Abstract
Molecular mechanisms transducing physical forces in the bone microenvironment to regulate bone mass are poorly understood. Here, we used mouse genetics, mechanical loading, and pharmacological approaches to test the possibility that polycystin-1 and Wwtr1 have interdependent mechanosensing functions in osteoblasts. We created and compared the skeletal phenotypes of control Pkd1flox/+;Wwtr1flox/+, Pkd1Oc-cKO, Wwtr1Oc-cKO, and Pkd1/Wwtr1Oc-cKO mice to investigate genetic interactions. Consistent with an interaction between polycystins and Wwtr1 in bone in vivo, Pkd1/Wwtr1Oc-cKO mice exhibited greater reductions of BMD and periosteal MAR than either Wwtr1Oc-cKO or Pkd1Oc-cKO mice. Micro-CT 3D image analysis indicated that the reduction in bone mass was due to greater loss in both trabecular bone volume and cortical bone thickness in Pkd1/Wwtr1Oc-cKO mice compared to either Pkd1Oc-cKO or Wwtr1Oc-cKO mice. Pkd1/Wwtr1Oc-cKO mice also displayed additive reductions in mechanosensing and osteogenic gene expression profiles in bone compared to Pkd1Oc-cKO or Wwtr1Oc-cKO mice. Moreover, we found that Pkd1/Wwtr1Oc-cKO mice exhibited impaired responses to tibia mechanical loading in vivo and attenuation of load-induced mechanosensing gene expression compared to control mice. Finally, control mice treated with a small molecule mechanomimetic, MS2 that activates the polycystin complex resulted in marked increases in femoral BMD and periosteal MAR compared to vehicle control. In contrast, Pkd1/Wwtr1Oc-cKO mice were resistant to the anabolic effects of MS2. These findings suggest that PC1 and Wwtr1 form an anabolic mechanotransduction signaling complex that mediates mechanical loading responses and serves as a potential novel therapeutic target for treating osteoporosis.
Subject terms: Bone, Bone quality and biomechanics
Introduction
In vivo and in vitro studies demonstrate that the polycystin-1(PC1)/polycystin-2 (PC2) heterotetrameric complex functions in osteoblasts and osteocytes to regulate bone mass1,2 and acts as a mechanosensor to transduce the bone anabolic response to mechanical loading in vivo.3,4 Genetic ablation of either PC1 or PC2 deficiency in osteoblasts or osteocytes has similar effects in reducing bone mass by decreasing osteoblast-mediated bone formation.3,5–8 PC1 and PC2 mechanosensing functions are mediated by heterotetrameric complex activation of common signal transduction pathways. In this regard, PC1 and PC2 conditional knockout models exhibit concordant effects in impairing osteoblast-mediated bone formation. However, PC1 and PC2 have discordant effects on bone marrow adipogenesis that implicates separate signaling mechanisms.4,6,9 In this regard, PC1 deficiency stimulates adipogenesis, leading to increased bone marrow adipose tissue (MAT) deposition,2,6 whereas PC2 loss-of-function inhibits adipogenesis.4 Additional in vitro and in vivo data show that PC1 activates Runx2 transcriptional activity to stimulate osteoblastogenesis but diminishes PPARγ signaling leading to reduced bone marrow fat.2,4 In agreement with the low turnover bone disorder in Pkd1 mouse models, not only bone-specific alkaline phosphatase was significantly lower in patients with ADPKD but also histomorphometric parameters of bone formation were decreased compared to control subjects without ADPKD.10–12 The molecular mechanisms underlying the different effects of PC1 and PC2 on osteoblastogenesis and adipogenesis are not clear. In the current studies, we sought to understand the mechanism for the apparent PC1-specific effect in reciprocally regulating transcriptional control of osteoblastogenesis and adipogenesis.
The Hippo-Yap/Wwtr1(also called TAZ) pathway is also regulated by mechanical forces.13–16 Alterations of matrix stiffness in cell culture modulates nuclear translocation of non-phosphorylated Wwtr1 resulting in co-activation of Runx2 and stimulation of osteoblastogenesis and in Wwtr1 binding to PPARγ to inhibit adipogenesis.2,17,18 The physiological importance of Wwtr1 in bone homeostasis is revealed by transgenic overexpression of Wwtr1 in osteoblasts in mice, which increases osteoblast-mediated bone formation and inhibits bone marrow adipogenesis19; depletion of Wwtr1 in zebrafish, which impairs bone development,18 and global knockout of Wwtr1 in mice, which have small stature and ossification defects.20 Based on these observations, we posit that PC1 dependent Wwtr1 signaling might explain the differential functions of PC1 and PC2 on adipogenesis.
Recent studies show crosstalk between PC1 and Wwtr1 signaling that is mediated by the binding of Wwtr1 to the PC1 C-terminal tail.2,17 The PC1/Wwtr1 complex is cleaved to allow nuclear translocation of Wwtr1, a mechanism of Wwtr1 regulation that differs from the canonical Hippo regulation of Yap/Wwtr1 signaling.21 Wwtr1 binds to the PC1 C-terminal tail (PC1-CTT) and undergoes nuclear translocation in response to changes in bone ECM microenvironment to stimulate osteoblastogenesis and inhibit adipogenesis through transcriptional co-activation of Runx2 and co-repression of PPARγ activity.2 Wwtr1 also binds to PC2, leading to PC2 degradation.22 These observations in bone parallel the interactions between PC1/PC2 and Wwtr1 in primary cilia in renal epithelial cells.20,23,24 In this regard, Wwtr1 knockout from the kidney result in cystic kidney disease in mice, similar to polycystin complex inactivation, suggesting that PC1/PC2 and Wwtr1 act through common pathways in the kidney as well as bone.20,23,24 Furthermore, a small molecule mechanomimetic (named as molecular staple, MS) that binds to the PC1:PC2 C-terminal tail in a presumptive coiled-coil region (e.g. PC1 residue Tyr4236 and PC2 residues Arg877, Arg878, and Lys874) has been shown to activate this complex and mimic the effects of physical forces to activate polycystins/Wwtr1 signaling and stimulate bone mass in mice.2 Collectively these observations suggest that Wwtr1 modulates polycystin’s mechanosensing functions to differentially regulate osteoblastogenesis and adipogenesis.
In this study, we examined the interaction between PC1 and Wwtr1 in mouse bone by using Osteocalcin (Oc)-Cre to conditionally delete both Pkd1 and Wwtr1 in osteoblasts. We compared skeletal phenotypes of double Pkd1/Wwtr1Oc-cKO mice with single conditional Pkd1 and Wwtr1 null mice under baseline conditions, after mechanical loading, and following treatment with a more potent mechanomimetic, MS2, that activates the PC1/PC2 complex. We found that genetic ablation of PC1 and Wwtr1 in osteoblasts results in additive loss of bone mass and anabolic responses to mechanical loading. Compound PC1 and Wwtr1 deficient mice were also resistant to the bone inductive effects of the MS2 mechanomimetic in vivo. Our findings provide a new mechanism whereby Wwtr1 regulates skeletal homeostasis through co-dependent functions with PC1 in osteoblasts.
Results
Wwtr1 has gene-dosage dependent effects on bone mass
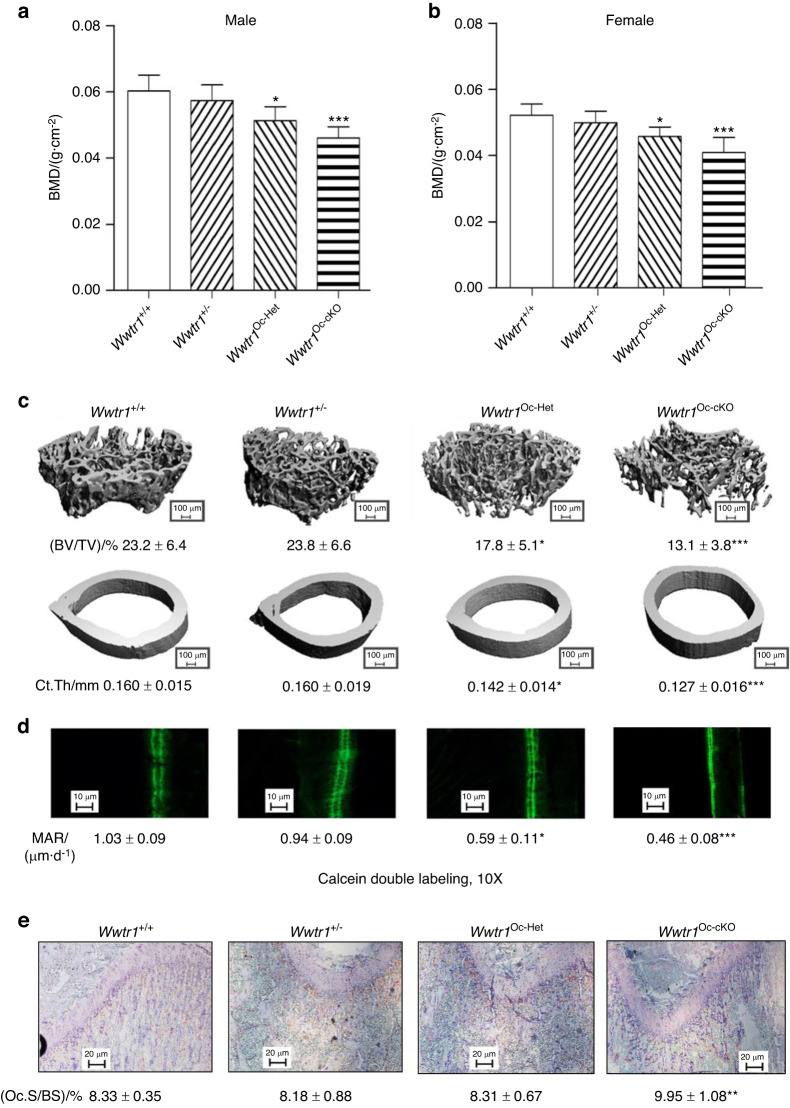
In the osteoblast specific conditional Wwtr1 knockout mouse model, we found a gene-dosage effect on osteoblast-mediated bone formation and bone mass (Figs. 1 and S1). Compared with control mice (Wwtr1+/+), heterozygous conditional Oc-Cre;Wwtr1flox/+ (Wwtr1Oc-Het), which has an approximately 37% reduction in Wwtr1 message expression in bone (Table 1), showed 15% and 12% reductions of BMD in male and female adult mice, respectively (Fig. 1a, b). Oc-Cre;Wwtr1flox/− (Wwtr1Oc-cKO) mice, which had a 64% reduction in Wwtr1 message expression in bone, showed an even greater loss of bone mass, with 24% and 22% reductions of BMD in male and female adult mice, respectively. Micro-CT 3D analysis showed that the reduction in bone mass in single conditional Wwtr1Oc-Het heterozygous mice arose from a 23% reduction in trabecular bone volume and a 11% reduction in cortical bone thickness in both male and female adult mice. Conditional Wwtr1Oc-cKO mice had a 44% loss in trabecular volume and 20% loss in cortical thickness.
Fig. 1.
Conditional deletion of Wwtr1 in mature osteoblasts on postnatal bone homeostasis. a, b Bone mineral density by DEXA scan in male and female mice. c Bone structure by micro-CT 3D images analysis from both male and female mice. d Periosteal mineral apposition rate (MAR) by calcein double labeling. There was a significant reduction in periosteal MAR in single Wwtr1Oc-Het heterozygous mice compared with age-matched control mice and an even greater decrement in Wwtr1Oc-cKO null mice, indicating a gene-dosage effect of Wwtr1 on osteoblast-mediated bone formation. e TRAP staining (red color) for osteoclast activity. Data are expressed as the mean ± S.D. from serum samples of individual mice (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001 compared with wild-type control mice. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
Table 1.
Gene-expression profiles in bone from Wwtr1-deficient mice
| Gene | Wild-type | Wwtr1+/− | Wwtr1Oc-Het | Wwtr1Oc-cKO | P-value |
|---|---|---|---|---|---|
| Osteoblast lineage | |||||
| Pkd1 | 1.00 ± 0.15 | 1.03 ± 0.22 | 0.99 ± 0.34 | 0.96 ± 0.32 | 0.971 8 |
| Wwtr1 | 1.00 ± 0.24 | 0.61 ± 0.10*** | 0.63 ± 0.14*** | 0.36 ± 0.09*** | <0.000 1 |
| Fgf23 | 1.00 ± 0.10 | 0.97 ± 0.22 | 1.61 ± 0.45* | 2.31 ± 0.61*** | <0.000 1 |
| Runx2-II | 1.00 ± 0.13 | 0.80 ± 0.21 | 0.82 ± 0.27 | 0.61 ± 0.10** | 0.017 7 |
| Osteopontin | 1.00 ± 0.26 | 0.83 ± 0.20 | 0.67 ± 0.11* | 0.62 ± 0.21* | 0.014 3 |
| Bsp | 1.00 ± 0.11 | 0.94 ± 0.28 | 0.68 ± 0.16* | 0.54 ± 0.15*** | 0.000 3 |
| Mepe | 1.00 ± 0.12 | 1.04 ± 0.22 | 1.43 ± 0.28** | 1.81 ± 0.22*** | <0.000 1 |
| Col1 | 1.00 ± 0.20 | 0.81 ± 0.23 | 0.67 ± 0.21* | 0.65 ± 0.18* | 0.030 4 |
| Alp | 1.00 ± 0.20 | 1.01 ± 0.29 | 0.69 ± 0.11* | 0.52 ± 0.15*** | 0.000 2 |
| Osteocalcin | 1.00 ± 0.19 | 0.80 ± 0.23 | 0.53 ± 0.19** | 0.57 ± 0.15** | 0.001 3 |
| OPG | 1.00 ± 0.13 | 0.97 ± 0.29 | 0.89 ± 0.31 | 0.96 ± 0.24 | 0.892 6 |
| RankL | 1.00 ± 0.17 | 1.13 ± 0.24 | 1.12 ± 0.11 | 1.52 ± 0.15*** | <0.000 1 |
| Fzd2 | 1.00 ± 0.25 | 1.02 ± 0.18 | 1.03 ± 0.18 | 0.96 ± 0.28 | 0.935 4 |
| Wnt10b | 1.00 ± 0.08 | 1.08 ± 0.28 | 0.97 ± 0.17 | 0.73 ± 0.06* | 0.016 4 |
| CYR61 | 1.00 ± 0.13 | 1.06 ± 0.10 | 1.54 ± 0.25** | 2.06 ± 0.55*** | <0.000 1 |
| CTGF | 1.00 ± 0.10 | 1.06 ± 0.28 | 1.18 ± 0.41 | 1.54 ± 0.31* | 0.022 1 |
| Osteoclast | |||||
| Trap | 1.00 ± 0.15 | 0.95 ± 0.19 | 1.10 ± 0.22 | 1.41 ± 0.19** | 0.002 1 |
| Mmp9 | 1.00 ± 0.14 | 0.85 ± 0.22 | 0.87 ± 0.15 | 0.86 ± 0.18 | 0.380 8 |
| Chondrocyte | |||||
| Collagen II | 1.00 ± 0.20 | 0.86 ± 0.20 | 0.79 ± 0.26 | 0.67 ± 0.15* | 0.045 8 |
| Collagen X | 1.00 ± 0.21 | 0.87 ± 0.36 | 0.74 ± 0.39 | 0.45 ± 0.23* | 0.033 9 |
| Adipocyte | |||||
| PPARγ2 | 1.00 ± 0.17 | 1.23 ± 0.14 | 1.57 ± 0.30** | 2.03 ± 0.42*** | <0.000 1 |
| aP2 | 1.00 ± 0.18 | 1.27 ± 0.31 | 1.64 ± 0.29** | 2.51 ± 0.36*** | <0.000 1 |
| Lpl | 1.00 ± 0.15 | 1.31 ± 0.37 | 1.63 ± 0.34* | 2.56 ± 0.47*** | <0.000 1 |
Data are mean ± S.D. from 6 tibias of wild-type control, Wwtr1+/−, Wwtr1Oc-Het, and Wwtr1Oc-cKO mice and expressed as the fold changes relative to the housekeeping gene β-actin subsequently normalized to control mice. *, **, *** indicates significant difference from wild-type control at P < 0.05, P < 0.01, P < 0.001, respectively. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
In agreement with the DEXA and micro-CT data, analysis of bone histology showed a Wwtr1 gene-dosage dependent reduction in trabecular bone volume (Figs. 1c and S1a) and cortical bone thickness (Figs. 1c and S1b) in the distal femur and a decrease in bone formation rate measured by double calcein labeling (Fig. 1d). There were 43% and 55% reductions of periosteal MAR in both conditional Wwtr1Oc-Het heterozygous and Wwtr1Oc-cKO null mice, respectively, compared with control mice (Wwtr1+/+). Unexpectedly, conditional deletion of Wwtr1 in osteoblasts resulted in enhanced osteoclast activity, as evidenced by increased TRAP immunostaining in the growth plate of conditional Wwtr1Oc-cKO null mice (Fig. 1e).
Real-time RT-PCR analysis revealed a Wwtr1 gene-dosage dependent changes in osteoblast markers including Runx2-II and Wnt10b, FGF-23, Mepe, RankL, CYR61, and CTGF and chondrocyte markers such as Collagen II and Collagen X. Reductions of Runx2-II and Wnt10b impairs osteoblast proliferation and differentiation. Increments of FGF-23 and Mepe as well as Yap signaling such as increased CYR61 and CTGF gene expressions inhibits osteoblast differentiation and mineralization. An increase in RankL and the RankL/OPG ratio promotes osteoclast differentiation, leading to greater TRAP staining and higher osteoclast activity in conditional Wwtr1Oc-cKO null mice compared with control mice (Wwtr1+/+). Conditional deletion of Wwtr1 also resulted in increased adipocyte markers such as PPARγ2, aP2 and Lpl gene expressions (Table 1).
Unexpectedly, global Wwtr1+/− heterozygous mice, which had a ~40% reduction in Wwtr1 message expression in bone, did not have significant changes in BMD or bone volume. Single-heterozygous Wwtr1+/– showed normal bone gene expression profiles as well (Table 1). The osteoblast specific reduction but not global reductions of Wwtr1 on the skeletal phenotype points to other important co-factors in osteoblasts, such as Wwtr1 interactions with PC1, or counteracting effects of Wwtr1 in non-osteoblastic cells.
Additive effects of combined Pkd1 and Wwtr1 deficiency to reduce bone mass
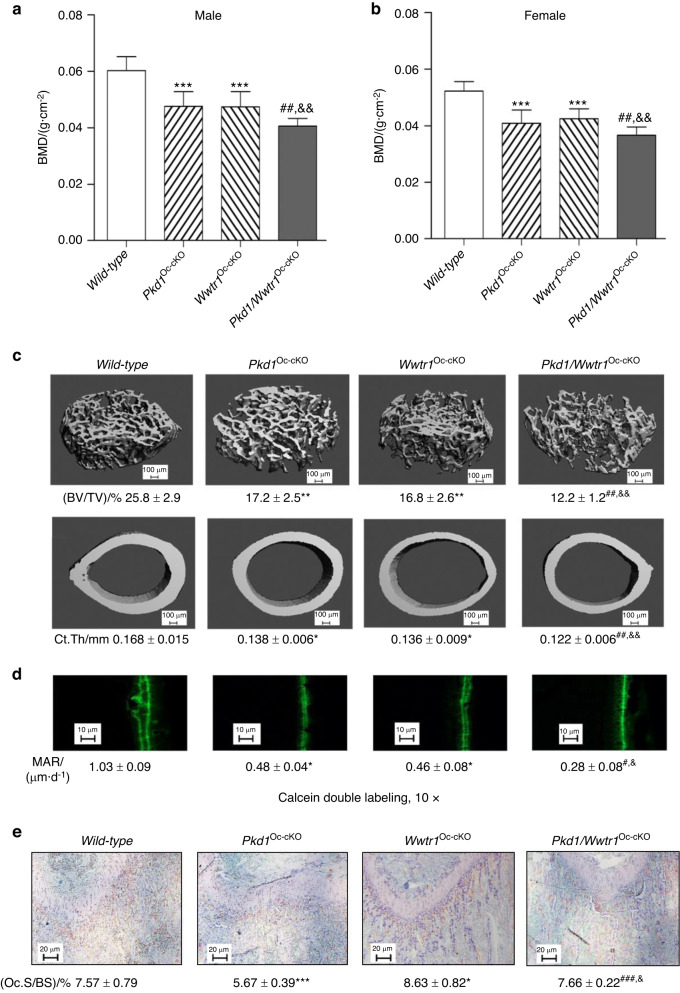
To test the functional effects of the interaction between Pkd1 and Wwtr1 in osteoblasts, we characterized Osteocalcin-Cre;Pkd1flox/−; Wwtr1flox/− (Pkd1/Wwtr1Oc-cKO) double null mice with osteoblast-specific conditional deletions of both Pkd1 and Wwtr1 in bone. In the conditional Pkd1/ Wwtr1 double knockout model, we found that Pkd1 and Wwtr1 have additive effects in reducing osteoblast-mediated bone formation. Compared with control mice (Pkd1flox/+;Wwtr1flox/+), both conditional Pkd1Oc-cKO and Wwtr1Oc-cKO null mice had similar reductions in bone mass, as evidenced by respective 22% and 21% reductions of BMD (Fig. 2a, b), 33% and 35% reductions in trabecular bone volume (Figs. 2c and S2a), and 18% and 19% reductions in cortical bone thickness (Figs. 2c and S2b) as determined by distal femur Micro-CT 3D image analysis and Goldner staining. Also, the reductions in bone mass were similar in male and female adult mice (Fig. 2a, b).
Fig. 2.
Conditional deletion of Pkd1 and Wwtr1 in mature osteoblasts on postnatal bone homeostasis. a, b Bone mineral density by DEXA scan in male and female mice. c Bone structure by micro-CT 3D images analysis from male mice. d Periosteal mineral apposition rate (MAR) by Calcein double labeling. There was a significant reduction in periosteal MAR in single Pkd1Oc-cKO or TAZOc-cKO mice compared with age-matched control mice and an even greater decrement in double Pkd1/TAZOc-cKO null mice, indicating an additive effect of PC1 and TAZ on osteoblast-mediated bone formation. e TRAP staining (red color) for osteoclast activity. Data are expressed as the mean ± S.D. from serum samples of individual mice (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001 compared with wild-type mice, #P < 0.05, ##P < 0.01, ###P < 0.001 compared with Wwtr1Oc-cKO mice, and &P < 0.05, &&P < 0.01, &&&P < 0.001 compared with Pkd1Oc-cKO mice, respectively. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
The skeletal phenotype of double Pkd1/Wwtr1Oc-cKO mice was more severe than either single Pkd1Oc-cKO or Wwtr1Oc-cKO null mice. Double Pkd1/Wwtr1Oc-cKO mice had greater losses in BMD, trabecular bone volume, and cortical bone thickness with 33%, 53%, and 27% reductions, respectively in both male and female adult mice. This indicates the additive effects of Pkd1 and Wwtr1 in postnatal bone homeostasis (Figs. 2 and S2). Consistent with lower bone mass, combined Pkd1 and Wwtr1 deficiency also resulted in additive reductions in osteoblast-related gene expressions, such as in Runx2-II, Osteocalcin, and Dmp1 (Table 2), as well as mechanosensing responsive genes such as in Wnt10b, c-Jun, and PTGS2 (Table 2). Periosteal MAR (Fig. 2d) was decreased by ~73% in conditional double Pkd1/Wwtr1Oc-cKO mice compared to controls, whereas Wwtr1Oc-cKO and Pkd1Oc-cKO single conditional knockout mice had reductions in periosteal MAR of 55% and 53%, respectively compared to control mice. Loss of either Pkd1 or Wwtr1 resulted in enhanced marrow adipogenesis, but no additive effects on adipocyte differentiation-related gene expressions were observed in the conditional double Pkd1/Wwtr1Oc-cKO mice (Table 2).
Table 2.
Gene-expression profiles in bone from Pkd1Oc-cKO or/and Wwtr1Oc-cKO mice
| Gene | Wild-type | Pkd1Oc-cKO | Wwtr1Oc-cKO | Pkd1/Wwtr1Oc-cKO | P-value |
|---|---|---|---|---|---|
| Osteoblast lineage | |||||
| Pkd1 | 1.00 ± 0.11 | 0.41 ± 0.10*** | 0.93 ± 0.31 | 0.45 ± 0.10&&& | <0.000 1 |
| Wwtr1 | 1.00 ± 0.24 | 0.98 ± 0.27 | 0.46 ± 0.10*** | 0.43 ± 0.15### | <0.000 1 |
| Pkd2 | 1.00 ± 0.12 | 1.05 ± 0.21 | 0.93 ± 0.25 | 1.09 ± 0.31 | 0.627 1 |
| Runx2-II | 1.00 ± 0.14 | 0.76 ± 0.11** | 0.75 ± 0.10* | 0.62 ± 0.21#,& | 0.001 7 |
| Osteocalcin | 1.00 ± 0.13 | 0.75 ± 0.15** | 0.78 ± 0.11** | 0.66 ± 0.11#,& | 0.001 0 |
| FGF-23 | 1.00 ± 0.13 | 0.55 ± 0.17* | 2.30 ± 0.56*** | 1.02 ± 0.25 | <0.000 1 |
| Dmp1 | 1.00 ± 0.20 | 0.80 ± 0.10* | 0.78 ± 0.14* | 0.65 ± 0.10#,& | 0.001 2 |
| Wnt10b | 1.00 ± 0.04 | 0.77 ± 0.20** | 0.73 ± 0.19** | 0.63 ± 0.11#,& | 0.001 4 |
| Wnt1 | 1.00 ± 0.21 | 0.70 ± 0.16* | 0.92 ± 0.35 | 0.52 ± 0.21#,&&& | 0.000 7 |
| FzD2 | 1.00 ± 0.33 | 0.92 ± 0.17 | 0.93 ± 0.24 | 0.55 ± 0.19#,& | 0.001 6 |
| c-Jun | 1.00 ± 0.24 | 0.95 ± 0.28 | 1.07 ± 0.22 | 0.62 ± 0.12#,&& | 0.000 9 |
| c-Fos | 1.00 ± 0.20 | 0.92 ± 0.21 | 0.74 ± 0.15* | 0.67 ± 0.13### | 0.001 1 |
| Axin-2 | 1.00 ± 0.31 | 0.77 ± 0.10* | 0.88 ± 0.20 | 0.67 ± 0.13& | 0.001 2 |
| PTGS2 | 1.00 ± 0.27 | 0.70 ± 0.15* | 0.68 ± 0.16** | 0.44 ± 0.19#,& | <0.000 1 |
| OPG | 1.00 ± 0.19 | 0.93 ± 0.26 | 1.08 ± 0.26 | 0.97 ± 0.18 | 0.708 0 |
| RankL | 1.00 ± 0.15 | 0.73 ± 0.14* | 1.45 ± 0.13*** | 0.89 ± 0.18#,&&& | <0.000 1 |
| CYR61 | 1.00 ± 0.13 | 1.40 ± 0.18* | 1.94 ± 0.51*** | 1.67 ± 0.33 | 0.000 5 |
| CTGF | 1.00 ± 0.10 | 1.38 ± 0.23* | 1.68 ± 0.30** | 1.55 ± 0.36 | 0.001 4 |
| Osteoclast | |||||
| Trap | 1.00 ± 0.15 | 0.63 ± 0.14** | 1.43 ± 0.20*** | 0.93 ± 0.14##,&&& | <0.000 1 |
| Mmp9 | 1.00 ± 0.14 | 0.63 ± 0.08* | 0.86 ± 0.18 | 0.95 ± 0.35# | 0.031 7 |
| Chondrocyte | |||||
| Collagen II | 1.00 ± 0.19 | 0.92 ± 0.24 | 0.73 ± 0.15* | 0.51 ± 0.07##,& | <0.000 1 |
| VegfA | 1.00 ± 0.38 | 1.74 ± 0.22* | 1.66 ± 0.49* | 1.70 ± 0.57 | 0.014 3 |
| Adipocyte | |||||
| PPARγ2 | 1.00 ± 0.20 | 1.78 ± 0.34** | 1.88 ± 0.51** | 1.61 ± 0.22 | 0.001 0 |
| aP2 | 1.00 ± 0.16 | 1.71 ± 0.33*** | 2.09 ± 0.31*** | 1.77 ± 0.38 | <0.000 1 |
| Lpl | 1.00 ± 0.13 | 1.62 ± 0.28** | 2.01 ± 0.51*** | 1.88 ± 0.38 | 0.000 4 |
Data are mean ± S.D. from 6 tibias of wild-type control, Pkd1Oc-cKO, Wwtr1Oc-cKO, and Pkd1/Wwtr1Oc-cKO mice and expressed as the fold changes relative to the housekeeping gene β-actin subsequently normalized to control mice. *P < 0.05, **P < 0.01, ***P < 0.001 compared with wild-type mice, #P < 0.05, ##P < 0.01, ###P < 0.001 compared with Wwtr1Oc-cKO mice, and &P < 0.05, &&P < 0.01, &&&P < 0.001 compared with Pkd1Oc-cKO mice, respectively. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
We found that the conditional deletion of Pkd1 or Wwtr1 in osteoblasts has opposite effects on osteoclast activities. There was a decreased RankL/OPG expression ratio and TRAP immunostaining in Pkd1Oc-cKO mice but an increased RankL/OPG expression ratio and TRAP immunostaining in Wwtr1Oc-cKO mice (Table 2 and Fig. 2e). In contrast, double Pkd1/Wwtr1Oc-cKO had similar RankL expression and TRAP immunostaining when compared to control, indicating a recovery of osteoclast activities in the double null mice (Table 2 and Fig. 2e). Changes in gene expression and immunostaining in bone correlated with alterations in serum biomarkers (Table 3). In this regard, further evidence for osteoblast and osteocyte dysfunction includes reductions in serum osteocalcin and FGF-23 from in single Pkd1Oc-cKO or Wwtr1Oc-cKO mice compared with age-matched control mice and an even greater decrement in double Pkd1/Wwtr1Oc-cKO null mice (Table 3). In contrast, serum levels of TRAP, a marker of bone resorption, were decreased in single Pkd1Oc-cKO mice, increased in single Wwtr1Oc-cKO mice, but restored in double Pkd1/Wwtr1Oc-cKO null mice compared with control littermates (Table 3). In addition, serum level of leptin was significantly higher in single Pkd1Oc-cKO or Wwtr1Oc-cKO mice than age-matched control mice. However, we did not observe further increase in double Pkd1/Wwtr1Oc-cKO null mice (Table 3). These findings suggest that Pkd1 and Wwtr1 have distinct functions among osteoblasts, adipocytes, and osteoclasts in bone in vivo.
Table 3.
Biochemistry analysis of serum from Pkd1Oc-cKO or/and Wwtr1Oc-cKO mice
| Genotype | Wild-type | Pkd1Oc-cKO | Wwtr1Oc-cKO | Pkd1/Wwtr1Oc-cKO | P-value |
|---|---|---|---|---|---|
| Urea nitrogen/(mg·dL−1) | 19 ± 5 | 22 ± 5 | 21 ± 6 | 22 ± 4 | 0.723 6 |
| Calcium/(mg·dL−1) | 10.3 ± 0.9 | 10.4 ± 1.3 | 10.2 ± 1.5 | 9.4 ± 0.9 | 0.399 0 |
| Phosphorus/(mg·dL−1) | 10.1 ± 1.9 | 10.4 ± 1.4 | 9.8 ± 2.4 | 10.3 ± 1.5 | 0.935 5 |
| Osteocalcin (ng/mL) | 17.5 ± 3.5 | 3.4 ± 0.7*** | 10.7 ± 2.6*** | 1.0 ± 0.8#,&&& | <0.000 1 |
| TRAP/(U·L−1) | 10.8 ± 1.3 | 7.6 ± 0.6*** | 12.2 ± 1.9* | 9.8 ± 1.1##,&& | <0.000 1 |
| FGF23/(pg·mL−1) | 117 ± 32 | 79 ± 16* | 116 ± 35 | 46 ± 16#,&&& | 0.070 4 |
| Leptin/(pg·mL−1) | 836 ± 200 | 2 636 ± 759*** | 2 649 ± 915*** | 1 793 ± 600###,&& | <0.000 1 |
Data are mean ± S.D. from 6 serum samples of wild-type control, Pkd1Oc-cKO, Wwtr1Oc-cKO, and Pkd1/Wwtr1Oc-cKO mice. *P < 0.05, **P < 0.01, ***P < 0.001 compared with wild-type mice, #P < 0.05, ##P < 0.01, ###P < 0.001 compared with Wwtr1Oc-cKO mice, and &P < 0.05, &&P < 0.01, &&&P < 0.001 compared with Pkd1Oc-cKO mice, respectively. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
Loss of mechanical loading response in conditional Pkd1 and Wwtr1 deficient mice
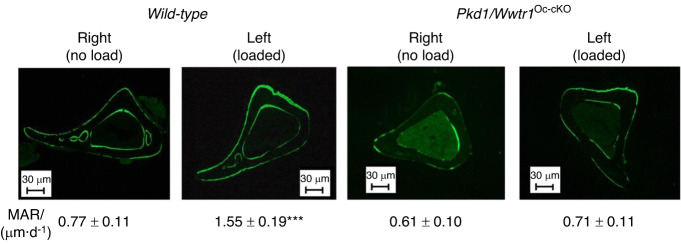
The cross-sections of tibiae from control and double Pkd1/Wwtr1Oc-cKO null mice after mechanical tibia loading studies in vivo are shown in Fig. 3. In wild-type control mice, loaded tibia showed a 2-fold increase in periosteal mineral apposition rate. In contrast, there was no measurable increase in periosteal mineral apposition in the loaded tibia from double Pkd1/Wwtr1 knockout mice (Fig. 3). In addition, a real-time RT-PCR analysis revealed that loaded tibia from the control mice had a dramatic response to mechanical stimulation, evidenced by significant increases of mechanosensing (e.g., Wnt10b, FzD2, Axin2, PTGS2, c-Jun, c-Fos, and Runx2-II) and osteogenic (e.g., Osteocalcin, Alp, Collagen I, and Dmp1) but decreases of adipogenic (e.g., PPARγ2, aP2, and Lpl) gene expressions when compared with no load control tibia. In contrast, even when using the same loading regimen, no changes of mechanosensing, osteogenic, and adipogenic gene expression profiles were observed in the loaded tibia from double Pkd1/ Wwtr1Oc-cKO null mice when compared with no load control tibia (Table 4). Thus, PC1 and Wwtr1 are important in mediating mechanotransduction in bone.
Fig. 3.
An impairment of anabolic response to mechanical loading in conditional Pkd1 and Wwtr1 deletion in bone. Representative images of midshaft tibia cross sections from no-load and loaded ulnae of wild-type control and compound Pkd1/Wwtr1Oc-cKO null mice after loading. Data are mean ± S.D. from 6 tibias of wild-type control and Pkd1/Wwtr1Oc-cKO mice. ***P < 0.001 compared with wild-type mice. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
Table 4.
Mechanosensing, osteogenic, and adipogenic gene-expression profiles that respond to mechanical loading in wild-type and double Pkd1/Wwtr1Oc-cKO mice
| Gene | Wild-type (no load) | Wild-type (loaded) | Pkd1/Wwtr1Oc-cKO (no load) | Pkd1/Wwtr1Oc-cKO (loaded) |
|---|---|---|---|---|
| Wnt10b | 1.00 ± 0.32 | 1.74 ± 0.62** | 1.00 ± 0.31 | 0.94 ± 0.31 |
| c-Jun | 1.00 ± 0.27 | 1.72 ± 0.41*** | 1.00 ± 0.18 | 0.95 ± 0.29 |
| c-Fos | 1.00 ± 0.22 | 2.04 ± 0.66*** | 1.00 ± 0.24 | 0.92 ± 0.29 |
| Axin-2 | 1.00 ± 0.16 | 1.54 ± 0.25*** | 1.00 ± 0.19 | 0.92 ± 0.33 |
| PTGS2 | 1.00 ± 0.16 | 3.34 ± 1.05*** | 1.00 ± 0.22 | 1.15 ± 0.17 |
| Runx2-II | 1.00 ± 0.28 | 1.94 ± 0.54*** | 1.00 ± 0.19 | 0.92 ± 0.19 |
| FzD2 | 1.00 ± 0.26 | 1.92 ± 0.47*** | 1.00 ± 0.26 | 0.93 ± 0.32 |
| Osteocalcin | 1.00 ± 0.34 | 5.26 ± 1.58*** | 1.00 ± 0.36 | 1.77 ± 0.41 |
| Alp | 1.00 ± 0.32 | 2.82 ± 1.05*** | 1.00 ± 0.30 | 0.92 ± 0.29 |
| Collagen I | 1.00 ± 0.25 | 4.93 ± 1.59*** | 1.00 ± 0.58 | 1.01 ± 0.33 |
| Dmp1 | 1.00 ± 0.32 | 1.83 ± 0.29*** | 1.00 ± 0.34 | 1.03 ± 0.27 |
| PPARγ2 | 1.00 ± 0.40 | 0.24 ± 0.12*** | 1.00 ± 0.21 | 0.86 ± 0.28 |
| aP2 | 1.00 ± 0.25 | 0.57 ± 0.14*** | 1.00 ± 0.18 | 0.89 ± 0.17 |
| Lpl | 1.00 ± 0.32 | 0.35 ± 0.11*** | 1.00 ± 0.30 | 0.88 ± 0.30 |
Data are mean ± S.D. from 6 tibiae of wild-type control and Pkd1/Wwtr1Oc-cKO mice and expressed as the fold changes relative to the housekeeping gene eEF1a1 subsequently normalized to control or Pkd1/Wwtr1Oc-cKO no load tibiae. *, **, *** indicates significant difference from wild-type or Pkd1/Wwtr1Oc-cKO no load control at P < 0.05, P < 0.01, P < 0.001
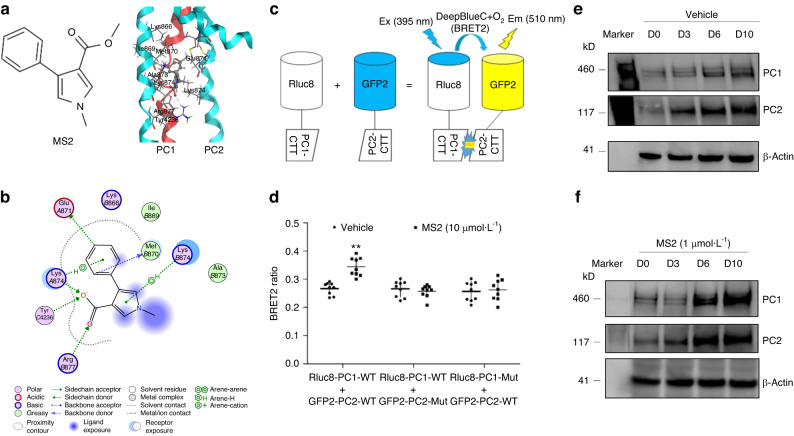
Validation of MS2 key binding to residues in PC1/PC2 C-terminus tails
We have previously showed that the small molecule MS2 activates PC1/PC2 complex signaling.2 Using computational modeling, we engaged in an induced fit docking campaign and predicted several potential ligand binding complexes. From these predicted poses, we identified key residues in the PC1 and PC2 C-terminus tail regions with which MS2 is predicted to interact. As shown in Fig. 4, the PC2-CTT binding site for MS2 is predicted to include Lysine874 and Arginine877, whereas the PC1-CTT binding site for MS2 involves Tyrosine4236 (Fig. 4a, b). To test these predictions, we performed site-mutagenesis of key residues in both PC1 and PC2 and tested the effects of MS2 on PC1 and PC2 assembly using a BRET2 assay. We observed that the compound MS2 markedly enhances the BRET2 signal in wild-type constructs, while mutagenesis of key residues in either PC1-CTT or PC2-CTT constructs completely abolished the BRET2 signal in the presence of compound MS2, confirming the role of MS2 in enhancing PC1 and PC2 interactions and lending support that MS2 is binding to the protein complex (Fig. 4c, d).
Fig. 4.
Cell-based BRET2 assays for MS2-target engagement assays. a Chemical structure of MS2 and an example of predicted 3D binding mode for MS2 (ball-and-sticks rendering in CPK colors) in PC2 (light blue). PC1 (red) as bound to PC2 in the homology structure is superimposed. b An example of calculated 2D binding mode and residues for MS2 in PC1/PC2 C-tails. c A diagram of BRET2 constructs and reactions in the presence of DeepBlue C with or without MS2 stimulation. d BRET2 signal changes from wild-type and mutant constructs with or without MS2 incubation. e, f Time-dependent changes of PC1 and PC2 proteins with or without MS2 incubation during osteogenic differentiation cultures in MC3T3-E1 cell line. Incubation Data are presented as the mean ± SD from 3 independent experiments (n = 3). **P < 0.01 compared with vehicle control
Next, we examined PC1/PC2 complex formation during MC3T3-E1 osteoblast differentiation in vitro. We observed culture duration dependent increase of PC1/PC2 complex formation by western blot analysis. Incubation with 1 μmol·L−1 of MS2 in osteogenic cultures markedly increased the amount of PC1 and PC2 protein as assessed by western blot analysis (Fig. 4e, f). These data suggests that MS2 may molecularly stabilize the PC1/PC2 complex in osteoblast culture in vitro.
Loss of MS2-mediated stimulated increase in bone mass in conditional Pkd1 and Wwtr1 deficiency mice
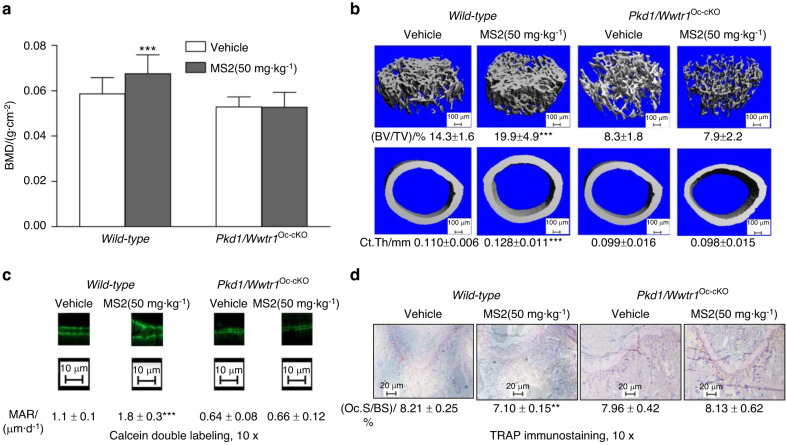
Finally, we treated wild-type and conditional double Pkd1/Wwtr1Oc-cKO null mice with vehicle or MS2 (50 mg·kg−1) i.p. daily and assessed their skeletal response. After only 2 weeks, we observed that wild-type control mice treated with MS2 had a 15% increment in femoral bone mineral density compared to vehicle control (Fig. 5). Micro-CT 3D images revealed that MS2 treated wild-type mice had a 39% increase in trabecular bone volume and 16% increase in cortical bone thickness.
Fig. 5.
Effects of MS2 on bone formation in wild-type and compound Pkd1/Wwtr1Oc-cKO null mice. a Bone mineral density by DEXA scan. b Bone structure by micro-CT 3D images analysis. c Periosteal mineral apposition rate (MAR) by Calcein double labeling. d Osteoclast activities by TRAP immunostaining after MS2 (50 mg·kg−1) treatment for 4 weeks compared to vehicle control. Data are mean ± S.D. from 6 tibias of wild-type control and compound Pkd1/Wwtr1Oc-cKO null mice. *P < 0.05, **P < 0.01, ***P < 0.001 compared with wild-type control mice. P values were determined by 1-way ANOVA with Tukey’s multiple-comparisons test
In contrast, administration of MS2 had no effects on bone mineral density and bone structure in double Pkd1/Wwtr1Oc-cKO null mice, suggesting specific-target dependent effects of MS2 on polycystins/Wwtr1 signaling (Fig. 5). We also observed that there were 1.6-fold increases in bone formation rate in wild-type mice treated with MS2 compared to the vehicle control, in agreement with enhanced osteoblastogenesis (e.g., Runx2-II, Osteocalcin, ALP and Dmp1) and suppressed marrow adipogenesis (e.g., PPARγ2, aP2, and Lpl) by a real-time RT-PCR analysis (Table 5 and Fig. 5). Again, administration of MS2 had no effects on bone formation rate and bone gene expression profiles in double Pkd1/Wwtr1Oc-cKO null mice. Furthermore, MS2 stimulated mechanosensing gene expressions, including Wnt1, Wnt10b, Axin-2, FzD2, c-Jun, c-Fos, eNOS, and PTGS2, consistent with MS2 acting as a small molecule “mechanomimetic”. MS2 treatment decreased RankL/OPG expression ratio and TRAP immunostained osteoclasts in the MS2-treated mice compared to vehicle control (Table 5 and Fig. 5). Administration of MS2 had no effects on osteoblast-mediated bone formation rate, marrow adipogenesis, and osteoclast activity in conditional double Pkd1/Wwtr1Oc-cKO null mice (Table 5 and Fig. 5). These data support that MS2 functions as anabolic drugs through the polycystins/Wwtr1 pathway to promote the bone remodeling process.
Table 5.
Gene expression profiles in bone from MS2-treated wild-type control and Pkd1/Wwtr1Oc-cKO mice
| Gene | Wild-type (Vehicle) | Wild-type (MS2) | Pkd1/Wwtr1Oc-cKO (Vehicle) | Pkd1/Wwtr1Oc-cKO (MS2) |
|---|---|---|---|---|
| Pkd1 | 1.00 ± 0.13 | 0.94 ± 0.15 | 1.00 ± 0.37 | 1.03 ± 0.28 |
| Wwtr1 | 1.00 ± 0.32 | 1.01 ± 0.32 | 1.00 ± 0.40 | 0.82 ± 0.19 |
| Runx2-II | 1.00 ± 0.19 | 1.79 ± 0.52** | 1.00 ± 0.32 | 1.03 ± 0.28 |
| Osteocalcin | 1.00 ± 0.24 | 1.68 ± 0.24*** | 1.00 ± 0.57 | 0.86 ± 0.61 |
| ALP | 1.00 ± 0.14 | 1.56 ± 0.23*** | 1.00 ± 0.52 | 1.09 ± 0.55 |
| Dmp1 | 1.00 ± 0.18 | 1.45 ± 0.17*** | 1.00 ± 0.33 | 1.02 ± 0.48 |
| OPG | 1.00 ± 0.38 | 1.10 ± 0.37 | 1.00 ± 0.30 | 1.18 ± 0.33 |
| RankL | 1.00 ± 0.30 | 0.57 ± 0.15* | 1.00 ± 0.50 | 0.99 ± 0.33 |
| Trap | 1.00 ± 0.26 | 0.56 ± 0.10* | 1.00 ± 0.58 | 0.93 ± 0.28 |
| PPARγ2 | 1.00 ± 0.43 | 0.59 ± 0.11* | 1.00 ± 0.47 | 1.06 ± 0.18 |
| aP2 | 1.00 ± 0.22 | 0.60 ± 0.26* | 1.00 ± 0.22 | 1.07 ± 0.12 |
| Lpl | 1.00 ± 0.28 | 0.63 ± 0.12* | 1.00 ± 0.40 | 0.95 ± 0.39 |
| Wnt1 | 1.00 ± 0.30 | 3.02 ± 0.98*** | 1.00 ± 0.60 | 0.97 ± 0.40 |
| Wnt10b | 1.00 ± 0.15 | 1.52 ± 0.25*** | 1.00 ± 0.40 | 1.12 ± 0.21 |
| FzD2 | 1.00 ± 0.26 | 3.36 ± 1.56*** | 1.00 ± 0.30 | 0.95 ± 0.18 |
| c-Fos | 1.00 ± 0.20 | 1.47 ± 0.34* | 1.00 ± 0.22 | 1.00 ± 0.27 |
| c-Jun | 1.00 ± 0.23 | 1.74 ± 0.24*** | 1.00 ± 0.29 | 0.84 ± 0.20 |
| Axin-2 | 1.00 ± 0.22 | 2.57 ± 1.02** | 1.00 ± 0.16 | 0.92 ± 0.17 |
| PTGS2 | 1.00 ± 0.35 | 2.25 ± 0.78** | 1.00 ± 0.26 | 0.90 ± 0.22 |
| eNOS | 1.00 ± 0.45 | 3.09 ± 0.68*** | 1.00 ± 0.13 | 1.03 ± 0.22 |
Data are mean ± S.D. from 6 tibias of wild-type control and Pkd1/Wwtr1Oc-cKO mice and expressed as the fold changes relative to the housekeeping gene eEF1a1 subsequently normalized to control or Pkd1/Wwtr1Oc-cKO no load tibias. *, **, *** indicates significant difference from wild-type control at P < 0.05, P < 0.01, P < 0.001
Discussion
In the current study, we found that conditional deletion of both Pkd1 and Wwtr1 in osteoblasts using Oc-Cre in double Pkd1/Wwtr1Oc-cKO null mice resulted in a more severe skeletal phenotype than loss of either PC1 or WWTR1 alone in the single Pkd1Oc-cKO or Wwtr1Oc-cKO null mice. Indeed, double Pkd1/Wwtr1Oc-cKO null mice had greater reductions of bone mineral density and periosteal mineral apposition rate compared to single Wwtr1Oc-cKO or Pkd1Oc-cKO null mice. Micro-CT 3D image analysis revealed that this was due to greater loss in both trabecular bone volume and cortical bone thickness in double Pkd1/ Wwtr1Oc-cKO null mice compared to single Pkd1Oc-cKO or Wwtr1Oc-cKO null mice. Double Pkd1/ Wwtr1Oc-cKO null mice also exhibited greater reductions in osteoblast-related message expression in bone compared to single Pkd1Oc-cKO or Wwtr1Oc-cKO null mice. Moreover, double Pkd1/Wwtr1Oc-cKO null mice in osteoblasts resulted in greater reductions in mechanosensing responsive gene expressions in physiological conditions and failed to respond to mechanical loading in vivo. Control mice treated with the PC1/PC2 agonist MS2 had significant increases in femoral BMD and periosteal MAR, whereas MS2 had no anabolic effects on double Pkd1/Wwtr1Oc-cKO mice. Thus, both loss-of-function genetic and gain-of-function pharmacological studies support co-dependent functions of PC1 and Wwtr1 in regulating osteoblast-mediated bone formation and post-natal bone homeostasis. These functional interactions are consistent with the fact that Wwtr1 binds to the C-terminal tail of PC1 and translocates to the nucleus in response to PC1/PC2 activation.2,17
The PC1/Wwtr1 signaling pathway may impact bone formation and bone mass by several cellular mechanisms. First, both PC1 and Wwtr1 have a direct role in osteoblast function and differentiation. In previous work we and others confirmed that conditional deletion of either Pkd1 or Wwtr1 in osteoblasts has a direct role in osteoblast-mediated bone formation.6,25 Our current studies extend these findings by demonstrating the additive osteopenia phenotypes in double Pkd1/Wwtr1Oc-cKO null mice are due to direct loss of both PC1 and/or Wwtr1 signaling in the osteoblast lineage. In addition, control mice treated with MS2 that targets the PC1/PC2/Wwtr1 complex had anabolic effects on osteoblast-mediated bone formation and increased bone mass. The magnitude of the anabolic bone response to MS2 was similar to increments in bone mass in mice treated with PTH analogues and RANKL blocking antibodies that were developed to treat osteoporosis.26 However, this response required PC1 and Wwtr1 co-dependent signaling, since the administration of MS2 had no effects on bone formation in double Pkd1/Wwtr1Oc-cKO null mice. These genetic loss-of-function and pharmacological gain-of-function studies indicate a functional interaction in osteoblast activity between PC1 and Wwtr1 in bone under normal physiological conditions.
Second, PC1 and Wwtr1 function as a mechanosensor in osteoblasts and osteocytes. We reported that conditional deletion of Pkd1 in osteocytes markedly attenuated mechanical loading-induced bone formation.3 The Wwtr1/Yap Hippo pathway is known to play an essential role in mechanosensing of alterations in cell stiffness and extracellular matrix.13–16 Recent studies demonstrated that mechanical loading activates the Wwtr1/Yap pathway27–30 and enhances Wwtr1/Yap nuclear translocation in response to shear stress in both bone marrow mesenchymal stem cells and MLO-Y4 osteocyte-like cell line.31,32 Thus, loss of mechanosensing responses in double Pkd1/Wwtr1Oc-cKO null mice can be explained by loss of Wwtr1 interactions with PC1 and their co-dependent signaling. Indeed, we confirmed that PC1-CTT interacted with PC2-CTT to produce BRET2 signal and that the small molecule MS2 enhanced this interaction and stimulated mechanosensing gene expressions like mechanical loading in wild-type control mice acting as a “mechanomimetic”. Prior in vitro studies showed that overexpression of either full-length PC1 or PC1-CTT along with full-length PC2 constructs markedly increased Wwtr1-induced activation of TEAD activity and that the cleavage of the PC1-C-Tail/Wwtr1 complex by mechanical stretch-induced activation of γ-secretase translocates to the nucleus to stimulate Runx2- and Wwtr1-mediated gene transcription, consistent with our proposition that PC1/Wwtr1 functions as a mechanosening complex in bone.2,17
Third, PC1 and Wwtr1 have opposite effects on osteoclast mediated bone resorption. Global Pkd1 heterozygous mice showed fewer osteoclasts in bone as evidenced by a lower number of TRAP positive osteoclasts,2,3,6 consistent with lower TRAP levels in ADPKD patients compared to non-ADPKD control.10–12,33 Conditional Pkd1Oc-cKO null mice in osteoblast lineages with one global null and one conditional knockout alleles had greater reductions in osteoclast activities,3,6 indicating a gene-dosage dependent effect of loss-of-PC1 in osteoblasts impacting osteoclast functions, likely through paracrine effects.34 Whereas global Wwtr1 heterozygous mice exhibited no changes in osteoclast activities compared to wild-type control mice,2 in our current study conditional Wwtr1oc-cKO null mice had higher osteoclast activities in bone as evidenced by increased number of TRAP positive osteoclasts. In contrast, double Pkd1/Wwtr1Oc-cKO mice had normal osteoclast parameters, consistent with offsetting effects of conditional deletion of Pkd1 and/or Wwtr1 in osteoblasts on osteoclasts. These findings agree with previous publications regarding the effects of osteoblast-specific deletion of Pkd12,3,6–8 or Wwtr125,27,35 on osteoclast activity. Indeed, osteocytes can regulate osteoclast activity through the RANKL/OPG paracrine pathway.36,37 PC1 and Wwtr1 deficiency resulted in respective decrease and increase in RANKL expression but no difference in OPG expression in the Pkd1Oc-cKO null and Wwtr1Oc-cKO null mice, this could account for the differential effects of PC1 and Wwtr1 on osteoclast activity in bone. Moreover, MS2 significantly decreased RANKL/OPG ratio bone and attenuated osteoclast activity. Our understanding of Wwtr1 regulation of osteoclast function is further supported by the observation by Yang et al. that either global or osteoclast-specific knockout of Wwtr1 led to a low-bone mass phenotype due to elevated osteoclast formation.38 Thus, PC1 and Wwtr1 signaling have divergent effects on osteoclasts and bone resorption.
Finally, PC1 and Wwtr1 regulate adipogenesis in bone marrow. We have previously reported that global PC1 deficiency in mice has an inverse effect, inhibiting osteoblastogenesis and enhancing adipogenesis.2,7 Similar to conditional Pkd1Oc-cKO null mice,3,6 we also observed conditional Wwtr1Oc-cKO null mice had greater increments in adipogenic markers than global or conditional Wwtr1 heterozygous mice in the current study, suggesting a gene-dosage dependent effect of loss-of-Wwtr1 in osteoblasts on bone marrow adipogenesis. Consistent with these loss-of-function results, administration of MS2 or mechanical loading in wild-type control mice suppressed marrow adipogenesis but had no effects in double Pkd1/Wwtr1Oc-cKO null mice. Interestingly, we found a similar increase in adipogenic markers in both PC1 and/or Wwtr1 osteoblast conditional knockout mice, consistent with the phenotypes of senile osteoporosis characterized by decreased osteoblastogenesis and increased adipogenesis leading to increased bone marrow fat.39–42 The increase of adipogenic markers could be theoretically explained by increased transdifferentiation of osteoblast precursors to adipocytes,43,44 or effects of PC1 and Wwtr1 in osteoblasts/osteocytes differentially releasing paracrine factors that modulate adipogenesis,45–47 analogous to paracrine factors that regulate osteoclastogenesis. Regardless, our studies revealed that double Pkd1/Wwtr1Oc-cKO null mice had no differences in adipogenic markers relative to single Pkd1Oc-cKO or Wwtr1Oc-cKO null mice, suggesting that PC1 and Wwtr1 regulate adipocyte differentiation through the common polycystins/Wwtr1/PPARγ pathway.2
In conclusion, polycystins and Wwtr1 have interdependent effects in mediating bone mechanotransduction, regulating osteoblast mediated bone formation, and the anabolic skeletal responses to a small molecule mechanomimetic and mechanical loading. These observations suggest that the PC1/PC2/ Wwtr1 complex functions through a common pathway and is a novel drug target to treat age-related osteoporosis.48,49
Materials and methods
Mice
All animal studies were conducted according to guidelines provided by the National Institutes of Health and the Institute of Laboratory Animal Resources, National Research Council. The University of Tennessee Health Science Center’s Animal Care and Use Committee approved all animal studies (Protocol number: 21-0301). We obtained the floxed Wwtr1 mice (Wwtr1flox/flox) which harbors two loxP sites flanking exon 2 of the Wwtr1 gene from Drs. Jeff Wrana and Helen McNeill.50 We obtained the floxed Pkd1 mice (Pkd1flox/flox) from Dr. Gregory Germino at Johns Hopkins University51 and Oc (Osteocalcin)-Cre transgenic mice from Dr. Thomas Clemens at the University of Alabama.52 These mice were bred and maintained on a C57BL/6 J background. At first, we used Oc-Cre; Wwtr1+/− mice and homozygous Wwtr1flox/flox mice to generate conditional Wwtr1 heterozygous Oc-Cre; Wwtr1flox/+ (Wwtr1Oc-Het) and homozygous Oc-Cre; Wwtr1flox/− (Wwtr1Oc-cKO) null mice as well as Wwtr1 heterozygous mice (Wwtr1+/−) and Oc-Cre negative control mice (Wwtr1flox/+ equivalent to wild-type). Second, we also used Oc-Cre;Pkd1+/− mice and homozygous Pkd1flox/flox mice to generate Oc-Cre;Pkd1flox/− (Pkd1Oc-cKO) mice. Then we used Oc-Cre;Pkd1+/−;Wwtr1+/− mice and homozygous Pkd1flox/flox;Wwtr1flox/flox mice to generate Oc-Cre;Pkd1flox/−;Wwtr1flox/− (Pkd1/Wwtr1Oc-cKO) mice as previously described.6,9 These mice were used for skeletal phenotype analysis. The mice were anesthetized with Ketamine (90 mg·kg−1) and Xylazine (10 mg·kg−1) for LUNARPIXIMUS bone densitometer scan, and the mice not useful for experimental purposes were sacrificed by CO2 inhalation followed by cervical dislocation. In addition, we used wild-type C57BL/6 J mice at 8 weeks of age to examine the effects of MS2 on osteogenesis and adipogenesis in vivo. The mice were treated with intraperitoneal injection of MS2 (50 mg·kg−1) i.p. or vehicle control (5% DMSO in PBS solution) once a day for 4 weeks. The bone samples were collected 4 h after the last dose administration.
Bone densitometry, histomorphometric and micro-CT analysis
BMD of femurs was assessed at 8 weeks of age using a LUNARPIXIMUS bone densitometer (Lunar Corp, Madison, WI). Calcein (Sigma, St. Louise, MO) double labeling of bone and histomorphometric analyses of periosteal mineral apposition rate (MAR) in tibias and osteoclast surface per bone surface (Oc.S/BS, %) in femurs by TRAP immunostaining were performed using the osteomeasure analysis system (Osteometrics). Bone Masson-Goldner staining was performed by the Histology and histomorphometry Core iLab service from Indiana University School of Medicine. The distal femoral metaphyses were also scanned using a Scanco μCT 40 (Scanco Medical AG, Brüttisellen, Switzerland). A 3D images analysis was done to determine bone volume (BV/TV) and cortical thickness (Ct.Th) as previously described.3,4,6
Real-time quantitative reverse transcription PCR (real-time qRT-PCR) and western blot analysis
For real-time qRT−PCR, 1.0 μg total RNA isolated from either the long bone of 6-week-old mice or 8-days cultured BMSCs in differentiation media was reverse transcribed as previously described.4,6 PCR reactions contained 20 ng template (cDNA), 375 nmol·L−1 each forward and reverse primers, and 1 X EvaGreen Supermix (Bio-Rad, Hercules, CA) in 10 μL. The threshold cycle (Ct) of tested-gene product from the indicated genotype was normalized to the Ct for cyclophilin A. Then the tested-gene product vs cyclophilin A is normalized to the mean ratio of wild-type or control group, which has been set to 1.
For Western blot analysis, protein concentrations of the supernatant were determined with a total protein assay kit (Bio-Rad, Hercules, CA). Equal quantities of protein were subjected to 4%–12% Bis-Tris or 3%–8% Tris-Acetate gradient Gels (Invitrogen, Carlsbad, CA) and were analyzed with standard western blot protocols as previously described.4,6 Polycystin-1 antibody (7E12, sc-130554), Polycystin-1 antibody (C-20, sc-10372), Polycystin-2 antibody (H-280, sc-25749), and Polycystin-2 antibody (YCE2, sc-47734) were purchased from Santa Cruz Biotechnology (Paso Robles, CA). Purified mouse Wwtr1 antibody (560236) was purchased from BD Biosciences (San Jose, CA). Phosphorylated p-Wwtr1 (Ser 89, sc-17610) and β-actin antibody (sc-47778) antibodies were from Santa Cruz Biotechnology (Paso Robles, CA). The intensity of the bands was quantified using Image J software (http://rsb.info.nih.gov/ij/).
BRET2 assays for target engagement assay in vitro
In collaboration with Oak Ridge National Laboratory and The University of Tennessee, Knoxville, we previously identified a compound that is thought to bind to the polycystin1 (PC1-CTT)/polycystin2 (PC2-CTT) complex in their C-terminus tails that we refer to as molecular staple two (MS2).2 Here, using computational ligand docking with an initial rigid receptor search using the proxy triangle algorithm and London dG scoring function53 and subsequent induced-fit refinement using a “free” receptor geometry, the Amber14SB:EHT force-field,54,55 and GBVI/WSA scoring function, as implemented in the Molecular Operating Environment (MOE, 2022.02 Chemical Computing Group ULC, 910–1010 Sherbrooke St. W., Montreal, QC H3A 2R7, Canada, 2023) software package, against our previously identified binding pocket (in a previously published homology model of the PC1/PC2 CTT domain) we predicted over 30 different potential ligand-protein complexes. Investigation of these complexes identified two classes of binding poses, those dominated by PC2-ligand contacts and poses with PC1/PC2 bridging contacts. From the poses containing the largest number of ligand-protein contacts, we identified key residues of MS2 interacting with PC2-CTT [e.g., Arg(R)877 and Lys(K)874] and PC1-CTT [e.g., Tyr(Y)4236]. To confirm the interaction between MS2 and PC1/PC2 C-tail complex (1:3), we used PC1-CTT and PC2 C-terminal tail (PC2-CTT) cDNAs to create Rluc8_PC1-CTT and GFP2_PC2-CTT constructs and develop BRET2 assays for their target engagement. The HEK-293T cells were transiently co-transfected with Rluc8_PC1-CTT (3.0 µg) and GFP2_PC2-CTT constructs (3.0 µg) by electroporation using a cell line optimal transfection kit according to the manufacturer’s protocol (Amaxa Inc, Gaithersburg, MD). The transfected cells were plated in 96-well black isoplate and cultured for 48 h after transfection. We used the Synergy H4 plate reader to monitor the BRET2 signal (Fluorescence/Luminescence ratio). The relative fluorescence (515/30 nm) and luminescence (410/80 nm) raw data were detected from each well after adding DeepBlue C (5 μmol·L−1) in the presence or absence of compound MS2 (10 μmol·L−1). In addition, based on the identification of crucial contact residues [e.g. Lys(K)874 and Arg(R)877 in PC2-CTT and Tyr(Y)4236 in PC1] that bind to MS2 in the computational modeling, we used a Q5 site-directed mutagenesis kit to generate amino-acid residue substitutions at the interaction sites in wild-type GFP2_PC2-CTT (K874E & R877P) and Rluc8_PC1-CTT (Y4236F) cDNAs to create mutant constructs (GFP2_PC2-CTT- K874E & R877P and Rluc8_PC1-CTT-Y4236F mutants) that disrupt the contact sites in MS2-PC1/PC2 binding pocket of wild-type PC1-CTT/PC2-CTT complex. Then we used the same approach to co-transfect GFP2_PC2-CTT-K874E & R877P mutant along with wild-type Rluc8_PC1-CTT or Rluc8_PC1-CTT-Y4236F mutant along with wild-type GFP2_PC2-CTT constructs into HEK293 cells to measure the changes of the BRET2 signal after treated with vehicle or compound MS2 (10 μmol·L−1).
Synthesis of MS2
A MS analogue, MS2 was synthesized to provide preliminary structure-activity relationship information based on the computational model shown in Figure 6A.2 The compound has a purity of greater than 98% and their structures were authenticated by standard analytical chemistry analyses.
Statistical analysis
We evaluated differences between two groups by unpaired t-test, multiple groups by one-way ANOVA with Turkey’s multiple comparison test. All values are expressed as means ± S.D. All computations were performed using a commercial biostatistics software (GraphPad Software Inc. La Jolla, CA).
Supplementary information
Acknowledgements
This work was supported by National Institutes of Health (NIH), National Institute of Arthritis and Musculoskeletal and Skin Diseases (Grant RO1 -AR071930) and National Institute of Diabetes and Digestive and Kidney Diseases (Grant RO1 DK121132). This research also used resources of the Compute and Data Environment for Science (CADES) at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725.
Author contributions
Z.X. and L.D.Q. designed the study, analyzed the data, and wrote the paper. L.C., H.L., and Z.X. performed the in vivo and in vitro experiments. M.D.S. and J.C.S. performed computational ligand docking. W.L., M.D.S., and J.C.S. revised the paper.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-023-00295-4.
References
- 1.Qin L, Liu W, Cao H, Xiao G. Molecular mechanosensors in osteocytes. Bone Res. 2020;8:23. doi: 10.1038/s41413-020-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao Z, et al. Polycystin-1 interacts with TAZ to stimulate osteoblastogenesis and inhibit adipogenesis. J. Clin. Investig. 2018;128:157–174. doi: 10.1172/JCI93725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao Z, et al. Conditional deletion of Pkd1 in osteocytes disrupts skeletal mechanosensing in mice. FASEB J. 2011;25:2418–2432. doi: 10.1096/fj.10-180299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Z, et al. Osteoblast-specific deletion of Pkd2 leads to low-turnover osteopenia and reduced bone marrow adiposity. PLoS One. 2014;9:e114198. doi: 10.1371/journal.pone.0114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu N, Xiao Z, Cao L, David V, Quarles LD. Conditional mesenchymal disruption of pkd1 results in osteopenia and polycystic kidney disease. PLoS One. 2012;7:e46038. doi: 10.1371/journal.pone.0046038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao Z, et al. Conditional disruption of Pkd1 in osteoblasts results in osteopenia due to direct impairment of bone formation. J. Biol. Chem. 2010;285:1177–1187. doi: 10.1074/jbc.M109.050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Z, Zhang S, Magenheimer BS, Luo J, Quarles LD. Polycystin-1 regulates skeletogenesis through stimulation of the osteoblast-specific transcription factor Runx2-II. J. Biol. Chem. 2008;283:12624–12634. doi: 10.1074/jbc.M710407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Z, et al. Cilia-like structures and polycystin-1 in osteoblasts/osteocytes and associated abnormalities in skeletogenesis and Runx2 expression. J. Biol. Chem. 2006;281:30884–30895. doi: 10.1074/jbc.M604772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu N, Cao L, David V, Quarles LD, Xiao Z. Kif3a deficiency reverses the skeletal abnormalities in Pkd1 deficient mice by restoring the balance between osteogenesis and adipogenesis. PLoS One. 2010;5:e15240. doi: 10.1371/journal.pone.0015240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evenepoel P, et al. A distinct bone phenotype in ADPKD patients with end-stage renal disease. Kidney Int. 2019;95:412–419. doi: 10.1016/j.kint.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Gitomer B, et al. Mineral bone disease in autosomal dominant polycystic kidney disease. Kidney Int. 2021;99:977–985. doi: 10.1016/j.kint.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rechter S, et al. Evidence for bone and mineral metabolism alterations in children with autosomal dominant polycystic kidney disease. J. Clin. Endocrinol. Metab. 2017;102:4210–4217. doi: 10.1210/jc.2017-01157. [DOI] [PubMed] [Google Scholar]
- 13.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 14.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 15.Dupont S. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res. 2016;343:42–53. doi: 10.1016/j.yexcr.2015.10.034. [DOI] [PubMed] [Google Scholar]
- 16.Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrick D, et al. Polycystin-1 regulates bone development through an interaction with the transcriptional coactivator TAZ. Hum. Mol. Genet. 2019;28:16–30. doi: 10.1093/hmg/ddy322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 19.Yang JY, et al. Osteoblast-targeted overexpression of TAZ increases bone mass in vivo. PLoS One. 2013;8:e56585. doi: 10.1371/journal.pone.0056585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hossain Z, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl. Acad. Sci. USA. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 22.Tian Y, et al. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol. Cell Biol. 2007;27:6383–6395. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin F, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kegelman CD, et al. Skeletal cell YAP and TAZ combinatorially promote bone development. FASEB J. 2018;32:2706–2721. doi: 10.1096/fj.201700872R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuya Y, et al. Increased bone mass in mice after single injection of anti-receptor activator of nuclear factor-kappaB ligand-neutralizing antibody: evidence for bone anabolic effect of parathyroid hormone in mice with few osteoclasts. J. Biol. Chem. 2011;286:37023–37031. doi: 10.1074/jbc.M111.246280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kegelman CD, Collins JM, Nijsure MP, Eastburn EA, Boerckel JD. Gone caving: roles of the transcriptional regulators YAP and TAZ in skeletal development. Curr. Osteoporos. Rep. 2020;18:526–540. doi: 10.1007/s11914-020-00605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarka M, et al. Mechanical loading activates the YAP/TAZ pathway and chemokine expression in the MLO-Y4 osteocyte-like cell line. Lab. Investig. 2021;101:1597–1604. doi: 10.1038/s41374-021-00668-5. [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Luo Q, Lin C, Song G. Simulated microgravity inhibits osteogenic differentiation of mesenchymal stem cells through down regulating the transcriptional co-activator TAZ. Biochem. Biophys. Res. Commun. 2015;468:21–26. doi: 10.1016/j.bbrc.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Luo Q, Lin C, Kuang D, Song G. Simulated microgravity inhibits osteogenic differentiation of mesenchymal stem cells via depolymerizing F-actin to impede TAZ nuclear translocation. Sci. Rep. 2016;6:30322. doi: 10.1038/srep30322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, X. et al. Stimulation of Piezo1 by mechanical signals promotes bone anabolism. Elife8, e49631 (2019). [DOI] [PMC free article] [PubMed]
- 32.Kim KM, et al. Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS One. 2014;9:e92427. doi: 10.1371/journal.pone.0092427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zubidat D, et al. Bone health in autosomal dominant polycystic kidney disease (ADPKD) patients after kidney transplantation. Bone Rep. 2023;18:101655. doi: 10.1016/j.bonr.2023.101655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, J. M., Lin, C., Stavre, Z., Greenblatt, M. B. & Shim, J. H. Osteoblast-osteoclast communication and bone homeostasis. Cells9, 2073 (2020). [DOI] [PMC free article] [PubMed]
- 35.Xiong J, Almeida M, O’Brien CA. The YAP/TAZ transcriptional co-activators have opposing effects at different stages of osteoblast differentiation. Bone. 2018;112:1–9. doi: 10.1016/j.bone.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong J, O’Brien CA. Osteocyte RANKL: new insights into the control of bone remodeling. J. Bone Miner. Res. 2012;27:499–505. doi: 10.1002/jbmr.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, H. N. et al. Osteocyte RANKL is required for cortical bone loss with age and is induced by senescence. JCI Insight5, e138815 (2020). [DOI] [PMC free article] [PubMed]
- 38.Yang W, et al. TAZ inhibits osteoclastogenesis by attenuating TAK1/NF-kappaB signaling. Bone Res. 2021;9:33. doi: 10.1038/s41413-021-00151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung DK, et al. Osteoporosis is associated with increased marrow fat content and decreased marrow fat unsaturation: a proton MR spectroscopy study. J. Magn. Reson. Imaging. 2005;22:279–285. doi: 10.1002/jmri.20367. [DOI] [PubMed] [Google Scholar]
- 40.Suchacki KJ, Cawthorn WP, Rosen CJ. Bone marrow adipose tissue: formation, function and regulation. Curr. Opin. Pharmacol. 2016;28:50–56. doi: 10.1016/j.coph.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al Saedi A, et al. Age-related increases in marrow fat volumes have regional impacts on bone cell numbers and structure. Calcif Tissue Int. 2020;107:126–134. doi: 10.1007/s00223-020-00700-8. [DOI] [PubMed] [Google Scholar]
- 42.Justesen J, et al. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2:165–171. doi: 10.1023/A:1011513223894. [DOI] [PubMed] [Google Scholar]
- 43.Salmi A, Quacquarelli F, Chauveau C, Clabaut A, Broux O. An integrative bioinformatics approach to decipher adipocyte-induced transdifferentiation of osteoblast. Genomics. 2022;114:110422. doi: 10.1016/j.ygeno.2022.110422. [DOI] [PubMed] [Google Scholar]
- 44.Clabaut A, et al. Adipocyte-induced transdifferentiation of osteoblasts and its potential role in age-related bone loss. PLoS One. 2021;16:e0245014. doi: 10.1371/journal.pone.0245014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao L, Gong FZ, Ma LY, Yang JH. Uncarboxylated osteocalcin promotes osteogenesis and inhibits adipogenesis of mouse bone marrow-derived mesenchymal stem cells via the PKA-AMPK-SIRT1 axis. Exp. Ther. Med. 2021;22:880. doi: 10.3892/etm.2021.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palhinha L, et al. Leptin induces proadipogenic and proinflammatory signaling in adipocytes. Front. Endocrinol. 2019;10:841. doi: 10.3389/fendo.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell. 2016;18:782–796. doi: 10.1016/j.stem.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 48.Xiao Z, Quarles LD. Physiological mechanisms and therapeutic potential of bone mechanosensing. Rev. Endocr. Metab. Disord. 2015;16:115–129. doi: 10.1007/s11154-015-9313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Xiao Z, Quarles LD, Li W. Osteoporosis: mechanism, molecular target and current status on drug development. Curr. Med. Chem. 2021;28:1489–1507. doi: 10.2174/0929867327666200330142432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reginensi A, et al. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013;9:e1003380. doi: 10.1371/journal.pgen.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piontek KB, et al. A functional floxed allele of Pkd1 that can be conditionally inactivated in vivo. J. Am. Soc. Nephrol. 2004;15:3035–3043. doi: 10.1097/01.ASN.0000144204.01352.86. [DOI] [PubMed] [Google Scholar]
- 52.Zhang M, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 53.Naim M, et al. Solvated interaction energy (SIE) for scoring protein-ligand binding affinities. 1. Exploring the parameter space. J. Chem. Inf. Model. 2007;47:122–133. doi: 10.1021/ci600406v. [DOI] [PubMed] [Google Scholar]
- 54.Maier JA, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerber PR, Muller K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.