Abstract
A complete and high-quality reference genome has become a fundamental tool for the study of functional, comparative, and evolutionary genomics. However, efforts to produce high-quality genomes for African taxa are lagging given the limited access to sufficient resources and technologies. The southern African dwarf chameleons (Bradypodion) are a relatively young lineage, with a large body of evidence demonstrating the highly adaptive capacity of these lizards. Bradypodion are known for their habitat specialization, with evidence of convergent phenotypes across the phylogeny. However, the underlying genetic architecture of these phenotypes remains unknown for Bradypodion, and without adequate genomic resources, many evolutionary questions cannot be answered. We present de novo assembled whole genomes for Bradypodion pumilum and Bradypodion ventrale, using Pacific Biosciences long-read sequencing data. BUSCO analysis revealed that 96.36% of single copy orthologs were present in the B. pumilum genome and 94% in B. ventrale. Moreover, these genomes boast scaffold N50 of 389.6 and 374.9 Mb, respectively. Based on a whole genome alignment of both Bradypodion genomes, B. pumilum is highly syntenic with B. ventrale. Furthermore, Bradypodion is also syntenic with Anolis lizards, despite the divergence between these lineages estimated to be nearly 170 Ma. Coalescent analysis of the genomic data also suggests that historical changes in effective population size for these species correspond to notable shifts in the southern African environment. These high-quality Bradypodion genome assemblies will support future research on the evolutionary history, diversification, and genetic underpinnings of adaptation in Bradypodion.
Keywords: Africa, chameleon genomics, HiFi data, Hi-C, reference genome, reptiles
Significance.
We generated high-quality reference genome assemblies for two southern African dwarf chameleons, Bradypodion pumilum and Bradypodion ventrale. These genomes are currently among the most complete and contiguous reptile assemblies and are the second and third on record for Chamaeleonidae. Chameleons show unique adaptive traits and ecology, and these assemblies presented here provide a novel resource for evolutionary research into this group.
Introduction
There has been a recent concerted effort to produce fully sequenced genomes for a range of vertebrate species. Initially centered around model organisms and charismatic groups, attention has now turned to include less studied groups (Blaxter et al. 2022; Ebenezer et al. 2022; Genome 10 K Community of Scientists 2009; Koepfli et al. 2015; Lewin et al. 2018). Reptiles are one of the most diverse clades of extant tetrapods, boasting nearly 12,000 species. However, the number of reptile genome assemblies is few relative to other tetrapod taxonomic groups. Furthermore, over 2,200 reptile species (18% of the total) occur in mainland Africa and associated landmasses (Uetz et al. 2023), yet only two full genome assemblies have yet been published (Paroedura picta, Hara et al. 2018; Hemicordylus capensis, Leitão et al. 2023). With finite resources, minimal access to sequencing technologies, and limited bioinformatic expertise, efforts to produce high-quality genome assemblies for African taxa are lagging (Ebenezer et al. 2022). However, given the efforts of genomic initiatives (Ebenezer et al. 2022; Lewin et al. 2022), the production of high-quality genomic resources for African taxa is gaining momentum (Ebenezer et al. 2022; Gupta 2022; Lewin et al. 2022; Ishengoma 2023).
An iconic group of African reptiles are the Chamaeleonidae—arguably one of the most specialized groups of reptiles (Bickel and Losos 2002; Tolley and Herrel 2013; Diaz et al. 2015). Chameleons have undergone evolutionary shifts from a terrestrial to an arboreal lifestyle (Tolley et al. 2013), exhibiting an array of adaptive functional traits presumably linked to habitat specialization (Hopkins and Tolley 2011; Herrel et al. 2013). The southern African dwarf chameleons, Bradypodion, are the youngest chameleon lineage, with multiple instances of adaptive responses to habitat shifts since the Miocene. Some clades have undergone rapid diversification, largely associated with changes in natural environments through ecological speciation (Tolley et al. 2008, 2022). Multiple convergence events have resulted in a few phenotypic morphs, or ecomorphs, in the genus. However, it is unclear whether the convergence of ecomorphs is due to standing genetic variation or if ecomorphs arise because of convergent novel mutations regardless of their phylogenetic history. Similarly, Anolis, a large clade of new world lizards occupying arboreal habitats broadly comparable to Bradypodion, display convergent ecomorphs across distantly related lineages. Given these parallels, anole and dwarf chameleon ecomorphs might employ similar genetic pathways to give rise to shared elements of their ecomorphological phenotypes. High-quality reference genomes from these clades are necessary to investigate this hypothesis.
Using long-read sequencing, we generated genome assemblies of high quality and completeness de novo for two Bradypodion species, the cape dwarf chameleon (Bradypodion pumilum) and the eastern dwarf chameleon (Bradypodion ventrale). We place these assemblies within a phylogenetic framework, identify syntenic relationships within Bradypodion and between Anolis sagrei, and explore the demographic history of B. pumilum and B. ventrale. These genomes provide an exciting opportunity to explore the genetic architecture that underlies the genotype–phenotype and genotype–environment associations in Bradypodion, which could extend to other chameleon genera.
Results and Discussion
Assembly Contiguity and Chromosome Size
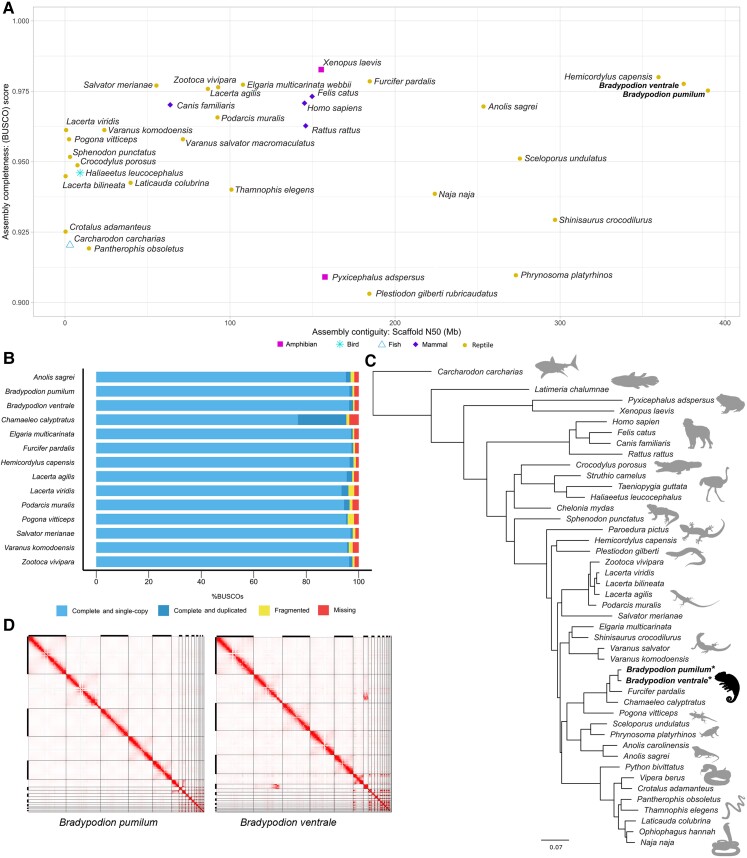
The assemblies for both dwarf chameleon species are the most contiguous reptile genomes produced to date, with a scaffold N50 of 389.6 Mb (B. pumilum, genome size: 2.43 Gb) and 374.9 Mb (B. ventrale, genome size: 2.40 Gb; fig. 1A). The next most contiguous published assembly is H. capensis (359.65 Mb, genome size: 2.29 Gb; Leitão et al. 2023), followed by Shinisaurus crocodilurus (296.95 Mb, genome size: 2.19 Gb; Xie et al. 2022). For each Bradypodion assembly, the three largest scaffolds together cover more than 50% of the genome. Bradypodion pumilum (BraPum1.0) compromises 90% of the genome within the seven largest scaffolds, while B. ventrale (BraVen1.1) holds 90% of the genome within the nine largest scaffolds similar to the contiguity of Sceloporus undulatus and Phrynosoma platyrhinos (supplementary table S1, Supplementary Material online). Both Bradypodion assemblies are highly correlated (supplementary fig. S1, Supplementary Material online; BraPum1.0: r2 = 0.991, P < 0.001; BraVen1.1: r2 = 0.982, P < 0.001) with chromosome sizes estimated based on the published karyotype for Bradypodion (Rovatsos et al. 2017).
Fig. 1.
(A) Scatterplot of recent high-quality vertebrate genomes comparing assembly completeness scores against scaffold N50. Study species are highlighted in bold. BUSCO scores presented only contain assemblies with scores above 0.9. (B) BUSCO assessment of assembly completeness for Bradypodion and closest nine available reptile assemblies based on BUSCO scores. (C) Maximum likelihood phylogenetic tree based on single-copy proteins from BUSCO analysis. Bradypodion assemblies are placed within the context of representative vertebrate taxa in all major groups. The assemblies presented here are indicated with an asterisk. Supported nodes (70% bootstrap) are indicated with a black dot. (D) Link density histograms depict associations between parts of the Bradypodion assemblies, B. pumilum and B. ventrale, depicting the mapping location of HiC read pairs. Increasing intensity of red indicated more read pairs map to a particular coordinate of the histogram. White shaded areas indicate no reads mapping to that coordinate in the histogram. Black bars on each axis indicate every other chromosome in the assembly.
Assembly Completeness and Phylogenetics
To estimate assembly completeness, Bradypodion assemblies were tested for the occurrence of a curated set of 3,354 protein-coding genes present in single copy across vertebrate genomes (vertebrata_odb10; Manni et al. 2021). BraPum1.0 contains 3,232 single-copy genes with 39 duplicated genes, and BraVen1.1 has 3,230 genes in single copy and 49 duplicated (fig. 1B). BraVen1.1 has a total of 3,279 genes present, behind H. capensis with 3,287 genes and Furcifer pardalis with 3,282 of the possible 3,354 coding genes that are assumed to be present in all vertebrates. Both BraPum1.0 and BraVen1.1 retain approximately 97% of the possible coding genes present in all vertebrates.
Using 3,354 single-copy orthologs from the vertebrata_odb10 Benchmarking Universal Single-Copy Orthologs (BUSCO) data set, we generated a phylogenetic tree to incorporate the placement of Bradypodion relative to other vertebrates with available assemblies (fig. 1C). Tree estimates using the concatenated sequence alignment produced a topology that mirrors the most complete squamate phylogeny (Burbrink et al. 2020). We also included the transcriptome of Chamaeleo calyptratus and recovered the placement of Chamaeleonidae as sister to the family Agamidae (Pinto et al. 2019), supporting the hypothesis that acrodonts are a monophyletic group. The phylogeny generated using representative genomes from a few groups contains a fraction of the power that could be harnessed using genomic data to address hypotheses relating to evolutionary histories. While the implementation of whole genomic data within the field of phylogenetics is still in its infancy (Simões and Pyron 2021; Card et al. 2023), the addition of these Bradypodion assemblies will be valuable in resolving relationships between lineages while uncovering the origins of traits within squamates (Card et al. 2023).
Hi-C and Inversion
Each link density histogram for the Bradypodion assemblies indicated the presence of six larger chromosomes, consistent with the six macrochromosomes observed in a karyotype of Bradypodion thamnobates (fig. 1D; Rovatsos et al. 2017). For B. ventrale, scaffolds seven and eight were not colinear; however, these scaffolds are homologous to each other with large inversion along scaffold eight (supplementary fig. S2, Supplementary Material online). Upon further investigation of the reduced coverage regions in both scaffolds, we detected breakpoints for an inversion on scaffold eight (position Sc8: 11,460–11,465 kb; position Sc8: 56,620–56,630 kb; supplementary fig. S3, Supplementary Material online). Furthermore, along the inversion breakpoints, we found that PacBio long reads span these sections of the assembly indicating the inversion is not an assembly artifact (supplementary fig. S3, Supplementary Material online). Subsequently, we retained scaffold seven while removing scaffold eight from the initial B. ventrale assembly. After removing this scaffold, HiC reads appropriately mapped along scaffold seven in the updated assembly (BraVen1.1; fig. 1D), with minor contacts corresponding to scaffold two indicative of homology in this region.
Repetitive Element Content
The total repetitive element content for B. pumilum was estimated to be 61% of the genome, with 52% comprised of known interspersed repeats and 5.62% unclassified repeats (supplementary table S2, Supplementary Material online). Comparatively, the B. ventrale assembly was shown to have a slightly lower repeat content estimated to be 59.54%, 50% of which were found to be interspersed repeats and 6.65% unclassified repeats. Both assemblies held a similar diversity in the type of repeat classes, including short interspersed elements (SINEs), long interspersed nuclear elements (LINEs), long terminal repeat (LTR) retrotransposons, and DNA transposons. However, B. pumilum contained a higher proportion of LTRs, whereas B. ventrale retained a higher proportion of LINEs. Overall, the repetitive element content appears to be relatively conserved within Bradypodion (supplementary fig. S4, Supplementary Material online), given that these two species are roughly 7 Ma divergent (Tolley et al. 2008).
Synteny Analysis
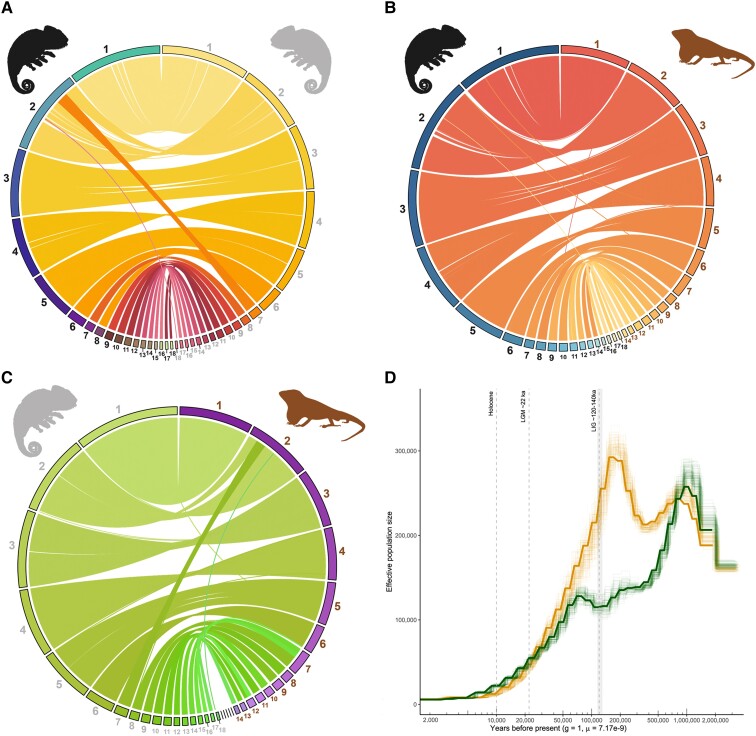
A synteny analysis of the two Bradypodion assemblies shows a conserved genome structure when comparing the 18 largest chromosomes (links > 1 kb), with synteny conserved primarily between the five largest chromosomes (fig. 2A). Similarly, this pattern is repeated between the two Bradypodion assemblies with A. sagrei, with rearrangements only on a few smaller chromosomes. Within B. pumilum, chromosomes six and eight both align with chromosome six of B. ventrale and A. sagrei. This may be due to a chromosomal fission in the B. pumilum lineage after the divergence of B. pumilum and B. ventrale. However, this pattern might instead suggest a failure to fully assemble chromosome six in B. pumilum, and this alternative explanation is supported by the substantial number of HiC links between these scaffolds, more than either scaffolds six or eight share with any other scaffold in the assembly (fig. 1D). Similarly, both chromosomes two and seven of B. ventrale are homologous to chromosome two in B. pumilum and A. sagrei. Chromosome seven in B. pumilum is syntenic with chromosome eight in B. ventrale, with both of these chromosomes aligning partly to chromosome seven in A. sagrei (fig. 2B). We were unable to conclusively distinguish between the alternatives—either a lineage-specific fission of chromosomes two and seven in B. ventrale or a failure to assemble these scaffolds into a single chromosome in B. pumilum. The latter, however, is more likely given the increased number of links between chromosomes two and seven in B. ventrale but additional data are required to confidently resolve between the alternatives.
Fig. 2.
Synteny Circos plots for whole genome alignment between (A) B. pumilum (black) and B. ventrale (gray), (B) B. pumilum and A. sagrei (brown), and (C) B. ventrale and A. sagrei. Each segment of the outer circle represents a unique scaffold in the genome alignment, with the largest scaffolds at the top of each plot. Links are drawn between scaffolds with similarity, where scaffolds with links of at least 1 kb in length were retained for each plot. (D) Changes in effective population size for B. pumilum (green) and B. ventrale (gold) over longer time periods inferred using the PSMC method. Solid lines indicate the model results using the average mutation rate (7.17 × 10−9). Clouded lines indicate unique bootstrapping results. The dotted lines indicate Holocene, the Last Glacial Maximum, and the Last Interglacial periods.
Anolis sagrei has an XY sex-determination system, and recently, chromosome seven in A. sagrei was identified as the X chromosome (Geneva et al. 2022). Despite these highly complete Bradypodion assemblies, it remains unclear which sex-determination system is present in Bradypodion. Within the chameleon genus Furcifer, the sex determination system was found to be a highly differentiated ZW system; however, similar patterns were inconclusive when tested across the Chamaeleonidae family (Rovatsos et al. 2017). The discovery of XX/XY sex chromosomes in C. calyptratus suggests that at least one transition between ZZ/ZW and XX/XY systems has occurred within the Chamaeleonidae (Nielsen et al. 2018). When comparing the synteny between Bradypodion and A. sagrei, chromosome seven in A. sagrei is related to chromosomes 7, 13, and 14 in B. pumilum and chromosomes 8, 14, and 15 in B. ventrale (fig. 2C). Given that there is no distinct pattern between Bradypodion and known X chromosome in A. sagrei, it is likely that the sex chromosomes for Bradypodion reside elsewhere.
Estimation of Population Size History
The trajectory of changes in the effective population size (Ne) of B. pumilum and B. ventrale was estimated using a pairwise sequentially Markovian coalescent (PSMC) approach (Li and Durbin 2011). We found the greatest change in predicted population size using the maximum mutation rate of 3.76 × 10−9 per base pair per year relative to the minimum and average in our models (supplementary fig. S5, Supplementary Material online). When using the minimum and average rates, there were similar trends between both predicted models, resulting in a conservative estimate for changes in the population size of these species. An increase in Ne for both species of Bradypodion was estimated for the early Pleistocene (1–2 Ma: fig. 2D), followed by a sharp decline in Ne of B. pumilum from 1 Ma to 100 kya. In contrast, there was a gradual reduction in Ne of B. ventrale between 1 Ma and 500 kya, after which a sharp increase led to a peak in Ne at ∼200 kya. Furthermore, the results suggest that both species of Bradypodion experienced a decrease in Ne along similar trajectories over the last ∼70 kya; however, PSMC estimates of Ne near present should be interpreted with caution as these estimates can be unreliable (i.e., <10 kya BP; Dussex et al. 2021). While the low estimates of Ne for the currently range-restricted B. pumilum are plausible, the predicted Ne over the last 70 kyr for B. ventrale may be underestimated given its widespread distribution at present (Tolley and Burger 2007), and Ne is unlikely to be extremely low. Nevertheless, some initial inferences can be made regarding the estimated trends in Ne in the Pleistocene. Over the last 120 kya, the biomes of South Africa underwent shifts in distribution before reaching their current extent roughly 10 kya (Tolley et al. 2014; Verboom et al. 2009). Similar to other species occurring in this region (Barlow et al. 2013; Péron and Altwegg 2015; Taft et al. 2022), favorable habitats for Bradypodion likely contracted as other biomes expanded. These range contractions could have resulted in a decline in Ne. Given that Bradypodion is largely affected by drastic changes in habitat type over longer time periods (da Silva and Tolley 2013; Tolley et al. 2008, 2022), it is possible that the decline in population size of these two species is related to recent contractions in their natural available habitats and is not due to recent anthropogenic habitat transformation.
Materials and Methods
Sampling and Sequencing
For long-read sequencing, a male B. pumilum from Cape Town (−34.03, 18.73) and a male B. ventrale from an introduced population in Johannesburg (−26.15, 28.07) were collected. High-molecular-weight DNA was extracted from muscle and liver tissues using the Qiagen Genomic-tip kit and sequenced on PacBio Sequel II 8M SMRT cells. Additional details regarding sampling, Omni-C library preparation, and sequencing are in the supplementary methods, Supplementary Material online.
Assembly Contiguity and Chromosome Size Analysis
Basic assembly statistics were generated using the stats tool in BBTools v38.9 to calculate contiguity statistics (Bushnell 2021). To compare contiguity between representative assemblies across all vertebrate groups, we downloaded 38 additional assemblies from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/) to compare contiguity between representative assemblies across all vertebrate groups, with 70% being reptiles (supplementary table S1, Supplementary Material online).
We estimated whether chromosome sizes in Bradypodion correlate with the length of scaffolds in our assemblies (Geneva et al. 2022) by estimating the size of each chromosome in the published Bradypodion karyotype (figure 1A in Rovatsos et al. 2017) by measuring the chromosome sizes using ImageJ (Schneider et al. 2012). The fraction of the total karyotype occupied by each chromosome was calculated and multiplied by the total size of each Bradypodion assembly to generate an estimate of nucleotide content. We then calculated the correlation between scaffold size and estimated chromosome size via linear regression using the lm function in R v4.2.2 (R Core Team 2022).
Assembly Completeness and Phylogenetics
A BUSCO (BUSCO v5.4.4; Simão et al. 2015) analysis was used to evaluate the completeness of the assemblies using the vertebrata odb10 data set. Subsequently, we constructed a phylogenetic tree using the single-copy BUSCOs for all assemblies downloaded from NCBI. Implementing the BUSCO phylogenomic pipeline (McGowan 2019), we generated multiple sequence alignments using MUSCLE for all protein sequences present in all assemblies. Alignments were trimmed with trimal v1.2rev59 (Capella-Gutiérrez et al. 2009). A maximum likelihood phylogenetic tree was generated using FastTree v2.1.11 (Price et al. 2010).
HiC Chromatin Conformation Mapping
Link density histograms of paired reads were generated from the Omni-C Libraries using Juicer v1.6 ad visualized in Juicebox v2.16.00. This revealed reduced read mapping on scaffolds seven and eight in the B. ventrale initial assembly (supplementary fig. S6, Supplementary Material online). With MUMmer v4.0.0 (Marçais et al. 2018), the initial assembly is aligned as a query and reference using nucmer to inspect whether scaffolds seven and eight are colinear duplicates of the same chromosome. Coverage across the genome assembly was estimated using SAMtools v1.7 (Li et al. 2009) to examine the points in the assembly with reduced mapping.
Repetitive Element Content
Estimating the repetitive landscape of the Bradypodion assemblies was performed by modeling repeats de novo on each assembly using RepeatModeler v v2.0.2a (Flynn et al. 2020) and annotated the repeat consensus sequences using RepeatMasker v4.1.2 (http://www.repeatmasker.org/). To maximize element identification, a custom bash script to specify the order of the four libraries was used as references for the masking process: 1) simple repeats, 2) Tetrapoda DFam library v3.7, 3) classified elements from the species-specific library, and 4) unknown elements from the species-specific library. For the age distribution of transposable elements in each genome, the divergence of an insert from its family consensus was used as a proxy for its age. Alignments for each repeat family were generated, and the Kimura-2 parameter divergence from consensus (correcting for CpG sites) was calculated using the calcDivergenceFromAlign.pl RepeatMasker tool.
Synteny Analysis
A whole genome alignment using SatsumaSynteny v3.1 was used to investigate synteny between the Bradypodion assemblies. The alignment was visualized in the R package circlize v0.4.15 to implement Circos. Both Bradypodion assemblies were aligned to A. sagrei v2.1 to investigate synteny between these 160–170 Ma divergent taxa (Zheng and Wiens 2016; Burbrink et al. 2020).
Estimation of Population Size History
We applied PSMC analysis to infer changes in effective population size history from the assembly sequence of B. pumilum and B. ventrale (Li and Durbin 2011). For each genome, the associated PacBio long-read data were mapped using Minimap2 (Li 2018). PSMC parameters were used as described in Bergeron et al. (2023), including -d 13 and -D 100. Next, we generated PSMC curves with 100 bootstraps using the parameters linked above. For the mutation rate, we used the minimum (9.45 × 10−9), average (7.17 × 10−9), and maximum (3.76 × 10−9) per base pair per year, estimated from the closest relative Pogona vitticeps (Bergeron et al. 2023) and a generation time of 1 year (Tolley and Burger 2007).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of South Africa (grant number 136381) and the National Science Foundation (grant numbers 1927194 and 2152059). Samples exported to the USA under CITES permit (#206297) issued by the South African Department of Fisheries, Forestry and Environment and provincial permits (CapeNature #CNN4459-5795 permit to K.A.T. and Gauteng Nature Conservation #000186 permit to G.J.A.) and ethics clearance from the University of Johannesburg (2019-10-10). Thanks to Matthew Adair, Marius Burger, Devon C. Main, and Bettine van Vuuren for their assistance with this project and to Cleo H. Falvey and Alyssa A. Vanerelli for their helpful comments on this manuscript.
Contributor Information
Jody M Taft, School of Animal, Plant and Environmental Sciences, University of the Witwatersrand, Johannesburg, South Africa; South African National Biodiversity Institute, Kirstenbosch Research Centre, Claremont, South Africa.
Krystal A Tolley, South African National Biodiversity Institute, Kirstenbosch Research Centre, Claremont, South Africa; Centre for Ecological Genomics and Wildlife Conservation, University of Johannesburg, Johannesburg, South Africa.
Graham J Alexander, School of Animal, Plant and Environmental Sciences, University of the Witwatersrand, Johannesburg, South Africa.
Anthony J Geneva, Department of Biology, Center for Computational and Integrative Biology, Rutgers University–Camden, Camden, New Jersey, USA.
Data Availability
Data presented in this article are available from NCBI under BioProject PRJNA986319, BioSample SAMN35825189 (Bradypodion pumilum), and SAMN35825190 (Bradypodion ventrale). Repetitive elements available at https://doi.org/10.7910/DVN/EPSA3O.
Literature Cited
- Barlow A, et al. 2013. Phylogeography of the widespread African puff adder (Bitis arietans) reveals multiple Pleistocene refugia in southern Africa. Mol Ecol. 22:1134–1157. [DOI] [PubMed] [Google Scholar]
- Bergeron LA, et al. 2023. Evolution of the germline mutation rate across vertebrates. Nature 615:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel R, Losos JB. 2002. Patterns of morphological variation and correlates of habitat use in chameleons. Biol J Linn. 76:91–103. [Google Scholar]
- Blaxter M, et al. 2022. Why sequence all eukaryotes? Proc Natl Acad Sci USA. 119:e2115636118. [Google Scholar]
- Burbrink FT, et al. 2020. Interrogating genomic-scale data for Squamata (lizards, snakes, and amphisbaenians) shows no support for key traditional morphological relationships. Syst Biol. 69:502–520. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2021. BBMap version 38.90. [cited 2021] Available from: https://sourceforge.net/projects/bbmap.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DC, Jennings WB, Edwards SV. 2023. Genome evolution and the future of phylogenomics of non-avian reptiles. Animals (Basel) 13:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva JM, Tolley KA. 2013. Ecomorphological variation and sexual dimorphism in a recent radiation of dwarf chameleons (Bradypodion). Biol J Linn. 109:113–130. [Google Scholar]
- Diaz RE, et al. 2015. The veiled chameleon (Chamaeleo calyptratus Duméril and Duméril 1851): a model for studying reptile body plan development and evolution. Cold Spring Harb Protoc. 2015:889–894. [DOI] [PubMed] [Google Scholar]
- Dussex N, van der Valk T, Morales HE, Wheat CW, Díez-del-Molino D, et al. 2021. Population genomics of the critically endangered kākāpō. Cell Genom. 1:100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer TE, et al. 2022. Africa: sequence 100,000 species to safeguard biodiversity. Nature 603:388–392. [DOI] [PubMed] [Google Scholar]
- Flynn JM, et al. 2020. Repeatmodeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA. 117:9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneva AJ, et al. 2022. Chromosome-scale genome assembly of the brown anole (Anolis sagrei), an emerging model species. Commun Biol. 5:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome 10 K Community of Scientists . 2009. Genome 10k: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J Hered. 100:659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK. 2022. Earth Biogenome Project: present status and future plans. Trends Genet. 38:811–820. [DOI] [PubMed] [Google Scholar]
- Hara Y, et al. 2018. Madagascar ground gecko genome analysis characterizes asymmetric fates of duplicated genes. BMC Biol. 16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrel A, et al. 2013. Slow but tenacious: an analysis of running and gripping performance in chameleons. J Exp Biol. 216:1025–1030. [DOI] [PubMed] [Google Scholar]
- Hopkins KP, Tolley KA. 2011. Morphological variation in the cape dwarf chameleon (Bradypodion pumilum) as a consequence of spatially explicit habitat structure differences. Biol J Linn. 102:878–888. [Google Scholar]
- Ishengoma E. 2023. Vertebrate genomics and adaptation—status and prospects in Africa. Mol Ecol. 32:3368–3381. [DOI] [PubMed] [Google Scholar]
- Koepfli KP, Paten B, O’Brien SJ; Genome 10K Community of Scientists . 2015. The Genome 10K Project: a way forward. Annu Rev Anim Biosci. 3:57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão HG, Diedericks G, Broeckhoven C, Baeckens S, Svardal H. 2023. Chromosome-level genome assembly of the cape cliff lizard (Hemicordylus capensis). Genome Biol Evol. 15:evad001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, et al. 2018. Earth Biogenome Project: sequencing life for the future of life. Proc Natl Acad Sci USA. 115:4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, et al. 2022. The earth BioGenome project 2020: starting the clock. Proc Natl Acad Sci USA. 119:e2115635118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, et al. 2018. Mummer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan J. 2019. BUSCO phylogenomics utility script. https://github.com/jamiemcg/BUSCO_phylogenomics.
- Nielsen SV, Banks J, Diaz RE Jr, Trainor P, Gamble T. 2018. Dynamic sex chromosomes in old world chameleons (Squamata: Chamaeleonidae). J Evol Biol. 31:484–490. [DOI] [PubMed] [Google Scholar]
- Péron G, Altwegg R. 2015. Departures from the energy-biodiversity relationship in South African passerines: are the legacies of past climates mediated by behavioral constraints on dispersal? PloS One 10:e0133992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto BJ, et al. 2019. The transcriptome of the veiled chameleon (Chamaeleo calyptratus): a resource for studying the evolution and development of vertebrates. Dev Dyn. 248:702–708. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2—approximately maximum-likelihood trees for large alignments. PloS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2022. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rovatsos M, et al. 2017. Evolution of karyotypes in chameleons. Genes (Basel) 8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simões TR, Pyron RA. 2021. The squamate tree of life. Bull Mus Comp Zool. 163:47–95. [Google Scholar]
- Taft JM, Maritz B, Tolley KA. 2022. Stable climate corridors promote gene flow in the cape sand snake species complex (Psammophiidae). Zoolo Scr. 51:58–75. [Google Scholar]
- Tolley KA, Bowie RC, Measey JG, Price BW, Forest F, et al. 2014. The shifting landscape of genes since the Pliocene: terrestrial phylogeography in the Greater Cape Floristic Region. In: Allsopp N, Colville JF, Verboom GA, et al. editors. Fynbos: ecology, evolution, and conservation of a megadiverse region. New York: Oxford University Press. p. 142–163. [Google Scholar]
- Tolley KA, Burger M. 2007. Chameleons of southern Africa. Cape Town: Struik Publishers. [Google Scholar]
- Tolley KA, Chase BM, Forest F. 2008. Speciation and radiations track climate transitions since the Miocene climatic optimum: a case study of southern African chameleons. J Biogeogr. 35:1402–1414. [Google Scholar]
- Tolley KA, Herrel A. 2013. The biology of chameleons. Los Angeles: University of California Press. [Google Scholar]
- Tolley KA, Tilbury CR, Burger M. 2022. Convergence and vicariance: speciation of chameleons in the Cape Fold Mountains, South Africa, and the description of three new species of Bradypodion fitzinger, 1843. Afr J Herpetol. 71:14–38. [Google Scholar]
- Tolley KA, Townsend TM, Vences M. 2013. Large-scale phylogeny of chameleons suggests African origins and Eocene diversification. Proc R Soc B Biol Sci. 280:20130184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., et al. 2023. The Reptile Database. http://www.reptile-database.org.
- Verboom GA, et al. 2009. Origin and diversification of the Greater Cape flora: ancient species repository, hot-bed of recent radiation, or both? Mol Phylogenet Evol. 51:44–53. [DOI] [PubMed] [Google Scholar]
- Xie HX, et al. 2022. Ancient demographics determine the effectiveness of genetic purging in endangered lizards. Mol Biol Evol. 39:msab359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol. 94:537–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this article are available from NCBI under BioProject PRJNA986319, BioSample SAMN35825189 (Bradypodion pumilum), and SAMN35825190 (Bradypodion ventrale). Repetitive elements available at https://doi.org/10.7910/DVN/EPSA3O.