Abstract
Autism spectrum disorder (ASD) is one of the common neurodevelopmental disorders in children. Its etiology and pathogenesis are poorly understood. Previous studies have suggested potential changes in the complement and coagulation pathways in individuals with ASD. In this study, using multiple reactions monitoring proteomic technology, 16 of the 33 proteins involved in this pathway were identified as differentially-expressed proteins in plasma between children with ASD and controls. Among them, CFHR3, C4BPB, C4BPA, CFH, C9, SERPIND1, C8A, F9, and F11 were found to be altered in the plasma of children with ASD for the first time. SERPIND1 expression was positively correlated with the CARS score. Using the machine learning method, we obtained a panel composed of 12 differentially-expressed proteins with diagnostic potential for ASD. We also reviewed the proteins changed in this pathway in the brain and blood of patients with ASD. The complement and coagulation pathways may be activated in the peripheral blood of children with ASD and play a key role in the pathogenesis of ASD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12264-023-01055-4.
Keywords: Autism spectrum disorder, Biomarker, Complement and coagulation cascade, Complement system, Machine learning, Multiple reaction monitoring
Introduction
Autism spectrum disorder (ASD) is a type of neurodevelopmental disorder characterized by social communication disorders, as well as repetitive and restricted behavior patterns. Worldwide, ASD affects 1% to 2% of children [1–4]. ASD is more than three times more common in males than in females [5]. The situation is similar in different ethnic groups [6]. In China, the incidence of ASD in children aged 6–12 is ~0.7% [7, 8].
ASD is a multifactorial disease and the interaction between genetic and environmental factors may play a critical role in its pathogenesis [9]. It is highly heterogeneous, has no precise diagnostic criteria, and is usually diagnosed using the Diagnostic and Statistical Manual of Mental Disorders (DSM-V) [10]. This is not only subjective but also often causes a diagnostic delay or misdiagnosis. However, it is known that early diagnosis and intervention in children with ASD improve their outcomes [11]. As a result, there is an urgent need to find potential biomarkers to diagnose ASD at an early stage [12].
Blood is one of the most easily obtained samples and one of the best sources of diagnostic markers of disease. Blood-based biomarkers are more convenient for clinical use. There have been some reports on blood protein diagnostic biomarkers for ASD, and they have been well reviewed [12–16]. Among them, complement-related proteins have been broadly reported to be altered in the blood of children with ASD [11–23], including our previous studies [11–14, 17] and the earliest ASD blood proteomic studies [18]. The immune system is composed of the innate immune system and the adaptive immune system. One of the main effector mechanisms of the innate immune system is the complement system, which serves to signal increased inflammation and clear pathogens and cell debris [24]. The complement cascade can be initiated by 3 major pathways: the classical pathway, the lectin pathway, and the alternate pathway. More than 40 proteins are now recognized as part of the complement system [25]. Of note, the relationship between complement proteins and neuropsychiatric disorders, such as ASD, schizophrenia, Alzheimer’s disease (AD), multiple sclerosis, and Huntington’s disease, has recently been reviewed [26–31]. Beyond immune function, they participate in brain architecture and are involved in the development of these diseases [31]. Consequently, it is of interest to simultaneously investigate whether these proteins are altered in the blood of children with ASD. Besides, the complement system and coagulation are interrelated. Indeed, several studies have shown the complement and coagulation cascade pathway to be associated with ASD [9, 13, 19, 24]. Thus, when detecting complement proteins, it is also important to include some proteins related to coagulation. By detecting the expression of these proteins in the blood of children with ASD, we can not only explore their association with ASD but also search for potential diagnostic markers. At present, the systematic investigation of protein changes in complement and coagulation pathways associated with ASD has not been reported.
The development of targeted proteomics technology has bridged the gap between screening and validation, as well as validation and transformation. Multiple reaction monitoring (MRM) is a classical method of target proteomics, using a triple quadrant or quadrant-ion trap mass spectrometer to detect the parent and daughter ion mass spectrometry response signals of target molecules, and can detect multiple target proteins simultaneously in one experiment [32]. This technology has been applied to the study of markers for various diseases due to its convenience, high throughput, and accuracy [32–35]. In this study, we used MRM technology to detect 33 proteins involved in the complement and coagulation pathways in the plasma of children with ASD and healthy controls. On this basis, we further used machine learning to identify a group of differentially-expressed complement and coagulation pathway proteins between these two groups, which may serve as a potential biomarker to assist clinicians in diagnosing ASD.
Materials and Methods
Study Population
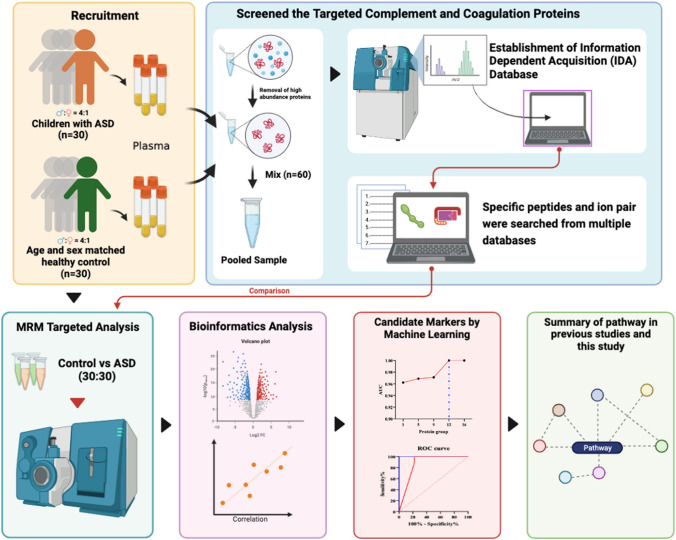
The workflow used in this study is shown in Fig. 1 from the website of BioRender (https://biorender.com/). A total of 30 children with ASD (24 males and 6 females) were recruited from Bao’an Maternal and Child Health Hospital in Shenzhen, as well as a gender and age-matched control group. ASD children were diagnosed by the same child neuropsychologist based on the ASD criteria defined in DSM-V [10]. Inclusion criteria: (1) meeting the DSM-V diagnostic criteria for ASD; (2) having detailed clinical data; and (3) having obtained written informed consent from the child’s caregiver. Exclusion criteria: (1) unclear diagnosis accompanied by other organic diseases of the nervous system; (2) serious physical disorders, such as heart, liver, and kidney disease; and (3) mental retardation, language developmental disorders, other mental disorders, and deafness. Children with ASD were also assessed using the Autism Behavior Checklist (ABC) and the Child Autism Rating Scale (CARS) [36]. The ABC score was 71.70 ± 19.37, while the CARS score was 35.57 ± 3.24 on average. There were no significant differences in age and body mass index between the ASD group and the control group (Table S1).
Fig. 1.
The experimental flow chart of this study.
Blood Sample Collection
Blood samples were collected by a pediatric nurse under the supervision of a child psychiatrist. Venous blood was collected into a 5 mL EDTA tube (vacuum collector system; Becton Dickinson Inc., Plymouth, UK) in the morning while the subjects were in the fasting state, then centrifuged at 1300 g at 4 ℃ for 10 min, and plasma was separated. Subsequently, the inhibitor mixture (30 μL per 1 mL plasma) was added to the resultant plasma sample (cocktail inhibitor solution: 2.0 mol/L Tris, 0.9 mol/L Na-EDTA, 0.2 mol/L Benzamidine, 92 μmol/L E-64, and 48 μmol/L Pepstatin; Sigma, St. Louis, MI, USA). The plasma was stored at −80 ℃.
Establishment of the Data Dependent Acquisition (DDA) Database
Before MRM analysis, a background library was established. For each sample, 20 µL plasma was used to remove high-abundance plasma proteins by using the Multiple Affinity Removal LC Column-Human 14 (Agilent, Santa Clara, CA, USA). The collected low-abundance protein was quantified by the BCA method [37], and 10 µg protein was taken from each sample and mixed to obtain DDA library samples. After reductive alkylation of the sample, digestion with trypsin (Promega, Madison, WI, USA) at a ratio of 1:30 at 37 ℃ overnight was carried out[9]. All enzyme lysis-treated samples were concentrated in a vacuum until dried.
The dried DDA database sample was reconstituted in 100 µL of Mili-Q water (Milli-Q System, Millipore Corp.) before injection into an Agilent high-performance liquid chromatograph (Agilent Technologies, Santa Clara, CA, USA) equipped with a high pH RP column (Durashell, C18, 250 mm × 4.6 mm, 5 μm; Bonna-Agela Technologies, Inc., Wilmington, DE, USA). The database sample was eluted, separated into 30 groups, lyophilized, and stored at –80 ℃. The DDA database was analyzed by using QTRAP 6500+ (AB SCIEX, Framingham, MA, USA). The specific peptide segments and mass spectrum (MS) information of these proteins were screened by Skyline (http://proteome.gs.washington.edu/software/skyline/) [38] and PeptideAtlas (https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/GetTransitions) [39]. 3–5 specific peptides were selected for each protein for qualitative and quantitative analysis.
MRM Analysis
For 60 samples (30 children with ASD and 30 controls), the protein concentration was quantified by the BCA method, and 100µg protein was taken from each sample for MRM analysis. The protein samples were reduced/alkylated, and digested with trypsin at a ratio of 1:30 at 37 ℃ overnight[9]. 2 μg protein from each MRM sample was mixed as a quality control sample. In optimized conditions the mobile phase consisted of solvent A (0.1% formic acid with water) and solvent B (acetonitrile with 0.1% formic acid) using the following gradient: 0 min 5% B, 0.5 min 6% B, 25 min 22% B, 31 min 35% B, 32 min 80% B, 36 min 5% B at a constant flow rate of 0.3 mL/min. The injection volume was 6 µL. The MS was operated in positive mode. Instrument parameters including collision energies were then optimized to yield the highest sensitivity for all peptides and transitions. The retention time of each peptide was identified using full scan data. The MRM detection window was 100 s and the cycle time was 0.7 s. The target scan time was 0.7 s.
Data Pre-processing and Quality Control
Raw MS files were processed using Skyline software. Peaks were manually checked, and peak integrations were adjusted accordingly where necessary. After automated integration, the chromatograms were controlled visually and then the integration results were exported to a Microsoft Excel spreadsheet.
Bioinformatics and Statistical Analysis
Principal component analysis (PCA) was applied using SIMCA-P 14.1 (V14.1, Sartorius Stedim Data Analytics AB, Umea, Sweden). OMICSBEAN online tools (http://www.omicsbean.cn/) were used for data standardization and statistical t-tests. The cutoff value for up-regulation was a 1.2-fold change and for down-regulation was a 0.83-fold change, and a false discovery rate-corrected P-value <0.05 was established for significantly differentially- expressed proteins (DEPs) between autistic children and controls [40]. The value of pathway activation intensity (PAS) was calculated by OMICSBEAN, which served as the activation profiles of the signaling pathways based on the expression of individual proteins [13]. The correlation matrix between age and DEPs was calculated using an online tool (https://www.omicsolution.org/). Pearson’s correlation analysis was applied to calculate the correlation between the CARS and ABC scores using the R packages ggplot2 (v3.3.5) (https://ggplot2.tidyverse.org) and ggpubr (v0.4.0) (https://CRAN.R-project.org/package=ggpubr). Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and the protein-protein interaction (PPI) network were analyzed by OMICSBEAN and STRING (https://string-db.org/). Statistical graphs were drawn using a statistical t-test in GraphPad Prism software 8.0 (GraphPad Software, Inc., San Diego, USA). The data are presented as the mean ± standard deviation. A P-value <0.05 was considered statistically significant.
Screening Potential Biomarkers by Machine Learning Techniques
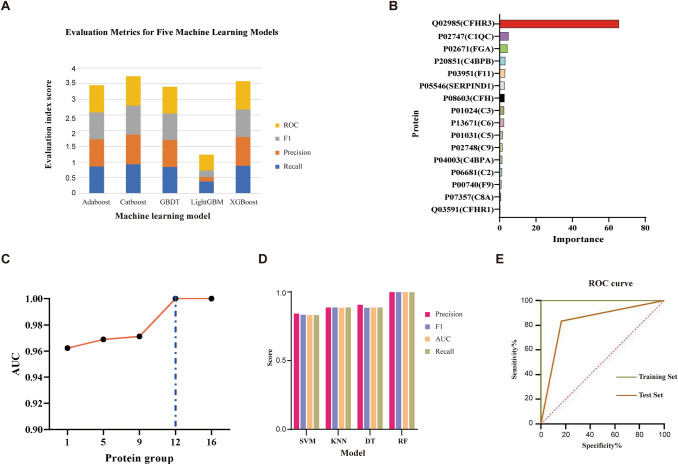
The most suitable algorithm for MRM data features was found among the five machine learning algorithms based on the decision tree model, including the adaptive boost classifier (Adaboost) [41], categorical boosting (Catboost) [42], gradient boosting decision tree (GBDT) [43], extreme gradient boosting (XGBoost) [44], and light gradient boosting machine (LightGBM) [45]. The four performance indexes of recall, precision, F1, and area under the curve (AUC) were used to select the best algorithm. The five-fold cross-validation strategy was used to reduce the model over-fitting caused by random local data. Finally, the average value of each evaluation index was used as the basis for screening. After obtaining the most suitable integrated machine learning algorithm, the importance of each protein under this condition could be calculated. A feature with a high importance value indicated that it made a significant contribution to grouping. After the combined screening of correlation coefficient and cumulative AUC, a group of biomarker panels with high information and the least number was obtained. To validate the correctness and classification performance of candidate markers, their classification ability on four classical models (decision tree (DT) [46], random forest (RF) [47], k-nearest neighbors (KNN) [48], and support vector machine (SVM)) [49] was evaluated.
Results
Establishment of the DDA Database
By examining the pooled blood samples from children with ASD and controls, the DDA database was established. More than 70,000 peptides were detected, which were involved in 1,747 proteins. Among these, 37 proteins related to the complement and coagulation pathway were found by background library database and website matching. We identified each protein using more than 3 specific peptides (Table S2). Twenty-seven proteins were associated with the complement pathway, and the other 10 proteins were involved in the coagulation cascade.
Identification of Differentially-Expressed Complement and Coagulation-Related Proteins by MRM Analysis
Further, through the identification and quantification of specific peptides unique to each protein, we quantitatively analyzed 33 proteins related to the complement and coagulation pathways (Tables S3 and S4). As confirmed by STRING database analysis, among these 33 proteins, 32 were involved in complement and coagulation cascades, 22 were involved in complement activation (C1QA, C1QC, C1RL, C1S, C2, C3, C4A, C4BPA, C4BPB, C5, C6, C7, C8A, C8B, C8G, C9, CFH, CFHR1, CFHR3, CLU, CR2, and MBL2), while 11 were involved in blood coagulation: C4BPB, F10, F11, F12, F13A1, F13B, F9, FGA, SERPINA5, SERPIND1, and PLG.
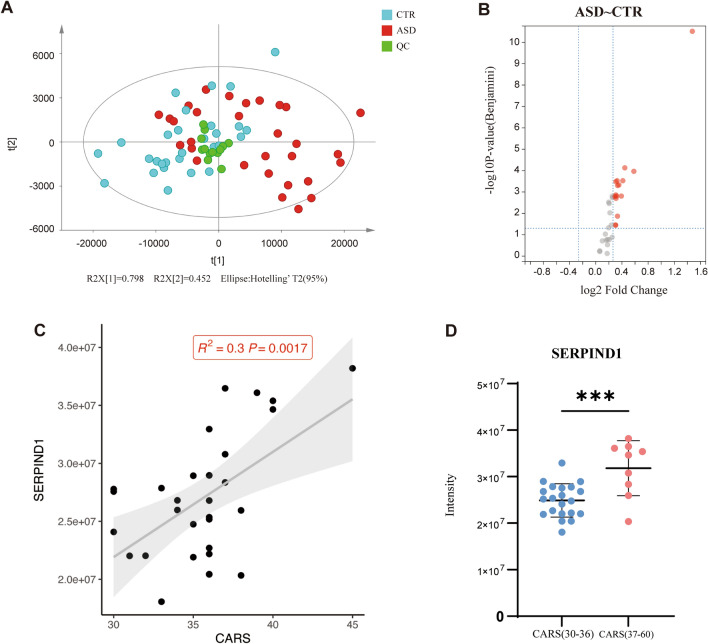
Interestingly, the PCA analysis showed that the expression pattern of the 33 quantified plasma proteins had a separation trend (Fig. 2A). Out of the 33 proteins, 16 were identified as DEPs between children with ASD and healthy controls (Table 1 and Fig. 2B): C1QC, C2, C3, C4BPA, C4BPB, C5, C6, C8A, C9, CFH, CFHR1, CFHR3, F11, F9, FGA, and SERPIND1. Compared with the controls, all of the proteins were up-regulated in the plasma of children with ASD. In addition, we analyzed the correlation between the expression of plasma DEPs and age in the ASD group, and the results were not statistically different. Similarly, there was no significant difference between the expression of plasma DEPs and age in the control group (Fig. S1). We also analyzed the correlation between the ABC and CARS scores of ASD children and the corresponding mass spectral intensity of each DEP. The results showed that there was no statistical difference between the correlation between the intensity of each DEP and the ABC score. In the correlation analysis with the CARS score, the corresponding strength of SERPIND1 was positively correlated with the CARS score (R2 = 0.3, P = 0.0017; Fig. 2C), while the rest showed no significant difference (Figs S2 and S3). There was a significant difference in the expression level of SERPIND1 between mild-moderate and severe ASD children (P < 0.05; Fig. 2D).
Fig. 2.
Screening of differentially-expressed proteins in the complement and coagulation pathways. A PCA analysis of complement and coagulation pathway-related proteins in plasma. B Volcano plot analysis and identification of differentially-expressed proteins. Red dots indicate P < 0.05 and fold change >1.2 (ASD vs control). C Correlation between the corresponding strength of SERPIND1 and CARS score in the ASD group. The y-axis represents the corresponding strength of the mass spectrum. D The corresponding strength of SERPIND1 between mild-moderate (30–36) and severe (37–60) ASD children. *** P <0.001.
Table 1.
The DEPs between children with ASD and healthy controls
| No. | Uniprot ID | Protein name | Gene name | Fold change | P-value a | Reference b |
|---|---|---|---|---|---|---|
| 1 | P04003 | C4b-binding protein alpha chain | C4BPA | 1.28 | 2.27E-04 | No |
| 2 | P20851 | C4b-binding protein beta chain | C4BPB | 1.24 | 6.72E-04 | No |
| 3 | P00740 | Coagulation factor IX | F9 | 1.33 | 1.44E-04 | No |
| 4 | P03951 | Coagulation factor XI | F11 | 1.23 | 2.36E-02 | No |
| 5 | P02747 | Complement C1q subcomponent subunit C | C1QC | 1.23 | 2.36E-02 | [15] |
| 6 | P06681 | Complement C2 | C2 | 1.26 | 8.90E-03 | [6] |
| 7 | P01024 | Complement C3 | C3 | 1.25 | 7.55E-04 | [10, 15, 20] |
| 8 | P01031 | Complement C5 | C5 | 1.36 | 3.60E-05 | [10, 11] |
| 9 | P13671 | Complement component C6 | C6 | 1.25 | 1.44E-04 | [84] |
| 10 | P07357 | Complement component C8 alpha chain | C8A | 1.24 | 1.11E-03 | No |
| 11 | P02748 | Complement component C9 | C9 | 1.5 | 5.24E-05 | No |
| 12 | P08603 | Complement factor H | CFH | 1.24 | 1.60E-04 | No |
| 13 | Q03591 | Complement factor H-related protein 1 | CFHR1 | 1.23 | 8.52E-04 | [15] |
| 14 | Q02985 | Complement factor H-related protein 3 | CFHR3 | 2.78 | 1.48E-11 | No |
| 15 | P02671 | Fibrinogen alpha chain | FGA | 1.31 | 7.55E-04 | [49] |
| 16 | P05546 | Heparin cofactor 2 | SERPIND1 | 1.26 | 2.42E-04 | No |
a FDR-corrected p-value < 0.05 vs. the control.
b No, not been reported.
In-depth Analysis of Pathways Related to the Differentially-expressed Proteins
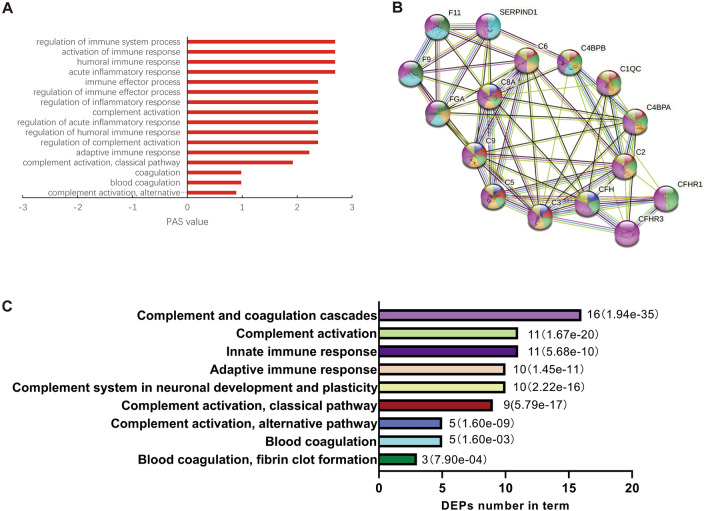
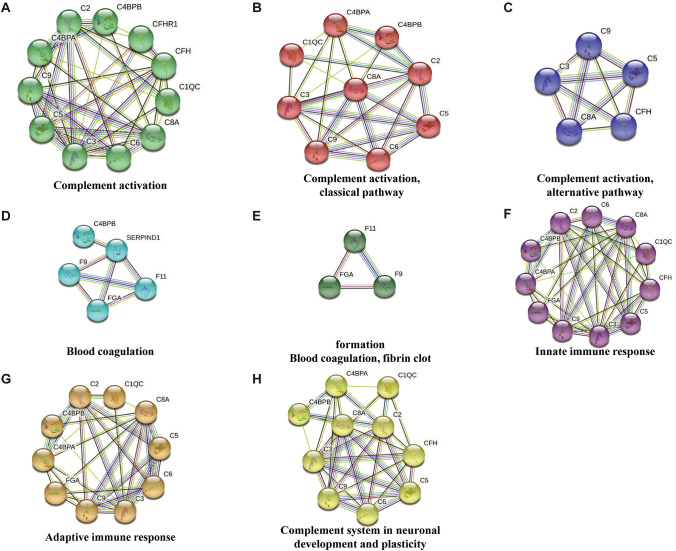
The results of GO analysis showed that the DEPs were mainly involved in the immune response and in the complement and coagulation cascades (Table S5); all these pathways were activated (Fig. 3A). PPI analysis revealed a remarkably significant enrichment of known interactions among these 16 proteins (Fig. 3B and C), including the complement and coagulation cascades (16 DEPs), complement activation (11 DEPs), innate immune response (11 DEPs), adaptive immune response (10 DEPs), complement system in neuronal development and plasticity (10 DEPs), complement activation, classical pathway (9 DEPs), complement activation, alternative pathway (5 DEPs), blood coagulation (5 DEPs), blood coagulation, and fibrin clot formation (3 DEPs). We further display the PPI of each pathway (Fig. 4). Eleven proteins were involved in complement activation: C1QC, C2, C3, C4BPA, C4BPB, C5, C6, C8A, C9, CFH, and CFHR1. Five proteins were involved in blood coagulation: F9, F11, FGA, C4BPB, and SERPIND1. Of note, 10 DEPs were associated with neuronal development and plasticity: C1QC, C2, C3, C4BPA, C4BPB, C5, C6, C8A, C9, and CFH.
Fig. 3.
Protein-protein interaction of differentially-expressed proteins in the complement and coagulation pathway. A Based on OMICSBEAN, the PAS values of the GO-BP term in which DEPs are involved. Positive PAS values indicate the upregulation of molecular pathways compared to controls. B, C PPI was analyzed by the STRING database (GO, KEGG, and Wiki databases). The colors represent the different pathways and the number of DEPs contained in them. The P-values are represented by the bar graph.
Fig. 4.
Details of the PPI network associated with DEPs.
Analysis of Candidate ASD Biomarkers Using Machine Learning Techniques
Among the five decisions tree-based machine learning models (Adaboost, Catboost, GBDT, LightGBM, and XGBoost), recall, precision, F1, and AUC were used as evaluation indexes, and the average value of each index was used as the basis for screening. We found that Catboost was most suitable for data features (Fig. 5A), so the importance of each protein was determined by using the Catboost model. The characteristics of CFHR3, C1QC, and FGA with large importance values were found from 16 DEPs, indicating that this characteristic contributes greatly to the grouping (Fig. 5B). Following the accumulation of the AUC, a group of biomarker panels with high information was obtained. We found that using the first 12 DEPs as a group, the AUC could reach 1, indicating that ASD and control groups could be effectively separated (Fig. 5C). To verify the correctness and classification performance of candidate markers, their classification ability on four classical models (DT, RF, KNN, and SVM) was evaluated (Fig. 5D). We randomly divided the data into two groups (training set and test set), with a 7:3 ratio between the two groups. The 12 proteins calculated above were used to identify ASD children from the control group, and AUC curves were obtained using the training and test sets. These 12 proteins are potential biomarkers for distinguishing ASD from the control group (Fig. 5E). The expression of marker proteins in each group is shown in Fig. 6.
Fig. 5.
Screening for potential biomarkers through machine learning. A Based on the decision tree model, the fit model is screened. B Using the Catboost model, the importance of each protein under this condition is obtained. C The AUCs of different potential biomarker combinations in the Catboost model. D There is a good trend of separation in various models. The classification performance of candidate biomarker combinations on the four classical models is evaluated. E Validation of potential marker combinations using ROC curves. The sensitivity of ROC in the training set is 1, and the sensitivity in the test set is 0.88.
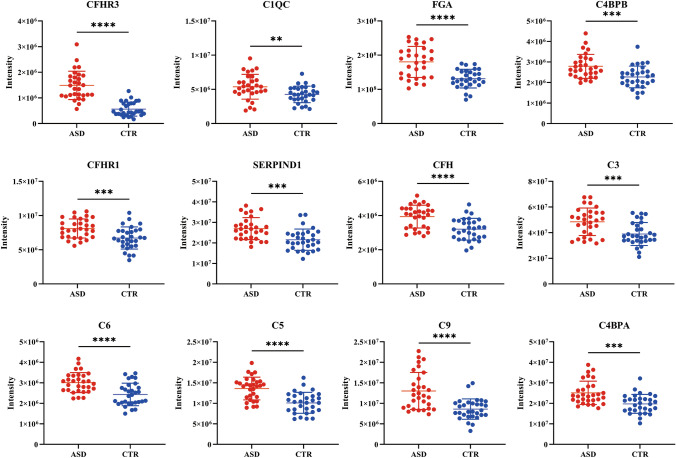
Fig. 6.
The expression of potential marker proteins identified in this study. The y-axis represents the corresponding strength of the mass spectrum. ** P <0.01; ***P < 0.001; ****P < 0.0001.
Discussion
Proteomic analysis of proteins in the complement and coagulation pathways was chosen for this study. Between children with ASD and control participants, 16 proteins implicated in the complement and coagulation pathways were identified as DEPs. All their levels were found to be higher in the plasma of children with ASD than in healthy controls. Apart from SERPIND1, there was no correlation between the expression levels of other proteins and the ABC and CARS scores. Of course, this needs to be verified with a larger sample size. Among the DEPs, 11 were associated with complement activation, 9 were related to activation of the classical pathway of complement, and 5 were related to the alternative pathway of complement activation. Consistent with this, C1QC, C2, C3, C5, C6, and CFHR1 have been reported to be increased in the blood of autistic children in previous studies [9, 11–13, 18, 50]. To the best of our knowledge, C4BPA, C4BPB, C8A, C9, CFH, and CFHR3 are first reported to be related to ASD and increased in the plasma of children with ASD. Indeed, in addition to the above proteins, other complement proteins have also been reported to change in the blood of children with ASD. For example, elevated levels of complement C1s subcomponent (C1S) [9], complement C1q subcomponent subunit A (C1QA), complement C1q subcomponent subunit B (C1QB) [18], and high complement factor I (CFI) [19] activity has been detected in the blood of children with ASD, while C4B levels in the plasma of children with ASD were significantly lower [51]. Another lectin pathway protein, collectin-10 (COLEC10), has also been found to be down-regulated in the plasma of children with ASD [9]. In the present study, 23 complement-related proteins were found to be altered in the blood of children with ASD. They are involved in the three activation pathways of complement. Of these, 18 are complement activators and components: C1QA, C1QB, C1QC, C1S, C2, C3, C4, C4B, C4BPA, C4BPB, C5, C6, C8A, C9, CFH, CFHR1, CFHR3, and COLEC10, and 6 are complement regulatory proteins: CFI, CFH, C4BPA, C4BPB, CFHR1, and CFHR3. The expression of most complement-related proteins is up-regulated in the peripheral blood of children with ASD, implying that the complement pathway may be activated in the periphery of children with ASD. Moreover, 5 proteins were associated with blood coagulation (C4BPB, F9, F11, FGA, and SERPIND1), of which, FGA has been reported to be increased in the blood of children with ASD [52], while C4BPB, F9, and F11 have not been reported to be associated with ASD. SERPIND1 has been reported to be increased in the peripheral blood mononuclear cells (PBMCs) in children with ASD [14]. From the perspective of genetic studies, this has also been reported to be associated with ASD [53] and developmental delay [54]. In the present study, the expression levels of SERPIND1 were positively correlated with CARS scores and were higher in children with severe ASD than in those with mild to moderate ASD, implying that it has the potential to classify the severity of diseases and deserves further investigation.
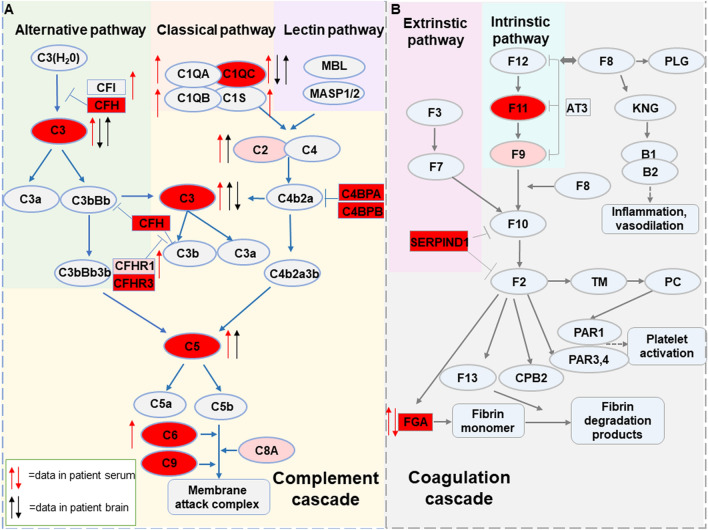
We then used the machine learning method to find a set of proteins with diagnostic potential from the 16 DEPs. We found that Catboost was the best fit for the data features. After the combined screening of the correlation coefficient and cumulative AUC, a group of biomarker panels that included 12 DEPs with high information content was obtained. Subsequently, we used four classical models to verify and evaluate the correctness and classification performance of these candidate markers, which achieved high indexes of recall, precision, F1, and AUC. Among these 12 DEPs, as noted above, C1QC, FGA, C3, C5, and C6 have been reported to be associated with ASD [13, 14, 18, 52], while CFHR3, C4BPB, C4BPA, CFH, C9, SERPIND1, and F11 have not been reported to be altered in ASD plasma. CFHR3, C4BPB, C4BPA, and CFH are regulatory proteins of the complement cascade that inhibit complement activation [55–58]. Their increased expression can inhibit complement activation. There is a balance between activation and inhibition of the complement system in vivo [58]. The activity of the complement system is tightly regulated to protect host cells from indiscriminate attack [50]. Therefore, except for CFHR3, C4BPB, C4BPA, and CFH, the increased expression of these proteins implies activation of the peripheral complement system in children with ASD. C9 is a component of the cell surface membrane attack complex (MAC), which is composed of C5–C9 and serves as the complement cascade’s final common pathway. The increase of its expression in plasma further supports the hypothesis that the peripheral complement system is activated in ASD children. The proteins involved in the complement and coagulation pathways in blood and associated with ASD are summarized in Fig. 7 [9, 11–14, 18, 19, 27, 51, 52, 59].
Fig. 7.
Summary of complement and coagulation pathway proteins associated with ASD identified in previous studies and this study. A Complement activation pathways and complement-related proteins associated with ASD. Red and pink mark DEPs, of which the red targets are 12 DEPs as potential markers. We compared the protein expression of the complement system in the brain and plasma in other articles. ↑ red, increased protein activity or expression in blood from patients; ↓ red, decreased protein activity or expression in blood from patients; ↑ black, increased RNA expression in brain tissue from patients; ↓ black, decreased RNA expression in brain tissue from patients. B Blood coagulation pathway and coagulation-related proteins associated with ASD.
Based on bioinformatics analysis, our results showed that among the 16 DEPs identified in this study, 11 were involved in the innate immune response and 9 in the adaptive immune response. Accumulating evidence suggests that the immune system plays a potential role in the pathophysiology of ASD. Indeed, the complement system, which was originally identified as a component of the innate immune system and is a major mediator of inflammation, is essential in the host’s defense against infection and serves as an immune surveillance system by clearing cellular debris and apoptotic cells. Activation of the complement pathway results in the release of inflammatory mediators. It has been reported that the increased levels of inflammatory markers and abnormal immune function in children with autism may be the potential mechanism of ASD [14, 60]. A previous study has found that inflammatory factors are present in the postmortem brain and are activated in ASD [61, 62]. Hence, our results suggest that the complement system and inflammatory reactions are involved in the pathophysiology of ASD.
Circulating complement proteins are mostly produced by liver cells, but many immune cells, especially macrophages and dendritic cells, produce complement locally when activated [21]. Due to the presence of the blood-brain barrier, the central nervous system (CNS) does not receive the same composition of circulating complement factors unless it is damaged [63]. In the brain, complement proteins can be locally synthesized by neurons, astrocytes, and microglia [25]. Therefore, peripheral and central complement production remains isolated [25]. Of note, in addition to their role in the innate immune response, complement proteins appear to play an important role in neurodevelopment, including neurogenesis, neuronal migration, and synapse pruning and remodeling [64]. The function of the complement system in the CNS is being under extensive investigation. Animal and in vitro cell models have shown that specific complement components play an important role in regulating neurogenesis in embryonic and adult brains [63]. Studies have shown that knockout of C1q and C3 in postnatal mice leads to increased brain wiring [59, 65, 66]. In mouse embryos, C1s and C3 knockout or knockdown results in reduced neuronal migration [67]. In mouse embryos, C3 gene knockout or knockdown causes increased proliferation of neural progenitor cells [67]. C3-, MaSp1-, or MASP2-deficient mice show radial migration disorder, resulting in improper localization of neurons and disorder of the cortical layers [67]. In addition, C3a and C5a are pro-inflammatory peptides that interact with and activate immune cells through their receptors (C3a and C5a receptors) [27]. Interestingly, knocking out, knocking down, or inhibiting these receptors has an effect similar to knocking out or knocking down the corresponding complement proteins, while activating them has the opposite effect [27]. Together, previous studies suggest that the complement cascade plays a role in the pathogenesis of neurodevelopmental disorders [21, 25, 68].
Indeed, altered complement proteins in the brain have been reported to be associated with ASD [21, 25, 69]. It has been shown that the levels of C2, C5, and MASP1 mRNA are increased, but C1q, C3, and C4 mRNA levels are decreased in the middle frontal gyrus of ASD subjects compared to controls [50]. Another study reported overexpression of C1q, C3, and CR3 genes in ASD brains [70]. More recently, a study has shown that C4 is reduced in the induced pluripotent stem cell-derived astrocytes of ASD subjects [22]. C1q, C3, and C4 participate in synapse elimination [71–74]. Their deficiency may limit the synaptic pruning process. Dendritic spine density is increased in the cortex [75], while a higher spine density has been reported in the temporal cortex of ASD patients [76]. C3 deficiency in mouse brains results in ASD-like behavior [50, 72]. In this study, bioinformatics analysis showed that 10 DEPs associated with the complement system were related to neuronal development and plasticity. The altered expression of complement RNA expression in the brains of ASD patients reported in the previous studies is summarized in Fig. 7 [21, 22, 50, 69, 70].
Overall, the results of this study and previous studies have shown that the levels of most complement-related proteins in the peripheral blood of ASD children are elevated. However, in the brain of ASD patients, conflicting results in the expression of C1q and C3 genes have been reported [27, 50, 70]. Likewise, a recent study that offspring whose mother has a low IL-10 level have decreased complement expression in the periphery and an increase in complement in the brain [77]. In addition, although a previous study found that the permeability of the blood-brain barrier is increased in some people with autism [78], it is unclear whether the changes of complement molecules in the CNS are related to peripheral changes [50]. Therefore, the involvement of the complement system in the pathogenesis of ASD is complex. Nevertheless, the existing evidence supports the conclusion that the complement system plays a key role in the pathogenesis of ASD. Dysregulation of complement proteins in peripheral blood may be a common feature in children with ASD [9, 11, 12].
Complement proteins are abundant in the blood, accounting for about 10% of serum protein. Consequently, they are suitable as potential diagnostic biomarkers for ASD. However, ASD is not only caused by changes in this pathway [11, 12, 79, 80]; many diseases also cause changes in the complement system [27–31, 81, 82]. Nevertheless, they still have the potential to become diagnostic markers. From the existing data, whether it is the brain or the periphery, the specific complement proteins that are changed in these diseases do not completely overlap [27, 28, 83–86]. For example, the types of changes in peripheral blood complement protein in patients with AD, schizophrenia, and multiple sclerosis are not completely consistent with those reported in the present study [27, 28, 82, 83]. Therefore, there may be differences in complement changes in different neurological diseases, and the similarities and differences between them deserve further study. The changes in complement proteins in different neuropsychiatric diseases may be different from those of ASD. Meanwhile, in practical applications, these proteins can be used as panels as described in this study, rather than as markers in the form of individual proteins. On the other hand, diseases with multiple factors or multiple pathogeneses may need to involve different types of markers, and complement proteins can be used as one or a class of them, which can be combined with other proteins to become markers. This may be a better strategy to improve the specificity of a diagnosis [11, 12]. This is easily achieved using the MRM technique used in the current study. Taken together, complement and coagulation system proteins have the potential to serve as independent biomarkers of ASD. However, it is important to note that this study is preliminary, with small sample size, and needs to be further validated by other cohorts as well as studies with larger sample sizes. Currently, while the data of this experiment conforms to the machine learning model, the 5-fold cross-validation effectively reduces the occurrence of over-fitting. Nevertheless, caution is needed as the AUC value for a panel consisting of 12 proteins is 1. Therefore, it is still crucial to validate another independent sample database in future experiments. More data could validate whether this group of proteins could provide an auxiliary diagnosis for children with ASD.
Moreover, post-translational modifications of complement proteins have also been investigated. Seven (C1QC, C1RL, C4BPB, C5, C8A, C8B, and CFHR2) and four (C1QA, C1QB, C8G, and CFH) glycosylated complement proteins have been found to be up-regulated and down-regulated in ASD patients, respectively [17, 52, 87]. The carbonyl levels of C8A are significantly higher in the plasma of autistic children than in healthy controls [17]. Complement C3 was also found to be up-regulated in the PBMCs of children with ASD in our previous study [14]. These changes deserve attention and further research. Together, the changes in the complement system in peripheral blood, PBMCs, and the brain of ASD patients highlight that this system may play a key role in the pathogenesis of ASD.
Furthermore, abnormal complement expression during brain development may be a causative factor for ASD that is independent of inflammation [27]. This is due to genetic factors more than anything else. Gene association studies have shown an increased frequency of a complement C4B gene null allele in patients with ASD [88–90]. On the other hand, abnormal complement activation systemically and in the CNS due to inflammatory insults or maternal immune activation during prenatal or early postnatal neurodevelopment also plays a role in the pathophysiology of ASD [27]. For example, maternal infection during pregnancy increases the risk of ASD in offspring [91]. This can be attributed to environmental factors more than anything else. The cross-sectional design of this study limits the strength of inferred causal relationships and may not reflect dynamic changes and exposure during the development of ASD. The relationship between complement and coagulation cascades and the development of ASD is worthy of further study.
In conclusion, we used MRM proteomics analysis to systematically investigate differences in peripheral blood complement and coagulation-related proteins between children with ASD and controls. The results identified 16 proteins as DEPs between these two groups, and they were all up-regulated in children with ASD. Using machine learning methods, 12 DEPs were identified as a group of potential biomarkers for ASD diagnosis. Among them, the expression of SERPIND1 was positively correlated with the CARS score and has the potential to classify the severity of the disease. We also summarized the complement and coagulation-related proteins associated with ASD, including those found to be altered in the brain and blood in the present and previous studies. These results support the conclusion that the complement and coagulation pathway is activated in the periphery of children with ASD and suggest that this pathway plays a critical role in ASD pathogenesis. Moreover, it would also be interesting to use MRM technology to study proteins from other pathways associated with children with ASD, or to select proteins that are significantly changed in different pathways to form a panel of diagnostic markers to be studied.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31870825), the Shenzhen Bureau of Science, Technology, and Information (JCYJ20170412110026229), the Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2022SHIBS0003), and 2019 Guiyang Science and Technology Bureau, and the Guiyang First People’s Hospital, Great Health Science and Technology Cooperation Project. We thank all the individuals who participated in the study and the Instrument Analysis Center of Shenzhen University.
Data Availability
All raw data have been deposited as an online resource to the Figshare database with the name: “A systematic investigation of complement and coagulation-related protein in autism spectrum disorder by multiple reaction monitoring technology” (https://doi.org/10.6084/m9.figshare.21830145.v1).
Conflict of interest
The authors claim that there are no conflicts of interest.
Ethical Approval
This research was approved by the Human Research Ethics Committees of Shenzhen University (M20220203) and the Maternal and Child Health Hospital of Baoan (20170801) and complies with the guidelines of the Helsinki Declaration. Written informed consent for study participation was given by the children’s guardians.
References
- 1.Masi A, DeMayo MM, Glozier N, Guastella AJ. An overview of autism spectrum disorder, heterogeneity and treatment options. Neurosci Bull. 2017;33:183–193.. doi: 10.1007/s12264-017-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora NK, Nair MKC, Gulati S, Deshmukh V, Mohapatra A, Mishra D, et al. Neurodevelopmental disorders in children aged 2–9 years: Population-based burden estimates across five regions in India. PLoS Med. 2018;15:e1002615.. doi: 10.1371/journal.pmed.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter AJ, Brugha TS, Erskine HE, Scheurer RW, Vos T, Scott JG. The epidemiology and global burden of autism spectrum disorders. Psychol Med. 2015;45:601–613.. doi: 10.1017/S003329171400172X. [DOI] [PubMed] [Google Scholar]
- 4.Maenner MJ, Shaw KA, Baio J, EdS, Washington A, Patrick M, et al. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveill Summ 2020, 69: 1–12. [DOI] [PMC free article] [PubMed]
- 5.Loomes R, Hull L, Mandy WPL. What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J Am Acad Child Adolesc Psychiatry. 2017;56:466–474.. doi: 10.1016/j.jaac.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Singer L. Thoughts about sex and gender differences from the next generation of autism scientists. Mol Autism. 2015;6:52.. doi: 10.1186/s13229-015-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Xu X, Yan W, Zou X, Wu L, Luo X, et al. Prevalence of autism spectrum disorder in China: A nationwide multi-center population-based study among children aged 6 to 12 years. Neurosci Bull. 2020;36:961–971.. doi: 10.1007/s12264-020-00530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang ZC, Han J. The first national prevalence of autism spectrum disorder in China. Neurosci Bull. 2020;36:959–960.. doi: 10.1007/s12264-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen L, Zhang H, Lin J, Gao Y, Chen M, Khan NU, et al. A Combined Proteomics and Metabolomics Profiling to Investigate the Genetic Heterogeneity of Autistic Children. Mol Neurobiol. 2022;59:3529–3545.. doi: 10.1007/s12035-022-02801-x. [DOI] [PubMed] [Google Scholar]
- 10.Vahia VN. Diagnostic and statistical manual of mental disorders 5: A quick glance. Indian J Psychiatry. 2013;55:220–223.. doi: 10.4103/0019-5545.117131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L, Zhao Y, Zhang H, Feng C, Gao Y, Zhao D, et al. Advances in biomarker studies in autism spectrum disorders. Adv Exp Med Biol. 2019;1118:207–233.. doi: 10.1007/978-3-030-05542-4_11. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Liu X, Zhang H, Lin J, Feng C, Iqbal J. Biomarkers in autism spectrum disorders: Current progress. Clin Chim Acta. 2020;502:41–54.. doi: 10.1016/j.cca.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Shen L, Zhang K, Feng C, Chen Y, Li S, Iqbal J, et al. iTRAQ-based proteomic analysis reveals protein profile in plasma from children with autism. Proteomics Clin Appl. 2018;12:e1700085.. doi: 10.1002/prca.201700085. [DOI] [PubMed] [Google Scholar]
- 14.Shen L, Feng C, Zhang K, Chen Y, Gao Y, Ke J, et al. Proteomics study of peripheral blood mononuclear cells (PBMCs) in autistic children. Front Cell Neurosci. 2019;13:105.. doi: 10.3389/fncel.2019.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristori MV, Mortera SL, Marzano V, Guerrera S, Vernocchi P, Ianiro G, et al. Proteomics and metabolomics approaches towards a functional insight onto AUTISM spectrum disorders: Phenotype stratification and biomarker discovery. Int J Mol Sci. 2020;21:6274.. doi: 10.3390/ijms21176274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham J, Szoko N, Natowicz MR. Proteomic investigations of autism spectrum disorder: Past findings, current challenges, and future prospects. Adv Exp Med Biol. 2019;1118:235–252.. doi: 10.1007/978-3-030-05542-4_12. [DOI] [PubMed] [Google Scholar]
- 17.Feng C, Chen Y, Pan J, Yang A, Niu L, Min J, et al. Redox proteomic identification of carbonylated proteins in autism plasma: Insight into oxidative stress and its related biomarkers in autism. Clin Proteomics. 2017;14:2.. doi: 10.1186/s12014-017-9138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett BA, Kantor AB, Schulman H, Walker WL, Lit L, Ashwood P, et al. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry. 2007;12:292–306.. doi: 10.1038/sj.mp.4001943. [DOI] [PubMed] [Google Scholar]
- 19.Momeni N, Brudin L, Behnia F, Nordström B, Yosefi-Oudarji A, Sivberg B, et al. High complement factor I activity in the plasma of children with autism spectrum disorders. Autism Res Treat. 2012;2012:868576.. doi: 10.1155/2012/868576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngounou Wetie AG, Wormwood K, Thome J, Dudley E, Taurines R, Gerlach M, et al. A pilot proteomic study of protein markers in autism spectrum disorder. Electrophoresis. 2014;35:2046–2054.. doi: 10.1002/elps.201300370. [DOI] [PubMed] [Google Scholar]
- 21.Ziabska K, Ziemka-Nalecz M, Pawelec P, Sypecka J, Zalewska T. Aberrant complement system activation in neurological disorders. Int J Mol Sci. 2021;22:4675.. doi: 10.3390/ijms22094675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansur F, Teles E Silva AL, Gomes AKS, Magdalon J, de Souza JS, Griesi-Oliveira K, et al. Complement C4 is reduced in iPSC-derived astrocytes of autism spectrum disorder subjects. Int J Mol Sci 2021, 22: 7579. [DOI] [PMC free article] [PubMed]
- 23.Momeni N, Bergquist J, Brudin L, Behnia F, Sivberg B, Joghataei MT, et al. A novel blood-based biomarker for detection of autism spectrum disorders. Transl Psychiatry. 2012;2:e91.. doi: 10.1038/tp.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masi A, Glozier N, Dale R, Guastella AJ. The immune system, cytokines, and biomarkers in autism spectrum disorder. Neurosci Bull. 2017;33:194–204.. doi: 10.1007/s12264-017-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garred P, Tenner AJ, Mollnes TE. Therapeutic targeting of the complement system: From rare diseases to pandemics. Pharmacol Rev. 2021;73:792–827.. doi: 10.1124/pharmrev.120.000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruno A, Dolcetti E, Rizzo FR, Fresegna D, Musella A, Gentile A, et al. Inflammation-associated synaptic alterations as shared threads in depression and multiple sclerosis. Front Cell Neurosci. 2020;14:169.. doi: 10.3389/fncel.2020.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magdalon J, Mansur F, Teles E Silva AL, de Goes VA, Reiner O, Sertié AL. Complement system in brain architecture and neurodevelopmental disorders. Front Neurosci 2020, 14: 23. [DOI] [PMC free article] [PubMed]
- 28.Krance SH, Wu CY, Zou Y, Mao H, Toufighi S, He X, et al. The complement cascade in Alzheimer’s disease: A systematic review and meta-analysis. Mol Psychiatry. 2021;26:5532–5541.. doi: 10.1038/s41380-019-0536-8. [DOI] [PubMed] [Google Scholar]
- 29.Morgan BP, Gommerman JL, Ramaglia V. An, “outside-In” and “inside-out” consideration of complement in the multiple sclerosis brain: Lessons from development and neurodegenerative diseases. Front Cell Neurosci. 2020;14:600656.. doi: 10.3389/fncel.2020.600656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellingacci L, Mancini A, Gaetani L, Tozzi A, Parnetti L, di Filippo M. Synaptic dysfunction in multiple sclerosis: A red thread from inflammation to network disconnection. Int J Mol Sci. 2021;22:9753.. doi: 10.3390/ijms22189753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Druart M, Le Magueresse C. Emerging roles of complement in psychiatric disorders. Front Psychiatry. 2019;10:573.. doi: 10.3389/fpsyt.2019.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat Methods. 2014;11:149–155.. doi: 10.1038/nmeth.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Wang Q, Chen J, Zhou J, Zhou X, Xie P. A multiple-reaction-monitoring mass spectrometric method for simultaneous quantitative analysis of five plasma apolipoproteins. Sci China Chem. 2014;57:723–731.. doi: 10.1007/s11426-013-5036-0. [DOI] [Google Scholar]
- 34.Li Y, Liang J, Gao JN, Shen Y, Kuang HX, Xia YG. A novel LC-MS/MS method for complete composition analysis of polysaccharides by aldononitrile acetate and multiple reaction monitoring. Carbohydr Polym. 2021;272:118478.. doi: 10.1016/j.carbpol.2021.118478. [DOI] [PubMed] [Google Scholar]
- 35.Lin TT, Zhang T, Kitata RB, Liu T, Smith RD, Qian WJ, et al. Mass spectrometry-based targeted proteomics for analysis of protein mutations. Mass Spectrom Rev. 2023;42:796–821.. doi: 10.1002/mas.21741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng FC, Xu XJ, Song TJ, Shou XJ, Wang XL, Han SP, et al. Development of an autism subtyping questionnaire based on social behaviors. Neurosci Bull. 2018;34:789–800.. doi: 10.1007/s12264-018-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du X, Shi Q, Zhao Y, Xie Y, Li X, Liu Q, et al. Se-methylselenocysteine (SMC) improves cognitive deficits by attenuating synaptic and metabolic abnormalities in alzheimer’s mice model: A proteomic study. ACS Chem Neurosci. 2021;12:1112–1132.. doi: 10.1021/acschemneuro.0c00549. [DOI] [PubMed] [Google Scholar]
- 38.Frewen B, MacCoss MJ. Using BiblioSpec for creating and searching tandem MS peptide libraries. Curr Protoc Bioinformatics 2007, Chapter 13: 13.7.1–13.713.7.12. [DOI] [PubMed]
- 39.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9.. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen L, Yang A, Chen X, Xiao S, Liu X, Lin J, et al. Proteomic profiling of Cerebrum mitochondria, myelin sheath, and synaptosome revealed mitochondrial damage and synaptic impairments in association with 3 × tg-AD mice model. Cell Mol Neurobiol. 2022;42:1745–1763.. doi: 10.1007/s10571-021-01052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freund Y, Schapire R. Experiments with a new boosting algorithm. ICML’96: Proceedings of the Thirteenth International Conference on International Conference on Machine Learning, 1996: 148–156.
- 42.Dorogush AV, Ershov V, Gulin A. CatBoost: Gradient boosting with categorical features support. 2018. 10.48550/arXiv.1810.11363.
- 43.Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Statist. 2001;29:1189–1232.. doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- 44.Chen Tianqi", Guestrin C. XGBoost: A scalable tree boosting system". 2016: arXiv: 1603.02754. https://arxiv.org/abs/1603.02754".
- 45.Ke G, Meng Q, Wang T, Chen W, Ma W, Liu TY. A highly efficient gradient boosting decision tree. NIPS’17: Proceedings of the 31st International Conference on Neural Information Processing Systems, 2017: 3149–3157.
- 46.Quinlan JR. Induction of decision trees. Mach Learn. 1986;1:81–106.. doi: 10.1007/BF00116251. [DOI] [Google Scholar]
- 47.Everson TM, Lyons G, Zhang H, Soto-Ramírez N, Lockett GA, Patil VK, et al. DNA methylation loci associated with atopy and high serum IgE: A genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7:89.. doi: 10.1186/s13073-015-0213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell HB, Schaefer PA. A? soft? K-nearest neighbor voting scheme. Int J Intell Syst. 2001;16:459–468.. doi: 10.1002/int.1018. [DOI] [Google Scholar]
- 49.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422.. doi: 10.1023/A:1012487302797. [DOI] [Google Scholar]
- 50.Fagan K, Crider A, Ahmed AO, Pillai A. Complement C3 expression is decreased in autism spectrum disorder subjects and contributes to behavioral deficits in rodents. Mol Neuropsychiatry. 2017;3:19–27.. doi: 10.1159/000465523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren RP, Burger RA, Odell D, Torres AR, Warren WL. Decreased plasma concentrations of the C4B complement protein in autism. Arch Pediatr Adolesc Med. 1994;148:180–183.. doi: 10.1001/archpedi.1994.02170020066011. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Chen Y, Xiong X, Zhou X, Han L, Ni L, et al. Peptidome analysis reveals novel serum biomarkers for children with autism spectrum disorder in China. Proteomics Clin Appl. 2018;12:e1700164.. doi: 10.1002/prca.201700164. [DOI] [PubMed] [Google Scholar]
- 53.Woodward KJ, Stampalia J, Vanyai H, Rijhumal H, Potts K, Taylor F, et al. Atypical nested 22q11.2 duplications between LCR22B and LCR22D are associated with neurodevelopmental phenotypes including autism spectrum disorder with incomplete penetrance. Mol Genet Genomic Med. 2019;7:e00507.. doi: 10.1002/mgg3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Liu X, Gao H, He R, Zhao Y. Identifying of 22q11.2 variations in Chinese patients with development delay. BMC Med Genomics. 2021;14:26.. doi: 10.1186/s12920-020-00849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halawa OA, Lin JB, Miller JW, Vavvas DG. A review of completed and ongoing complement inhibitor trials for geographic atrophy secondary to age-related macular degeneration. J Clin Med. 2021;10:2580.. doi: 10.3390/jcm10122580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Józsi M, Tortajada A, Uzonyi B, de Goicoechea Jorge E, de Rodríguez Córdoba S. Factor H-related proteins determine complement-activating surfaces. Trends Immunol. 2015;36:374–384.. doi: 10.1016/j.it.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Blom AM, Villoutreix BO, Dahlbäck B. Functions of human complement inhibitor C4b-binding protein in relation to its structure. Arch Immunol Ther Exp (Warsz) 2004;52:83–95.. [PubMed] [Google Scholar]
- 58.Sjöberg AP, Trouw LA, Blom AM. Complement activation and inhibition: A delicate balance. Trends Immunol. 2009;30:83–90.. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Comer AL, Jinadasa T, Sriram B, Phadke RA, Kretsge LN, Nguyen TPH, et al. Increased expression of schizophrenia-associated gene C4 leads to hypoconnectivity of prefrontal cortex and reduced social interaction. PLoS Biol. 2020;18:e3000604.. doi: 10.1371/journal.pbio.3000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu VW. Frontiers in Autism Research: New Horizons for Diagnosis and Treatment. 1. Singapore: World Scientific Publishing; 2014. Chapter 17: Maternal Autoantibodies in Autism Spectrum Disorder; pp. 429–449.. [Google Scholar]
- 61.Greene RK, Walsh E, Mosner MG, Dichter GS. A potential mechanistic role for neuroinflammation in reward processing impairments in autism spectrum disorder. Biol Psychol. 2019;142:1–12.. doi: 10.1016/j.biopsycho.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Chauhan A, Sheikh AM, Patil S, Chauhan V, Li XM, et al. Elevated immune response in the brain of autistic patients. J Neuroimmunol. 2009;207:111–116.. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleine TO, Benes L. Immune surveillance of the human central nervous system (CNS): Different migration pathways of immune cells through the blood-brain barrier and blood-cerebrospinal fluid barrier in healthy persons. Cytometry A. 2006;69:147–151.. doi: 10.1002/cyto.a.20225. [DOI] [PubMed] [Google Scholar]
- 64.Carpanini SM, Torvell M, Morgan BP. Therapeutic inhibition of the complement system in diseases of the central nervous system. Front Immunol. 2019;10:362.. doi: 10.3389/fimmu.2019.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705.. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bialas AR, Stevens B. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci. 2013;16:1773–1782.. doi: 10.1038/nn.3560. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Gorelik A, Sapir T, Haffner-Krausz R, Olender T, Woodruff TM, Reiner O. Developmental activities of the complement pathway in migrating neurons. Nat Commun. 2017;8:15096.. doi: 10.1038/ncomms15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.English JA, Lopez LM, O’Gorman A, Föcking M, Hryniewiecka M, Scaife C, et al. Blood-based protein changes in childhood are associated with increased risk for later psychotic disorder: Evidence from a nested case-control study of the ALSPAC longitudinal birth cohort. Schizophr Bull. 2018;44:297–306.. doi: 10.1093/schbul/sbx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahman MR, Petralia MC, Ciurleo R, Bramanti A, Fagone P, Shahjaman M, et al. Comprehensive analysis of RNA-seq gene expression profiling of brain transcriptomes reveals novel genes, regulators, and pathways in autism spectrum disorder. Brain Sci. 2020;10:747.. doi: 10.3390/brainsci10100747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nardone S, Sams DS, Reuveni E, Getselter D, Oron O, Karpuj M, et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 2014;4:e433.. doi: 10.1038/tp.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chu Y, Jin X, Parada I, Pesic A, Stevens B, Barres B, et al. Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc Natl Acad Sci U S A. 2010;107:7975–7980.. doi: 10.1073/pnas.0913449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Alcazar M, Daborg J, Stokowska A, Wasling P, Björefeldt A, Kalm M, et al. Altered cognitive performance and synaptic function in the hippocampus of mice lacking C3. Exp Neurol. 2014;253:154–164.. doi: 10.1016/j.expneurol.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 73.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183.. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang C, Garbuzova-Davis S, Tan J, Obregon D. C1q as a regulator of brain development: Implications for autism spectrum disorders. Brain Disord Ther 2015, 4. doi: 10.4172/2168-975X.10001.
- 75.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94.. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 76.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143.. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jaini R, Wolf MR, Yu Q, King AT, Frazier TW, Jr, Eng C. Maternal genetics influences fetal neurodevelopment and postnatal autism spectrum disorder-like phenotype by modulating in-utero immunosuppression. Transl Psychiatry. 2021;11:348.. doi: 10.1038/s41398-021-01472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiorentino M, Sapone A, Senger S, Camhi SS, Kadzielski SM, Buie TM, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol Autism. 2016;7:49.. doi: 10.1186/s13229-016-0110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X, Lin J, Zhang H, Khan NU, Zhang J, Tang X, et al. Oxidative stress in autism spectrum disorder-current progress of mechanisms and biomarkers. Front Psychiatry. 2022;13:813304.. doi: 10.3389/fpsyt.2022.813304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Li X, Shen L, Khan NU, Zhang X, Chen L, et al. Trace elements in children with autism spectrum disorder: A meta-analysis based on case-control studies. J Trace Elem Med Biol. 2021;67:126782.. doi: 10.1016/j.jtemb.2021.126782. [DOI] [PubMed] [Google Scholar]
- 81.Lee JD, Coulthard LG, Woodruff TM. Complement dysregulation in the central nervous system during development and disease. Semin Immunol. 2019;45:101340.. doi: 10.1016/j.smim.2019.101340. [DOI] [PubMed] [Google Scholar]
- 82.Aiyaz M, Lupton MK, Proitsi P, Powell JF, Lovestone S. Complement activation as a biomarker for Alzheimer’s disease. Immunobiology. 2012;217:204–215.. doi: 10.1016/j.imbio.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 83.Rezeli M, Végvári A, Ottervald J, Olsson T, Laurell T, Marko-Varga G. MRM assay for quantitation of complement components in human blood plasma - a feasibility study on multiple sclerosis. J Proteomics. 2011;75:211–220.. doi: 10.1016/j.jprot.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 84.Neueder A, Bates GP. A common gene expression signature in Huntington’s disease patient brain regions. BMC Med Genomics. 2014;7:60.. doi: 10.1186/s12920-014-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watkins LM, Neal JW, Loveless S, Michailidou I, Ramaglia V, Rees MI, et al. Complement is activated in progressive multiple sclerosis cortical grey matter lesions. J Neuroinflammation. 2016;13:161.. doi: 10.1186/s12974-016-0611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin P, Nicholls L, Assareh H, Fang Z, Amos TG, Edwards RJ, et al. Transcriptome analysis of human brain tissue identifies reduced expression of complement complex C1Q Genes in Rett syndrome. BMC Genomics. 2016;17:427.. doi: 10.1186/s12864-016-2746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin Y, Chen Y, Yang J, Wu F, Zhao L, Yang F, et al. Serum glycopattern and Maackia amurensis lectin-II binding glycoproteins in autism spectrum disorder. Sci Rep. 2017;7:46041.. doi: 10.1038/srep46041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warren RP, Singh VK, Cole P, Odell JD, Pingree CB, Warren WL, et al. Increased frequency of the null allele at the complement C4b locus in autism. Clin Exp Immunol. 1991;83:438–440.. doi: 10.1111/j.1365-2249.1991.tb05657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Odell D, Maciulis A, Cutler A, Warren L, McMahon WM, Coon H, et al. Confirmation of the association of the C4B null allelle in autism. Hum Immunol. 2005;66:140–145.. doi: 10.1016/j.humimm.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 90.Mostafa GA, Shehab AA. The link of C4B null allele to autism and to a family history of autoimmunity in Egyptian autistic children. J Neuroimmunol. 2010;223:115–119.. doi: 10.1016/j.jneuroim.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 91.Smith SEP, Hsiao E, Patterson PH. Activation of the maternal immune system as a risk factor for neuropsychiatric disorders. In: In: Maternal Influences on Fetal Neurodevelopment. 1st ed. Springer New York, 2010: 97–115.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data have been deposited as an online resource to the Figshare database with the name: “A systematic investigation of complement and coagulation-related protein in autism spectrum disorder by multiple reaction monitoring technology” (https://doi.org/10.6084/m9.figshare.21830145.v1).