Abstract
The function of a protein within a cell critically depends on its interaction with other proteins as well as its subcellular localization. The expression of mutants of a particular protein that have selective perturbation of specific protein interaction motifs is a very useful strategy for resolving a protein’s mechanism of action in a cellular process. In addition, expression of fluorescent protein fusions is a key strategy for determining the subcellular localization of a protein. These strategies require tight regulation to avoid potential alterations in protein interactions or localizations that can result from protein overexpression. Previous work led to the development of a Sleeping Beauty transposon system that allows doxycycline-inducible expression of protein mutants or fusions; titration of doxycycline allows expression of protein fusions or mutants at near endogenous levels. When used in combination with siRNA gene silencing, this strategy allows for knockdown-rescue experiments to assess the function of specific protein mutants. In this protocol, we describe the use of this Sleeping Beauty strategy for expression of eGFP fusion or mutant proteins in ARPE-19 and MDA-MB-231 cells. This includes design of expression plasmids, transfection, and selection to obtain stable engineered cells, as well as doxycycline treatment for controlled induction of protein expression, either alone or in combination with siRNA silencing for knockdown-rescue experiments. This strategy is advantageous as it allows rapid generation of stable cells for controlled protein expression, suitable for functional studies that require knockdown-rescue as well as various forms of live cell fluorescence imaging.
Key features
• Highly versatile doxycycline-inducible expression system that can be used in various mammalian cell lines.
• Stable integration of transgene allows for sustained and stable expression.
• Titration of doxycycline levels allows expression of transgene at near endogenous levels.
Keywords: Stable cells, Doxycycline inducible, Mammalian cell lines, Sleeping beauty transposon system, Fluorescent protein fusions
Background
Cell biology studies often require the expression of exogenous or modified proteins (through the use of transgenes) in cells in culture or other model systems, including expression of specific mutants that each selectively perturb a single aspect of that protein’s function. In addition, the study of protein localization often requires expression of the protein of interest fused to a fluorescent protein such as eGFP (Snapp, 2005) or to an enzyme suitable for proximity biotinylation (Gingras et al., 2019). Hence, optimal strategies for expression of transgenes are essential for a wide range of cell biology approaches.
Transient transfection to introduce plasmids encoding these various transgenes into mammalian cells in culture are commonly used for this purpose. However, this strategy can lead to artificial localization or function of the expressed proteins, as the level of expression of the exogenous proteins using this approach often dramatically exceeds that of the endogenous. Furthermore, transient transfection often leads to expression restricted to a minor fraction of cells (5%–20%), which limits the use of this method for cell population–based assays. In contrast, retroviral and lentiviral systems allow for efficient integration of a cassette for expression of a transgene into the genome. When combined with a selection stage, this approach allows for expression of the transgene in a large majority of cells in a population, as well as relatively stable expression levels of the transgene over time. However, the utility of lentiviral or retroviral strategies may be limited by size constraints for the transgene imposed by the packaging process, the time required to generate viral particles, as well as the need for specific biosafety protocols for work with these virus-like particles.
The modification of endogenous genes with CRISPR/Cas9 has also emerged as a compelling strategy to allow expression of specific mutants or fusions of a specific gene from its endogenous promoter, thus avoiding artifacts of protein overexpression (Bukhari and Müller, 2019). While powerful, this strategy also has potential limitations. Generation of loss-of-function mutants of a particular gene using CRISPR/Cas9 involves passaging of these modified cells and may lead to cellular adaptation that may limit the study of the direct effects of the protein being studied. Furthermore, many genes are present in multiple copies in many cultured cell lines, in particular aneuploid cell lines such as those derived from tumors (e.g., the breast cancer cell line MDA-MB-231 used here). Hence, it can be challenging to use CRISPR/Cas9 to modify all copies of a gene within the genome of these cells in culture, limiting the use of this approach for expression of loss-of-function mutants.
The use of transposons for stable integration of transgenes into the genome of cultured cells is an effective strategy that parallels the use of viral transduction strategies, without the need for packaging of transgene material into viral particles. The Sleeping Beauty transposon method enables stable integration of a transgene found within a vector containing inverted terminal repeats (ITRs) when co-transfected into cells with a vector encoding a transposase enzyme. This approach can be used in combination with selection to achieve integration of the transgene into a large majority of cells in a population (Kowarz et al., 2015).
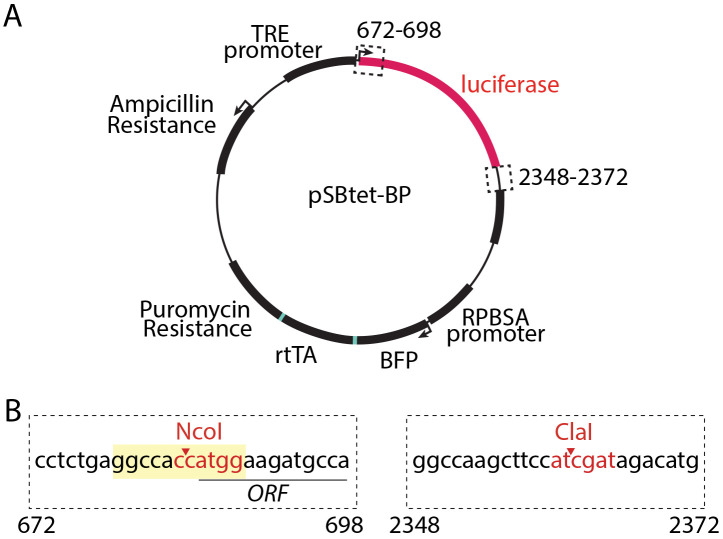
While the Sleeping Beauty system has been used extensively, a re-engineered transposase enzyme with enhanced activity—SB100X (Mátés et al., 2009)—is ideally suited for this approach, and achieves highly efficient and random integration into TA dinucleotide sites within the target cell genome (30,000 such sites in the human genome) (Kowarz et al., 2015). Kowarz & col. developed a suite of plasmids for use of the Sleeping Beauty transposon system that includes plasmids to allow either inducible (pSBtet) or constitutive (e.g., pSBbi) expression and a range of fluorescent protein and antibiotic selection markers and demonstrated the use of these strategies for stable gene expression in HeLa and HEK293T cells (Kowarz et al., 2015). The pSBtet series of plasmids developed by Kowarz & col. allows doxycycline-inducible expression of a transgene alongside constitutive expression of one of several selection markers (puromycin, hygromycin, neomycin, or blasticidin) and a fluorescent protein (BFP, eGFP, or RFP). We focus here on the use of pSBtet-BP (constitutive expression of BFP and puromycin selection, Figure 1A). Notably, the selection marker, fluorescent protein, and reverse tetracycline transactivator (rtTA) in the pSBtet plasmids are expressed as a self-cleaving polyprotein (Figure 1A).
Figure 1. pSBtet-BP plasmid and cloning strategy.
(A) Map of the pSBtet-BP plasmid as obtained from Addgene, plasmid number 60496 and developed by Kowarz & col. (Kowarz et al., 2015). Shown are the Tetracycline responsive element (TRE) promoter, the open reading frame encoding luciferase (which is replaced by the transgene of interest during the cloning stage, Procedure A), as well as the expression of a polyprotein from the constitutive synthetic RPBSA promoter; this polyprotein undergoes cleavage to generate blue fluorescent protein (BFP), reverse tetracycline transactivator (rtTA), and puromycin resistance protein. Also shown is the expression of ampicillin resistance for bacterial selection. Also shown (dashed line boxes) are the regions relevant to subcloning of the transgene, as described in Procedure A. (B) Shown is the cloning strategy for excision of the luciferase-encoding sequence and subcloning of the transgene of interest using NcoI and ClaI sites within the pSBtet-BP plasmid. NcoI and ClaI cut sites are shown in red text; the Kozak sequence is shown in yellow and the open reading frame (ORF) of the luciferase is underlined.
We recently used the Sleeping Beauty transposon strategy developed by Kowarz & col. (Kowarz et al., 2015) in ARPE-19, MDA-MB-231, and SUM149-PT cells for generation of stable cells that allow doxycycline-inducible expression of protein fusions or mutants (Cabral-Dias et al., 2022; Rahmani et al., 2023; Sugiyama et al., 2023) or short hairpin RNA (shRNA) (Lo et al., 2023). Here, we first describe subcloning of a transgene of interest into the pSBtet plasmid (Procedure A). We also note that the use of these Sleeping Beauty plasmids requires an initial transfection and selection process that is unique to some cell lines. We thus report the protocol optimized for the generation of stable cells in ARPE-19 and MDA-MB-231 (Procedure B). Finally, the use of doxycycline for expression of a transgene by the Sleeping Beauty strategy can allow for control of expression to approximate the endogenous level of expression of the protein. This is achieved through titration of doxycycline, allowing determination of an optimal range of doxycycline for each stable cell line generated. This approach allows expression of the transgene at near-endogenous levels, either alone or in combination with siRNA gene silencing of the endogenous protein for knockdown-rescue approaches (Cabral-Dias et al., 2022; Rahmani et al., 2023; Sugiyama et al., 2023). Thus, we also describe the use of doxycycline to induce expression of the protein of interest, either alone to determine optimal doxycycline condition for expression of the transgene (Procedure C), or in combination with siRNA gene silencing of the endogenous protein for knockdown-rescue approaches (Procedure D).
Materials and reagents
Restriction enzymes, e.g., NcoI and ClaI (New England Biolabs, catalog numbers: R0193L and R0197L, respectively)
Competent DH5α E. coli (Thermo Fisher, catalog number: 18265017)
D-MEM/F-12 (1×), liquid 1:1, with L-glutamine and HEPES buffer, 500 mL (Gibco, catalog number: 11330032)
RPMI-1640 medium, liquid; with L-glutamine and sodium bicarbonate, 500 mL (Sigma-Aldrich, catalog number: R8758-500ML)
Tissue culture flasks, 75 cm sq (250 mL, vented, polystyrene) (Sarstedt, catalog number: 83.3911.002)
6-well tissue culture plates, sterile, polystyrene (Sarstedt, catalog number: 86.1254.001)
Fetal bovine serum (Canada), 500 mL (Thermo Fisher Scientific, catalog number: 12483020)
Tetracycline-Free FBS (Wisent, catalog number: 081-150)
Penicillin-Streptomycin liquid 100 mL (Gibco, catalog number: 15070063)
Trypsin-EDTA (0.25% Trypsin with EDTA 1×, 500 mL (Gibco, catalog number: 25200072)
Dulbecco’s phosphate-buffered saline (dPBS), without calcium chloride and magnesium chloride, liquid, 500 mL (Sigma-Aldrich, catalog number: D8537-500ML)
Multiple PCR microtube, 0.5 mL (thin wall, neutral) (Sarstedt, catalog number: 72.735.002)
Ampicillin, sodium salt, 25 g (BioShop, catalog number: AMP201.25)
Chloramphenicol, 25 g (BioShop, catalog number: CLR201.25)
LB Broth (Miller), liquid microbial growth medium (Sigma-Aldrich, catalog number: L2542-500ML)
NucleoBond Xtra Midi EF, Midi kit for endotoxin-free plasmid DNA (Macherey-Nagel, catalog number: 740420.10)
FuGENE HD Transfection Reagent, 1 mL (Promega, catalog number: E2311)
Opti-MEM reduced serum medium (Thermo Fisher Scientific, catalog number: 31985070)
Puromycin dihydrochloride (Sigma Aldrich, catalog number: P7255-25MG)
pCMV(CAT)T7-SB100 plasmid (Addgene, Plasmid #34879)
-
pSBtet-BP plasmid (Addgene, Plasmid #60496)
Note: Various other plasmids described in Kowarz et al. (2015) allow alternatives for generation of stable cells to express the transgene using the Sleeping Beauty transposon system.
siRNA in RNAse-free water reconstituted at 20 μM
Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, catalog number: 13778150)
MDA-MB-231 cells (ATCC, catalog number: HTB-26)
ARPE-19 cells (ATCC, catalog number: CRL-2302)
ARPE-19 growth medium (see Recipes)
Tet-free ARPE-19 growth medium (see Recipes)
MDA-MB-231 growth medium (see Recipes)
Tet-free MDA-MB-231 growth medium (see Recipes)
Equipment
Shaking 37 °C platform incubator (Thermo Fisher Scientific, model: SHKE6000)
Cell incubator set to 37 °C and 5% CO2 (Thermo Fisher Scientific, model: HERAcell 150i)
Centrifuge (unrefrigerated) for 50 mL Falcon tubes (Thermo Fisher Scientific, model: Sorval ST16R)
Procedure
-
Plasmid design and isolation
Subclone transgene of interest into the pSBtet-BP plasmid, using standard molecular biology techniques. We suggest synthesizing the gene of interest using commercial services for subcloning into NcoI (base pair 684) and ClaI (base pair 2361) within the pSBtet-BP plasmid (see Figure 1B). This strategy will result in excision of the open reading frame for luciferase, so that it can be replaced with the transgene of interest. Please note that it is important to maintain the integrity of the Kozak sequence within the final plasmid (Figure 1B, sequence with yellow shading). It is also important to ensure that the synthesized transgene does not contain internal NcoI and ClaI cleavage sequences; these should be removed by silent point mutations if present. Alternatively, SfiI sites can be used to subclone a gene of interest within the pSBtet-BP plasmid (Kowarz et al., 2015).
-
Isolate plasmid using midi-prep or maxi-prep procedure.
Inoculate 100 mL of LB broth culture supplemented with 100 μg/mL of ampicillin with a single clone of competent DH5α E. coli transformed with the pSBtet-BP transgene plasmid.
Inoculate 100 mL of LB broth culture supplemented with 25 μg/mL of chloramphenicol with a single clone of competent DH5α E. coli transformed with the pCMV(CAT)T7-SB100 plasmid.
Grow bacterial liquid cultures in a shaking 37 °C incubator overnight.
Perform plasmid isolation using a commercial kit available for this purpose, ensuring that these are certified to be endotoxin-free. We suggest using NucleoBond Xtra Midi EF, and several other alternatives are available. This procedure is described as per the manufacturer’s instructions.
-
Transfection of Sleeping Beauty plasmids and puromycin selection
-
Thaw fresh batch of ARPE-19 cells and passage twice (see Note 1). The protocol for growth and one passage of cells (see Note 3) is as follows:
Grow cells in a T-75 flask in ARPE-19 growth medium at 37 °C and 5% CO2 (see Recipes below for medium composition).
When cells reach 60%–80% confluence, remove medium and wash with dPBS.
Incubate cells in 1.5 mL of trypsin solution for 3–5 min at 37 °C or until cells detach from the surface of the T-75 flask.
Add 8.5 mL of ARPE-19 growth medium and use a pipette to wash medium solution over the surface of the flask to ensure that all cells are detached.
Centrifuge cells at 200× g for 5 min and discard the supernatant.
Carefully resuspend the pellet into 10 mL of fresh ARPE-19 growth medium.
Transfer 2–3 mL of this cell suspension to a fresh T-75 flask, to which add an additional 7–8 mL of ARPE-19 growth medium.
The remaining 7–8 mL of cell suspension from the previous step can be used for seeding (e.g., step B2 below) or discarded.
Seed ARPE-19 cells into wells of a 6-well plate. Seeding should be done at approximately 5.0 × 105 cells per well (in 2 mL of medium per well), leading to ~20%–30% confluence at time of seeding and 40%–50% confluence on the day of transfection (step B3).
-
Transfect cells with plasmids using FuGENE HD transfection reagent. Each plasmid transfection condition should be performed in triplicate (3 wells of the 6-well plate per condition).
-
Prepare the following transfection mixture in a sterile microfuge tube per well of a 6-well plate to be transfected (see Note 4), in the following order:
i. Opti-MEM medium for a total volume of 150 μL
ii. 3 μg of pSBtet-BP transgene plasmid
iii. 0.3 μg of the pCMV(CAT)T7-SB100 plasmid
iv. 9 μL of FuGENE transfection reagent
Vortex for 30 s and incubate this transfection mixture for 15 min at room temperature.
Remove growth medium from cells and wash once with 2 mL of dPBS. After removal of dPBS, add 1.9 mL of Opti-MEM medium to each well. Then, add 100 μL of transfection mixture (from step B3) to each well, dropwise.
-
Incubate cells with transfection mixture in Opti-MEM for 24 h. Then, remove this medium, wash cells once in dPBS, and replace medium with ARPE-19 growth medium. Incubate cells for another 24 h at 37 °C with 5% CO2.
Replace medium with ARPE-19 growth medium and incubate for 24 h at 37 °C with 5% CO2. This step is essential to allow cells to recover and maintain viability following transfection.
After 24 h, replace regular growth medium with ARPE-19 growth medium supplemented with 2 μg/mL of puromycin and incubate cells for 2–3 days at 37 °C with 5% CO2. Some cell death can be observed during this period.
-
When cells in the 6-well plate reach ~50%–60% confluence, passage cells into a T-25 flask, as follows:
Remove medium from cells and wash 2× in dPBS.
Add 0.3 mL of Trypsin to each cell and incubate for 5 min at 37 °C and 5% CO2.
When cells begin to detach, add 2 mL of ARPE-19 growth medium, ensuring that cells are detached from the bottom of the well.
Combine the cell suspension mixture from each well of triplicate transfection condition into a single tube.
Centrifuge cells at 200× g for 5 min and discard the supernatant.
Carefully resuspend the pellet in 5 mL of fresh ARPE-19 growth medium.
Transfer cell suspension into a single T-25 flask and incubate for 24 h at 37 °C with 5% CO2.
Replace medium with ARPE-19 growth medium supplemented with 2 μg/mL of puromycin and incubate at 37 °C with 5% CO2, while changing medium supplemented with puromycin every 2–3 days.
When cells reach confluence, passage cells as in step B1 and transfer into a T-75 flask.
When cells reach confluence again, passage cells again as in step B1. Ensure that cells have been passaged approximately 3–4 times over the course of 2–3 weeks with puromycin selection medium. If expression of multiple transgenes using this strategy is required, please see Note 5.
-
-
Doxycycline induction of transgene expression—transgene expression alone
Culture ARPE-19 cells stably transfected with pSBtet-BP transgene (produced following Procedure B, called ARPE-19-transgene cells henceforth) in tet-free ARPE-19 growth medium. This should require 2–3 passages (approximately 7–8 days) of culture in this medium, as per step B1 above. This stage ensures that there is minimal expression of the transgene prior to silencing the expression of the endogenous protein.
Seed ARPE-19-transgene cells into wells of a 6-well plate. Seeding should be done at approximately 5 × 105 cells per well (in 2 mL of medium per well), leading to ~20%–30% confluence at time of seeding. For initial experiments, seeding ~6 wells worth of cells is recommended, as this will allow testing of a range of doxycycline concentrations.
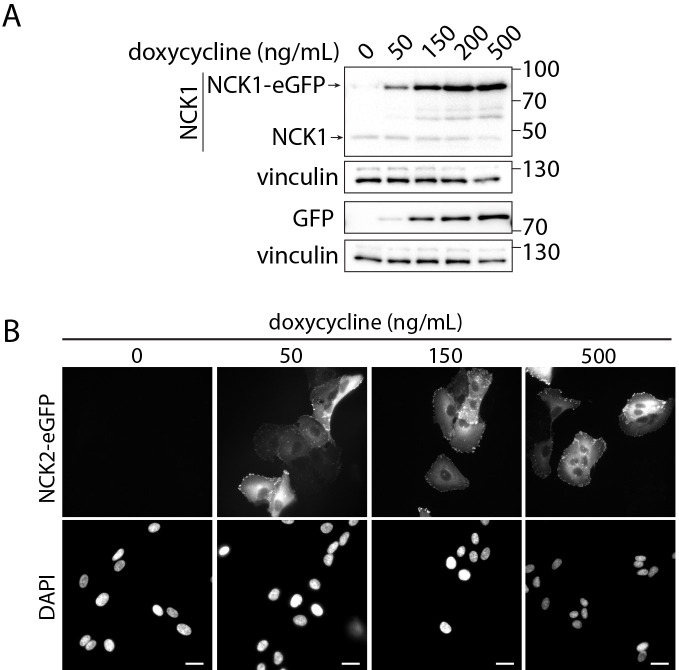
At 24 h post-seeding, replace medium with tet-free ARPE-19 growth medium supplemented with 0–2,000 ng/mL of doxycycline (e.g., 0, 50, 100, 250, 500, and 2,000 ng/mL doxycycline) and incubate for 24 h at 37 °C with 5% CO2 before proceeding to downstream experimental applications such as fluorescence microscopy or western blotting experiments. Examples of this approach, with detection of fluorescent protein fusion expression by western blotting, are seen in Figure 2A, and by fluorescence microscopy in Figure 2B. Detection of transgene expression by western blotting allows comparison of the expression of the transgene to that of the endogenous protein (Figure 2A, compare expression of NCK1-eGFP to that of endogenous NCK1), while detection of transgene expression by microscopy allows estimation of the proportion of cells that harbor the transgene. Additional examples of detection by western blotting are found in Cabral-Dias et al. (2022), Figure S2B and S2C (see Note 2).
-
Doxycycline induction of the transgene for knockdown-rescue experiments
Culture ARPE-19 cells stably transfected with pSBtet-BP transgene (as per Procedure B, called ARPE-19-transgene cells henceforth) in tet-free ARPE-19 growth medium. This should require 2–3 passages (approximately 7–8 days) of culture in this medium, as per step B1 above. This stage ensures that there is minimal expression of the transgene prior to silencing the expression of the endogenous protein. Please see Note 6 when designing plasmids for knockdown-rescue approaches.
Seed ARPE-19-transgene cells into wells of a 6-well plate. Seeding should be done at approximately 2.5 × 105 cells per well (in 2 mL of medium per well), leading to ~10%–15% confluence at time of seeding and 20%–30% confluence on the day of transfection (first transfection is 24 h after seeding).
-
Transfect cells with target siRNA using Lipofectamine RNAiMAX reagent (transfection is performed twice, separated by 24 h). Time (in hours) is relative to the time of seeding at t = 0 h, with transfections performed at t = 24 h and 48 h, as follows:
-
Prepare the following transfection mixture in a sterile microfuge tube for each well of a 6-well plate to be transfected:
i. 100 μL of Opti-MEM medium
ii. 3.25 μL of Lipofectamine RNAiMAX reagent
iii. 2.75 μL of 20 μM siRNA solution
Vortex for 10 s and incubate this transfection mixture for 15 min.
Wash ARPE-19 cells in a 6-well plate 2–3 times with dPBS and remove medium.
Add 900 μL of Opti-MEM to each well. Then, add 100 μL of transfection mixture to each well in a dropwise motion.
Let cells incubate in the incubator for 3 h at 37 °C with 5% CO2.
Wash cells with PBS after 3 h and replace with tet-free ARPE-19 growth medium.
Allow the cells to rest while incubating in tet-free ARPE-19 growth medium for 24 h at 37 °C with 5% CO2 and repeat this protocol (steps D3a–D3f) the next day, followed by another 24 h rest (incubation in tet-free ARPE-19 growth medium at 37 °C with 5% CO2) before proceeding to doxycycline induction.
-
At 72 h post-seeding (24 h after the last siRNA transfection), replace medium with tet-free ARPE-19 growth medium supplemented with 200 ng/mL of doxycycline to each well (or another optimal, experimentally determined doxycycline concentration) and incubate for 24 h at 37 °C with 5% CO2 before proceeding to downstream experimental applications such as immunofluorescence microscopy or western blotting experiments. Examples of this approach for immunofluorescence microscopy are found in (Cabral-Dias et al., 2022), Figures 4 and 5.
Figure 2. Detection of NCK1/2 transgenes in ARPE-19 cells stably engineered with pSBtet-BP-transgene plasmids.
ARPE-19 cells were subjected to transfection to generate cells that stably carry the transgene for inducible expression of eGFP-tagged NCK1 or 2, as in Procedure B. Then, induction of NCK1-eGFP (A) or NCK2 -eGFP (B) was performed as in Procedure C, using doxycycline concentrations (ng/mL) as indicated for 24 h. (A) Whole-cell lysates of NCK1-eGFP stable cells were resolved by SDS-PAGE followed by transfer to PVDF membranes and then immunoblotted using antibodies as indicated. Also shown are the approximate molecular weights (kDa) for each blot panel. (B) Cells were subjected to PFA fixation, and DAPI labeling using standard fluorescence microscopy sample preparation, and then imaged using a Quorum Diskovery instrument comprised of a Leica DMi8 microscope. Imaging was done using a 40× objective with 405- and 488-nm laser illumination, 450/55 and 525/50 emission filters, and acquired using a Zyla 4.2Plus sCMOS camera (Andor). Scale bars: 40 μm.
Notes
Ensure use of freshly thawed, low passage number cells when making stable cell lines (Procedure B).
The determination of optimal doxycycline conditions for induction will be specific to each experiment and protein being studied. Expression within ~2–3 fold of that of the endogenous protein, for example as determined by western blotting, is a reasonable initial condition to test for downstream applications; however, the appropriate expression level should be tested for each protein.
The protocol described here is well suited for ARPE-19 cells. The same protocol is also effective for other cell lines such as MDA-MB-231 cells, while noting differences in MDA-MB-231 growth medium and tet-free MDA-MB-231 growth medium (see Recipes).
The ratio of pSBtet-BP plasmids containing the transgene of interest and the transposase enzyme (pCMV(CAT)T7-SB100) used for transfection (step B3) may require optimization for each plasmid. We report transfection of a 10:1 ratio of pSBtet-BP transgene plasmid to pCMV(CAT)T7-SB100 integrase plasmid; other proportions may also be considered if stable transfection is not initially successful. The 10:1 ratio of pSBtet-BP transgene plasmid to the SB100 integrase plasmid is optimized to enhance the likelihood of a single integration event per cell; however, further reduction of the amount of pCMV(CAT)T7-SB100 integrase plasmid may be considered should further optimization be required to achieve a single integration event per cell (Grabundzija et al., 2010; Kowarz et al., 2015).
It is expected that the pCMV(CAT)T7-SB100 integrase plasmid is not retained following the selection process, as the latter takes two weeks or longer, and this integrase-expressing plasmid is not itself integrated into the host cell genome. Should an experimental design require expression of a second transgene using an additional pSBtet or pSBbi plasmid (e.g., using a different selection marker to express a different transgene), this will again require co-transfection of the pCMV(CAT)T7-SB100 alongside the appropriate pSBtet/pSBbi plasmid.
To allow for knockdown-rescue experiments, it is essential that the transgene used to generate the pSBtet plasmid also has a silent mutation in the region of the gene corresponding to the siRNA sequence.
Recipes
-
ARPE-19 growth medium
DMEM/F12 medium (500 mL)
10% FBS (50 mL)
Penicillin-Streptomycin (5,000 U/mL) (5 mL)
-
Tet-free ARPE-19 growth medium
DMEM/F12 medium (500 mL)
10% tetracycline-free FBS (50 mL)
Penicillin-Streptomycin (5,000 U/mL) (5 mL)
-
MDA-MB-231 growth medium
RPMI-1640 medium, liquid; with L-glutamine and sodium bicarbonate (500 mL)
10% FBS (50 mL)
Penicillin-Streptomycin (5,000 U/mL) (5 mL)
-
Tet-free MDA-MB-231 growth medium
RPMI-1640 medium, liquid; with L-glutamine and sodium bicarbonate (500 mL)
10% tetracycline-free FBS (50 mL)
Penicillin-Streptomycin (5,000 U/mL) (5 mL)
Acknowledgments
This work was supported by a Project Grant from the Canadian Institutes of Health Research (PJT156355) to C.N.A. We thank Laura A. Orofiamma for the critical reading of this manuscript. The Sleeping Beauty plasmids described here were developed by Kowarz & colleagues (Kowarz et al., 2015). We have previously used this method to develop stable cell lines in ARPE-19, MDA-MB-231, and SUM149-PT cells (Cabral-Dias et al., 2022; Lo et al., 2023; Rahmani et al., 2023; Sugiyama et al., 2023).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Bukhari H. and Müller T.(2019). Endogenous Fluorescence Tagging by CRISPR. Trends Cell Biol. 29(11): 912-928. [DOI] [PubMed] [Google Scholar]
- 2.Cabral-Dias R., Lucarelli S., Zak K., Rahmani S., Judge G., Abousawan J., DiGiovanni L. F., Vural D., Anderson K. E., Sugiyama M. G., et al.(2022). Fyn and TOM1L1 are recruited to clathrin-coated pits and regulate Akt signaling. J. Cell Biol. 221(4): e201808181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingras A. C., Abe K. T. and Raught B.(2019). Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48: 44-54. [DOI] [PubMed] [Google Scholar]
- 4.Grabundzija I., Irgang M., Mátés L., Belay E., Matrai J., Gogol-Döring A., Kawakami K., Chen W., Ruiz P., Chuah M. K., et al.(2010). Comparative Analysis of Transposable Element Vector Systems in Human Cells. Mol. Ther. 18(6): 1200-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowarz E., Löscher D. and Marschalek R.(2015). Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 10(4): 647-653. [DOI] [PubMed] [Google Scholar]
- 6.Lo L., Uchenunu O., Botelho R. J., Antonescu C. N. and Karshafian R.(2023). AMPK is required for recovery from metabolic stress induced by ultrasound microbubble treatment. iScience 26(2): 105883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mátés L., Chuah M. K. L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D. P., Schmitt A., Becker K., Matrai J., et al.(2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41(6): 753-761. [DOI] [PubMed] [Google Scholar]
- 8.Rahmani S., Ahmed H., Ibazebo O., Fussner-Dupas E., Wakarchuk W. W. and Antonescu C. N.(2023). O-GlcNAc transferase modulates the cellular endocytosis machinery by controlling the formation of clathrin-coated pits. J. Biol. Chem. 299(3): 102963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snapp E.(2005). Design and Use of Fluorescent Fusion Proteins in Cell Biology. Curr. Protoc. Cell Biol. 27(1): ecb2104s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama M. G., Brown A. I., Vega-Lugo J., Borges J. P., Scott A. M., Jaqaman K., Fairn G. D. and Antonescu C. N.(2023). Confinement of unliganded EGFR by tetraspanin nanodomains gates EGFR ligand binding and signaling. Nat. Commun. 14(1): 2681. [DOI] [PMC free article] [PubMed] [Google Scholar]