is dispensable for aminoacylation but mediates binding to other nucleic acids, for example genomic DNA (45). Through EMSA, we determined that the UNE-S domain is also necessary for SerRS to bind the VEGFA mRNA, which in turn mediates TR (Figure 2C). Consistently, expression of the catalytic domain alone had no effect on TR (Figure 2B). To investigate whether the catalytic activity of SerRS is required for SerRS-mediated TR, we mutated threonine 429 to alanine in full-length SerRS (SerRST429A). The mutation is located near the active site in the catalytic domain (Supplementary Figure S2), which disrupts tRNA charging (45,48). Despite the intact binding of SerRST429A to the VEGFA stem-loop (Figure 2D), overexpression of SerRST429A did not lead to increased TR (Figure 2E), demonstrating that enzymatic activity is essential for SerRS-mediated TR. These experiments suggested that to stimulate TR, not only the catalytic activity and tRNA-binding capacity of SerRS are required, but also the vertebrate-specific UNE-S domain, possibly through its capacity to bind nucleic acids other than tRNA.
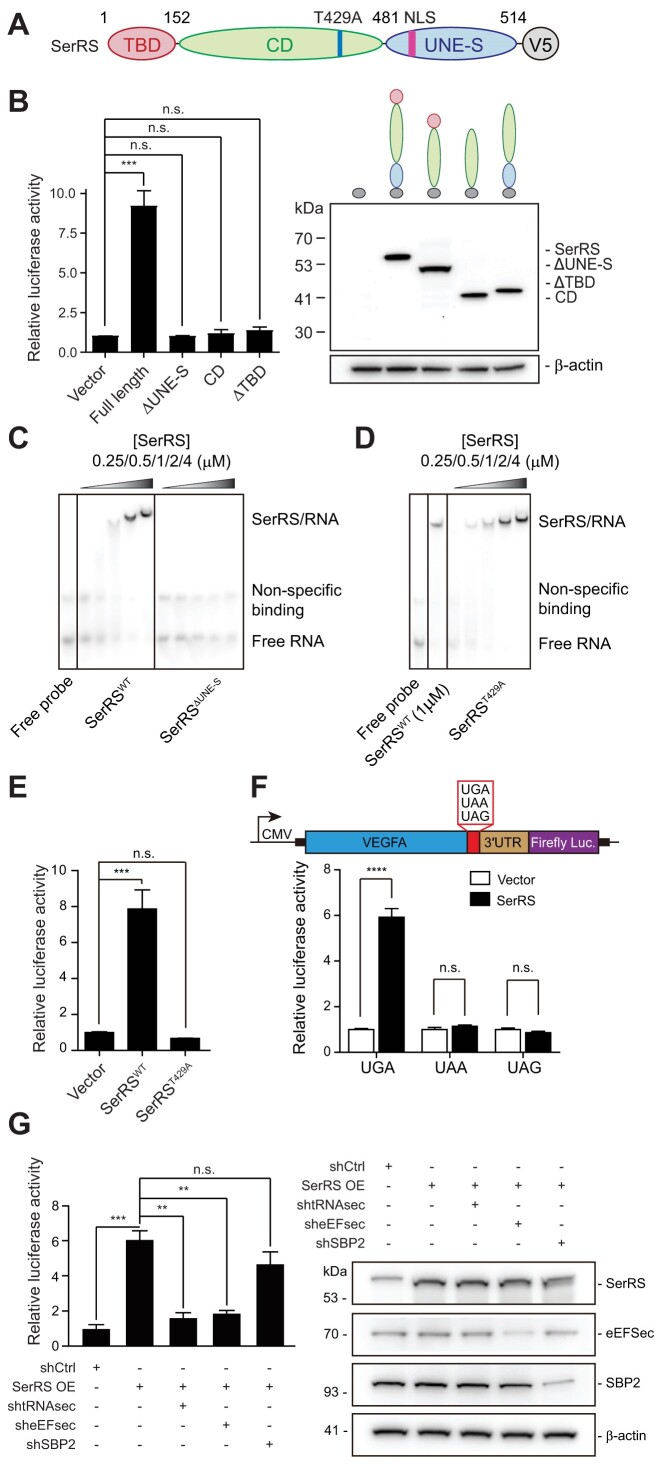
Figure 2.
SerRS-mediated translational readthrough is dependent on SerRS catalytic activity and selenocysteine incorporation elements. (A) Scheme of SerRS domain structure. (B) Domain mapping for SerRS-mediated translational readthrough. SerRS contains a tRNA-binding domain (TBD), a catalytic domain (CD) and a domain unique to SerRS (UNE-S) involved in nucleic acid binding. Expression of V5-tagged SerRS domains was confirmed by western blot. (C) EMSA showing binding of SerRSWT but not SerRSΔUNE-S to the VEGFA mRNA. (D) EMSA showing binding of the catalytic mutant SerRST429A to the VEGFA mRNA. (E) A point mutation in the SerRS catalytic site (T429A), which renders SerRS catalytically inactive, abolished SerRS translational readthrough activity. (F) Mutation of the UGA stop codon to UAA or UAG abrogated increased translational readthrough upon SerRS overexpression in a VEGFA-based luciferase reporter assay. (G) SerRS-mediated TR measured by a VEGFA reporter assay with SerRS overexpression. TR is dependent on tRNASec and eEFSec, as their knockdown abrogates the increase in translational readthrough by SerRS overexpression. SBP2 knockdown does not affect TR. Knockdown was verified by western blot. (A–G) (n.s., not significant; ** P< 0.01; *** P< 0.001; **** P< 0.0001)
SerRS-mediated translational readthrough utilizes tRNASec and eEFSec for stop codon suppression
As we could establish that SerRS binding directly to mRNA and its catalytic activity were required for TR, we next investigated the mechanism. VEGFA contains a UGA stop codon, so we tested whether SerRS-dependent TR would tolerate any of the three stop codons. Mutating UGA to either of the other two stop codons, UAA or UAG, abrogated SerRS-induced TR in our system (Figure 2F). This suggests that SerRS-mediated TR is highly specific to UGA and likely dependent on stop codon suppression by specific tRNAs recognizing the UGA stop codon.
SerRS initiates the first step in the incorporation of selenocysteine by charging tRNASec, which decodes UGA stop codons. If the mechanism behind SerRS-dependent TR is shared with selenocysteine incorporation, reduction of tRNASec should severely impair SerRS-dependent TR. We used a shRNA directed against tRNASec to reduce the tRNA level (49) (Supplementary Figure S3A), which indeed strongly reduced SerRS-dependent TR (Figure 2G). Selenocysteine incorporation is driven by a complex machinery of factors that enables stop codon suppression by tRNASec. A specific elongation factor, eEFSec, competes with termination factors and employs a mechanism distinct from other elongation factors (50). Knockdown of eEFSec also significantly impaired TR, suggesting that SerRS-dependent TR does rely on the same components as selenocysteine incorporation (Figure 2G). Knockdown of either tRNASec or eEFSec also significantly reduced the level of TR in the absence of SerRS overexpression (Supplementary Figure S3B), further confirming that the mechanism also occurs in cells with an endogenous level of SerRS.
During selenocysteine incorporation, the specific site of stop codon suppression is determined by SBP2, which recognizes the SECIS, an in-cis RNA stem-loop motif in the 3′UTR of selenoprotein mRNAs. SBP2 also interacts with the ribosome stalled at the upstream UGA stop codon, which brings the eEFSec/Sec-tRNASec complex via its binding to the SECIS to the proximity of the UGA codon (51). However, knockdown of SECISBP2 (SBP2 gene) did not reduce TR significantly as opposed to knockdown of tRNASec or EEFSEC (eEFSec gene) (Figure 2G and Supplementary Figure S3B). Therefore, SerRS-mediated TR shares some but not all key factors for selenocysteine incorporation.
SerRS rescues full-length protein expression by suppressing a pathogenic nonsense mutation in MSH2
To further explore the applicability of our observations to disease-causing nonsense mutations, we focused on a family with an autosomal dominant nonsense mutation in MSH2 (16) (Figure 3A). The MSH2 gene encodes for a DNA mismatch repair protein, and the loss of one copy of this gene is sufficient to cause microsatellite instability and increase cancer risk, especially of hereditary nonpolyposis colorectal cancer (HNPCC) or Lynch syndrome (52). The proband (III-M1) and the proband's mother (II-P) both carry a heterozygous (c.1835C > G) mutation, introducing a stop codon where a serine should be (p.Ser612X, annotated as S611X in the original case report (16)). We confirmed the mutation independently by Sanger sequencing, while sequencing of the proband's siblings (III-F1, III-M2) revealed no mutation (Supplementary Figure S4A). The nonsense mutation falls within the MSH3/MSH6 interaction domain and truncation of the protein would likely result in loss-of-function (Figure 3B). qRT-PCR analysis of the affected proband and parent compared to the proband's unaffected siblings revealed no significant differences in MSH2 mRNA levels (Figure 3C), suggesting that nonsense-mediated decay is not engaged. However, western blotting of MSH2 from immortalized patient-derived B cells showed reduced full-length protein in the individuals with the mutation (Figure 3D). Truncated MSH2 could not be detected in patient B cells, possibly due to degradation caused by instability. RNA structure prediction of the nucleotides directly following the aberrantly introduced stop codon revealed the formation of a G–C-containing stem-loop structure (Supplementary Figure S4B), and an EMSA performed with purified SerRS and the 30 nucleotides of MSH2 mRNA containing the stem-loop structure confirmed binding (Figure 3E). We developed a luciferase reporter system for MSH2 readthrough, similar to the one described for VEGFA: the N-terminal sequence of MSH2 including the p.Ser612X mutation and the 30 nucleotides downstream of mutation were placed under a CMV promoter, followed by a luciferase reporter (Figure 3F). This was transfected into 293AD cells along with SerRS or other aaRSs as controls. Overexpression of SerRS, but not of the other aaRSs, led to increased translational readthrough of the MSH2 reporter (Figure 3F). This was confirmed through western blot, where overexpression of SerRS increased full-length MSH2 expression and simultaneously reduced the truncated form (Figure 3G). These experiments suggest that SerRS overexpression could rescue full-length MSH2 translation in this single model of a clinically relevant nonsense mutation.
SerRS translational readthrough affects a specific gene set
As SerRS recognized stem-loop structures, we asked whether TR of other genes could be similarly regulated by SerRS. We enriched SerRS-bound mRNAs using cross-linking immunoprecipitation (CLIP) in human breast cancer MDA-MB-231 cells and manually identified genes bound by SerRS through PCR and Sanger sequencing (Supplementary Table S2). Enrichment of the so identified 9 genes and VEGFA could be verified by qRT-PCR (Supplementary Figure S5A). Of these genes (including VEGFA), four contained a UGA stop codon. To verify that SerRS overexpression could lead to increased readthrough, we developed the luciferase reporter assay for one of the identified target genes with a UGA stop codon, BAK1. BAK1, a mitochondrial outer membrane protein, also possesses a stem-loop motif in its 3′UTR (Supplementary Figure S5B). In line with our findings for VEGFA, SerRS overexpression increased TR of BAK1 over 14-fold while overexpressing other aaRSs did not affect TR (Supplementary Figure S5B). These findings reinforced that SerRS binding to mRNA stem-loop motifs is a common mechanism to promote TR in different genes.
While the CLIP method was able to identify SerRS-bound mRNAs, it could not pinpoint where SerRS bound and was limited in the number of genes that could feasibly be identified. To unbiasedly search for mRNAs that are potentially regulated by SerRS and their binding sites, we performed enhanced CLIP followed by next-generation sequencing of SerRS-bound RNA targets (eCLIP-seq).
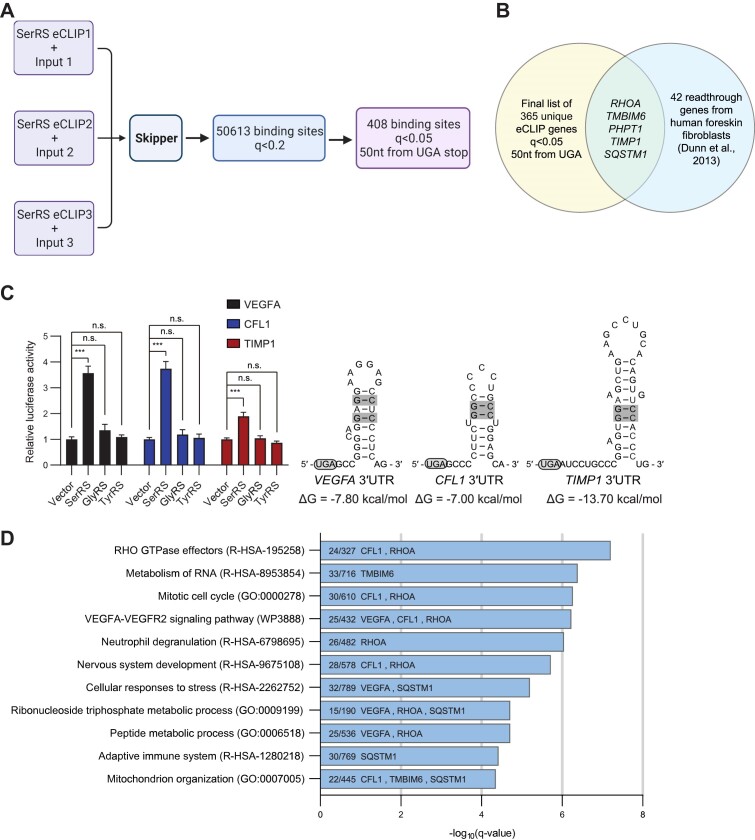
Reads from triplicate eCLIP-seq experiments in MDA-MB-231 cells were passed through a CLIP-seq analysis pipeline, Skipper (31), to obtain a set of 50613 sites with significant SerRS binding over size-matched input (q < 0.2), corresponding to 7102 unique RNAs (Figure 4A and Supplementary Table S3). Most of the genes identified in the original CLIP experiment (Supplementary Table S2) were also found in this new eCLIP set (6/9 genes). All tRNAs for serine and selenocysteine were found as top hits when calling repeat elements, confirming the suitability of the eCLIP technique and Skipper analysis pipeline (Supplementary Figure S6A and Supplementary Table S4). Although most binding sites were found within the coding sequence of mRNAs, including three binding sites in MSH2, many sites were also found within 5′UTRs and 3′UTRs (Supplementary Figure S6B). Further refinement to include only sites within 50 nucleotides of a protein-coding UGA stop codon and with q < 0.05 led to the identification of 408 sites, corresponding to 365 unique mRNAs (Figure 4A and Supplementary Table S5). Of note, VEGFA appeared in this list. As further validation that SerRS mediates physiological TR events we compared our gene set with previously reported readthrough events. We saw a sizeable overlap with a previous set of genes with TR confirmed through ribosome profiling by Dunn et al. (6), with 27/42 genes appearing in our eCLIP set and 5/27 of those genes having SerRS occupancy within 50 nucleotides of the UGA stop codon (RHOA, TMBIM6, PHPT1, TIMP1, SQSTM1) (Figure 4B and Supplementary Table S6).
Figure 4.
eCLIP-seq identifies a set of SerRS-bound RNAs that includes other translational readthrough genes. (A) A schematic showing the eCLIP-seq analysis workflow and filtering steps to reach the final list of 408 SerRS-bound windows within 50 nucleotides of a UGA stop codon (q < 0.05). Created with BioRender.com. (B) Venn diagram showing the overlap between the final 365 unique genes found in our eCLIP set (from 408 windows) with the confirmed translational readthrough genes identified in human foreskin fibroblasts by Dunn et al. (6). (C) Luciferase reporter assays were performed for the hits CFL1 and TIMP1, using VEGFA as a positive control. All three RNAs possess a G–C base pair-containing stem-loop after the canonical UGA stop codon. (D) Gene Ontology (GO) analysis was performed on RNAs from the final list of 408 windows. The number of genes enriched in each pathway are shown along with the overlapping translational readthrough genes from Figure 4B and our experimentally confirmed translational readthrough genes in Figure 4C. (A–D) (n.s., not significant; *** P< 0.001).
To test whether other mRNAs that appear in our eCLIP list beside VEGFA are subject to TR, we selected CFL1 and TIMP1 to perform the same luciferase readthrough reporter assay that we did for VEGFA. Both CFL1 and TIMP1 contain a stem-loop structure containing G-C base pairs immediately after the UGA stop codon (Figure 4C). We saw a similar increase in CFL1 readthrough as we did for VEGFA, and a smaller but significant increase in TR for TIMP1 (Figure 4C). Using Metascape (36), we performed a gene ontology (GO) analysis to determine cellular pathways that may be enriched by bound RNAs (q < 0.05, window within 50 nucleotides of UGA). We found that Rho GTPase effectors, metabolism of RNA, cell cycle, and VEGFA–VEGFR2 signaling pathways were enriched (Figure 4D and Supplementary Table S7). We highlighted VEGFA, CFL1, and the 5 readthrough genes from Dunn et al. (6) that overlapped with our set (Figure 4B) in each related GO pathway (Figure 4D).
Collectively, our data indicate a role for SerRS in controlling translational readthrough for a subset of genes defined by their mRNA structure and stop codon usage.
DISCUSSION
We showed that SerRS can bind and mediate TR of specific mRNAs by recruiting certain components of the selenocysteine incorporation machinery, including tRNASec and the corresponding elongation factor, eEFSec, for ribosomal delivery. Our data suggests that SerRS binds to specific stem-loop motifs in the 3′UTR that mimic the long variable loop (a major identity element) of tRNASer and tRNASec (Figure 1) and that the catalytic activity and UNE-S domain of SerRS are necessary for mediating TR (Figures 2B and C).
Not all components of the selenocysteine incorporation machinery are needed for SerRS-mediated TR. SerRS performs the same function in both mechanisms to initiate the charging of tRNASec with serine (53). For selenocysteine incorporation, this is followed by the conversion of Ser-tRNASec to Sec-tRNASec through PSTK and SepSecS (11). SepSecS forms a complex with the enzymes that provide activated selenium (54), suggesting a supramolecular hub for the conversion of Ser-tRNASec to Sec-tRNASec. To avoid nonspecific extension of off-target proteins for selenocysteine incorporation, mRNAs containing a SECIS are recognized by SBP2, which allows recruitment of Sec-tRNASec-bound eEFSec (55). While we showed that eEFSec and tRNASec are important for SerRS to mediate TR, SBP2 is not, suggesting that SerRS and SBP2, with their corresponding mRNA binding motifs, dictate mRNA selectivity and allow the separation of the two TR mechanisms.
The SECIS is quite complex in humans, consisting of a lower stem, central core, upper stem, and apical loop (55). In contrast, SerRS recognizes a comparatively simple G–C-containing stem-loop motif mimicking the variable loop of tRNASer and tRNASec, suggesting the need for further safeguard mechanisms. The RNA sequence surrounding the stop codon impacts the efficiency of TR, partly due to interactions between the translational machinery and structural features of the mRNAs (56). mRNAs with a stem-loop must be accessible to SerRS, so we speculate that SerRS competes with other RNA binding proteins for these structures or that exposure of these stem-loops during translation only occurs under specific conditions. SerRS is active as a homodimer (Supplementary Figure S2), so tethering one subunit to the mRNA close to the UGA stop codon would still allow the other subunit to recognize and charge tRNASec. The resulting high local concentration of Ser-tRNASec may enable suppression of the UGA stop codon and TR of specific mRNAs. Having high local concentrations of an aaRS for the charging of suppressor tRNAs has been shown to be favorable for efficient stop codon suppression (44). In addition, while binding (kon) is comparable between eEFSec and Sec-tRNASec versus Ser-tRNASec, the eEFSec-Ser-tRNASec complex dissociates faster (koff) than Sec-tRNASec, further suggesting that high local concentrations of Ser-tRNASec might be crucial (57). Recently, it was shown by another group that eEFSec can enable UGA readthrough using Ser-tRNASec, supporting our mechanism (51).
Based on our data, readthrough efficiency is determined by more factors than just the strength of SerRS binding to target mRNA. SerRS also needs to be catalytically competent (Figure 2E) and the binding of SerRS needs to occur close to the stop codon, as demonstrated by reduced TR efficiency when spacers are introduced between the UGA stop codon and the SerRS-binding stem-loop structure (Figure 1F). However, binding strength is still important as shown by the GC > AU and structural mutations that reduce SerRS binding and consequently TR efficiency (Figures 1E and G). SerRS binds to many distinct RNA species, and having TR efficiency be modified by binding strength, binding location, and catalytic activity allows greater regulation of SerRS-mediated TR.
Other RNA binding factors have been identified as regulators of VEGFA TR, such as HNRNPA2/B1 (25). Eswarappa et al. discovered that HNRNPA2/B1 is critical for VEGFA TR through interactions with translating ribosomes that prevent recruitment of eukaryotic translation termination factor 1 (eRF1) to the stop codon. The consensus binding sequence for hnRNPA2/B1 (5′-GCCAAGGAGCC-3′) falls within the stem-loop structure of VEGFA that we also identified as critical for binding by SerRS. Mutations in the hnRNPA2/B1 region which abrogated TR could also affect stem-loop formation for SerRS binding. It is possible that hnRNPA2/B1 and SerRS bind to VEGFA mRNA at different stages of TR (blocking recruitment of the termination factor versus increasing local tRNA concentrations) or that hnRNPA2/B1 and SerRS promote TR through cooperative binding. In contrast to other RNA-binding proteins, only SerRS directly generates the necessary aminoacylated suppressor tRNA, thereby enabling translation to continue through the stop codon.
The 3′UTR stem-loops in target mRNAs must compete with high endogenous levels of tRNASer for SerRS binding. In addition, tRNASec must be both locally available and aminoacylated over tRNASer despite a slight preference of SerRS for tRNASer over tRNASec (47). We speculate that reducing the availability of tRNASer could contribute to the success of SerRS-mediated TR. As tRNA levels are regulated during stress conditions by selective cleavage (58) and retroactive nuclear import (59), SerRS-mediated TR might offer an additional pathway for cells to adapt to stress by altering protein variant production. Strict regulation is necessary to allow SerRS-regulated TR to be both gene-specific and potentially responsive to the present state of the cell despite the ubiquitous expression of SerRS as an integral part of the translation machinery.
Recently, studies into aaRSs and their noncanonical roles have expanded the involvement of aaRSs in regulatory processes, with several of these new functions being mechanistically involved in the regulation of translation (60–63). SerRS is especially suitable to enable stop codon suppression as it does not use the anticodon of its cognate tRNA as an identity element (64,65), thus it can be repurposed to recognize tRNAs with different anticodons with more ease than other aaRSs. In addition, aaRSs acquired additional domains during evolution which allow for new interactions and functionalities (66). SerRS contains a unique domain at its C-terminus (UNE-S), which arose in vertebrates (45) and is necessary for TR, presumably by enabling SerRS to interact with target mRNAs (Figure 2B and Supplementary Figure S2). This suggests that the regulation of TR by SerRS is evolutionarily new, even though prototypes of the selenocysteine machinery trace back to archaea. It is possible that the pre-existing involvement of SerRS in the selenocysteine biosynthesis machinery and the emergence of the UNE-S domain enabled vertebrate SerRS to develop the function in mediating TR as described herein. It is worth noting that the appearance of the UNE-S domain has already been linked to the emergence of a closed circulatory system (45) and indeed SerRS is necessary for functional vascular development (45,67). Previous findings linked SerRS to the transcriptional regulation of VEGFA, the master regulator of angiogenesis (45,67). We suggest that vertebrate SerRS may further regulate angiogenesis via its ability to influence VEGFA translational readthrough.
To obtain additional evidence for SerRS-mediated readthrough in MDA-MB-231 cells, we performed ribosome profiling on control MDA-MB-231 cells and SerRS-overexpressing MDA-MB-231 cells. However, no significant difference in ribosome occupancy with SerRS overexpression was detected for most genes, including VEGFA (Supplementary Table S8), possibly due to insufficient read depth.
To determine whether our eCLIP-seq hits overlapped with previously published mechanisms of TR, we looked at the overlap between our set and the set described in Loughran et al. (68). This study identified UGA_CUAG as a vertebrate-conserved motif that enabled TR of a set of 23 human genes. Some of the hits identified in Loughran et al. appeared in our broader set of eCLIP hits (q < 0.2, CDKN3, CGGBP1, DCTN3, PHF19), but none of these appeared in our more stringent list (q < 0.05, binding within 50 nucleotides of UGA). We also do not see an enrichment of the UGA_CUAG motif in our dataset. Therefore, it appears that the TR mediated by SerRS and components of the selenocysteine incorporation machinery acts distinctively from the phenomenon described in Loughran et al.
Taken together we here describe a novel mechanism for the regulation of physiological translational readthrough of specific mRNAs through direct provision of suppressor tRNA by a tRNA synthetase. TR has powerful implications for therapeutics designed to counteract PTCs. Most current approaches to promote TR of genes with nonsense mutations rely on engineered RNAs or chemical modifiers, potentially promoting TR of undesired mRNAs. While our findings here might be more limited in their direct applicability as a therapeutic option due to the need for naturally occurring RNA motifs, the mechanism makes use of endogenous components. The findings discussed here uncover another aspect of how cells developed complex translational control by coordinating mRNA and protein features together to achieve functional readthrough. Insights such as these may improve therapeutics by leveraging the natural systems already in place to manage TR in cells, making them more efficient and less prone to off-target effects.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Scripps Research next generation sequencing and bioinformatics core for eCLIP sequencing and preliminary data analysis and the antibody core for the generation of SerRS antibody used for CLIP. We thank Dr. Paul Schimmel for valuable input and support and Dr. Paul Fox for providing material for preliminary experiments and discussion. We thank Dr. Kristopher W. Brannan for guidance on performing and analyzing the eCLIP-seq experiments. Scripts for the calculation of expected/observed were provided by Dr. Scott Adamson and we thank him for his advice on ribosome profiling analysis.
Notes
Present address: Haissi Cui, Department of Chemistry, University of Toronto, Toronto, ON M5S 3H6, Canada.
Contributor Information
Ze Liu, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA.
Justin Wang, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA.
Yi Shi, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA; Department of Biochemistry, School of Medicine, Nankai University, Tianjin, China.
Brian A Yee, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Markus Terrey, Howard Hughes Medical Institute, Department of Cellular and Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Qian Zhang, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA.
Jenq-Chang Lee, Department of Surgery, National Cheng Kung University Medical College and Hospital, Taiwan.
Kuo-I Lin, Genomics Research Center, Academia Sinica, Taiwan.
Andrew H-J Wang, The Ph.D. Program for Translational Medicine, College of Medical Science and Technology, Taipei Medical University and Academia Sinica, Taipei 110, Taiwan.
Susan L Ackerman, Howard Hughes Medical Institute, Department of Cellular and Molecular Medicine, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA; Department of Neurobiology, University of California San Diego, La Jolla, CA 92093, USA.
Gene W Yeo, Department of Cellular and Molecular Medicine, University of California San Diego, La Jolla, CA 92093, USA.
Haissi Cui, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA.
Xiang-Lei Yang, Department of Molecular Medicine, Scripps Research Institute, La Jolla, CA 92037, USA.
DATA AVAILABILITY
eCLIP-seq data was deposited at the GEO (GSE206674). UCSC Browser tracks for the analyzed data can be accessed at: https://genome.ucsc.edu/s/jwang95070/hg38_eCLIP_skipper_2.24.23. Ribosome profiling data can be accessed at the GEO (GSE230349).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R35 GM139627 to X.-L.Y., HG004659, HG009889 to G.W.Y.]; Deutsche Forschungsgemeinschaft [327097878 to H.C.]; Human Frontier Science Program [LT000207 to H.C.]; Z.L. and J.W. were supported by a fellowship from the National Foundation for Cancer Research; G.W.Y. was partially supported by an Allen Distinguished Investigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation. S.L.A. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: National Institutes of Health [R35 GM139627].
Conflict of interest statement. G.W.Y. is a co-founder, member of the Board of Directors, on the scientific advisory board, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. The authors declare no other competing financial interests.
REFERENCES
- 1. Schimmel P.R., Söll D.. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu. Rev. Biochem. 1979; 48:601–648. [DOI] [PubMed] [Google Scholar]
- 2. Jackson R.J., Hellen C.U.T., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schuller A.P., Green R.. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 2018; 19:526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G.et al.. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayr C. Regulation by 3’-untranslated regions. Annu. Rev. Genet. 2017; 51:171–194. [DOI] [PubMed] [Google Scholar]
- 6. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Driscoll D.M., Copeland P.R.. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003; 23:17–40. [DOI] [PubMed] [Google Scholar]
- 8. Leinfelder W., Zehelein E., Mandrand-Berthelot M.A., Böck A.. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988; 331:723–725. [DOI] [PubMed] [Google Scholar]
- 9. Wang C., Guo Y., Tian Q., Jia Q., Gao Y., Zhang Q., Zhou C., Xie W.. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015; 43:10534–10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson B.A., Xu X.-M., Kryukov G.V., Rao M., Berry M.J., Gladyshev V.N., Hatfield D.L.. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12848–12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palioura S., Sherrer R.L., Steitz T.A., Söll D., Simonovic M.. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009; 325:321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lescure A., Fagegaltier D., Carbon P., Krol A.. Protein factors mediating selenoprotein synthesis. Curr. Protein Pept. Sci. 2002; 3:143–151. [DOI] [PubMed] [Google Scholar]
- 13. Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M.. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000; 19:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A.. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008; 29:1037–1047. [DOI] [PubMed] [Google Scholar]
- 15. Atkinson J., Martin R.. Mutations to nonsense codons in human genetic disease: implications for gene therapy by nonsense suppressor tRNAs. Nucleic Acids Res. 1994; 22:1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen W.-C., Lin S.-C., Lee J.-C.. A novel nonsense mutation of MSH2 gene in a Taiwanese family with hereditary nonpolyposis colorectal cancer. Kaohsiung J. Med. Sci. 2011; 27:68–71. [DOI] [PubMed] [Google Scholar]
- 17. Keeling K.M., Xue X., Gunn G., Bedwell D.M.. Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 2014; 15:371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manuvakhova M., Keeling K., Bedwell D.M.. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000; 6:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porter J.J., Heil C.S., Lueck J.D.. Therapeutic promise of engineered nonsense suppressor tRNAs. Wiley Interdiscip. Rev. RNA. 2021; 12:e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Zhang Y., Mendonca C.A., Yukselen O., Muneeruddin K., Ren L., Liang J., Zhou C., Xie J., Li J.et al.. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature. 2022; 604:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lueck J.D., Yoon J.S., Perales-Puchalt A., Mackey A.L., Infield D.T., Behlke M.A., Pope M.R., Weiner D.B., Skach W.R., McCray P.B.et al.. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 2019; 10:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wangen J.R., Green R.. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. Elife. 2020; 9:e52611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat. Rev. Cancer. 2002; 2:795–803. [DOI] [PubMed] [Google Scholar]
- 24. Ferrara N., Gerber H.-P., LeCouter J.. The biology of VEGF and its receptors. Nat. Med. 2003; 9:669–676. [DOI] [PubMed] [Google Scholar]
- 25. Eswarappa S.M., Potdar A.A., Koch W.J., Fan Y., Vasu K., Lindner D., Willard B., Graham L.M., DiCorleto P.E., Fox P.L.. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014; 157:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eswarappa S.M., Fox P.L.. Antiangiogenic VEGF-Ax: a new participant in tumor angiogenesis. Cancer Res. 2015; 75:2765–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xin H., Zhong C., Nudleman E., Ferrara N.. Evidence for pro-angiogenic functions of VEGF-Ax. Cell. 2016; 167:275–284. [DOI] [PubMed] [Google Scholar]
- 28. Morgenstern J.P., Land H.. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990; 18:3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi Y., Wei N., Yang X.-L.. Studying nuclear functions of aminoacyl tRNA synthetases. Methods. 2017; 113:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darnell R. CLIP (cross-linking and immunoprecipitation) identification of RNAs bound by a specific protein. Cold Spring Harb Protoc. 2012; 2012:1146–1160. [DOI] [PubMed] [Google Scholar]
- 31. Boyle E.A., Her H.-L., Mueller J.R., Nguyen G.G., Yeo G.W.. Skipper analysis of RNA-protein interactions highlights depletion of genetic variation in translation factor binding sites. Cell Genomics. 2022; 3:100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang H., Lei R., Ding S.-W., Zhu S.. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014; 15:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu D. Algorithms for efficiently collapsing reads with Unique Molecular Identifiers. Peer J. 2019; 7:e8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.-L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L.. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014; 345:455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lauria F., Tebaldi T., Bernabò P., Groen E.J.N., Gillingwater T.H., Viero G.. riboWaltz: optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018; 14:e1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hellman L.M., Fried M.G.. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007; 2:1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reinkemeier C.D., Girona G.E., Lemke E.A.. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science. 2019; 363:eaaw2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu X., Shi Y., Zhang H.-M., Swindell E.C., Marshall A.G., Guo M., Kishi S., Yang X.-L.. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 2012; 3:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lenhard B., Orellana O., Ibba M., Weygand-Durasević I.. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 1999; 27:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Holman K.M., Puppala A.K., Lee J.W., Lee H., Simonović M.. Insights into substrate promiscuity of human seryl-tRNA synthetase. RNA. 2017; 23:1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fukui H., Hanaoka R., Kawahara A.. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ. Res. 2009; 104:1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kirchner S., Cai Z., Rauscher R., Kastelic N., Anding M., Czech A., Kleizen B., Ostedgaard L.S., Braakman I., Sheppard D.N.et al.. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 2017; 15:e2000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dobosz-Bartoszek M., Pinkerton M.H., Otwinowski Z., Chakravarthy S., Söll D., Copeland P.R., Simonović M.. Crystal structures of the human elongation factor eEFSec suggest a non-canonical mechanism for selenocysteine incorporation. Nat. Commun. 2016; 7:12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hilal T., Killam B.Y., Grozdanović M., Dobosz-Bartoszek M., Loerke J., Bürger J., Mielke T., Copeland P.R., Simonović M., Spahn C.M.T.. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science. 2022; 376:1338–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ligtenberg M.J.L., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y.H., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J.B.et al.. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat. Genet. 2009; 41:112–117. [DOI] [PubMed] [Google Scholar]
- 53. Gonzalez-Flores J.N., Shetty S.P., Dubey A., Copeland P.R.. The molecular biology of selenocysteine. Biomol Concepts. 2013; 4:349–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oudouhou F., Casu B., Dopgwa Puemi A.S., Sygusch J., Baron C.. Analysis of novel interactions between components of the selenocysteine biosynthesis pathway, SEPHS1, SEPHS2, SEPSECS, and SECp43. Biochemistry. 2017; 56:2261–2270. [DOI] [PubMed] [Google Scholar]
- 55. Howard M.T., Copeland P.R.. New directions for understanding the codon redefinition required for selenocysteine incorporation. Biol Trace Elem Res. 2019; 192:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E.. Translational readthrough potential of natural termination codons in eucaryotes–the impact of RNA sequence. RNA Biol. 2015; 12:950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paleskava A., Konevega A.L., Rodnina M.V.. Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB. J. Biol. Chem. 2010; 285:3014–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwenzer H., Jühling F., Chu A., Pallett L.J., Baumert T.F., Maini M., Fassati A.. Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress response. Cell Rep. 2019; 26:3416–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jeong S.J., Park S., Nguyen L.T., Hwang J., Lee E.-Y., Giong H.-K., Lee J.-S., Yoon I., Lee J.-H., Kim J.H.et al.. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat. Commun. 2019; 10:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mukhopadhyay R., Jia J., Arif A., Ray P.S., Fox P.L.. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 2009; 34:324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Putney S.D., Schimmel P.. An aminoacyl tRNA synthetase binds to a specific DNA sequence and regulates its gene transcription. Nature. 1981; 291:632–635. [DOI] [PubMed] [Google Scholar]
- 63. Jones J.A., Wei N., Cui H., Shi Y., Fu G., Rauniyar N., Shapiro R., Morodomi Y., Berenst N., Dumitru C.D.et al.. Nuclear translocation of an aminoacyl-tRNA synthetase may mediate a chronic “integrated stress response". Cell Rep. 2023; 42:112632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Normanly J., Ogden R.C., Horvath S.J., Abelson J.. Changing the identity of a transfer RNA. Nature. 1986; 321:213–219. [DOI] [PubMed] [Google Scholar]
- 65. Biou V., Yaremchuk A., Tukalo M., Cusack S.. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994; 263:1404–1410. [DOI] [PubMed] [Google Scholar]
- 66. Guo M., Schimmel P., Yang X.L.. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010; 584:434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shi Y., Xu X., Zhang Q., Fu G., Mo Z., Wang G.S., Kishi S., Yang X.-L.. tRNA synthetase counteracts c-Myc to develop functional vasculature. Elife. 2014; 3:e02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Loughran G., Jungreis I., Tzani I., Power M., Dmitriev R.I., Ivanov I.P., Kellis M., Atkins J.F.. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018; 293:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
eCLIP-seq data was deposited at the GEO (GSE206674). UCSC Browser tracks for the analyzed data can be accessed at: https://genome.ucsc.edu/s/jwang95070/hg38_eCLIP_skipper_2.24.23. Ribosome profiling data can be accessed at the GEO (GSE230349).