Abstract
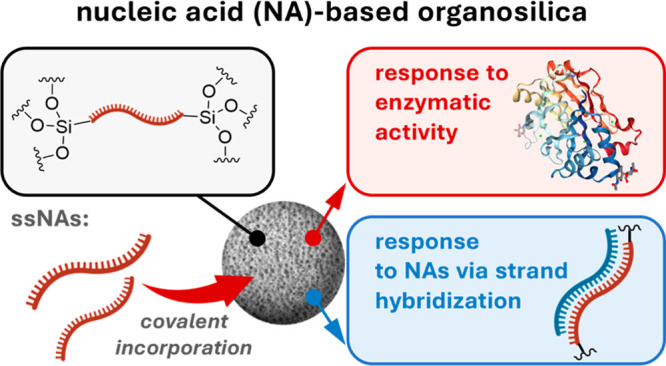
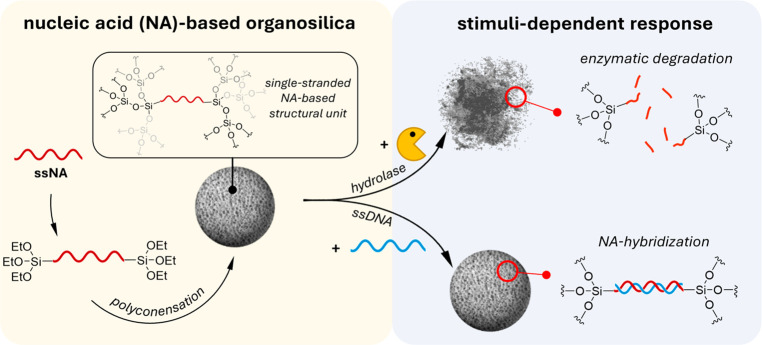
The development of smart nanoparticles (NPs) that encode responsive features in the structural framework promises to extend the applications of NP-based drugs, vaccines, and diagnostic tools. New nanocarriers would ideally consist of a minimal number of biocompatible components and exhibit multiresponsive behavior to specific biomolecules, but progress is limited by the difficulty of synthesizing suitable building blocks. Through a nature-inspired approach that combines the programmability of nucleic acid interactions and sol–gel chemistry, we report the incorporation of synthetic nucleic acids and analogs, as constitutive components, into organosilica NPs. We prepared different nanomaterials containing single-stranded nucleic acids that are covalently embedded in the silica network. Through the incorporation of functional nucleic acids into the organosilica framework, the particles respond to various biological, physical, and chemical inputs, resulting in detectable physicochemical changes. The one-step bottom-up approach used to prepare organosilica NPs provides multifunctional systems that combine the tunability of oligonucleotides with the stiffness, low cost, and biocompatibility of silica for different applications ranging from drug delivery to sensing.
Introduction
Nanomaterials have enormous potential for advancing disease detection, prevention, and treatment.1−6 Despite past achievements in the preparation of stimuli-responsive nanomaterials, it remains relatively difficult to encode information for specific functions into nanoparticles (NPs) such as the biomolecule recognition and on-demand degradation in the presence of biological agents. In the pursuit of obtaining NPs with tailored responsiveness, controlled radical polymerization7,8 can be used to synthesize polymers with a variety of functional groups, which are used to prepare self-assembled micelles and vesicular structures.9−12 Other soft materials, such as engineered supramolecular polymers13,14 or liposomes,15−17 have also been reported as functional nanomaterials for biomedical applications. Stimuli-responsive features have also been introduced into hard nanomaterials by embedding organic functional groups into the framework of silica NPs.18−20 The latter approach led to the development of hybrid systems that contain reactive and cleavable components which enable the stimuli-responsive release of cargo molecules and the degradation of the nanocarrier.21−23 In these examples, the feasibility of introducing a stimuli-responsive property depends on the synthetic accessibility of a variety of organic building blocks or multistep functionalization processes, all of which are tedious and potential pitfalls for the design of funtional materials. In the case of hard NPs (e.g., organosilicas), incorporating the stimuli-responsive functional group into the silica framework relies on covalent bridges.
The need to form covalently bonded organosilica is related to the stability of the particles during condensation of the silica precursors. In this respect, introducing building blocks that are held together by reversible supramolecular interactions into a rigid silica framework appears to be difficult because the structural integrity of the nanoparticles can be significantly affected. Double-stranded DNA is a unique example of a stable and rigid biopolymer that can be used to self-assemble 3D structures with nanometer-precise features and potential applications in biomedical technologies.24−26 The introduction of synthetic DNA aptamers,27 strand displacement reactions,28 and structure-switching probes29,30 as responsive modules in DNA-based self-assembled 3D structures has already been demonstrated for developing sensors29,31 and smart drug delivery vehicles.24 Despite their promising potential, purely synthetic DNA-based materials have several drawbacks, and there are still major obstacles on the way to real-world nanomedicine applications.
First, the production of large quantities of synthetic DNA is expensive, and second, long-term DNA stability in vitro or in vivo is a significant obstacle for biomedical applications.32 Therefore, the use of resistant oligonucleotide analogs for preparing NPs has become increasingly important.25 Peptide nucleic acids (PNAs), polyamide analogs of DNA,33 are more stable and easier to produce, and their derivatives can be particularly useful in nanotechnology. PNAs have a high affinity and sequence selectivity for DNA and RNA,34 are able to form highly stable PNA:DNA:PNA triplexes and PNA:PNA duplexes, are compatible with both aqueous and organic solvents,35 and, most importantly for in vivo applications, are completely resistant to peptidases and nucleases.36
Combining what we have learned from hard and soft materials with the advanced information-encoded world of oligonucleotides, we aimed to develop a strategy for making hybrid nucleic acid (NA)-based organosilica NPs in a one-step reaction using a bottom-up approach. Until now, the possibility of using organoalkoxysilane derivatives of NAs to prepare DNA- or PNA-bridged organosilica NPs has remained partially explored. Previous reports on the design of hybrid NA-based silica composites simply relied on the strategy of silicification of DNA origami, which served as a template structure.37,38 So far, only the loading of porous particles with oligonucleotides, the functionalization of the surface of NPs with nucleic acids for drug delivery,39−42 or sensing applications have been investigated.43−45
In this work, we capitalize on the principles of sol–gel chemistry, in which organo-alkoxysilanes enable the preparation of responsive organosilica NPs.18 We show a variety of rationally designed NPs that respond to biological, physical, and chemical stimuli by using NA components (i.e., DNA or PNA) to construct covalent or, more interestingly, noncovalent silica networks. We also show that NAs embedded in the hybrid material are still accessible to recognize complementary NA strands or small molecules, which leads to morphological changes and the disruption of the nanomaterial.
The results highlight new and sought-after possibilities for designing responsive NPs from nontoxic starting materials, which can be used in a wide range of applications from drug delivery to sensing. The oligonucleotide is an active component that can be covalently bridged between the silicon oxide moieties, leading to an interesting responsive silica framework in which the functionality of the NA remains retained and can noncovalently bind a complementary strand. These components can also be used for the noncovalent assembly of the stimuli-responsive nanoparticles, as described in the subsequent companion paper (see “Supramolecular Nucleic Acid-Based Organosilica Nanoparticles Responsive to Physical and Biological Inputs”).
Results and Discussion
ssDNA-Based Organosilica Nanoparticles
To merge the properties of DNA-based and sol–gel chemistry,46 we used alkoxysilane derivatives of single-stranded DNA (ssDNA) and single-stranded peptide nucleic acids (ssPNA)47,48 as building blocks for the preparation of ssNA-based organosilica NPs (Figure 1). The functionalization of the oligonucleotides at the 5′- and 3′-ends with 3-(triethoxysilyl)propyl isocyanate (ICPTES) prior to their hydrolysis and polycondensation with the secondary silica source tetraethyl orthosilicate (TEOS) in the presence of the surfactant cetyltrimethylammonium bromide (CTAB) allowed us to covalently embed ssNAs into the silicate framework of the final NPs. This approach leads to the formation of NA-based organosilica nanoparticles containing single-stranded NA-based structural units in their framework.
Figure 1.
The presence of a covalently embedded nucleic acid (DNA or PNA) building block endows nucleic acid-based organosilica NPs with sequence-specific chemical, physical, and biological responsiveness.
The covalent embedding of ssDNA as an organic bridging group into the silica scaffold (Figure 1, left) was achieved in two steps. First, we prepared a bis-alkoxysilane oligonucleotide upon reacting the 5′- and 3′-diamino-functionalized 10mer ssDNA with ICPTES (Supporting Information Section I.I) obtaining the corresponding bis-alkoxysilane derivative (Si-ssDNA-Si; Figure 2a, bottom). Then the Si-ssDNA-Si was mixed with TEOS in the presence of the surfactant CTAB in a basic (pH = 8.5) medium (Supporting Information Section II.II). The resulting ssDNA-bridged organosilica NPs (ssDNA-OSPs) from which the surfactant was extracted were spherical and had an average size of 387 ± 88 nm, as revealed by scanning electron microscopy (SEM, Figure 2a and Figure S2). The synthetic parameters for preparing ssDNA-OSPs were carefully adjusted to ensure DNA stability during particle formation. In particular, milder conditions based on the use of a pH of 8.5 and a low EtOH content (Supporting Information Section II.I, Figure S1) were selected by modifying an earlier procedure for the preparation of mesoporous silica NP.23 Electron microscopy analysis of ssDNA-OSPs indicates that the presence of DNA alters the formation pathway, as particles larger than the 100 nm model silica particles were obtained (Figure S1b). The different morphology of ssDNA-OSPs compared with model particles can be explained by the presence of the negatively charged and bulky NA linkers interacting with the positively charged CTAB micelles. This alters the formation pathway that normally occurs when no DNA or a shorter DNA strand is used (vide infra).49 The successful covalent embedding of DNA oligonucleotides into the inorganic silica framework was demonstrated by standard characterization techniques for organosilica particles, such as attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR, Figure S3a), UV–vis absorption spectroscopy (Figure S3b), and X-ray photoelectron spectroscopy (XPS, Figure S3c–f). The amount of DNA incorporated into the NPs, relative to the original amount of Si-ssDNA-Si (0.7 μmol), was >97%, as determined by UV–vis absorption spectroscopy (Supporting Information Section II.II, Figure S4), leading to DNA incorporation of 0.029 μmol/mg or 11.6% (w/w).
Figure 2.
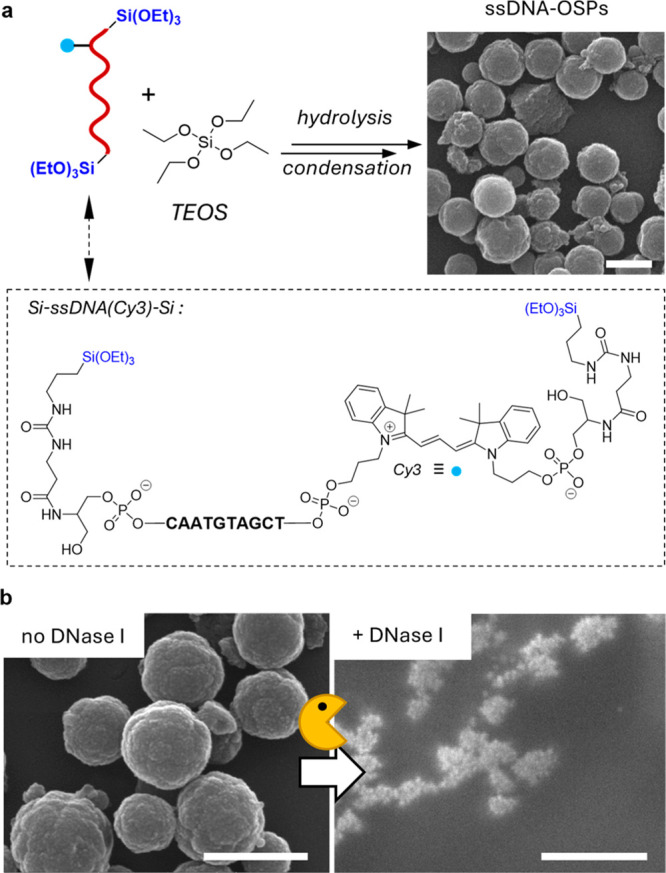
(a) The DNA bis-alkoxysilane (Si-ssDNA-Si) is hydrolyzed in basic aqueous solutions (pH = 8.5) in the presence of the surfactant CTAB to obtain nanometer-sized ssDNA-OSPs as visualized by SEM. (b) SEM images of ssDNA-OSPs before (left image) and after mixing (2 h, right image) with DNase I. All scale bars = 500 nm.
SAXS measurements on ssDNA-OSPs revealed the presence of a disordered mesoporous phase with a hexagonal arrangement, as indicated by the appearance of a broad signal at q = 1.3 nm–1 (Figure S5). The disordered arrangement of the pores was also confirmed by transmission electron microscopy (TEM), as the visibility of the pores was low (Figure S2b). The low order of the mesoporous phase can be explained by the presence of structurally flexible and highly negatively charged 10mer oligonucleotides, which interfere with the formation of a highly ordered hexagonal structure of CTAB micelles during particle formation. To demonstrate that the length of the ssNA affects the order of the mesopores, organosilica particles containing a 4mer oligonucleotide (Supporting Information Section I.I) were prepared by using the same synthetic procedure as for ssDNA-OSPs. The TEM images of the latter particles reveal the presence of clearly visible mesopores (Figure S6), confirming the effect of oligonucleotide length in the formation of an ordered mesophase in the final particle.
Next, the reactivity and hybridization properties of ssDNA-OSPs, namely, the degradation of ssDNA-OSPs in the presence of hydrolytic enzymes (Supporting Information Section SII.III) or the possibility of binding with a complementary DNA (Supporting Information Section SII.IV), were investigated, respectively. As shown in the SEM images in Figure 2b and Figure S7c, ssDNA-OSPs disintegrate into small debris due to enzyme-catalyzed hydrolysis of the NA bridge when a dispersion of ssDNA-OSPs is mixed with the enzyme DNase I, a hydrolase that catalyzes the hydrolysis of phosphodiester bonds of DNA. In the absence of the enzyme, however, the spherical morphology of the particles was preserved (Figure S7b). Enzyme-triggered degradation was further confirmed by dynamic light scattering (DLS) analysis, in which degradation induced by the enzyme resulted in the formation of smaller fragments (Figure S8). As shown in the electron micrographs, ssDNA-OSPs break up into smaller silica-based degradation products, which can be detected in the DLS analysis by the disappearance of the micrometer-sized aggregates of ssDNA-OSPs and the appearance of a new particulate aggregate (average Dh = 200 nm).
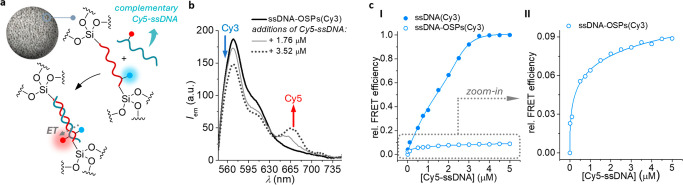
To evaluate if the DNA can recognize a complementary strand even when it is a structural component of ssDNA-OSPs, Förster resonance energy transfer (FRET)-based experiments (Supporting Information Section II.IV) were performed. The 10mer and Cy3-labeled Si-ssDNA-Si bisalkoxysilane were used to prepare the hybrid particles (i.e., ssDNA-OSPs(Cy3)). The complementary Cy5-labeled ssDNA (Cy5-ssDNA) was synthesized (Supporting Information Section I.I) and added in the form of aliquots to a dispersion of ssDNA-OSPs(Cy3). After light excitation (λex = 520 nm) of the Cy3 energy donor, energy transfer to the excited state of the acceptor, Cy5, occurs as proven by the emission at low energy (Cy5, λem = 665 nm; see Figure S9 for band identification). As FRET is regulated by distance, the evidence of FRET confirms the formation of a duplex between the immobilized DNA strand and the added complementary strand (Figure 3a). Indeed, successive addition of the Cy5-labeled ssDNA strand to the dispersion of ssDNA-OSPs(Cy3) resulted in a concentration-dependent sensitization of the Cy5 emission (λem = 665 nm) upon Cy3 excitation (Figure 3b). The ζ-potential measurements showed that ssDNA-OSPs had an overall negative surface charge of −27 ± 2 mV when dispersed in deionized water at pH = 7.4, ruling out the possibility of electrostatic interaction between the particles and the complementary Cy5-labeled ssDNA strand.
Figure 3.
(a) The ssDNA embedded in the silica framework of ssDNA-OSPs is accessible and can hybridize with its complementary strand. When the complementary and Cy5-labeled strand binds to Cy3-labeled ssDNA-OSPs, a partial energy transfer (ET) from the excited state of the donor, Cy3 (blue dot), to the acceptor, Cy5 (red dot), occurs. (b) Fluorescence spectra obtained upon addition of Cy5-ssDNA to a dispersion containing ssDNA-OSPs(Cy3) (0.06 mg·mL–1 ≙ 1.76 μM of Cy3-labeled DNA, bold line). (c) Relative FRET efficiencies obtained from energy-transfer experiments with the free Cy3-ssDNA strand (3.52 μM) or with the ssDNA-OSPs(Cy3) (0.12 mg·mL–1 ≙ 3.52 μM) in the presence of Cy5-ssDNA (0–5 μM). All of the experiments were performed in Tris-HCl buffer (10 mM, containing 3 mM MgCl2 at pH 7.5). All fluorescence spectra and intensities were recorded upon excitation at λex,Cy3 = 520 nm. The FRET intensities were recorded at λem,Cy3 = 570 nm and λem,Cy5 = 665 nm.
The observed FRET efficiency of ssDNA-OSP with Cy5-ssDNA was 8% of what was obtained with the same free oligonucleotides (Cy3-ssDNA and Cy5-ssDNA) in solution; this can be explained by considering that only about 11% of the materials are constituted by the oligonucleotides, and most of them are not on the surfaces but embedded in the silica framework that has a tile structure. In addition, some of the DNA could have a wrong conformation in the silica, therefore limiting the accessibility of the DNA binding sites in the particles.
However, as shown above, the apparently small recognition ability does not prevent the DNase from degrading the particles, suggesting the possible use of such particles for biomedical applications and for sensing purposes, as described in the subsequent companion paper (“Supramolecular Nucleic Acid-Based Organosilica Nanoparticles Responsive to Physical and Biological Inputs”).
The kinetic properties of the FRET process were found to be slow, as might be expected for relatively large objects when capturing small molecules. The energy transfer process increased over a sustained period and did not reach a steady state even after 120 min of mixing (Figure S10), unlike what was observed with free Cy3-ssDNA and Cy5-ssDNA, for which it already reached its maximum after 30 min (Figure 3 c).
ssPNA-Based Organosilica Nanoparticles
As mentioned earlier, the presence of highly charged bridging groups such as DNA can affect the control of the morphology of NPs. ssPNA has the same nucleobases as DNA, but its backbone is neutral48 and therefore can lead to a different morphology. Furthermore, using PNA instead of DNA can produce enzyme-resistant nanostructures with a high affinity for target DNA and RNA. To address these issues, we prepared NPs with uncharged ssPNA (Supporting Information Section I.II) under the same experimental conditions used for ssDNA-OSPs (Figure 4a; Supporting Information Section II.V).
Figure 4.
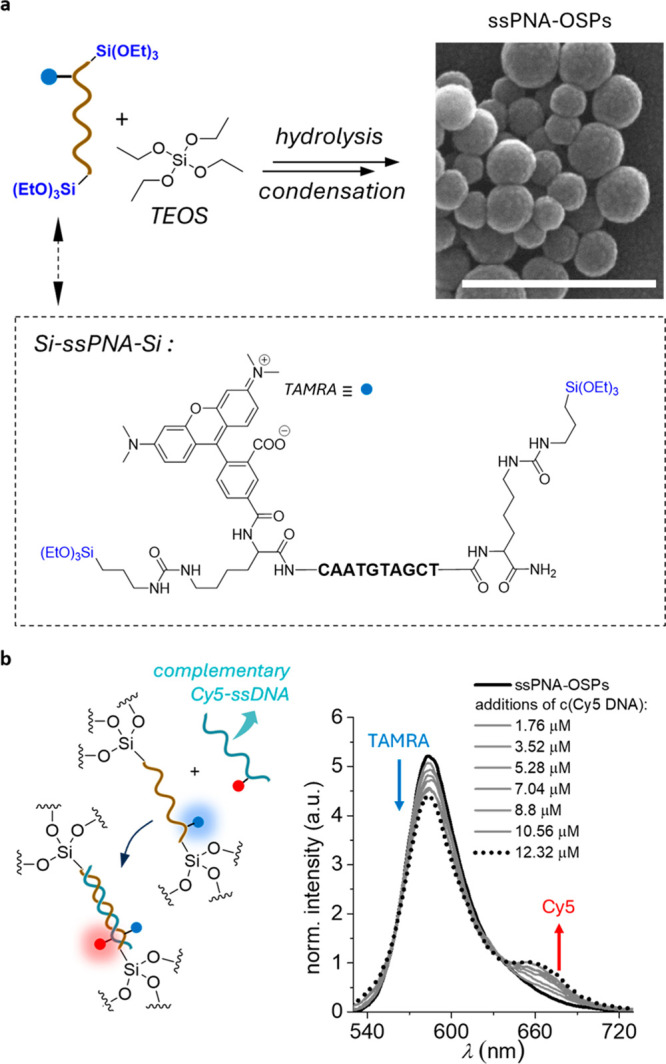
(a) Synthesis scheme for the preparation of ssPNA-OSPs shown in the SEM image (top right). Scale bar: 500 nm. (b) ssPNA-OSPs based FRET study in the presence of the Cy5-labeled and the PNA complementary ssDNA strand.
The ssPNA-bridged organosilica particles (ssPNA-OSPs) showed a more regular spherical shape and a narrower size distribution (103 ± 27 nm, Figure 4a and Figure S11a,b) compared with the ssDNA-OSPs. The PNA sequence was chosen to be the same as for ssDNA-OSP, which is complementary to the “seed region” of micro-RNA 221, an important target for cancer cell regulation.50 It is thus of interest to evaluate the ability of ssPNA-OSPs to recognize complementary NA sequences (using DNA as used for ssDNA-OSPs for comparison). UV–vis absorption spectroscopy confirmed the successful integration of the 5(6)-carboxytetramethylrhodamine (TAMRA)-labeled ssPNA (Figure 4a) into the final NPs (ssPNA-OSPs(TAMRA), Figure S11c), as indicated by the presence of the characteristic absorption band of the nucleobases at 250–300 nm and the presence of the absorption band of the fluorophore at 500–605 nm.
FRET experiments demonstrate that the PNA present in ssPNA-OSPs(TAMRA) is still accessible and can hybridize with complementary ssDNA-Cy5 (Figure 4b). To this end, aliquots of the Cy5-labeled ssDNA strand solution were added to a dispersion of ssPNA-OSPs(TAMRA) (Supporting Information Section II.VI). Upon subsequent TAMRA excitation (λex = 500 nm), sensitized Cy5 emission (λem = 667 nm) was recorded (Figure 4b).
As with ssDNA-OSPs, the observed FRET efficiency is also low for ssPNA-OSPs if compared to that derived from dsDNA in solution, which can be explained by the same reasons as those for ssDNA-OSPs. Moreover, due to the lack of negative charges on the PNA during particle formation, the PNA is less likely to be incorporated into regions where it is directly exposed to the silica–water interface than its charged DNA counterpart. Therefore, the PNA molecules will be located in even less-accessible sites for strand hybridization.
In contrast to ssDNA-OSPs, the addition of DNase I does not lead to the degradation of the particles, as shown by the DLS measurements (Figure S12). The size distribution of the micrometer-sized aggregates of ssPNA-OSPs does not change significantly upon incubation with the enzyme, contrary to what was observed for ssDNA-OSPs. This can be explained by the fact that DNase I enzymes cannot hydrolyze the PNA’s N-(2-aminoethyl)-glycine units and therefore do not degrade the corresponding ssPNA-containing hybrid nanoparticles.
Conclusions
This work shows that nucleic acid-derived alkoxysilanes can be prepared and used for a one-step bottom-up process to design multifunctional organosilica NPs. From a material chemistry point of view, the presence of Watson–Crick–Franklin-type supramolecular bonds leads to responsive and programmable behavior that can be explored for various applications. For example, when ssDNA is used as a bridging group within the inorganic framework of silica particles, enzymatically degradable particles capable of hybridizing with complementary DNA are obtained. Such NPs represent attractive nanomaterials for biomedical applications, as the NPs can carry therapeutically active oligonucleotides, an important aspect in developing safe nanomedicines and gene therapies. In addition, non-biodegradable NPs can be obtained using PNA building blocks, conserving the ability to bind to cognate NAs. The ability to incorporate these biocomponents can also be exploited to create new hybrid structures held together by noncovalent interactions as shown in the following companion paper (see “Supramolecular Nucleic Acid-Based Organosilica Nanoparticles Responsive to Physical and Biological Inputs”).
Acknowledgments
The authors acknowledge financial support from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement “Nano-Oligo Med” (no. 778133). This work has also benefited from the infrastructure and framework of the COMP-HUB Initiative, funded by the Departments of Excellence program of the Italian Ministry for Education, University, and Research (MIUR, 2018–2022).
Glossary
Abbreviations
- CTAB
cetyltrimethylammonium bromide
- Cy3
cyanine 3
- Cy5
cyanine 5
- ET
energy transfer
- FRET
Förster resonance energy transfer
- ICPTES
3-(triethoxysilyl)propyl isocyanate
- NAs
nucleic acids
- NPs
nanoparticles
- OSPs
organosilica particles
- ssDNA
single-stranded DNA
- ssDNA-OSPs
ssDNA-bridged DNA organosilica particles
- ssPNA
single-stranded peptide nucleic acids
- ssPNA-OSPs
ssPNA-bridged organosilica particles
- TAMRA
5(6)-carboxytetramethylrhodamine
- TEOS
tetraethyl orthosilicate
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c00393.
Details about instruments, materials, methods, supporting figures, and chemical synthesis (PDF)
Author Present Address
∇ M.D.D.: Department of Chemistry, Northwestern University, Evanston, Illinois 60208, United States
Author Present Address
• T.T.: Department of Radiology, Stanford University, Stanford, California 94305, United States
Author Contributions
° P.P. and S.V. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Mitchell M. J.; Billingsley M. M.; Haley R. M.; Wechsler M. E.; Peppas N. A.; Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discovery 2021, 20, 101–124. 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlerding E. B.; Grodzinski P.; Cai W.; Liu C. H. Big potential from small agents: Nanoparticles for imaging-Based companion diagnostics. ACS Nano 2018, 12, 2106–2121. 10.1021/acsnano.7b07252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konvalina G.; Haick H. Sensors for breath testing: From nanomaterials to comprehensive disease detection. Acc. Chem. Res. 2014, 47, 66–76. 10.1021/ar400070m. [DOI] [PubMed] [Google Scholar]
- Rosi N. L.; Mirkin C. A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Ventola C. L. Progress in nanomedicine: Approved and investigational nanodrugs. P&T 2017, 42, 742–755. [PMC free article] [PubMed] [Google Scholar]
- Chung Y. H.; Beiss V.; Fiering S. N.; Steinmetz N. F. COVID-19 Vaccine frontrunners and their nanotechnology design. ACS Nano 2020, 14, 12522–12537. 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- Szwarc M.; Levy M.; Milkovich R. Polymerization initiated by electron transfer to monomer. A new method of formation of block polymers. J. Am. Chem. Soc. 1956, 78, 2656–2657. 10.1021/ja01592a101. [DOI] [Google Scholar]
- Warren N. J.; Armes S. P. Polymerization-induced self-assembly of block copolymer nano-objects via RAFT aqueous dispersion polymerization. J. Am. Chem. Soc. 2014, 136, 10174–10185. 10.1021/ja502843f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K.; Harada A.; Nagasaki Y. Block copolymer micelles for drug delivery: Design, characterization and biological significance. Adv. Drug Delivery Rev. 2012, 64, 37–48. 10.1016/j.addr.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Lutz J.-F.; Lehn J.-M.; Meijer E. W.; Matyjaszewski K. From precision polymers to complex materials and systems. Nat. Rev. Mater. 2016, 1, 16024. 10.1038/natrevmats.2016.24. [DOI] [Google Scholar]
- O’Reilly R. K.; Joralemon M. J.; Hawker C. J.; Wooley K. L. Facile syntheses of surface-functionalized micelles and shell cross-linked nanoparticles. J. Polym. Sci. A Polym. Chem. 2006, 44, 5203–5217. 10.1002/pola.21602. [DOI] [Google Scholar]
- Discher D. E.; Eisenberg A. Polymer vesicles. Science 2002, 297, 967–973. 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- Brunsveld L.; Folmer B. J. B.; Meijer E. W.; Sijbesma R. P. Supramolecular polymers. Chem. Rev. 2001, 101, 4071–4097. 10.1021/cr990125q. [DOI] [PubMed] [Google Scholar]
- Álvarez Z.; Kolberg-Edelbrock A. N.; Sasselli I. R.; Ortega J. A.; Qiu R.; Syrgiannis Z.; Mirau P. A.; Chen F.; Chin S. M.; Weigand S.; Kiskinis E.; Stupp S. I. Bioactive scaffolds with enhanced supramolecularmotion promote recovery from spinal cord injury. Science 2021, 374, 848–856. 10.1126/science.abh3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane E. L.; Belessiotis-Richards A.; Backlund C.; Wang J.; Hidaka K.; Milling L. E.; Bhagchandani S.; Melo M. B.; Wu S.; Li N.; Donahue N.; Ni K.; Ma L.; Okaniwa M.; Stevens M. M.; Alexander-Katz A.; Irvine D. J. STING agonist delivery by tumour-penetrating PEG-lipid nanodiscs primes robust anticancer immunity. Nat. Mater. 2022, 21, 710–720. 10.1038/s41563-022-01251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Cheng Q.; Wei T.; Yu X.; Johnson L. T.; Farbiak L.; Siegwart D. J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 2021, 20, 701–710. 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. M.; Cullis P. R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug. Delivery Rev. 2013, 65, 36–48. 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Croissant J. G.; Fatieiev Y.; Almalik A.; Khashab N. M. Mesoporous silica and organosilica nanoparticles: Physical chemistry, biosafety, delivery strategies, and biomedical applications. Adv. Healthcare Mater. 2018, 7, 1700831 10.1002/adhm.201700831. [DOI] [PubMed] [Google Scholar]
- Inagaki S.; Guan S.; Ohsuna T.; Terasaki O. An ordered mesoporous organosilica hybrid material with a crystal-like wall structure. Nature 2002, 416, 304–307. 10.1038/416304a. [DOI] [PubMed] [Google Scholar]
- Quignard S.; Masse S.; Laurent G.; Coradin T. Introduction of disulfide bridges within silica nanoparticles to control their intra-cellular degradation. Chem. Commun. 2013, 49, 3410–3412. 10.1039/c3cc41062e. [DOI] [PubMed] [Google Scholar]
- Maggini L.; Cabrera I.; Ruiz-Carretero A.; Prasetyanto E. A.; Robinet E.; De Cola L. Breakable mesoporous silica nanoparticles for targeted drug delivery. Nanoscale 2016, 8, 7240–7247. 10.1039/C5NR09112H. [DOI] [PubMed] [Google Scholar]
- Picchetti P.; Moreno-Alcántar G.; Talamini L.; Mourgout A.; Aliprandi A.; De Cola L. Smart Nanocages as a Tool for Controlling Supramolecular Aggregation. J. Am. Chem. Soc. 2021, 143, 7681–7687. 10.1021/jacs.1c00444. [DOI] [PubMed] [Google Scholar]
- Travaglini L.; Picchetti P.; Totovao R.; Prasetyanto E. A.; De Cola L. Highly degradable imine-doped mesoporous silica particles. Mater. Chem. Front. 2019, 3, 111–119. 10.1039/C8QM00438B. [DOI] [Google Scholar]
- Hu Q.; Li H.; Wang L.; Gu H.; Fan C. DNA Nanotechnology-enabled drug delivery systems. Chem. Rev. 2019, 119, 6459–6506. 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- Lacroix A.; Sleiman H. F. DNA nanostructures: current challenges and opportunities for cellular delivery. ACS Nano 2021, 15, 3631–3645. 10.1021/acsnano.0c06136. [DOI] [PubMed] [Google Scholar]
- Kimna C.; Lieleg O. Molecular micromanagement: DNA nanotechnology establishes spatio-temporal control for precision medicine. Biophys. Rev. 2020, 1, 011305. 10.1063/5.0033378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H. M.; Liu H.; Kuai H.; Peng R.; Mo L.; Zhang X. B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016, 45, 2583–602. 10.1039/C5CS00645G. [DOI] [PubMed] [Google Scholar]
- Simmel F. C.; Yurke B.; Singh H. R. Principles and applications of nucleic acid strand displacement reactions. Chem. Rev. 2019, 119, 6326–6369. 10.1021/acs.chemrev.8b00580. [DOI] [PubMed] [Google Scholar]
- Leung K.; Chakraborty K.; Saminathan A.; Krishnan Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 2019, 14, 176–183. 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy N.; Chakraborty K.; Saminathan A.; Zeichner E.; Leung K.; Devany J.; Krishnan Y. A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat. Methods. 2019, 16, 95–102. 10.1038/s41592-018-0232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo S.; Porchetta A.; Ricci F. DNA-Based scaffolds for sensing applications. Anal. Chem. 2019, 91, 44–59. 10.1021/acs.analchem.8b05009. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran A. R. Nuclease resistance of DNA nanostructures. Nat. Rev. Chem. 2021, 5, 225–239. 10.1038/s41570-021-00251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P.Peptide Nucleic Acids, 3rd ed.; Springer: 2020. [Google Scholar]
- Saarbach J.; Sabale P. M.; Winssinger N. Peptide nucleic acid (PNA) and its applications in chemical biology, diagnostics, and therapeutics. Curr. Opin. Chem. Biol. 2019, 52, 112–124. 10.1016/j.cbpa.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Sen A.; Nielsen P. E. On the stability of peptide nucleic acid duplexes in the presence of organic solvents. Nucleic Acids Res. 2007, 35, 3367–3374. 10.1093/nar/gkm210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov V. V.; Potaman V. N.; Frank-Kamenetskii M. D.; Egholm M.; Buchard O.; Sönnichsen S. H.; Nielsen P. E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Liu X.; Zhang F.; Jing X.; Pan M.; Liu P.; Li W.; Zhu B.; Li J.; Chen H.; Wang L.; Lin J.; Liu Y.; Zhao D.; Yan H.; Fan C. Complex silica composite nanomaterials templated with DNA origami. Nature 2018, 559, 593–598. 10.1038/s41586-018-0332-7. [DOI] [PubMed] [Google Scholar]
- Nguyen L.; Döblinger M.; Liedl T.; Heuer-Jungemann A. DNA-Origami-Templated Silica Growth by Sol–Gel Chemistry. Angew. Chem., Int. Ed. 2019, 58, 912–916. 10.1002/anie.201811323. [DOI] [PubMed] [Google Scholar]
- Lülf H.; Bertucci A.; Septiadi D.; Corradini R.; De Cola L. Multifunctional Inorganic Nanocontainers for DNA and Drug Delivery into Living Cells. Chem.—Eur. J. 2014, 20, 10900–10904. 10.1002/chem.201403232. [DOI] [PubMed] [Google Scholar]
- Dhar S.; Gu F. X.; Langer R.; Farokhzad O. C.; Lippard S. J. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA–PEG nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 17356–17361. 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.; Sun S.; Varner J. A.; Howell S. B.; Ruoslahti E.; Sailor M. J. Securing the payload, finding the cell, and avoiding the endosome: Peptide-targeted, fusogenic porous silicon nanoparticles for delivery of siRNA. Adv. Mater. 2019, 31, 1902952 10.1002/adma.201902952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T.; Kovochich M.; Liong M.; Meng H.; Kabehie S.; George S.; Zink J. I.; Nel A. E. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano 2009, 3, 3273–3286. 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov V.; Xiao Y.; Shlyahovsky B.; Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- Song Y.; et al. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem., Int. Ed. 2019, 58, 2236–2240. 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- Cutler J. I.; Auyeung E.; Mirkin C. A. Spherical nucleic acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- Livage J.; Sanchez C.; Henry M.; Doeuff S. The chemistry of the sol-gel process. Solid State Ion. 1989, 32, 633–638. 10.1016/0167-2738(89)90338-X. [DOI] [Google Scholar]
- Egholm M.; Buchardt O.; Nielsen P. E.; Berg R. H. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral peptide backbone. J. Am. Chem. Soc. 1992, 114, 1895–1897. 10.1021/ja00031a062. [DOI] [Google Scholar]
- Shakeel S.; Karim S.; Ali A. Peptide nucleic acid (PNA) — a review. J. Chem. Technol. Biotechnol. 2006, 81, 892–899. 10.1002/jctb.1505. [DOI] [Google Scholar]
- Picchetti P.; DiMarco B. N.; Travaglini L.; Zhang Y.; Ortega-Liebana M. C.; De Cola L. Breaking with Light: Stimuli-Responsive Mesoporous Organosilica Particles. Chem. Mater. 2020, 32, 392–399. 10.1021/acs.chemmater.9b03937. [DOI] [Google Scholar]
- Sun T.; Wang Q.; Balk S.; Brown M.; Lee G. -S. M.; Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009, 69, 3356–3363. 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.