Abstract
Background
Tumor microenvironment (TME) characteristics are potential biomarkers of response to immune checkpoint inhibitors in metastatic melanoma. This study developed a method to perform unsupervised classification of TME of metastatic melanoma.
Methods
We used multiplex immunohistochemical and quantitative pathology-derived assessment of immune cell compositions of intratumoral and peritumoral regions of metastatic melanoma baseline biopsies to classify TME in relation to response to anti-programmed cell death protein 1 (PD-1) monotherapy or in combination with anti-cytotoxic T-cell lymphocyte-4 (ipilimumab (IPI)+PD-1).
Results
Spatial profiling of CD8+T cells, macrophages, and melanoma cells, as well as phenotypic PD-1 receptor ligand (PD-L1) and CD16 proportions, were used to identify and classify patients into one of three mutually exclusive TME classes: immune-scarce, immune-intermediate, and immune-rich tumors. Patients with immune-rich tumors were characterized by a lower proportion of melanoma cells and higher proportions of immune cells, including higher PD-L1 expression. These patients had higher response rates and longer progression-free survival (PFS) than those with immune-intermediate and immune-scarce tumors. At a median follow-up of 18 months (95% CI: 6.7 to 49 months), the 1-year PFS was 76% (95% CI: 64% to 90%) for patients with an immune-rich tumor, 56% (95% CI: 44% to 72%) for those with an immune-intermediate tumor, and 33% (95% CI: 23% to 47%) for patients with an immune-scarce tumor. A higher response rate was observed in patients with an immune-scarce or immune-intermediate tumor when treated with IPI+PD-1 compared with those treated with PD-1 alone.
Conclusions
Our study provides an automatic TME classification method that may predict the clinical efficacy of immunotherapy for patients with metastatic melanoma.
Keywords: PATHOLOGY, Immunotherapy, Tumor Microenvironment, Melanoma
WHAT IS ALREADY KNOWN ON THIS TOPIC
There is a significant knowledge gap regarding the interplay between specific immune populations and the effectiveness of immunotherapy in metastatic melanoma. In particular, the contribution of non-T-cell constituents, such as tumor-associated macrophages, within the tumor microenvironment (TME) has not been fully explored. As a result, a comprehensive TME classification approach incorporating both T cell and macrophage populations is needed to improve our ability to predict treatment responses and progression-free survival in metastatic melanoma.
WHAT THIS STUDY ADDS
Using multiplex immunofluorescence staining and quantitative image analysis, we characterize the TME into three distinct classes based on their immune profiles (immune-scarce, immune-intermediate, and immune-rich). This study incorporates the importance of not only CD8+T cells but also peritumoral CD16+CD68− non-macrophages and CD16+ cells in the TME, which are associated with improved clinical outcomes following immune checkpoint therapy. Our innovative classification methodology integrates the analysis of multiple markers (CD8, CD68, PD-L1, CD16, and SOX10) to optimize the associations between TME classifications and the progression-free survival of patients treated with immune checkpoint inhibitors.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study introduces a novel approach to classifying the TME using multidimensional high-plex tissue imaging and tests their possible associations with clinical outcomes. This study offers new perspectives on the roles of various cell types, particularly CD8+T cells and peritumoral CD16+CD68− non-macrophages. It suggests potential avenues for future research to delve deeper into these cellular components and their impact on tumor progression and treatment response. The classification model provides a framework to better understand patient responses to programmed cell death protein 1 (PD-1) and ipilimumab+PD-1 therapies in the context of their TME profiles.
Introduction
Significant advancements in the treatment of metastatic melanoma have been made possible with immune checkpoint-blocking (ICB) antibodies. Patients with metastatic melanoma have improved outcomes when treated with anti-programmed cell death protein 1 (PD-1) monotherapy or in combination with anti-cytotoxic T-cell lymphocyte-4 (CTLA-4).1–5 However, despite these significant improvements, approximately half of the patients with metastatic melanoma do not initially respond to checkpoint-based immunotherapies or respond but later develop resistance.6 Therefore, accurate predictive biomarkers are urgently needed to enable patient selection and identify patients who require alternative treatment strategies that lead to improved outcomes.7
Several studies have identified tumor microenvironment (TME) characteristics as potential biomarkers of response to ICB treatment in metastatic melanoma.8–11 The TME is composed of many cell types, including tumor cells, immune cells, stromal cells, blood vessels, and other mesenchymal cells, and plays a crucial role in tumor progression and metastasis.8–10 12 Differences in the compositions of resident cell types, such as cytotoxic T cells, tumor-associated macrophages, and inflammatory pathways within the TME, have been associated with the response of patients to ICB treatment.13–17 Additionally, the expression of the PD-1 receptor ligand, PD-L1, has been proposed as a predictive biomarker, although its utility is inconsistent across studies and cancer types.18 19
Traditionally, three major TME classes have been identified based on the compositions of resident cell types and their association with clinical outcomes in metastatic melanoma and other cancers.20–24 The general terminology includes inflamed (or “hot”), excluded (or “altered”) and ignored (“cold” or “desert”) TME classes.24 For example, the Tumor Profiler Consortium has defined thresholds for CD8+T-cell densities within the intratumoral (IT) or surrounding peritumoral (PT) regions to classify melanomas into inflamed, excluded, and ignored tumors.23 Inflamed tumors have overall high densities of IT and PT lymphocytes and are associated with a favorable response to ICB therapies.23 Excluded tumors have high PT lymphocytes at the tumor margin, but low IT lymphocytes and ignored tumors are characterized by the lack of lymphocytes and are associated with poor response to ICB therapies.23 However, such approaches are based on CD8+T cells alone.
The methodology for profiling the TME has expanded with the use of multiplex tissue imaging technologies to profile the immune cell population more comprehensively in patients’ biopsies. The use of multiplex immunofluorescence staining and quantitative pathology has enabled the assessment of multiple cell types and phenotypic markers (PD-L1) to generate a broader overview of the TME beyond T-cells alone.25 26 Quantifying each immune cell population within the TME for its association with response to ICB is at the core of developing more accurate TME-based biomarkers. Other populations are known to be important in relation to response to ICB therapies.27 28 Therefore, a novel approach is required to identify combinations of cell populations that result in clinically significant TME classes.
There is growing interest in the macrophage phenotype and composition of the TME because of their prominent roles in cancer progression and responses to therapy.13 29 Macrophages, particularly tumor-associated macrophages (TAMs), have been shown to contribute to the establishment and maintenance of an immunosuppressive TME, promote angiogenesis, and support tumor cell invasion and metastasis.30 31 Furthermore, emerging evidence suggests that macrophages can directly influence the efficacy of immunotherapies including checkpoint inhibitors.32 33 Our recent study by Lee et al 34 showed that intratumoral CD16+ macrophages are associated with clinical outcomes in patients with metastatic melanoma treated with a combination of anti-PD-1 and anti-CTLA-4 therapies. TME classification, which includes TAM beyond T-cells alone, along with phenotypic markers CD16/PD-L1, may improve the clinical utility of TME classification.
Here, we used multiplex tissue image-based immune profiling of baseline biopsies from 188 patients with metastatic melanoma, divided into a discovery cohort of 155 patients and a validation cohort of 33 patients, to determine TME classes that were associated with response to anti-PD-1 alone (PD-1) or combined with ipilimumab (IPI+PD-1). We developed a methodology that combined cytotoxic T-cell, macrophage and melanoma cell types, as well as phenotypic markers, PD-L1 and CD16 (FcγRIII) cell proportions to classify tumors. The study developed a novel algorithm that optimized the combination of these immune cell phenotypes and locations to maximize associations of TME classes with treatment response and progression-free survival.
Methods
Patient cohorts
A retrospective cohort of patients with unresectable stage III or IV melanoma treated with anti-PD-1 alone (PD-1) or in combination with IPI (IPI+PD-1) ICB therapy. Patients were required to have a baseline formalin-fixed, paraffin-embedded (FFPE) melanoma tissue biopsy taken prior to treatment with immune checkpoint inhibitors. Samples were collected as part of the Personalized Immunotherapy Program (Protocol Number: MIA2020/283). This study was conducted in accordance with the Declaration of Helsinki. Samples were acquired with written informed consent from all patients. Patient responses were determined using standard Response Evaluation Criteria in Solid Tumors (RECIST) v.1.1 criteria.35 In line with the previous studies,16 36 37 patient responses were determined using standard RECIST v.1.1 criteria. Tumor response to immunotherapy was assessed with regular scans as per the standard of care and according to each institution’s protocols (in general, 3-monthly CT or CT/positron emission tomography imaging). Patients who achieved a complete response, partial response (PR), or stable disease (SD) of greater than 6 months were classified as responders, while non-responders were classified as those with progressive disease, PR, or SD of less than or equal to 6 months PFS. PFS was defined as the interval between the start of treatment and disease progression.
Multiplex immunofluorescence staining
Multiplex immunofluorescence staining was performed on a single 4 µm FFPE melanoma section as previously described.11 16 25 37 Briefly, slides were heated in the oven at 65°C for 20 min, deparaffinized in xylene and rehydrated in graded ethanols. Antigen retrieval was performed in pH9 HIER buffer in the Decloaking Chamber (Biocare Medical) at 110°C for 10 min. Staining was performed using an Autostainer plus (Dako). Tissue sections were blocked with 3% hydrogen peroxide in Tris-buffered saline (TBST) for 5 min and then incubated with the antibody for CD68 (Cell Marque, clone KP-1, mouse, dilution 1:500) for 30 min. The antibody was detected using the Opal Polymer HRP Ms+Rb (Onestep) (Akoya Biosciences) detection system and visualized using Opal520 TSA (1:100) for 5 min. Antigen retrieval was then conducted again to prepare the slides for the next antibody. Using this method, all samples were stained sequentially with CD16a (Abcam, clone EPR16784, rabbit, dilution 1:400) visualized with Opal620 TSA (1:100), SOX10 (Biocare Medical, clone BD34, mouse, dilution 1:200) visualized with Opal690 TSA (1:100), PD-L1 (Cell Signaling Technology, clone E1L3N, rabbit, dilution 1:1000) visualized with Opal650 TSA (1:100), and CD8 (Dako, clone C8/144B, mouse, dilution 1:1500) visualized with Opal570 TSA (1:100). Slides were counterstained with DAPI (1:2000) for nuclei visualization, and coverslipped using the ProLong Diamond Antifade Mountant (Invitrogen).
Multispectral image analysis
Slides were imaged using the Vectra 3.0 microscope. Multispectral fluorescent images were visualized in Phenochart V.1.0.8 (Akoya Biosciences), and the tumor and surrounding peritumoral regions were selected for high-power whole slide imaging (20×). A spectral library for each fluorophore was generated in inForm V.2.4.1 (Akoya Biosciences), and multiplex images were subsequently spectrally unmixed. The individual 20× images were then stitched into a single multispectral image for each tumor specimen for analysis in HALO V.2.2 (Indica Labs). The Random Forest tissue classifier algorithm was trained to recognize IT and PT based on the presence or absence of SOX10. The positivity for each marker was determined by optimized thresholds based on the staining intensity, and the analysis settings were run across all samples.
Statistical analysis
Patients’ baseline clinical and blood count characteristics were summarized through frequencies and proportions for categorical variables and median and range for continuous variables. The baseline characteristics were compared between responders and non-responders using the Wilcoxon rank-sum test for continuous variables and Pearson’s χ2 test for categorical variables, with Yates’s continuity correction when appropriate. The immune cell proportions were uniformly scaled to the interval (0, 1) by fitting the empirical distribution functions of the cells.38 We employed unsupervised hierarchical clustering to group immune cells within IT and PT regions into relevant TME classes. The optimal number of TME classes was obtained across the entire cohort using the NbClust R package.39 We assessed immune cell compositions to identify the most represented cells within each TME class using a v-test based on the hypergeometric distribution.40 41
PFS was calculated from the date of commencement of treatment to the date of progression or death from any cause. PFS differences between TME classes were evaluated using the Kaplan-Meier method and tested with the log-rank statistics. The association between TME classes and PFS was assessed using multivariable Cox regression adjusted with baseline characteristics. All statistical analyses were performed in R.42 P values<0.05 were considered statistically significant for all analyses.
Results
Patient characteristics
This discovery cohort included pretreatment biopsies from 155 patients with metastatic melanoma treated with either PD-1 monotherapy (n=80; table 1) or IPI+PD-1 combination therapy (n=75; table 2). The median age of the PD-1 therapy cohort was 68 years (table 1). Responding patients tended to be older (p value=0.007) and with a lower neutrophil-to-lymphocyte ratio (p value=0.03). There was also a higher rate of brain metastases in the non-responding patients (p value=0.03). However, no associations were observed between response and sex, primary site, line of ICB, primary melanoma subtype (cutaneous vs non-cutaneous acral or mucosal), previous treatment with targeted kinase therapy, baseline lactate dehydrogenase (LDH), American Joint Committee on Cancer (AJCC) v8 stage, Eastern Cooperative Oncology Group (ECOG) status, biopsy site, hemoglobin and presence of lung or liver metastases. Median PFS for the PD-1 therapy cohort was 6.24 months (95% CI: 3.42 to 21.6 months), and 1-year PFS was 44% (95% CI: 34% to 56%).
Table 1.
Patients clinical and blood characteristics of the PD-1 therapy cohort
| Characteristics | Responders (n=39) | Non-responders (n=41) | Total (n=80) | P value |
| Age (years) | ||||
| Median (range) | 74 (37–93) | 65 (37–95) | 68 (37–95) | 0.007* |
| Gender, n (%) | ||||
| Female Male |
12 (30.8%) 27 (69.2%) |
13 (31.7%) 28 (68.3%) |
25 (31.2%) 55 (68.8%) |
0.928† |
| Primary site, n (%) | ||||
| Scalp, face, neck Occult Other |
8 (20.5%) 5 (12.8%) 26 (66.7%) |
10 (24.4%) 3 (7.3%) 28 (68.3%) |
18 (22.5%) 8 (10.0%) 54 (67.5%) |
0.6884‡ |
| Line of ICB, n (%) | ||||
| 1st line >1st line |
32 (82.1%) 7 (17.9%) |
30 (73.2%) 11 (26.8%) |
62 (77.5%) 18 (22.5%) |
0.4946† |
| Cutaneous primary, n (%) | ||||
| Yes No |
37 (94.9%) 2 (5.1%) |
38 (92.7%) 3 (7.3%) |
75 (93.8%) 5 (6.2%) |
1.000† |
| Previous MAPKi, n (%) | ||||
| Yes No |
2 (5.1%) 37 (94.9%) |
9 (22.0%) 32 (78.0%) |
11 (13.8%) 69 (86.2%) |
0.06299† |
| Baseline LDH, n (%) | ||||
| Elevated Normal |
11 (28.2%) 28 (71.8%) |
18 (43.9%) 23 (56.1%) |
29 (36.2%) 51 (63.7%) |
0.2197† |
| AJCC staging v8, n (%) | ||||
| III/M1A/M1B M1C/M1D |
18 (46.2%) 21 (53.8%) |
11 (26.8%) 30 (73.2%) |
29 (36.2%) 51 (63.7%) |
0.1177‡ |
| ECOG, n (%) | ||||
| 0 ≥1 Unknown |
23 (59.0%) 16 (41.0%) 0 (0.0%) |
15 (36.6%) 24 (58.5%) 2 (4.9%) |
38 (47.5%) 40 (50.0%) 2 (2.5%) |
0.07289‡ |
| Biopsy region, n (%) | ||||
| Brain Lung Lymph node Others Subcutaneous |
4 (10.3%) 0 (0.0%) 4 (10.3%) 3 (7.7%) 28 (71.8%) |
5 (12.2%) 2 (4.9%) 7 (17.1%) 3 (7.3%) 24 (58.5%) |
9 (11.2%) 2 (2.5%) 11 (13.8%) 6 (7.5%) 52 (65.0%) |
0.5267‡ |
| Hemoglobin | ||||
| Median (range) Nine missing details were removed |
140 (109–161) | 140.5 (72–159) | 140 (72–161) | 0.5039* |
| Neutro lympho ratio | ||||
| Median (range) Nine missing details were removed |
2.6 (0.6–19.5) | 3.35 (1.2–22.5) | 2.9 (0.6–22.5) | 0.02954* |
| Brain metastasis, n (%) | ||||
| Yes No |
5 (12.8%) 34 (87.2%) |
15 (36.6%) 26 (63.4%) |
20 (25.0%) 60 (75.0%) |
0.02814† |
| Lung metastasis, n (%) | ||||
| Yes No |
19 (48.7%) 20 (51.3%) |
22 (53.7%) 19 (46.3%) |
41 (51.2%) 39 (48.8%) |
0.8273† |
| Liver metastasis, n (%) | ||||
| Yes No |
6 (15.4%) 33 (84.6%) |
10 (24.4%) 31 (75.6%) |
16 (20.0%) 64 (80.0%) |
0.4673† |
| Median PFS (months) | Not yet reached | 2.27 | 6.24 (95% CI: 3.42 to 21.6) | |
| 1-year PFS (%) | 89 | 0 | 44 (95% CI: 34 to 56) |
Bold p-values indicate statistical significance at p < 0.05 level.
*Wilcoxon test.
†Pearson’s χ2 test with Yate’s correction was used for categorical variables.
‡Pearson’s χ2 test.
AJCC, American Joint Committee on Cancer; ECOG, Eastern Cooperative Oncology Group; ICB, immune checkpoint-blocking; LDH, lactate dehydrogenase; MAPKi, mitogen-activated protein kinases inhibitors; PD-1, anti-programmed cell death protein 1; PFS, progression-free survival.
Table 2.
Patients clinical and blood characteristics of the IPI+PD-1 therapy cohort
| Characteristics | Responders (n=48) | Non-responders (n=27) | Total (n=75) | P value |
| Age (years) | ||||
| Median (range) | 61 (19–79) | 57 (27–75) | 57 (19–79) | 0.2060* |
| Gender, n (%) | ||||
| Female Male |
18 (37.5%) 30 (62.5%) |
9 (33.3%) 18 (66.7%) |
27 (36.0%) 48 (64.0%) |
0.9122† |
| Primary site, n (%) | ||||
| Scalp, face, neck Occult Other |
8 (16.7%) 9 (18.8%) 31 (64.6%) |
3 (11.1%) 6 (22.2%) 18 (66.7%) |
11 (14.7%) 15 (20.0%) 49 (65.3%) |
0.7869‡ |
| Line of ICB, n (%) | ||||
| 1st line >1st line |
48 (100.0%) 0 (0.0%) |
24 (88.9%) 3 (11.1%) |
72 (96.0%) 3 (4.0%) |
0.0813† |
| Cutaneous primary, n (%) | ||||
| Yes No |
47 (97.9%) 1 (2.1%) |
24 (88.9%) 3 (11.1%) |
71 (94.7%) 4 (5.3%) |
0.2564† |
| Previous MAPKi, n (%) | ||||
| Yes No |
3 (6.2%) 45 (93.8%) |
5 (18.5%) 22 (81.5%) |
8 (10.7%) 67 (89.3%) |
0.2068† |
| Baseline LDH, n (%) | ||||
| Elevated Normal |
9 (18.8%) 39 (81.2%) |
13 (48.1%) 14 (51.9%) |
22 (29.3%) 53 (70.7%) |
0.0155† |
| AJCC staging v8, n (%) | ||||
| III/M1A/M1B M1C/M1D |
18 (37.5%) 30 (62.5%) |
8 (29.6%) 19 (70.4%) |
26 (34.7%) 49 (65.3%) |
0.6638‡ |
| ECOG, n (%) | ||||
| 0 ≥1 |
40 (83.3%) 8 (16.7%) |
18 (66.7%) 9 (33.3%) |
58 (77.3%) 17 (22.7%) |
0.1715‡ |
| Biopsy region, n (%) | ||||
| Brain Lung Lymph node Others Subcutaneous |
5 (10.4%) 3 (6.2%) 13 (27.1%) 4 (8.3%) 23 (47.9%) |
2 (7.4%) 2 (7.4%) 11 (40.7%) 2 (7.4%) 10 (37.0%) |
7 (9.3%) 5 (6.7%) 24 (32.0%) 6 (8.0%) 33 (44.0%) |
0.7920‡ |
| Hemoglobin | ||||
| Median (range) Nine missing details were removed§§ |
139.5 (87–183) | 140 (79–163) | 140 (79–183) | 0.5754* |
| Neutro lympho ratio | ||||
| Median (range) Nine missing details were removed§§ |
2.45 (0.8–11.7) | 3.78 (0.87–26.90) | 2.7 (0.8–26.9) | 0.05197* |
| Brain metastasis, n (%) | ||||
| Yes No |
8 (16.7%) 40 (83.3%) |
8 (29.6%) 19 (70.4%) |
16 (21.3%) 59 (78.7%) |
0.3069† |
| Lung metastasis, n (%) | ||||
| Yes No |
31 (64.6%) 17 (35.6%) |
19 (29.6%) 8 (70.4%) |
50 (66.7%) 25 (33.3%) |
0.7986† |
| Liver metastasis, n (%) | ||||
| Yes No |
9 (18.8%) 39 (81.2%) |
8 (29.6%) 19 (70.4%) |
17 (22.7%) 58 (77.3%) |
0.4278† |
| Median PFS (months) | Not yet reached | 2.37 | 48.7 (95% CI: 23 to NA) | – |
| 1-year PFS (%) | 96 | 0 | 61 (95% CI: 51 to 73) | – |
Bold p-values indicate statistical significance at p < 0.05 level.
*Wilcoxon test.
†Pearson’s χ2 test with Yate’s correction was used for categorical variables.
‡Pearson’s χ2 test.
§Nine missing hemoglobin and neutro lympho ratio samples were removed.
AJCC, American Joint Committee on Cancer; ECOG, Eastern Cooperative Oncology Group; ICB, immune checkpoint-blocking; IPI+PD1, anti-cytotoxic T lymphocyte antigen-4; MAPKi, mitogen-activated protein kinases inhibitors; PD-1, programmed cell death protein 1; PFS, progression-free survival.
The median age of patients treated with IPI+PD-1 therapy was 57 years (table 2). A higher proportion of responding patients had a normal baseline LDH compared with non-responders (p value=0.0155). However, no association was observed between response and age, sex, primary melanoma subtype, line of ICB, primary site, previous treatment with targeted kinase therapy, M stage at entry, ECOG, biopsy region, hemoglobin, neutrophil to lymphocyte ratio, presence of lung, brain or liver metastasis. Median PFS and 1-year PFS were 48.7 months (95% CI: 23 months to not yet achieved) and 61% (95% CI: 51% to 73%), respectively. The clinical and blood characteristics of all 155 patients are provided in online supplemental table 2.
jitc-2023-007144supp001.pdf (630.1KB, pdf)
An initial TME classification based on all 16 cell phenotypes showed associations with response and PFS in patients treated with PD-1 monotherapy but not with IPI+PD-1
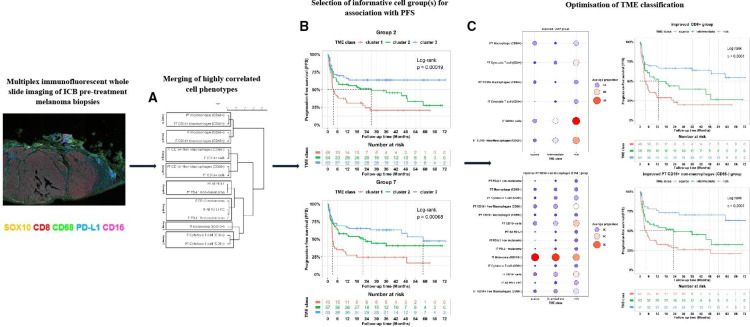
An initial TME classification was obtained using 16 cell phenotypes (online supplemental table 1). These included proportions of cytotoxic T-cells, macrophages and melanoma cells, as well as their PD-L1 and CD16 expressions within IT and PT regions. The optimal number of classes to classify the TME was determined to be three based on NbClust analysis (figure 1A). Unsupervised clustering revealed distinct content of these three TME classes (figure 1B).
Figure 1.
Initial TME classification using all cell phenotypes as individuals. (A) Nbclust analysis of the optimal number of classes with the metrics (indices) ranking three clusters as the most selected number. (B) Descriptive heatmap showing proportions of immune cells within TME classes identified via unsupervised clustering of all immune cells. (C) The response rates of all treated patients within the TME classes with higher response rate in TME class III and response rate (p value=0.0021). (D) Kaplan-Meier curves for PFS of the 155 patients showing the association between TME class and PFS (log-rank test, p value=0.0029). (E) Proportions of responders and non-responders treated with PD-1 in different TME classes (p value=0.02). (F) Kaplan-Meier curves showing the association between TME classes and PFS in the PD-1 cohort (log-rank test, p value=0.039). (G) Proportions of responders and non-responders treated with IPI+PD-1 in each TME class (p value=0.15). (H Kaplan-Meier curves show no association between TME classes and PFS in the IPI+PD-1 cohort (log-rank, p value=0.14). The table below each Kaplan-Meier plot shows patients at risk at 0, 6, 12, 18, 24, 30, 36, 42,48, 54, 60, 66, and 72 months mark. ECOG, Eastern Cooperative Oncology Group; IPI, ipilimumab; IT, intratumoral; PD1, programmed cell death protein 1; PD-L1, PD-1 receptor ligand; PFS, progression-free survival; PT, peritumoral; TME, tumor microenvironment.
While these TME classes were associated with response and PFS across the entire cohort (n=155, p value=0.0021 for response and log-rank test, p value=0.0029 for PFS; figure 1C,D) and within patients that received PD-1 monotherapy (p value=0.02 for response and log-rank test, p value=0.039 for PFS; figure 1E,F), no association was observed with either response (p value=0.15; figure 1G) or PFS (log-rank test, p value=0.14; figure 1H) for patients treated with IPI+PD-1.
TME classifications based on CD8+ and CD16+ cells improve association with PFS
As there was no association between the TME classes obtained across all phenotypes and PFS or response in the group of patients treated with IPI+PD-1, we reasoned that the presence of highly correlated cell phenotypes could result in suboptimal clustering. Hence, we sought to identify homogeneous43–47 (ie, highly correlated) cell phenotypes that, when grouped, would result in TMEs that maximized the association with PFS and response (see online supplemental material and figure 2). We identified eight cell groups: melanoma cells (group 1), CD8+T cells (group 2), IT CD16 macrophages (group 3), IT CD16+ cells (group 4), IT PD-L1 cells (group 5), PT CD16 macrophages (group 6), PT CD16+ non-macrophages (CD68−) cells (group 7), and PT PD-L1 cells (group 8) with distinct cell contents (figure 2A). Each cell group was then used separately to develop a TME classification (online supplemental figure 1). Next, we assessed the relationship between TME classes from each group with treatment response and PFS (online supplemental table 3 and online supplemental figures 2A,B). Our data showed that the TME classes from groups 2 and 7 were best associated with PFS and response. Group 2 was characterized by IT and PT cytotoxic (CD8+) T-cells, and group 7 was characterized by PT CD16+ non-macrophages (CD68−) and PT CD16+ cells.
Figure 2.
Workflow of the study methodology. We identified three TME classes (clusters I, II, and III) using all 16 cell phenotypes, but they did not achieve optimal stratification of PFS nor showed significant associations with the treatment-specific response (see figure 1A-H). To improve the TME classification, we developed a new method to cluster the 16 immune cells into k (we found k=8) homogeneous groups (A), from which we derived three TME classes (cluster 1, cluster 2 and cluster 3) (online supplemental figure 1). The most clinically divergent PFS and response associations were observed for TME classes from cell groups 2 and 7 (B). To further enhance TME classification, we identified other cell groups that could be efficiently combined with cells in groups 2 and 7, resulting in improved groups 2 and 7 (C). We then compared the TME classes (scarce, intermediate, and rich) derived from these two improved groups. The TME classes from the enhanced group 7 demonstrated a stronger association with PFS and response and were thus chosen as the final TME classes for the manuscript. ICB, immune checkpoint-blocking; IT, intratumoral; PD-L1, PD-1 receptor ligand; PT, peritumoral; TME, tumor microenvironment.
Optimal combinations of CD8+ and CD16+ cells with other cell groups resulted in TME classes that better stratify PFS than CD8+ and CD16+ cells
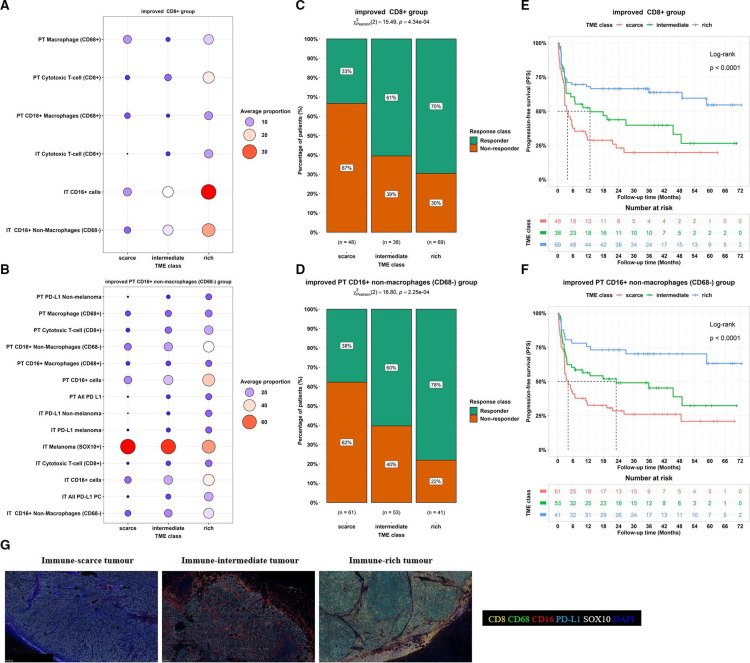
To further identify TME classes that improve the TME classes from groups 2 and 7 and associations with response and PFS, we reasoned that the combinations of the eight uncorrelated cell groups could result in TME classes that better stratify patients, as these groups capture unique information. To achieve this, we sought to further improve the TME classifications based on groups 2 and 7 by identifying optimal combinations of each of these groups with others via an exhaustive search algorithm (online supplemental table 4). Two classifiers were built based on groups 2 and 7 with the addition of linear combinations of cells in other groups to maximize the association with PFS. The subsequent classifiers were termed the improved CD8+ group (figure 3A) and the improved PT CD16+ non-macrophages (CD68−) group (figure 3B), respectively. These improved group combinations resulted in three TME classes labeled: immune-scarce, immune-intermediate and immune-rich.
Figure 3.
Improved TME clusters based on the integration of groups 2 and 7 with the remaining cell groups. (A,B) Circle plots of the average proportion of cells in different TME classes for the improved CD8 (A) and the improved PT CD16+ non-macrophages (CD68−) (B) groups. (C,D) Proportions of responders and non-responders in different TME classes; a significant association was found between treatment response and TME classes from the improved CD8 (p value=0.000434; (C) and the improved PT CD16+ non-macrophages (CD68−) (p value=0.000225; (D) groups, (E,F). Kaplan-Meier curves for PFS showing an association between TME class and PFS TME classes for the improved CD8 (log-rank test, p value<0.0001; (E) and the improved PT CD16+ non-macrophages (CD68−) (log-rank test, p value<0.0001; (F) groups. (G) Representative multiplex immunofluorescence staining images of immune cell phenotypes at the IT and PT regions of each TME class (CD16+: red, CD8+: yellow, CD68+: green, DAPI: blue, PD-L1: cyan, SOX10: white). The table below each Kaplan-Meier plot shows patients at risk at 0, 6, 12, 18, 24, 30, 36, 42,48, 54, 60, 66, and 72 months mark. IT, intratumoral; PD-L1, PD-1 receptor ligand; PT, peritumoral; TME, tumor microenvironment.
jitc-2023-007144supp002.xls (64KB, xls)
TME classification based on the improved CD16+ cell group maximised the associations with outcomes compared with the improved CD8+ cell group
Next, we assessed the association between the improved TME classes (immune-scarce, immune-intermediate and immune-rich) and response for the improved CD8+ group (figure 3C) and the improved PT CD16+ non-macrophages (CD68−) group (figure 3D). A higher proportion of responders had immune-rich tumors (improved CD8+ group: 70% vs 30%; improved PT CD16+ non-macrophages (CD68−) group: 78% vs 22%) and immune-intermediate tumors (improved CD8+ group: 61% vs 39%; improved PT CD16+ non-macrophages (CD68−) group: 60% vs 40%) compared with non-responders. Conversely, there was a lower incidence of responders than non-responders in the immune-scarce tumors (improved CD8+ group: 33% vs 67%; improved PT CD16+ non-macrophages (CD68−) group: 38% vs 62%).
Comparison of the PFS stratified by the TME classes from these two improved cell groups showed that the improved PT CD16+ non-macrophages (CD68−) group led to longer 1-year and median PFS (immune-rich: 76% and not reached, respectively, immune-intermediate: 56% and 23.0 months, respectively, and immune-scarce: 33% and 4.1 months, respectively; log-rank test p values<0.00001; figure 3E) compared with the improved CD8+ group (immune-rich: 68% and not reached, respectively, immune-intermediate: 53% and 12.7 months, respectively, and immune-scarce: 29% and 3.8 months, respectively; log-rank test p values<0.00001; figure 3F). Hence the improved PT CD16+ non-macrophages (CD68−) group-based TME classes (figure 3G) were chosen as the final (optimal) classification.
Immune microenvironment characterization of the improved CD16+-based TME classes
Immune cell content was assessed to understand the compositions of immune-scarce, immune-intermediate and immune-rich tumors (figure 4A–E). The immune-scarce tumor was characterized or defined by the contribution of all cell phenotypes, with PT cytotoxic T-cell (CD8+), IT CD16+, IT cytotoxic T-cell (CD8+), PT T-cell (CD8+), and IT CD16+ non-macrophages (CD68−) positive cells being the strongest contributing cells in labeling this class (figure 4B). This class had lower proportions of cytotoxic T-cells, macrophages, PD-L1 and CD16 expression within IT and PT regions compared with the other two TME classes (figure 4E). The immune-intermediate tumor class was mainly characterized by IT PD-L1 non-malignant cells, which were lower than the immune-rich tumor (figure 4C,E). The immune-rich tumor also had contributions from all cell phenotypes besides the IT macrophage (CD68+) positive cells (figure 4D). The class had the lowest proportions of melanoma (SOX10+) cells and the highest proportions of other cells compared with the immune-intermediate and immune-scarce phenotypes (figure 4E). Differences in CD8+ cell proportions in the IT and PT regions were also observed across all TME classes (figure 4F).
Figure 4.
Cell composition and contributing immune cell features to the improved TME group 7. (A) Heatmap showing proportions of immune cells in the final TME classes (ie, the improved PT CD16+ non-macrophages (CD68−) group-based TME class). (B–D) Immune cell compositions within each TME class: immune-scarce (B), immune-intermediate (C), and immune-rich (D) tumors, were ranked according to their representations (using v-test statistics). Cell phenotype with a v-test less than −2 (red line) or greater than 2 (red line) was significantly represented (corresponds to p values<0.05); the bar length signifies the cell phenotype representation order within TME class (E) Plot comparing the average proportions of immune cells across spatial phenotypes. All comparisons showed significant differences (p value<0.0001). (F) Plot comparing average proportions of immune cells in the IT and PT regions for each spatial phenotype. The symbols *, ** and **** imply significant differences at p value<0.05, 0.01 and 0.0001, respectively. ECOG, Eastern Cooperative Oncology Group; IT, intratumoral; PD-L1, PD-1 receptor ligand; PT, peritumoral.
CD16+ cell-based TME classification associated with treatment-specific PFS
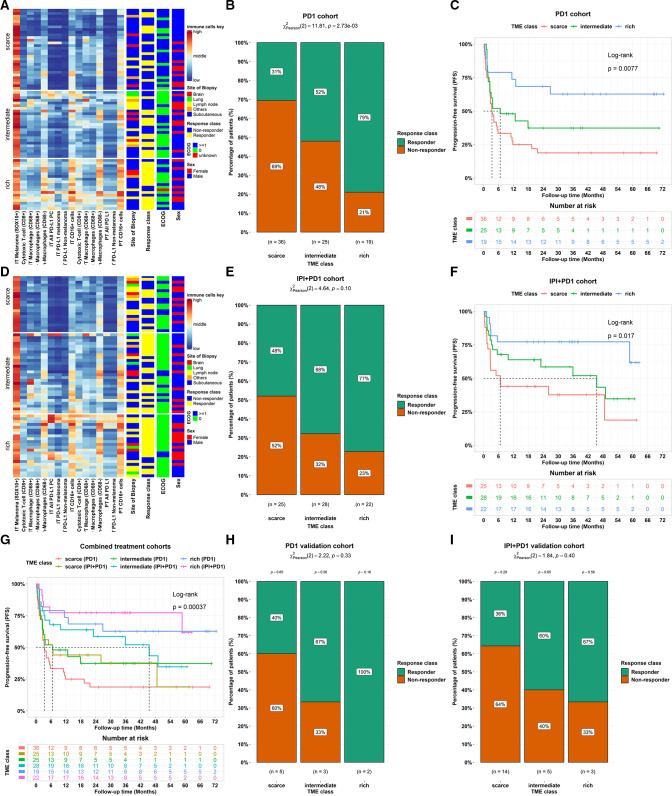
Next, we assessed the association between the optimal TME classes (ie, immune-scarce, immune-intermediate and immune-rich tumors) and response and PFS after stratifying by treatment class: PD-1 monotherapy (figure 5A) or IPI+PD-1 combination therapy (figure 5D).
Figure 5.
Associations between the final improved TME classifier and patient outcomes. (A) Heatmap showing proportions of immune cells in TME classes for the PD-1. (B) Proportions of responders and non-responders in different TME classes for the PD-1 (p value=0.00273). (C) Kaplan-Meier curves for PFS showing an association between TME class in PD-1-treated patients (log-rank test, p value=0.0077). (D) Heatmap showing proportions of immune cells in TME classes for the IPI+PD-1 treated patients. (E) Proportions of responders and non-responders in different TME classes for the IPI+PD-1 (p value=0.10). (F) Kaplan-Meier curves for PFS showing an association with TME class for IPI+PD-1-treated patients (log-rank test, p value=0.017). (G) Kaplan-Meier curves for PFS showing the association with TME class stratified by treatment types (combined cohorts: log-rank test, p value=0.00037). The table below each Kaplan-Meier plot shows patients at risk at 0, 6, 12, 18, 24, 30, 36, 42,48, 54, 60, 66, and 72 months mark. (H–I) Comparison of treatment response rates across TME classes within the validation cohort. (H) Proportions of responders and non-responders across TME classes in PD-1-treated patients. (I) Proportions of responders and non-responders across TME Classes in IPI+PD-1-treated patients. ECOG, Eastern Cooperative Oncology Group; IPI, ipilimumab; IT, intratumoral; PD1, programmed cell death protein 1; PD-L1, PD-1 receptor ligand; PT, peritumoral; TME, tumor microenvironment.
For patients treated with PD-1, higher proportions of responders were found in immune-intermediate and immune-rich classes compared with non-responders (immune-rich: 79% vs 21%, immune-intermediate: 52% vs 48%, and immune-scarce: 31% vs 69%; p value=0.00273; figure 5B). The TME classes were significantly associated with PFS (log-rank test, p value=0.0077; figure 5C). Similarly, higher proportions of responders were observed in both the immune-intermediate and immune-rich classes in patients treated with IPI+PD-1 (immune-rich: 77% vs 23%, immune-intermediate: 68% vs 32%, and immune-scarce: 48% vs 52%; figure 5E), though this association did not reach statistical significance (p value=0.10). The TME classes were significantly associated with PFS (log-rank test, p value=0.017; figure 5F).
Patients treated with IPI+PD-1 had a longer 1-year and median PFS compared with those treated with PD-1 (log-rank p value=0.00037; figure 5G) for each TME class. The 1-year PFS rate and median PFS in the IPI+PD-1 versus PD-1 were: 77% versus 74% and no median was achieved in both treatment groups in the immune-rich class, 64% versus 48% and 45.5 months versus 6.67 months in the immune-intermediate class, and 44% versus 25% and 6.67 versus 3.35 months in the immune-scarce class.
CD16+ cell-based TME classification associated with baseline clinical features
The associations between clinical outcomes and the TME classes combined with baseline clinical characteristics were investigated (online supplemental table 5). The immune-rich and immune-intermediate classes were associated with higher proportions of patients with an ECOG status of 0 (p value=0.0005). There was no association between the TME classes and gender, age at the start of treatment, primary site, lines of ICB, primary cutaneous status, previous targeted therapies, baseline LDH, M stage at entry, hemoglobin, neutrophil-to-lymphocyte ratio, or the presence of brain, lung, or liver metastases. Also, there were no associations between response and site of biopsy for any of the TME classes (p values: = 0.53 (immune-scarce), 0.63 (immune-intermediate), and 0.42 (immune-rich) (online supplemental figure 3A).
Multivariable regression analysis using response as an endpoint combining baseline clinical features with the TME classes revealed the immune-rich class (adjusted OR (AOR)=5.817; 95% CI: 1.932 to 17.518, p value=0.0017) was associated with a higher likelihood of responders compared with the scarce TME class (online supplemental table 6). Even though the association between the immune-intermediate TME class and the odds of response was not significant, the odds of responders were still higher in this group compared with the scarce TME class (AOR=2.127; 95% CI: 0.828 to 5.466, p value=0.1169). Also, the association between odds of response and gender, ECOG, baseline LDH, M stage at entry, site of biopsy, previously targeted therapies, lines of ICB, neutrophil-to-lymphocyte ratio, the presence of lung or liver metastasis was not significant.
Also, multivariable Cox regression analysis using PFS as an endpoint (online supplemental figure 3B) revealed that the immune-rich group had a lower risk of progression (adjusted HR=0.29; 95% CI: 0.14 to 0.60, p value<0.001) compared with patients with immune-scarce groups. However, immune-intermediate versus immune-scarce tumors, age at the start of treatment, sex, ECOG, biopsy regions, previous BRAF or MEK inhibitors, line of ICB and M stage at entry were not associated with PFS.
Immunotherapy response patterns in the validation cohort
This cohort comprised 23 patients treated with IPI+PD-1 immunotherapies and 10 with PD-1 monotherapy. While a small cohort, similar patterns to those observed in the larger discovery cohort were identified for the association between TME classes and treatment response in patients treated with PD-1 (p value=0.33; figure 5H) and IPI+PD-1 (p value=0.40; figure 5I), though these associations did not reach statistical significance. For patients treated with PD-1 monotherapy, the immune-rich and immune-intermediate classes had higher proportions of responders than the immune-scarce group. Specifically, the immune-rich class exhibited a complete response rate of 100%, with no non-responders. Similarly, the immune-intermediate class had a larger proportion of responders (67%) than of non-responders (33%). In contrast, the immune-scarce class had fewer responders (40%) than non-responders (60%). Likewise, IPI+PD-1-treated patients presented similar trends, with both the immune-rich and immune-intermediate classes having more responders than the non-responders. The response rate was 67% in the immune-rich group and 60% in the immune-intermediate group. In contrast, fewer patients (33%) in the immune-scarce group responded to treatment. This consistency across both cohorts underscores the potential relevance of TME classes in influencing response to immunotherapy.
Discussion
In this study, we quantitatively classified the TME of patients with metastatic melanoma treated with PD-1 therapy or IPI+PD-1 immunotherapy in relation to the clinical outcomes. Spatial profiling of cytotoxic T-cells, macrophages and melanoma cells, along with phenotypic PD-L1 and CD16 proportions within the IT and surrounding PT regions, were used to identify three TME classes: immune-scarce, immune-intermediate, and immune-rich.
Associations between TME classes and clinical outcomes have been investigated in many different tumor types.20 21 24 48 49 In concordance with previous studies, our data revealed that the TME classes were associated with response and PFS in patients with metastatic melanoma treated with PD-1 monotherapy and IPI+PD-1. The immune-scare tumor was associated with poor response and shorter PFS. The immune-intermediate tumor had a better response rate and longer PFS than the immune-scarce tumor. The immune-rich tumor was associated with the best response and longest PFS. Our findings were consistent with previous studies1 3 5 50 51 demonstrating that patients treated with IPI+PD-1 had a better response and PFS than those treated with PD-1. The response rates were about 1.5 times higher in patients with immune-scarce and immune-intermediate tumors treated with IPI+PD-1 immunotherapy compared with PD-1 monotherapy. PD-1+IPI has been shown to increase T-cell recognition of tumor antigens, with increased T-cell receptor diversity following IPI treatment, while PD-1 is suggested to predominantly expand existing clones.52 53 Therefore, patients with immune-scarce tumors may require IPI+PD-1 to induce tumor recognition and are the subgroup most likely to benefit from combination therapies.
Cytotoxic CD8+ tumor-infiltrating T-cells have been identified as one of the most relevant predictive tumor biomarkers in TME of metastatic melanoma.48 54–56 Elevated cytotoxic CD8+T cells in the TME have been linked with positive anti-tumor effects in breast,57 colorectal, glioblastoma,58 and cervical cancers.59 Our findings confirmed that CD8+T cells within IT and PT regions are essential biomarkers of ICB response. Our data further revealed PT CD16+CD68− non-macrophages and PT CD16+ cells as important combinations of biomarkers in TME, the presence of which are associated with improved outcomes. We observed a comparable PFS from the CD8+ and PT CD16+CD68− non-macrophages cell-based TME classes. High CD8+T cell and PT CD16+ non-macrophage (CD68−) levels within the TME were associated with improved immunotherapeutic response and prolonged PFS. High densities of CD16+ tumor-infiltrating cells have been associated with enhanced responsiveness to chemotherapy and improved survival in ovarian cancer60 61 and colorectal carcinoma.62 CD16 is expressed in a range of immune cells, such as natural killer (NK) cell monocytes, macrophages and gamma delta T-cells.63–65 The CD16 receptor is known to bind to the ipilimumab antibody (IgG166), which can induce antibody-dependent cellular cytotoxicity (ADCC) and depletion of regulatory T-cells (Treg).64 67 Therefore, the inclusion of CD16 within the TME classification of ipilimumab-treated patients is in line with functional assays, which suggests a direct interaction between the theory and the CD16-expressing cells.57 58
Our data revealed that the optimal TME classes (immune-scarce, immune-intermediate, and immune-rich) based on the integration of PT CD16+ non-macrophages (CD68−) cells with the linear combinations of other cells led to prolonged PFS compared with the classification based on PT CD16+ non-macrophages (CD68−) cells or CD8+ cells. The immune-scarce tumor had the highest proportions of melanoma cells and the lowest proportions of cytotoxic T-cells and macrophages, along with reduced PD-L1 and CD16 expression. The immune-intermediate tumor showed lower proportions of melanoma cells and higher proportions of other IT and PT immune cells than the immune-scarce class; it was mainly characterized by IT PD-L1 non-melanoma cells. The immune-rich tumor had the lowest proportions of melanoma cells and the highest proportions of other IT and PT immune cells compared with both immune-intermediate and immune-rich tumors. PT CD16+, PT cytotoxic T-cell (CD8+), IT PD-L1 non-melanoma, IT CD16+, and IT cytotoxic T-cell (CD8+) positive cells were the most represented cells of the immune-rich tumor.
Emerging evidence suggests that macrophages can directly influence the efficacy of immunotherapies including checkpoint inhibitors.32 33 A recent study by Lee et al 34 showed that intratumoral CD16+ macrophages are associated with clinical outcomes in patients with metastatic melanoma treated with a combination of anti-PD-1 and anti-CTLA-4 therapies. Moreover, CD16 is expressed on NK cells and some subsets of macrophages and mediates ADCC.68 69 Numerous studies have underscored the potential of using ADCC in cancer immunotherapy, including in melanoma, highlighting the significance of this immune mechanism.70 71 CD16 has also been shown to play a role in Treg clearance, which can significantly affect the immunotherapy efficacy.72 73 However, further investigation is required to comprehensively understand the therapeutic implications of CD16 in antibody-based immunotherapy.
In our study, multiplex immunofluorescence staining was performed using digital image analysis to identify immune cell phenotypes. Consequently, computed cell phenotypes may result in a certain degree of misclassification. Improvements in automated cell phenotyping, which can accommodate for batch effect and staining intensity changes, are required and may improve these classifications of the TME. Additionally, the findings presented in this study were obtained from a TME classification method that combined cell clustering and selection using optimal PFS stratification. While the proposed classification algorithm showed an encouraging degree of efficacy, it still has room for further refinements to optimize its performance. The cell selection component of the proposed method was specific to optimizing PFS; however, it could be optimized for other clinical outcomes, such as overall survival. Furthermore, as not all patients with immune-rich tumors will benefit from treatment with standard-of-care immunotherapies, clinical (as in74) or multiomics models should be incorporated into prediction models for improved patient outcomes. However, the findings here provide essential insights into the predictive significance of the tumor-immune profiles.
Moreover, when we analyzed the combined data set (online supplemental table 2), we found significant associations between the response to checkpoint inhibitor immunotherapy and age at the start of treatment, line of ICB, previous Mitogen-activated protein kinases inhibitor (MAPKi) treatment, baseline LDH, ECOG status, neutro–lympho ratio, and the presence of brain metastasis. However, after stratifying the data by treatment type (PD-1 alone or IPI+PD-1), most of these associations were no longer apparent (tables 1 and 2). This outcome could be largely attributed to the reduced sample size in the stratified analysis, which potentially affected the statistical power to detect the significant associations. While we acknowledge that our sample size might have limited our ability to detect associations between the clinical variables and response to checkpoint inhibitor immunotherapy when separated by treatment type, our study provides valuable insight towards developing a more comprehensive and accurate model for response and PFS using the identified TME classes. This would undoubtedly contribute to personalized treatment strategies and improve the outcomes of patients with melanoma. Other limitations include the focus on cytotoxic T-cells and CD68+ macrophages. Future studies that consider other influential cell types alone and in combination75 such as polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) and neutrophils, will shed light on their role in TME-based classification and their influence on immunotherapy response.76 77 As high-dimensional imaging technologies become more assessable, broader cell and marker panels expand the classification of the TME.
In conclusion, the comprehensive evaluation of the cells within IT and PT tissue regions associated with TME infiltration patterns yielded several insights that shed light on how tumors respond to PD-1 monotherapy or IPI+PD-1 combination immunotherapy. Quantitative spatial profiling of immune markers via multiplex immunofluorescence could be a valuable tool for classifying TME into relevant classes based on the levels of different immune populations. Our study has accurately assigned tumors from patients with metastatic melanoma into three TME classes with distinct IT and PT cell patterns and clinical features. The TME classes were associated with patient response to PD-1 monotherapy or IPI+PD-1 immunotherapy and PFS. Critically, this TME classification, which is performed prior to the commencement of therapy and is correlated to outcomes, will be invaluable in stratifying patients to receive standard-of-care therapies (predicted good outcome) versus novel strategies and clinical trials (predicted poor outcome). Our study provides an additional diagnostic or immune-oncology tool for immunotherapy biomarkers in precision medicine by categorizing quantitative pathology data into a functional and clinically relevant immune phenotype.
Acknowledgments
We would like to thank MIA patients and their families for their contributions to our study. Ongoing support from colleagues at the Melanoma Institute Australia and the Charles Perkins Centre, University of Sydney, is also greatly acknowledged. Support from the CLEARbridge Foundation as well as from colleagues at Melanoma Institute Australia and Royal Prince Alfred Hospital is also gratefully acknowledged.
Footnotes
Twitter: @SerineLo, @Twitter @ProfRScolyerMIA
Contributors: Study design: NAA, TNG, RAS, GL and JSW. Sample and clinical data acquisition: IPdS, TNG, YM, AMM, MSC, RAS, GL and JSW. Experiments and data analysis: NAA, TNG and JSW. Statistical models and analysis: NAA. Data interpretation: NAA, TNG, IAV, SNL and JSW. Writing—original draft: NAA, TNG, IAV, CQ, EP and JSW. Writing—review and editing: all authors. Supervision: RAS, GL and JSW. Funding: TNG, RAS, GL and JSW. All authors have read and agreed to the published version of the manuscript. JSW acts as the guarantor of this study.
Funding: This work was supported by a National Health and Medical Research Council of Australia (NHMRC) Program Grant (APP2006415 and APP2018514) (to RAS and GL). RAS, GL and AMM are supported by NHMRC Investigator grants. GL is also supported by the University of Sydney Medical Foundation. JSW is supported by an NHMRC Fellowship (APP1174325), Melanoma Research Alliance young investigator fellowship (#700455), Cancer Council NSW project grant (RG19-15), and CINSW Translational Program Grant (TPG 2021/TPG2114). TNG, CQ and IPdS are supported by a CINSW Early Career Fellowship (2020/ECF1153, 2020/ECF1244, and 2021/ECF1376).
Competing interests: GL is a consultant advisor for Agenus, Amgen, Array Biopharma, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Evaxion, Hexal AG (Sandoz Company), Highlight Therapeutics S.L., Innovent Biologics USA, Merck Sharpe & Dohme, Novartis, OncoSec, PHMR Ltd, Pierre Fabre, Provectus, Qbiotics, Regeneron. IPdS reports personal fees from Bristol Myers Squibb, personal fees from Merck Sharpe & Dohme, and personal fees from Roche. AMM is a consultant advisor for BMS, Merck MSD, Novartis, Roche, Pierre Fabre, and QBiotics. MSC is a consultant advisor for Amgen, BMS, Eisai, Ideaya, MSD, Nektar, Novartis, Oncosec, Pierre Fabre, QBiotics, Regeneron, Roche, Merck, and Sanofi and received honoraria from BMS, MSD, and Novartis. RAS has received fees for professional services from Evaxion, Provectus Biopharmaceuticals Australia, QBiotics, Novartis, MSD, NeraCare, AMGEN, BMS, Myriad Genetics, and GlaxoSmithKline.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Data are available upon reasonable request from the corresponding author.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Sydney Local Health District Human Research Ethics Committee (Protocol No. X20-0086 and 2020/ETH00426). Participants gave informed consent to participate in the study before taking part.
References
- 1. Ascierto PA, Long GV, Robert C, et al. Survival outcomes in patients with previously untreated BRAF wild-type advanced Melanoma treated with Nivolumab therapy: three-year follow-up of a randomized phase 3 trial. JAMA Oncol 2019;5:187–94. 10.1001/jamaoncol.2018.4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schachter J, Ribas A, Long GV, et al. Pembrolizumab versus Ipilimumab for advanced Melanoma: final overall survival results of a Multicentre, randomised, open-label phase 3 study. The Lancet 2017;390:1853–62. 10.1016/S0140-6736(17)31601-X [DOI] [PubMed] [Google Scholar]
- 3. Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or monotherapy in untreated Melanoma. N Engl J Med 2015;373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus Docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. The Lancet 2016;387:1540–50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 5. Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-year survival with combined Nivolumab and Ipilimumab in advanced Melanoma. N Engl J Med 2019;381:1535–46. 10.1056/NEJMoa1910836 [DOI] [PubMed] [Google Scholar]
- 6. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 7. Garutti M, Bonin S, Buriolla S, et al. n.d. Find the flame: predictive biomarkers for immunotherapy in melanoma. Cancers;13:1819. 10.3390/cancers13081819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423–37. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009;9:239–52. 10.1038/nrc2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human Melanoma. J Clin Invest 2016;126:3447–52. 10.1172/JCI87324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gide TN, Silva IP, Quek C, et al. Close proximity of immune and tumor cells underlies response to anti-PD-1 based therapies in metastatic melanoma patients. OncoImmunology 2020;9. 10.1080/2162402X.2019.1659093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scott DW, Gascoyne RD. The tumour Microenvironment in B cell Lymphomas. Nat Rev Cancer 2014;14:517–34. 10.1038/nrc3774 [DOI] [PubMed] [Google Scholar]
- 13. Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated Macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of Stromal cells in the tumour Microenvironment. Nat Rev Immunol 2015;15:669–82. 10.1038/nri3902 [DOI] [PubMed] [Google Scholar]
- 15. Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune Contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 16. Gide TN, Quek C, Menzies AM, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019;35:238–55. 10.1016/j.ccell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 17. Somasundaram R, Connelly T, Choi R, et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat Commun 2021;12:346. 10.1038/s41467-020-20600-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carlino MS, Long GV, Schadendorf D, et al. Outcomes by line of therapy and programmed death ligand 1 expression in patients with advanced Melanoma treated with Pembrolizumab or Ipilimumab in KEYNOTE-006: A randomised clinical trial. Eur J Cancer 2018;101:236–43. 10.1016/j.ejca.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 19. Hodi FS, Chiarion-Sileni V, Gonzalez R, et al. Nivolumab plus Ipilimumab or Nivolumab alone versus Ipilimumab alone in advanced Melanoma (Checkmate 067): 4-year outcomes of a Multicentre, randomised, phase 3 trial. Lancet Oncol 2018;19:1480–92. 10.1016/S1470-2045(18)30700-9 [DOI] [PubMed] [Google Scholar]
- 20. Gruosso T, Gigoux M, Manem VSK, et al. Spatially distinct tumor immune Microenvironments stratify triple-negative breast cancers. J Clin Invest 2019;129:1785–800. 10.1172/JCI96313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination Immunotherapies. Nat Rev Drug Discov 2019;18:197–218. 10.1038/s41573-018-0007-y [DOI] [PubMed] [Google Scholar]
- 22. Sharma P, Allison JP. The future of immune Checkpoint therapy. Science 2015;348:56–61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 23. Sobottka B, Nowak M, Frei AL, et al. Establishing standardized immune Phenotyping of metastatic Melanoma by Digital pathology. Lab Invest 2021;101:1637. 10.1038/s41374-021-00676-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammerl D, Martens JWM, Timmermans M, et al. Spatial Immunophenotypes predict response to anti-Pd1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat Commun 2021;12:5668. 10.1038/s41467-021-25962-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yaseen Z, Gide TN, Conway JW, et al. Validation of an accurate automated Multiplex Immunofluorescence method for Immuno-profiling Melanoma. Front Mol Biosci 2022;9:810858. 10.3389/fmolb.2022.810858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taube JM, Roman K, Engle EL, et al. Multi-institutional TSA-amplified Multiplexed Immunofluorescence reproducibility evaluation (MITRE) study. J Immunother Cancer 2021;9:e002197. 10.1136/jitc-2020-002197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dummer R, Long GV, Robert C, et al. Randomized phase III trial evaluating Spartalizumab plus Dabrafenib and Trametinib for BRAF V600-mutant Unresectable or metastatic Melanoma. J Clin Oncol 2022;40:1428–38. 10.1200/JCO.21.01601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daud AI, Wolchok JD, Robert C, et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody Pembrolizumab in Melanoma. J Clin Oncol 2016;34:4102–9. 10.1200/JCO.2016.67.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noy R, Pollard JW. Tumor-associated Macrophages: from mechanisms to therapy. Immunity 2014;41:49–61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39–51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cassetta L, Pollard JW. Targeting Macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 2018;17:887–904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- 32. Arlauckas SP, Garris CS, Kohler RH, et al. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci Transl Med 2017;9:eaal3604. 10.1126/scitranslmed.aal3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Veglia F, Perego M, Gabrilovich D. Myeloid-derived Suppressor cells coming of age review-article. Nat Immunol 2018;19:108–19. 10.1038/s41590-017-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H, Ferguson AL, Quek C, et al. Intratumoral CD16+ macrophages are associated with clinical outcomes of patients with metastatic melanoma treated with combination anti-PD-1 and anti-CTLA-4 therapy. Clinical Cancer Research 2023;29:2513–24. 10.1158/1078-0432.CCR-22-2657 [DOI] [PubMed] [Google Scholar]
- 35. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 36. Kluger HM, Tawbi HA, Ascierto ML, et al. Defining tumor resistance to PD-1 pathway blockade: recommendations from the first meeting of the SITC Immunotherapy resistance Taskforce. J Immunother Cancer 2020;8:e000398. 10.1136/jitc-2019-000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gide TN, Pires da Silva I, Quek C, et al. Clinical and molecular heterogeneity in patients with innate resistance to anti-PD-1 +/- anti-CTLA-4 Immunotherapy in metastatic Melanoma reveals distinct therapeutic targets. Cancers (Basel) 2021;13:3186. 10.3390/cancers13133186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao XH, Stojkovic I, Obradovic Z. A robust data Scaling algorithm to improve classification Accuracies in BIOMEDICAL data. BMC Bioinformatics 2016;17. 10.1186/s12859-016-1236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charrad M, Ghazzali N, Laval U, et al. Nbclust: an R package for determining the relevant number of clusters in a data set Véronique Boiteau. 2014. Available: http://www.jstatsoft.org/
- 40. Iseas S, Sendoya JM, Robbio J, et al. Prognostic impact of an integrative landscape of clinical, immune, and molecular features in non-metastatic Rectal cancer. Front Oncol 2022;11. 10.3389/fonc.2021.801880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verdonschot JAJ, Merlo M, Dominguez F, et al. Phenotypic clustering of dilated cardiomyopathy patients highlights important pathophysiological differences. Eur Heart J 2021;42:162–74. 10.1093/eurheartj/ehaa841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. R Core Team . R: A language and environment for statistical computing. 2021. [Google Scholar]
- 43. Kiselev VY, Andrews TS, Hemberg M. Challenges in Unsupervised clustering of single-cell RNA-Seq data. Nat Rev Genet 2019;20:273–82. 10.1038/s41576-018-0088-9 [DOI] [PubMed] [Google Scholar]
- 44. Kiers HAL. Simple structure in component analysis techniques for mixtures of qualitative and quantitative variables. Psychometrika 1991;56:197–212. 10.1007/BF02294458 [DOI] [Google Scholar]
- 45. Chavent M, Kuentz-Simonet V, Saracco J. Orthogonal rotation in PCAMIX. Adv Data Anal Classif 2012;6:131–46. 10.1007/s11634-012-0105-3 [DOI] [Google Scholar]
- 46. Vandenbon A, Diez D. A clustering-independent method for finding Differentially expressed genes in single-cell Transcriptome data. Nat Commun 2020;11:4318. 10.1038/s41467-020-17900-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu C, Su Z. Identification of cell types from single-cell Transcriptomes using a novel clustering method. Bioinformatics 2015;31:1974–80. 10.1093/bioinformatics/btv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Desbois M, Udyavar AR, Ryner L, et al. Integrated Digital Pathology and Transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat Commun 2020;11:5583. 10.1038/s41467-020-19408-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao Y, Ma D, Zhao S, et al. Multi-Omics profiling reveals distinct Microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res 2019;25:5002–14. 10.1158/1078-0432.CCR-18-3524 [DOI] [PubMed] [Google Scholar]
- 50. Zimmer L, Apuri S, Eroglu Z, et al. Ipilimumab alone or in combination with Nivolumab after progression on anti-PD-1 therapy in advanced Melanoma. Eur J Cancer 2017;75:47–55. 10.1016/j.ejca.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 51. Munhoz RR, Postow MA. Clinical development of PD-1 in advanced Melanoma. Cancer J 2018;24:7–14. 10.1097/PPO.0000000000000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gangaev A, Rozeman EA, Rohaan MW, et al. Differential effects of PD-1 and CTLA-4 blockade on the Melanoma-reactive Cd8 T cell response. Proc Natl Acad Sci U S A 2021;118:e2102849118. 10.1073/pnas.2102849118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amaria RN, Reddy SM, Tawbi HA, et al. Neoadjuvant immune Checkpoint blockade in high-risk Resectable Melanoma. Nat Med 2018;24:1649–54. 10.1038/s41591-018-0197-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Attrill GH, Owen CN, Ahmed T, et al. Higher proportions of Cd39+ tumor-resident cytotoxic T cells predict recurrence-free survival in patients with stage III Melanoma treated with adjuvant Immunotherapy. J Immunother Cancer 2022;10:e004771. 10.1136/jitc-2022-004771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wong PF, Wei W, Smithy JW, et al. Multiplex quantitative analysis of tumor-infiltrating lymphocytes and Immunotherapy outcome in metastatic Melanoma. Clin Cancer Res 2019;25:2442–9. 10.1158/1078-0432.CCR-18-2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Attrill GH, Lee H, Tasker AT, et al. Detailed spatial Immunophenotyping of primary Melanomas reveals immune cell subpopulations associated with patient outcome. Front Immunol 2022;13:979993. 10.3389/fimmu.2022.979993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim PS, Ahmed R. Features of responding T cells in cancer and chronic infection. Curr Opin Immunol 2010;22:223–30. 10.1016/j.coi.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kmiecik J, Poli A, Brons NHC, et al. Elevated Cd3+ and Cd8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor Microenvironment and at the systemic level. J Neuroimmunol 2013;264:71–83. 10.1016/j.jneuroim.2013.08.013 [DOI] [PubMed] [Google Scholar]
- 59. Piersma SJ, Jordanova ES, van Poelgeest MIE, et al. High number of intraepithelial Cd8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage Cervical cancer. Cancer Res 2007;67:354–61. 10.1158/0008-5472.CAN-06-3388 [DOI] [PubMed] [Google Scholar]
- 60. Lalos A, Neri O, Ercan C, et al. High density of Cd16+ tumor-infiltrating immune cells in recurrent ovarian cancer is associated with enhanced responsiveness to chemotherapy and prolonged overall survival. Cancers (Basel) 2021;13:5783. 10.3390/cancers13225783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poznanski SM, Nham T, Chew MV, et al. Expanded Cd56Superbrightcd16+ NK cells from ovarian cancer patients are cytotoxic against Autologous tumor in a patient-derived Xenograft murine model. Cancer Immunol Res 2018;6:1174–85. 10.1158/2326-6066.CIR-18-0144 [DOI] [PubMed] [Google Scholar]
- 62. Sconocchia G, Zlobec I, Lugli A, et al. Tumor infiltration by FciγRIII (Cd16)+ myeloid cells is associated with improved survival in patients with colorectal carcinoma. Int J Cancer 2011;128:2663–72. 10.1002/ijc.25609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee H, Quek C, Silva I, et al. Integrated molecular and Immunophenotypic analysis of NK cells in anti-PD-1 treated metastatic Melanoma patients. Oncoimmunology 2019;8:e1537581. 10.1080/2162402X.2018.1537581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yeap WH, Wong KL, Shimasaki N, et al. Cd16 is indispensable for Antibodydependent cellular cytotoxicity by human monocytes. Sci Rep 2016;6:34310. 10.1038/srep34310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Braakman E, van de Winkel JG, van Krimpen BA, et al. Cd16 on human Γδ T lymphocytes: expression, function, and specificity for Mouse IgG isotypes. Cell Immunol 1992;143:97–107. 10.1016/0008-8749(92)90008-d [DOI] [PubMed] [Google Scholar]
- 66. Romano E, Kusio-Kobialka M, Foukas PG, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by Nonclassical monocytes in Melanoma patients. Proc Natl Acad Sci U S A 2015;112:6140–5. 10.1073/pnas.1417320112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Arce Vargas F, Furness AJS, Litchfield K, et al. Fc Effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell 2018;33:649–663. 10.1016/j.ccell.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ravetch JV, Lanier LL. Immune inhibitory receptors. Science 2000;290:84–9. 10.1126/science.290.5489.84 [DOI] [PubMed] [Google Scholar]
- 69. Veillette A, Chen J. SIRPα-Cd47 immune Checkpoint blockade in anticancer therapy. Trends Immunol 2018;39:173–84. 10.1016/j.it.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 70. Gandhi MK, Arpon D, Keane C, et al. A novel anti-lymphoma immune evasion mediated by the interaction between PD-1 enriched NK-cells and Cd163+PD-L1+PD-L2+ tumor associated Macrophages, that is more prominent in Hodgkin lymphoma than diffuse large B-cell lymphoma. Blood 2016;128:22. 10.1182/blood.V128.22.918.918 [DOI] [Google Scholar]
- 71. Wang W, Erbe AK, Hank JA, et al. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer Immunotherapy. Front Immunol 2015;6:368. 10.3389/fimmu.2015.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer Immunotherapy. Nat Immunol 2016;17:1025–36. 10.1038/ni.3518 [DOI] [PubMed] [Google Scholar]
- 73. Melero I, Rouzaut A, Motz GT, et al. T-cell and NK-cell infiltration into solid tumors: A key limiting factor for efficacious cancer Immunotherapy. Cancer Discov 2014;4:522–6. 10.1158/2159-8290.CD-13-0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Inˆ I, Silva I, Ahmed T, et al. Clinical models to define response and survival with anti-PD-1 antibodies alone or combined with Ipilimumab in metastatic Melanoma. J Clin Oncol 2022;40:1068–80. 10.1200/JCO.21 [DOI] [PubMed] [Google Scholar]
- 75. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune Microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bronte V, Brandau S, Chen S-H, et al. Recommendations for myeloid-derived Suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Coffelt SB, de Visser KE. Immune-mediated mechanisms influencing the efficacy of anticancer therapies. Trends Immunol 2015;36:198–216.:S1471-4906(15)00037-X. 10.1016/j.it.2015.02.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007144supp001.pdf (630.1KB, pdf)
jitc-2023-007144supp002.xls (64KB, xls)
Data Availability Statement
Data are available upon reasonable request. Data are available upon reasonable request from the corresponding author.