Abstract
The basis of joint tolerance to β-lactam and fluoroquinolone antibiotics in Escherichia coli mediated by hipA was examined. An antibiotic tolerance phenotype was produced by overexpression of hipA under conditions that did not affect the growth rate of the organism. Overexpressing hipA probably decreases the period in which bacteria are susceptible to the antibiotics by temporarily affecting some aspect of chromosome replication or cell division.
β-Lactam and fluoroquinolone antibiotics exhibit a bactericidal action against growing cultures of most pathogenic bacteria (16). However, mutants that are as sensitive to growth inhibition by the antibiotics as the wild-type parent strains but which undergo only a slow loss of viability in the presence of the antibiotics have been described previously (9, 12, 14, 15, 17). The phenomenon has been called tolerance or high persistence (9, 12). Mutants tolerant to β-lactam antibiotics have been described in both laboratory mutant strains and clinical isolates (9). However, mutants tolerant to fluoroquinolones have so far only been described in laboratory mutants of Escherichia coli (12, 14, 17). These mutants also display tolerance to β-lactam antibiotics (3, 4, 12, 17). Tolerance to β-lactam antibiotics may have clinical significance (11), but it is not known whether joint tolerance to β-lactams and fluoroquinolones has clinical relevance.
In E. coli, joint tolerance to both peptidoglycan and DNA synthesis inhibitors is under the control of the hip (high persistence) locus (14, 17). Thus, strains containing chemically mutagenized hip exhibit a 1,000-fold reduction in the rate of killing by β-lactam and fluoroquinolone antibiotics (14, 17). The hip locus in E. coli consists of two genes, hipA (1,320 bp), which encodes a weakly expressed 50-kDa protein, and hipB (264 bp), which encodes a Cro-like protein which is a hipA repressor and responsible for low-level hipA expression (3). HipA is found exclusively in a tight complex with HipB, and the stop codon of hipB overlaps the start codon of hipA by one base (3, 4). This close relationship is essential since hipB mutant strains are nonviable, indicating that nonregulated expression of hipA might be lethal (4).
An understanding of antibiotic tolerance mediated by mutations in the hipA gene may provide the key to a link between β-lactam and quinolone mechanisms of action. In this paper, we report on the distribution of hip in bacteria and the role of hipA in tolerance of E. coli through studies on overexpression of hipA.
Distribution of hipA.
A search for hipA and hipB homologues was performed with a range of gram-negative and -positive bacteria. By using standard techniques (13), PCR-amplified E. coli hipA and hipB were used to probe restriction digests of chromosomal DNA from Shigella sonnei, Salmonella typhimurium, Klebsiella aerogenes, Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus hominis, Bacillus subtilis, Providencia vulgaris, and Serratia marcescens. To determine the distribution of the hip locus in E. coli, the hip operon was amplified by PCR with two sets of primers designed to amplify the entire hipA gene and the entire hipB gene (3). Although both hipA and hipB were identified in S. sonnei, homologues could not be identified, even with low-stringency hybridization, in any of the other bacteria examined, including organisms such as Salmonella typhimurium and Klebsiella pneumoniae, which are closely related to E. coli and Shigella sonnei. However, three homologues were identified by amino acid database searches, one in Haemophilus influenzae and two in the Rhizobium symbiosis plasmid pNGR234 (7, 8). In H. influenzae, the gene (HI0665) is significantly disrupted and would be unlikely to express a protein with similar function to HipA. In Rhizobium, the genes (y4mE and y4dM) encode proteins with 28 and 27% identity to HipA, respectively. Significantly, as for HipB, these genes are under the control of strong transcriptional regulators (y4mF and y4dL).
Chromosomal DNA from 40 clinical isolates of E. coli, from different locations worldwide, was PCR amplified with two sets of primers designed for hipA and hipB. Approximately 20% of the strains tested were negative with both primer sets, and the remaining strains were amplification positive with both sets.
Chromosomal deletion of hipA in E. coli.
The E. coli LN2666 (1) hipA gene was replaced with a copy which expressed only the first 25 amino acids of the protein, resulting in strain IC4. This was done by homologous recombination (6) with the plasmid pHp100, a pFC24 (6) derivative in which a 618-bp BssHII fragment of hipA had been excised. To determine if hipA deletion or disruption exhibited a specific phenotype, strains IC4 and LN3559 (hipA::tetA), a gift from J.-M. Louarn, were compared to the parent strain, LN2666, with respect to growth rate, morphology, antimicrobial susceptibility, and total cellular protein profiles. For all these characteristics, there were no differences between the three strains (data not shown).
Overexpression of hipA.
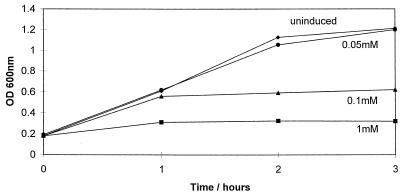
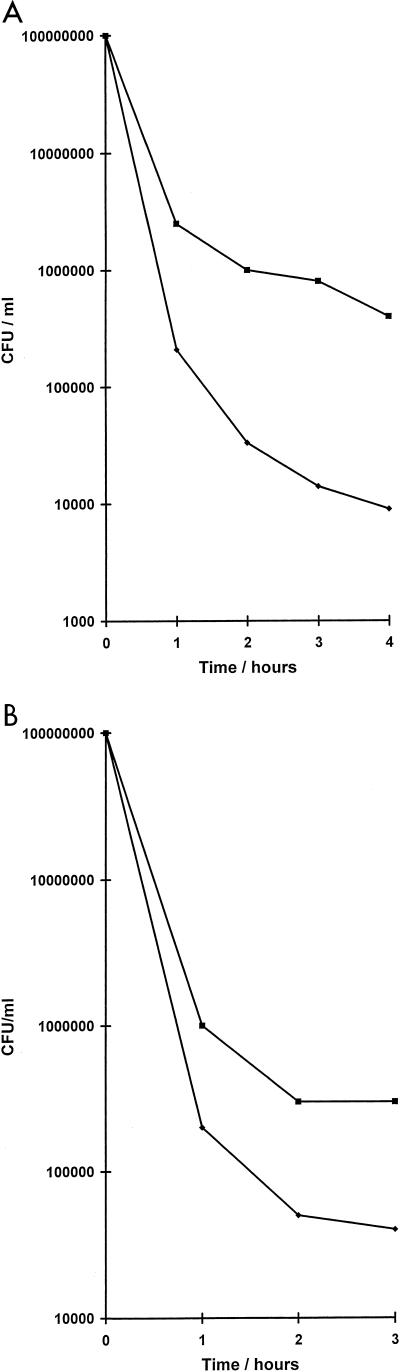
Growth of E. coli BL21 (DE3) carrying pLysS (Promega) and pHp200, a pET30b (Novagen) construct containing the entire hipA open reading frame, was induced with 0.05 to 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Induction produced dose-dependent inhibition of cell division in E. coli BL21 (Fig. 1). The growth rates of cells exposed to IPTG concentrations of less than 0.03 mM were unaffected. However, when challenged with 100 μg of ampicillin per ml, E. coli containing pHp200, induced with 0.01 mM IPTG, exhibited significantly reduced killing in comparison to cells containing the vector alone (Fig. 2A). To determine if this phenomenon was joint tolerance as identified in the original mutants (14), survival studies were performed for cells containing pHp200 that had been exposed to the quinolone ciprofloxacin. In this case, there was a 10-fold difference in the killing of cells containing pHp200 compared to that of cells containing pET30b when exposed to 100 μg of the drug per ml (Fig. 2B). Expression of hipA was confirmed by S•Tag Western blot (Novagen), which showed HipA to be a protein of 49 kDa (data not shown), the size predicted by its sequence.
FIG. 1.
Growth curve of E. coli BL21 (DE3) carrying pLysS and pHp200 with and without IPTG induction. OD600, optical density at 600 nm.
FIG. 2.
Killing of E. coli BL21 (DE3) containing plasmid pHp200 (■) or pET30b (⧫) with 100 μg of ampicillin per ml (A) or 100 μg of ciprofloxacin per ml (B). Cultures contained 0.01 mM IPTG.
The ability to overexpress hipA has allowed us to demonstrate that joint tolerance to β-lactam and quinolone antibiotics in E. coli is due to expression of hipA in excess of hipB and that HipA is toxic to E. coli. Failure to isolate HipA unbound to HipB (4) and the observation made here that overexpression of hipA produced a tolerance phenotype similar to that observed in an earlier work (14) indicate that in the original mutants, reduced binding of HipA to HipB probably resulted in the observed tolerance. However, since neither cassette insertion inactivation (4) nor chromosomal deletion of hipA (this study) conferred tolerance, at least part if not all of HipA is required for the phenotype.
Since the hip locus is restricted to relatively few bacterial species, including not even all strains of E. coli, antibiotic tolerance resulting from hip mutations is unlikely to be clinically significant. In addition, persistence identified in species other than E. coli, e.g., the persistence of β-lactams in staphylococci (2), is unlikely to be related to hip. However, although the role of hip in E. coli tolerance to β-lactam and quinolone antibiotics has been established in this study, the mechanism of tolerance has not. Bigger (2) suggested that persisting bacteria are cells briefly existing in the nondividing phase of their life cycle and survive because penicillin kills only dividing cells. If bacteria are susceptible to β-lactam and quinolone antibiotics only during specific phases of their life cycle, then any phenomenon that reduced the window of opportunity for killing would enable more bacteria to survive the cidal effects of both antibiotics. It is tempting to speculate that HipA may decrease the period of time during which bacteria are susceptible to these antibiotics by affecting some aspect of chromosome replication or cell division. This may relate to the location of hip in the terminal domain of the chromosome within 100 bp of the dif locus and close to terC. These loci are involved in chromosome partitioning and termination of chromosome replication, respectively (5, 10).
Acknowledgments
This work was supported by a grant to I.C. from SmithKline Beecham Pharmaceuticals.
REFERENCES
- 1.Berg C M, Curtiss R. Transposition derivatives of an Hfr strain of Escherichia coli K12. Genetics. 1967;56:503–525. doi: 10.1093/genetics/56.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigger J W. Treatment of staphylococcal infections with penicillin. Lancet. 1944;ii:497–500. [Google Scholar]
- 3.Black D S, Kelly A J, Mardis M J, Moyed H S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black D S, Irwin B, Moyed H S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakely G, Colloms S D, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 6.Cornet F, Mortier I, Patte J, Louarn J M. Plasmid pSC101 harbors a recombinant site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J Bacteriol. 1994;176:3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischmann R D, Adams M D, White O, Clayton R A, Kerlavage E F, Bult C J, Tomb T J, Dougherty B A, Merrick J M, McKenney K, Sutton G, Fitzhugh W, Fields C, Gocayne D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394–401. [DOI] [PubMed]
- 9.Handwerger S, Tomasz A. Antibiotic tolerance among clinical isolates of bacteria. Annu Rev Pharmacol Toxicol. 1985;25:349–380. doi: 10.1146/annurev.pa.25.040185.002025. [DOI] [PubMed] [Google Scholar]
- 10.Hill T M, Hensen J M, Kuempel P L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci USA. 1987;84:1754–1758. doi: 10.1073/pnas.84.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holtje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 12.Hooper D C, Wolfson J S. Mechanisms of quinolone action and bacterial killing. In: Hooper D C, Wolfson J S, editors. Quinolone antimicrobial agents. 2nd ed. Washington, D.C: American Society for Microbiology; 1993. pp. 53–75. [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 14.Moyed H S, Bertrand K P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyed H S, Broderick S H. Molecular cloning and expression of hipA, gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell A D, Chopra I. Understanding antibacterial action and resistance. 2nd ed. New York, N.Y: Ellis Horwood; 1996. [Google Scholar]
- 17.Wolfson J S, Hooper D C, McHugh G L, Bozza M A, Swartz M N. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and β-lactam antimicrobial agents. Antimicrob Agents Chemother. 1990;34:1938–1943. doi: 10.1128/aac.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]