Abstract
Quinolone resistance in clinical isolates of Campylobacter jejuni in Sweden increased more than 20-fold at the beginning of the 1990s. Resistance to 125 μg of ciprofloxacin per ml in clinical isolates was associated with chromosomal mutations in C. jejuni leading to a Thr-86-Ile substitution in the gyrA product and a Arg-139-Gln substitution in the parC product.
Quinolones have been used extensively for the treatment of diarrhea caused by Campylobacter jejuni. Quinolones are also frequently used as prophylaxis for travellers. The frequency of resistance to quinolones among C. jejuni strains was low (1 to 2%) in Sweden until the beginning of the 1990s (16), when it rapidly increased to almost 25% within a few years (17, 23). The mechanisms for quinolone resistance in pathogenic bacteria have been shown to involve chromosomal mutations that modify DNA gyrase (18, 20, 22, 24) or DNA topoisomerase IV (1, 21), the targets of quinolone action. Decreased outer membrane permeability (2, 5, 13) and export through an active efflux system (3, 8) have also been linked to quinolone resistance. Recently, transferable plasmid-borne resistance to quinolones has also been inferred (10). In this study, we investigated mutations of gyrase and topoisomerase IV genes in C. jejuni.
Campylobacter strains were grown at 37°C in a microaerobic atmosphere (5% O2, 10% CO2, 85% N2) for 48 h on blood-free, nonselective nutrient agar (Oxoid Ltd., Basingstoke, United Kingdom). Determination of MICs by broth dilution was performed as described previously (16).
Characteristics of the clinical isolates studied are summarized in Table 1. A quinolone-susceptible reference strain (8382), originally isolated from a fecal sample and susceptible to tetracyclines, erythromycin, and clindamycin, was also included.
TABLE 1.
Antibiotic resistance in clinical isolates of C. jejuni
| Straina | Serogroup (HS:HL)b | MIC (μg/ml) of:
|
||
|---|---|---|---|---|
| Norfloxacin | Ciprofloxacin | Nalidixic acid | ||
| 33324 | 2:4 | 500 | 64 | 1,000 |
| 33325 | 1:9 | 500 | 34 | 1,000 |
| 33328 | 4:1 | 1,000 | 125 | 2,000 |
| 27611 | NT:NT | 500 | 125 | 1,000 |
| 8382 | NT:2 | 0.50 | 0.13 | 34 |
| 34156 | 1:2 | 2 | 0.19 | 8 |
| 34157 | 1:2 | 500 | 125 | >256 |
| 34158 | 1:2 | 1,000 | 125 | >256 |
Strains 33324, 33325, and 33328 are fecal isolates from patients with severe diarrhea; 27611 is a blood isolate from a patient with diarrhea; 8382 is a quinolone-susceptible fecal isolate; and 34156 to 34158 are isolates from a patient with diarrhea who was treated with 400 mg of norfloxacin orally twice daily for 0, 4, and 24 days, respectively. All strains are from an earlier investigation (23).
In order to characterize mutations associated with resistance, the quinolone resistance-determining region (QRDR) of the gyrA gene in the clinical isolates was amplified by PCR. The primers and the method were described previously (22). The templates for PCR amplification were prepared by the boiling method (19). Products of PCR were purified as described previously (11) and digested with EcoRI and BamHI, generating a fragment of 219 bp, which was cloned into M13mp18 or M13mp19 as described previously (14).
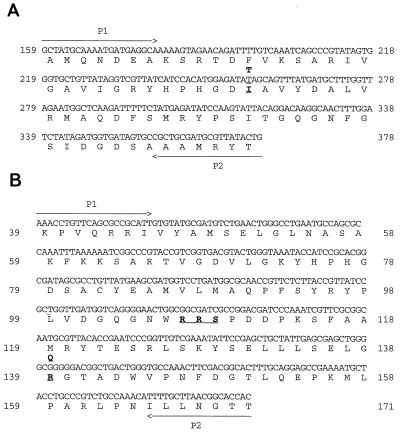
The QRDR sequence of the gyrA gene obtained for the eight isolates in Table 1 is shown in Fig. 1A. The sequence for the quinolone-susceptible strains 8382 and 34156 (with one exception; see below) was identical to that previously reported (22). The QRDRs of the quinolone-resistant isolates (Table 1) all showed the C-to-T transition at nucleotide 256 (Fig. 1A), leading to a Thr-86-Ile substitution, which was earlier observed to mediate high quinolone resistance (22). All eight isolates also had a silent C-to-T change at nucleotide 242 (Fig. 1A) which was not previously reported.
FIG. 1.
Nucleotide and deduced amino acid sequences of the QRDR of the gyrA gene (A) and the corresponding region of the parC gene (B) of the quinolone-resistant isolates (Table 1). The primers for PCR amplification in each case are indicated. (A) The Thr-86-Ile substitution, associated with high-level resistance to quinolones, is shown. (B) Numbering refers to the amino acid residues. Differences in amino acids in comparison to the product of the corresponding gene of E. coli are shown.
In order to identify further mutations associated with resistance, the QRDR of the parC gene of C. jejuni, corresponding to nucleotides 140 to 534 in Escherichia coli parC (6, 21), was amplified. Two primers were used, 5′-TGGGATCCAAACCTGTTCAGCGCCGCATT-3′ (P1) and 5′-CGGAATTCGTGGTGCCGTTAAGCAAA-3′ (P2). BamHI and EcoRI sites were added to the 5′ ends of P1 and P2, respectively. The PCR amplification was as described previously (20), except that 1.0 mM MgCl2 and an annealing temperature of 58°C were used.
The sequence of the parC gene of C. jejuni downstream of the QRDR (sequence between positions 538 and 1238 [Fig. 2]) was determined for the quinolone-susceptible isolates (8382 and 34156 [Table 1]) and for one resistant strain (33324 [Table 1]) by three sequential runs of the capture PCR (CPCR) technique, as described previously (7). Controls without templates were included.
FIG. 2.
Nucleotide sequence of a part of the C. jejuni parC gene starting from the QRDR. The extra bases (TCG) that result in the insertion of a serine residue are in boldface type and underlined. Differences from the nucleotide sequence of the corresponding E. coli parC gene are indicated below the sequence.
The first run of CPCR was carried out with a biotinylated primer based on the known sequence of the QRDR of C. jejuni parC (positions 412 to 437; in Fig. 2, positions 140 to 538 correspond to the QRDR). An amplified fragment of about 355 bp was obtained. The identity of the amplified DNA was confirmed by carrying out two independent PCR runs with two specific primers located downstream of the biotinylated one (nucleotides 438 to 457 and 492 to 514 [Fig. 2]). Three independent clones of the amplified DNA were sequenced as described previously (15). Based on the sequence detected by the first CPCR run (nucleotides 538 to 890 [Fig. 2]), two other CPCR runs were carried out one after the other with two biotinylated primers representing positions 807 to 835 and positions 1094 to 1116 (Fig. 2).
The amplified QRDR of C. jejuni parC was found to be slightly larger (398 bp) than that observed by Vila et al. for E. coli (395 bp) (21). The two identical sequences of this amplification product from the two quinolone-susceptible strains (8382 and 34156 in Table 1) are shown in Fig. 1B. The amino acid sequence is identical to that of the corresponding fragment in E. coli, with four exceptions. The most conspicuous of these are the two arginine residues substituted for glycine and alanine at residues 107 and 108, respectively, immediately followed by the insertion of an extra serine (Fig. 2) flanked by a sequence corresponding to a 5-bp direct repeat (CGATC). The fourth exception is an arginine instead of a glutamine at residue 139. The resistant strains all show a mutation substituting glutamine for arginine at this position. The differences at residues 107 to 109 could be endogenous to C. jejuni. An alternative interpretation could be that because of the intensive exposure to quinolones used in medicine and agriculture, C. jejuni has adapted by mutations at residues 107 to 109, resulting in a protein with a lower affinity for quinolones (resistance) but a lower enzymatic efficiency. The Gln-139-Arg substitution detected in the susceptible isolates could be regarded as a compensatory mutation resulting in an increase in enzyme activity with a concomitant loss of resistance (4).
The nucleotide sequence of the part of C. jejuni parC analyzed here (about 50% of the whole parC gene, compared to the size of the corresponding gene in E. coli) showed 94% similarity (93% after translation) to the corresponding gene in E. coli. A higher degree of similarity was found near the part of the gene corresponding to the N terminus, while the part corresponding to the C terminus was generally found to be more variable (Fig. 2).
In this study, quinolone-resistant isolates of C. jejuni, representing the increase in resistance frequency mentioned above, were analyzed for mutational changes in both the gyrA and parC genes. A single mutation in gyrA, leading to a Thr-86-Ile substitution, in combination with a single mutation in parC, leading to an Arg-139-Gln substitution, had been found in clinical isolates of C. jejuni resistant to high concentrations of ciprofloxacin. The gyrA mutation seen in this study is identical to that observed earlier in C. jejuni (22), but the C. jejuni strains examined were found to display a higher resistance level to ciprofloxacin (34 to 125 μg/ml) than what was observed in the previously mentioned study (22). The interpretation that the combination of mutations in gyrA and parC could be associated with the high resistance is supported by the data presented in Table 1, which show that isolates of the same serogroup from a patient under treatment with ciprofloxacin showed high resistance to quinolones already on the fourth day of treatment and that a comparison of susceptible isolates from the initiation of treatment with resistant isolates from the fourth day showed the two mutations. However, the gyrA and parC mutations observed did not explain why the MICs of ciprofloxacin were 34 μg/ml for strain 33325, 64 μg/ml for strain 33324, and 125 μg/ml for others (27611 and 34157). One possibility could be that there was an additional mutation(s) in another location(s) of the gyrA or gyrB gene. Mutations that affect drug permeation and efflux could also alter the ultimate MICs for these isolates.
Nucleotide sequence accession number.
The sequence of the parC gene has been deposited in the EMBL database under accession number Y18300.
Acknowledgments
We thank Maria Lagerström-Fermér for generously providing the information about the CPCR technique and her kind assistance in determining the parC sequence. The competent assistance of Elsy Johnson is gratefully acknowledged.
REFERENCES
- 1.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charvalos E, Tselentis Y, Hamzehpour M M, Köhler T, Pechere J-C. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob Agents Chemother. 1995;39:2019–2022. doi: 10.1128/aac.39.9.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen S P, Hooper D C, Wolfson J S, Souza K S, McMurry L M, Levy S B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988;32:1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fermér C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J Bacteriol. 1997;179:831–837. doi: 10.1128/jb.179.3.831-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper D C, Wolfson J S, Souza K S, Tung C, McHugh G L, Swartz M N. Genetic and biochemical characterization of norfloxacin resistance in Escherichia coli. Antimicrob Agents Chemother. 1986;29:639–644. doi: 10.1128/aac.29.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in Escherichia coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 7.Lagerström-Fermér M, Parik J, Malmgren H, Stewart J, Pettersson U, Landegren U. Capture PCR: efficient amplification of DNA fragments adjacent to a known sequence in human and YAC DNA. PCR Methods Appl. 1991;1:111–119. doi: 10.1101/gr.1.2.111. [DOI] [PubMed] [Google Scholar]
- 8.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Martinez L, Pascual A, Jacoby G A. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 11.Öfverstedt L G, Hammarström K, Balgobin N, Hjertén S, Pettersson U, Chattopadyaya J. Rapid and quantitative recovery of DNA fragments from gels by displacement electrophoresis (isotachophoresis) Biochim Biophys Acta. 1984;782:120–126. doi: 10.1016/0167-4781(84)90014-9. [DOI] [PubMed] [Google Scholar]
- 12.Penner J L, Hennessy J N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robillard N J, Scarpa A L. Genetic and physiological characterization of ciprofloxacin resistance in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1988;32:535–539. doi: 10.1128/aac.32.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjögren E, Kaijser B, Werner M. Antimicrobial susceptibilities of Campylobacter jejuni and Campylobacter coli isolated in Sweden: a 10-year follow-up report. Antimicrob Agents Chemother. 1992;36:2847–2849. doi: 10.1128/aac.36.12.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjögren E, Lindblom G-B, Kaijser B. Norfloxacin resistance in Campylobacter jejuni and Campylobacter coli from Swedish patients. J Antimicrob Chemother. 1997;40:257–261. doi: 10.1093/jac/40.2.257. [DOI] [PubMed] [Google Scholar]
- 18.Stein D C, Danaher R J, Cook T M. Characterization of a gyrB mutation responsible for low-level nalidixic acid resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1991;35:622–626. doi: 10.1128/aac.35.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Eys G J J M, Gravekamp C, Gerritsen M J, Quint W, Cornelissen M T E, Schegget J T, Terpstra W J. Detection of leptospires in urine by polymerase chain reaction. J Clin Microbiol. 1989;27:2258–2262. doi: 10.1128/jcm.27.10.2258-2262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila J, Ruiz J, Goñi P, Marcos A, Jimenez De Anta T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995;39:1201–1203. doi: 10.1128/aac.39.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vila J, Ruiz J, Goñi P, Jimenez De Anta M T. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Huang W M, Taylor D E. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother. 1993;37:457–463. doi: 10.1128/aac.37.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wretlind B, Strömberg A, Östlund L, Sjögren E, Kaijser B. Rapid emergence of quinolone resistance in Campylobacter jejuni in patients treated with norfloxacin. Scand J Infect Dis. 1992;24:685–686. doi: 10.3109/00365549209054659. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]