Abstract
Fast, high-throughput methods for measuring the level and duration of protective immune responses to SARS-CoV-2 are needed to anticipate the risk of breakthrough infections. Here we report the development of two quantitative PCR assays for SARS-CoV-2-specific T cell activation. The assays are rapid, internally normalized and probe-based: qTACT requires RNA extraction and dqTACT avoids sample preparation steps. Both assays rely on the quantification of CXCL10 messenger RNA, a chemokine whose expression is strongly correlated with activation of antigen-specific T cells. On restimulation of whole-blood cells with SARS-CoV-2 viral antigens, viral-specific T cells secrete IFN-γ, which stimulates monocytes to produce CXCL10. CXCL10 mRNA can thus serve as a proxy to quantify cellular immunity. Our assays may allow large-scale monitoring of the magnitude and duration of functional T cell immunity to SARS-CoV-2, thus helping to prioritize revaccination strategies in vulnerable populations.
Infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, leads to a broad range of clinical symptoms, ranging from asymptomatic infection to severe pneumonia and acute respiratory distress syndrome (ARDS)1,2. As the deployment of vaccines attenuates the pandemic3, vaccine effectiveness and the duration of protective immunity will need to be systematically assessed and monitored at a global level. Long-term protection from viral infection is mediated by both humoral (antibodies) and cellular immunity4. Quantification of SARS-CoV-2-specific IgG and neutralizing antibodies is often used as a marker of immune protection5, but measurement of T cell responses is rarely performed because of the associated technical challenges. Many recent studies point to the importance of determining T cell function in convalescent and vaccinated individuals for the management of vaccination campaigns. Data from animal models and human patients show that T cells protect against severe COVID-19 disease and reduce viral spread6–8. Because of the heterogeneity of individual immune responses, humoral immune measurements do not always correlate with the magnitude of the T cell response9–11, as seen in nonseroconverters or low-neutralizer patients12. SARS-CoV-2-specific T cells may be present in the absence of antibodies13,14, and antibody and T cell responses are independent in magnitude and in persistence during the memory phase of the immune response9,15.
Several groups, including ours, have been quantifying SARS-CoV-2-specific T cells using synthetic peptides (15-mers long) to activate T cells in vitro following overnight incubation with whole blood. These peptides are either presented directly by HLA-class II or processed by proteases present in the blood and presented by HLA-class I. We and others have previously demonstrated that these peptides activate SARS-CoV-2-specific CD4 and CD8 T cells16–19. More recently, we demonstrated that the same peptides used in the present study activate SARS-CoV-2 T cells in peripheral blood mononuclear cells (PBMCs) and in whole blood14. In ref. 14, we demonstrated that the quantity of cytokines (IL-2 and IFN-γ) measured by enzyme-linked immunosorbent assay in the whole blood after overnight stimulation correlates with the number of SARS-CoV-2-specific T cells quantified with ELISpot, which was further confirmed by intracellular cytokine staining. These data collectively confirm that the addition of peptides to whole blood allows precise quantification of SARS-CoV-2-specific T cells11,14,20,21.
Despite the recognized need to quantify the levels of cellular immunity, the complexity and lack of scalability of these traditional methods (that is, ELISpot and flow cytometry), has so far prevented large-scale studies of the cellular immune response to COVID-19 in recovered and vaccinated individuals. Thus, most studies using ELISpot or flow cytometry assess only 10–40 participants, with larger clinical trials assessing around 200 (refs. 22–27). Furthermore, the process of freezing/thawing PBMCs, often used for testing T cell response, can introduce high variability in the results28,29, which can be bypassed by using whole blood.
Results
CXCL10 mRNA levels can be measured as a proxy for T cell activation.
To address these problems, we have implemented a probe-based quantitative PCR (qPCR) rapid T cell activation (qTACT) assay (Fig. 1a), based on ex vivo stimulation of whole-blood samples with a pool of viral peptides covering spike [S] or other SARS-CoV-2 viral proteins (that is, nucleoprotein)14,30, followed by direct amplification of IFNG (directly produced by SARS-CoV-2 antigen-specific T cells) or CXCL10, a molecule expressed by monocytes31,32 in response to T cell activation (Fig. 2a). A further technical implementation of the assay allows quantification of T cell immunity directly from blood, bypassing the need for red blood cell lysis and RNA purification, thus reducing labor time and minimizing operator-induced errors. To this end, following overnight incubation with a dimethylsulfoxide (DMSO) control, or SARS-CoV-2 peptide pools, 50 μl of blood are diluted (1:4) to avoid PCR inhibition by anticoagulants (that is, heparin) and 2 μl are directly loaded onto a qPCR instrument (dqTACT). We call this latter assay direct qPCR-based rapid T cell activation (dqTACT) (Fig. 1a).
Fig. 1 |. CXCL10 mRNA levels can be measured as a proxy for T cell activation.

a, Schematic of workflow for the T cell activation assays. All assays begin with whole-blood collection followed by overnight stimulation with DMSO, nucleocapsid (NP) or SpG peptide pools. Next, supernatants are collected for ELLA or Olink; RNA is extracted and used for probe-based qPCR (qTACT) or next-generation sequencing (TACTseq) or whole blood is diluted and used directly for qPCR (dqTACT). Figure created with Biorender.com. b, CXCL10 is upregulated in response to spike peptide pool activation of whole blood. Volcano plot displaying differentially expressed genes stimulated by the spike peptide pool versus DMSO, grouped by the participant’s COVID-19 or vaccination status, displayed as red (upregulated) or blue (downregulated). Significantly differentially expressed genes were defined as having an adjusted P value <0.05 and |log2FC| > 1. P values were calculated using DESeq2 (two-sided) and adjusted using the Benjamini–Hochberg method. c, CXCL10 and IFNG mRNA induction correlate. Scatter plot displaying each gene’s correlation to IFNG expression across samples (x axis), and the corresponding log2FC (calculated by DESeq2) for the spike peptide pool versus DMSO, grouped by COVID-19 or vaccination status. Correlation calculated using Spearman’s index. d, Venn diagram displaying overlap in significantly upregulated genes in convalescent and vaccinated participants for spike peptide-stimulated samples compared to DMSO control samples. Significantly upregulated genes were defined as having P < 0.05 and log2FC > 1. e, GSEA plot displaying significantly positive enrichment for IFN-γ response genes among the upregulated genes in spike peptide simulate samples compared to DMSO control samples in COVID-19 convalescent and vaccinated cohorts.
Fig. 2 |. CXCL10 is upregulated by monocytes in response to IFN-γ released by antigen-specific T cells.

a, Schematic of proposed mechanism of CXCL10 transcript upregulation. On spike stimulation of whole blood, antigen presenting cells (APCs) present spike peptides to antigen-specific T cells that subsequently release IFN-γ. IFN-γ release can be quantified by ELLA, ELISpot or flow cytometry. Next, IFN-γ stimulates monocytes, which, in turn, upregulate CXCL10 mRNA, which can be detected by the qTACT/dqTACT assays. Figure created with Biorender.com. b, Only monocytes upregulate CXCL10 in response to IFN-γ and TNF-α. Whole blood was stimulated with DMSO (Neg) or IFN-γ + TNF-α in the presence of brefeldin/Monensin (BFA/Mon). CXCL10/IP-10 positive T cells (T), B cells (B), natural killer cells (NK), natural killer T cells (NK-T), monocytes (Mono) and neutrophils (Neutro) were quantified using flow cytometry. P values were calculated using a two-sided Wilcoxon matched-pairs signed rank test. n = 4 biologically independent samples. c, Both monocytes and neutrophils release CXCL10/IP-10 on SpG stimulation. Whole blood was stimulated with DMSO (Neg) or a spike peptide pool (SpG) in the absence (left panel) or presence (right panel) of BFA/Mon. CXCL10 positive T, B, NK, NK-T, Mono and Neutro were quantified using flow cytometry. P values were calculated using a two-sided Wilcoxon matched-pairs signed rank test. n = 4 biologically independent samples. d, Monocytes, and not neutrophils, upregulate CXCL10/IP-10 in response to SpG. Whole blood was simulated with DMSO (Neg) or a spike peptide pool (SpG) overnight with BFA/Mon added for the last 4 h. CXCL10 positive Monocytes and Neutrophils were quantified using flow cytometry. P values were calculated using a two-sided Wilcoxon matched-pairs signed rank test (P = 0.015625). n = 7 biologically independent samples. e, CXCL10 mRNA is upregulated in monocytes on stimulation with spike peptides. Monocytes and neutrophils were sorted from whole blood after overnight stimulation with spike peptides or DMSO control. RNA was extracted from the cells and the relative CXCL10 mRNA expression was determined using the qTACT assay. P values were calculated using a two-sided Wilcoxon matched-pairs signed rank test (P = 0.03125). n = 7 biologically independent samples. For b–e, the box bounds represent the first quartile (bottom), median (center) and the third quartile (top). The whiskers represent the range of samples up to 1.5 times the interquartile range. Beyond this point, samples are shown as outliers.
To select genes whose induction would correlate with the presence and activation of antigen-specific T cells, we first evaluated the transcriptional profile of whole blood after overnight stimulation with SARS-CoV-2 peptide pools by RNA-sequencing (TACTseq, Fig. 1a). This initial cohort consisted of 11 naïve, eight participants who had recovered from COVID-19 and 16 vaccinated participants (13 collected at 2–3 months and five collected at 5–8 months after the second BNT162b2 dose) (Supplementary Table 1).
Briefly, whole blood was incubated overnight with either DMSO or Spike-Gold (SpG) peptides. The latter is a refined set of peptides covering immunodominant spike peptides (previously described and validated in ref. 14). RNA was extracted from the cell pellet and subjected to Illumina single-end sequencing33. We identified genes activated by viral peptides by performing a differential expression analysis between peptide-stimulated samples and untreated controls, across all participants (Fig. 1b and Supplementary Table 1). Treatment with SpG robustly induced a largely overlapping set of genes (121 genes, FDR < 0.05, log2 fold change (FC) > 1) in both SARS-CoV-2 convalescent and vaccinated participants (Fig. 1d). Genes associated with the ‘cellular response to interferon gamma signaling’ pathway are significantly enriched among these upregulated genes, consistent with antigen-specific T cell activation in convalescent or vaccinated participants (Fig. 1e). To narrow down a shortlist of candidates for further investigation by qPCR, we selected a panel of the top six genes that were significantly upregulated following SpG stimulation and that correlated with IFNG expression, as a proxy for antigen-specific T cell activation (Fig. 1c, Extended Data Fig. 1a,b and Supplementary Table 1). Of note, correlation between IFNG mRNA levels and IFN-γ protein (detected by the ELLA automated assay system), were high, supporting our initial screen (Extended Data Fig. 1c and Supplementary Table 2).
We validated transcriptional induction of shortlisted genes by qPCR in 13 COVID-19 convalescent and 16 naïve participants (Supplementary Table 3), and included stimulation with a refined sets of peptides covering immunodominant SARS-CoV-2 nucleoprotein peptides (NP2), in addition to the SpG pool14,30. The former was used as an additional control to differentiate between naïve and convalescent participants. For each gene, the same participant was run between two and ten times to determine how results varied with multiple replicates. Out of all genes tested (Extended Data Fig. 1d and Supplementary Table 3), CXCL10 displayed the lowest variance (Extended Data Fig. 1e and Supplementary Table 3), and is thus the most reproducible and reliable biomarker for our PCR-based assay.
CXCL10 is upregulated in monocytes in response to IFN-γ released by SARS-CoV-2 antigen-specific T cells.
Our working hypothesis (Fig. 2a) is that CXCL10 mRNA is a reliable proxy for IFN-γ secreted by antigen-specific T cells. CXCL10/IP-10 protein is less so20, as it is abundantly stored by neutrophils and monocytes before IFN-γ stimulation. To formally prove this, we have performed the following analysis: first, we showed which immune cell subsets produce CXCL10/IP-10 in response to stimulation with IFN-γ and TNF-α in the presence of brefeldin/monensin (BFA/Mon) to inhibit protein secretion. Monocytes and neutrophils are the main immune cells that increase their CXCL10/IP-10 production in response to IFN-γ and TNF-α stimulation. Monocytes and, in particular, neutrophils, have elevated CXCL10/IP-10 levels at baseline (before stimulation) (Fig. 2b, Extended Data Fig. 2a and Supplementary Table 4).
Next, we determined whether monocytes and neutrophils produce CXCL10/IP-10 on stimulation with SARS-CoV-2-specific spike peptides. For this purpose, we set up three conditions: (1) BFA/Mon was not added, allowing cytokine release from immune cells and (2) a negative control in which BFA/Mon was added from the beginning blocking IFN-γ production and CXCL10 mRNA induction in neighboring cells (Fig. 2c and Supplementary Table 4); (3) BFA/Mon was added in the last 4 hours of an overnight incubation (delayed BFA/Mon), which prevents CXCL10/IP-10 secretion in the cell culture media, but should not prevent CXCL10 mRNA induction (Fig. 2d, Extended Data Fig. 2b and Supplementary Table 5). Our flow cytometric results indicated an accumulation of CXCL10/IP-10 in monocytes and not in neutrophils on stimulation with spike peptides (Fig. 2c,d). Last, we sorted cells in the BFA/Mon delayed condition. mRNA was extracted and CXCL10 levels were quantified by qTACT (Fig. 2e and Supplementary Table 5), confirming a significant induction only in monocytes.
We conclude that monocytes can produce CXCL10 in a tightly regulated manner and in response to the IFN-γ secreted by antigen-specific T cells, and thus serves as a proxy of T cell activation on spike peptide stimulation of whole blood (Fig. 2a).
TACT assays are concordant with gold standard ELLA and ELISpot assays.
We next performed a series of validation experiments in a larger cohort of naïve participants, who were COVID-19 convalescent and SARS-CoV-2 vaccinated, to assess the correlation between CXCL10 mRNA expression and IFN-γ level. First, CXCL10 measured by both qTACT and dqTACT is in robust concordance with both IFN-γ protein quantification by ELLA (Fig. 3a,b and Supplementary Tables 6 and 7) and ELISpot (Fig. 3c,d and Supplementary Tables 8 and 9). We additionally show a robust correlation between TACTseq and either ELLA or ELISpot (Extended Data Fig. 3a,b and Supplementary Tables 10 and 11). In addition to TACTseq, we used an independent and complementary approach to assess the cytokines/chemokines induced by both lymphoid and myeloid cells in whole blood. Following stimulation with SARS-CoV-2 peptide pools, the supernatant from naïve (n = 7) and BNT162b2 vaccinated (n = 19) participants was collected and analyzed using the Olink multiplex assay (Olink Bioscience). This technology can accurately quantify secretion of a panel of 45 inflammatory cytokines/chemokines. In addition to IFN-γ and IL-2, stimulation with spike (SpG), but not NP2 peptides nor DMSO, induced CXCL10/IP-10, CCL2, CCL4, CCL8, CXCL8 and CXCL9 in vaccinated participants (Extended Data Fig. 4a,b and Supplementary Table 12). qTACT CXCL10 quantification correlates well with Olink IFN-γ and IL-2 levels, and less so with CXCL10/IP-10, consistent with the fact that CXCL10 mRNA but not protein is the best proxy for T cell activation (Extended Data Fig. 4c–e and Supplementary Table 12). This discrepancy is most likely due to the prestimulation reservoir of CXCL10/IP-10 in neutrophils (Fig. 2c and Supplementary Table 4), which are highly abundant in whole blood34. Finally, we confirmed previously published results by demonstrating a high degree of correlation between ELLA and ELISpot quantified IFN-γ (Extended Data Fig. 3c and Supplementary Table 13).
Fig. 3 |. TACT assays are concordant with gold standard ELLA and ELISpot assays.
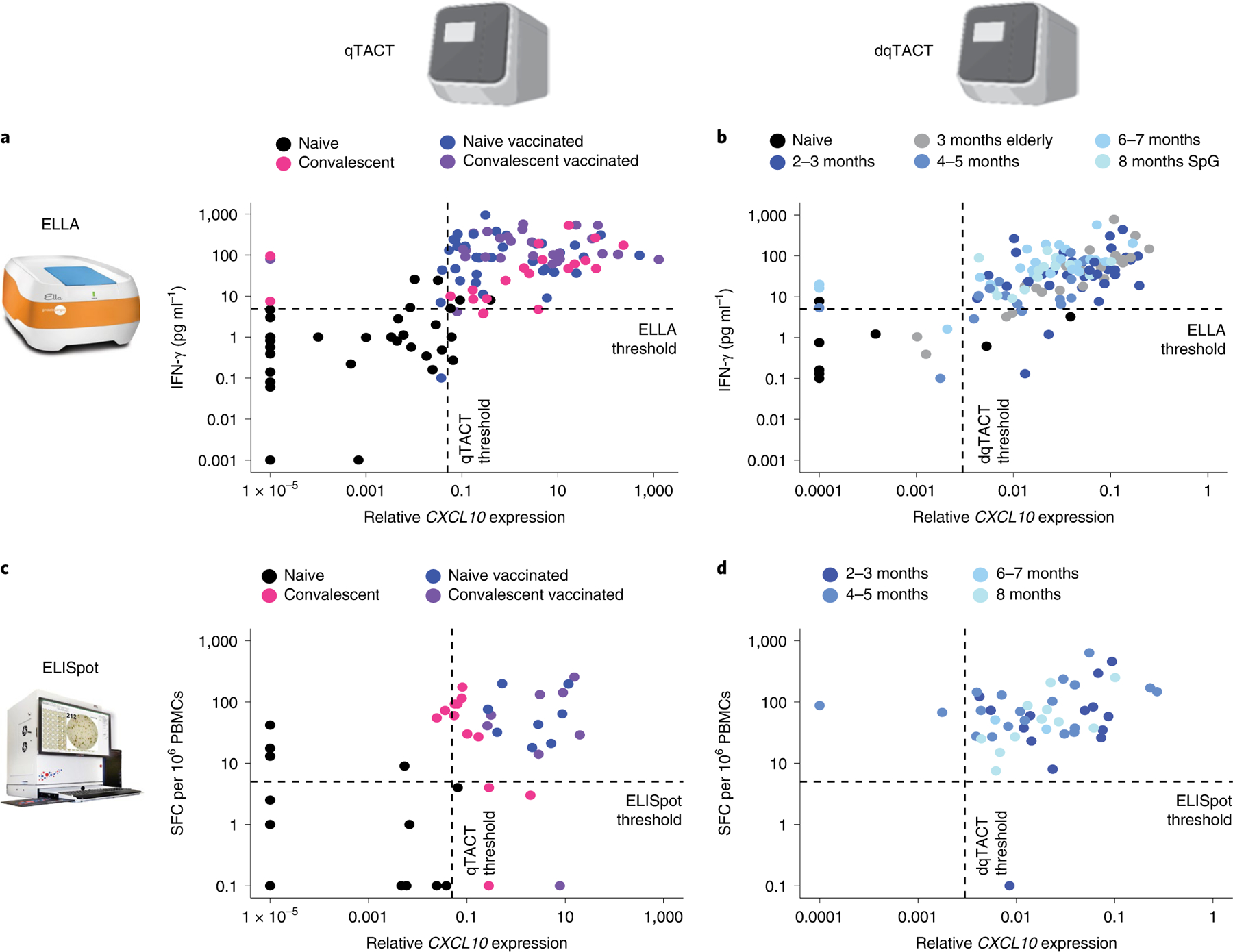
a–d, Concordance between assays to quantify cellular immunity. The quantification shown is with the DMSO control subtracted from the spike peptide-stimulated sample. Each dot represents a unique participant color coded based on their COVID-19 and vaccination statuses (legend). The dashed line represents thresholds for each assay. a, Concordance between ELLA and qTACT assays. For 117 participants, IFN-γ protein secretion was quantified by ELLA (y axis) and CXCL10 mRNA by qTACT (x axis). b, Concordance between ELLA and dqTACT assays. For 133 participants, IFN-γ protein secretion was quantified by ELLA (y axis) and CXCL10 mRNA by dqTACT (x axis). c, Concordance between ELISpot and qTACT assays. For 50 participants, IFN-γ producing cells were quantified by ELISpot (y axis) and CXCL10 mRNA by qTACT (x axis). d, Concordance between ELISpot and dqTACT assays. For 46 participants, IFN-γ producing cells were quantified by ELISpot (y axis) and CXCL10 mRNA by dqTACT (x axis).
Analytical validation and comparison of available T cell assays.
Second, each method has a different dynamic range, but we were able to use our large set of data to establish robust thresholds to call true positives and true negatives, and to assess specificity, sensitivity and accuracy of the different assays. We also performed standard receiver operating characteristic (ROC) curve analysis to calculate the area under the curve (AUC)35. Overall, all assays have high accuracy and AUC > 0.90 (Fig. 4a,b and Supplementary Tables 6–13). We computed two-sided Fisher’s exact tests for each assay, reporting the associated P values and odds ratios (with corresponding 95% CIs) in Supplementary Table 14. These additional metrics confirm the high standards of the qTACT and dqTACT tests, which are comparable to the gold standard ELLA and ELISpot assays. These assays will be useful for the evaluation of cellular immunity toward future vaccines and variants, each with their advantages and limitations. Traditional flow cytometry and ELISpot have the highest specificity and sensitivity, but are difficult to scale given the intense labor time needed to isolate PBMCs. Whole-blood-based assays are very comparable in terms of specificity, sensitivity and accuracy. The strongest downside of ELLA, which directly quantifies IFN-γ secretion, is the price per sample, with the additional requirement of specialized equipment and/or plates. Finally, both qTACT and dqTACT were optimized on multiple qPCR machines, including the 7500 Fast System (Applied Biosystems), CFX96 (BioRad), CFX384 (BioRad) and bCUBE 2.0 (Hyris), with comparable results (Extended Data Fig. 3d,e and Supplementary Tables 15 and 16), thus allowing a wide application of these studies across multiple diagnostic laboratories worldwide. The Hyris bCUBE is a portable, two-channel qPCR machine that is able to quantify up to 36 samples at a time. Unlike BioRad’s CFX machines, the bCUBE uses a patented detection technology based on a solid-state complementary metal-oxide-semiconductor sensor with a peculiar optical stack to capture and quantify fluorescence, coupled with a disposable cartridge specifically designed for this setup. It features a reaction chamber design with reagents lying directly on the optical window, in conjunction with the efficient heat transfer given by an aluminum plate integrated in the cartridge. As opposed to the BioRad or AB machines, this is a portable, cheap and easy to use instrument that might enable point of care implementation of the assay.
Fig. 4 |. Analytical validation and comparison of available T cell assays.
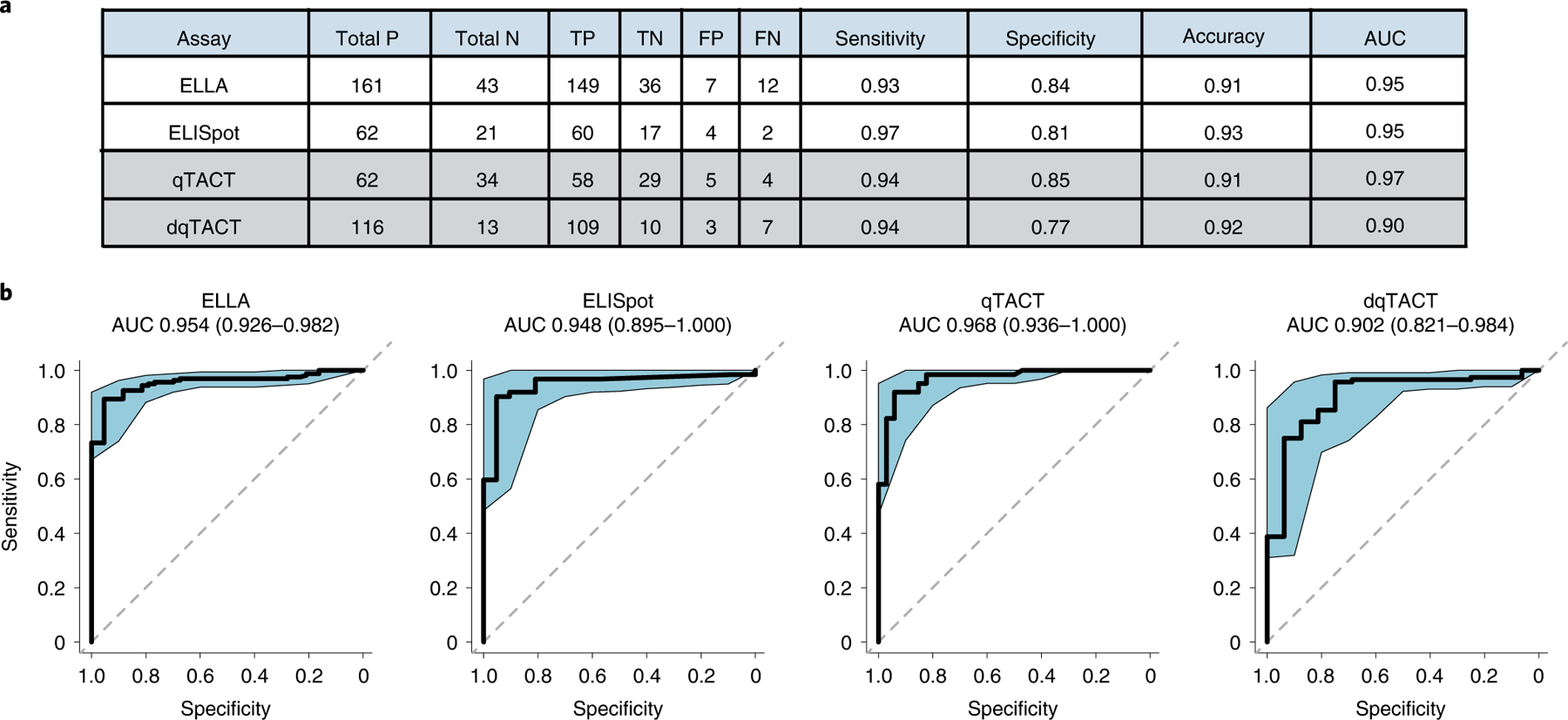
a, Comparison of the various assays used to determine T cell response to spike peptides. Total P/N = total positives/negatives (that is, above or below threshold, respectively). TP/TN = true positives/negatives (that is, correctly above or below threshold, respectively, according to the participant’s COVID-19/vaccination status). FP/FN = false positives/negatives. Sensitivity = true positives/(true positives + false negatives). Specificity = true negatives/(true negatives + false positives). Diagnostic accuracy = (true positives + true negatives)/all samples). b, ROC curves, AUC values and associated 95% confidence intervals for each assay.
Using qTACT and dqTACT assays to monitor cellular immunity in large cohorts.
Finally, we used our scalable qTACT and dqTACT tests, in parallel with ELLA and ELISpot, across multiple cohorts of participants. First, we used a cohort of 91 participants (45 naïve and 46 COVID-19 convalescent, Supplementary Tables 17–19), for which we have previously published the corresponding ELLA results19. For this validation cohort, we chose to quantify IFNG mRNA, despite its inferiority relative to CXCL10, to include a gene that is expressed by antigen-specific T cells and to directly correlate mRNA expression (qTACT) with IFN-γ protein secretion (ELLA). The cohort was recruited before vaccination and followed at days 10 and 20 after the first and second dose of the BNT162b2 vaccine and the data on IFN-γ an IL-2 cytokine secretion have been described in a separate paper19. Consistent with our previously published data, compared to naïve participants, COVID-19 recovered individuals had a higher median expression of CXCL10 before vaccination (Fig. 5a and Supplementary Table 17). A similar trend was observed for IFNG levels, but the difference was not statistically significant (Fig. 5b, Extended Data Fig. 5a and Supplementary Table 17). Notably, quantification of the Spike-specific T cell response by both CXCL10 and IFNG qPCR (qTACT) 10 and 20 days after the first and second dose, confirmed similar findings obtained by more traditional ELLA (Fig. 5c and Supplementary Table 18) and ELISpot (Fig. 5d and Supplementary Table 19) assays, with the clear advantage of cost effectiveness and scalability. The technical advantage of our test over ELLA or ELISpot is the ease of use of qPCR and the internal normalization standard (that is, ACTIN levels), which is absent in other more laborious methods of quantifying cellular immunity.
Fig. 5 |. Using qTACT to monitor cellular immunity in vaccinated participants.

a, Quantification of CXCL10 mRNA (qTACT) prevaccination and at 10 and 20 days after the first and second dose of an mRNA-based vaccine. Time points are indicated on the x axis and relative CXCL10 expression (minus DMSO control) on the y axis. The dashed line represents the qTACT threshold (0.05). The number of participants for each time point is indicated above the box plots along with the percentage of participants who fall above the threshold. Colors represent the participant’s COVID-19/vaccination status (legend). b, Quantification of IFNG mRNA (qTACT) prevaccination and at 10 and 20 days after the first and second dose of an mRNA-based vaccine. Time points are indicated on the x axis and relative IFNG expression (minus DMSO control) on the y axis. The number of participants for each time point is indicated above the box plots. Colors represent the participant’s COVID-19/vaccination status (legend). c, Quantification of IFN-γ protein secretion (ELLA) prevaccination and at 10 and 20 days post the first and second dose of an mRNA-based vaccine. Time points are indicated on the x axis and IFN-γ protein secretion (minus DMSO control) on the y axis. The dashed line represents the ELLA threshold (5). The number of participants for each time point is indicated above the box plots along with the percentage of participants who fall above the threshold. Colors represent the participants’ COVID-19/vaccination statuses (legend). d, Quantification of IFN-γ producing cells (ELISpot) prevaccination and at 10 and 20 days after the first and second dose of an mRNA-based vaccine. Time points are indicated on the x axis and number of IFN-γ producing cells (minus DMSO control) on the y axis. The dashed line represents the ELISpot threshold (5). The number of participants for each time point is indicated above the box plots along with the percentage of participants who fall above the threshold. Colors represent the participants’ COVID-19/vaccination statuses (legend). For a–d, the box bounds represent the first quartile (bottom), median (center) and the third quartile (top). The whiskers represent the range of samples up to 1.5 times the interquartile range. Beyond this point, samples are shown as outliers.
We next used the dqTACT assay to quantify the levels of CXCL10, as a proxy for cellular immunity in vaccinated participants at different time points after the second dose. We observed a consistent detection of T cells above threshold by dqTACT (Fig. 6a and Supplementary Table 20), ELLA (Fig. 6b and Supplementary Table 21) and ELISpot (Fig. 6c and Supplementary Table 22), up to 8 months postvaccination.
Fig. 6 |. Using dqTACT to monitor the persistence of cellular immunity and cross reactivity with spike epitopes from VOC in vaccinated participants.

a–c, Detection of CXCL10 mRNA (dqTACT (a)), IFN-γ protein secretion (ELLA (b)) or IFN-γ producing cells (ELISpot (c)) in vaccinated participants over time. Time points (postvaccination) are indicated on the x axis and relative CXCL10 expression (a), IFN-γ protein secretion (b) or IFN-γ producing cells (c) on the y axis (all values minus DMSO). The dashed lines represent the dqTACT (0.003), ELLA (5) or ELISpot (5) thresholds. d, Schematic to show how T cell responses against the VOC can be evaluated using the delta variant as an example. Orange regions refer to amino acid mutations present in the delta variant compared to the wildtype (WT) SARS-CoV-2 strain. Pool HS (hotspot) contains peptides covering the nonconserved Spike-Wuhan regions affected by mutations present in the delta variant (24 peptides). Pool delta MT contains peptides from Pool HS with the amino acid mutations present in the Spike protein of the delta variant. e–g, Quantification of CXCL10 mRNA (dqTACT (e)), IFN-γ protein secretion (ELLA (f)) or IFN-γ producing cells (ELISpot (g)) in vaccinated participants stimulated with spike peptides covering the hotspot wildtype (HS WT) or delta variant region. The peptide pool used for stimulation, either the wildtype (HS) or delta (delt) sequence, is indicated on the x axis and relative CXCL10 expression (e), IFN-γ protein secretion (f) or IFN-γ producing cells (g) on the y axis (all values minus DMSO). The dashed lines represent the dqTACT (0.003), ELLA (5) or ELISpot (5) thresholds. P values were calculated using a two-sided Wilcoxon rank sum test. h,i, Quantification of relative CXCL10 mRNA expression using dqTACT (h) or IFN-γ protein section using ELLA (i) in an elderly cohort. For a–c and e–i, the box bounds represent the first quartile (bottom), median (center) and the third quartile (top). The whiskers represent the range of samples up to 1.5 times the interquartile range. Beyond this point, the samples are shown as outliers. The number of participants for each time point is indicated above the box plots along with the percentage of participants who fall above the threshold.
Finally, we used the dqTACT assay to assess cellular immunity in: (1) a population of elderly individuals at 3 months after the second dose. We did not observe any appreciable difference compared to a younger population, and results were consistent with data obtained by ELLA (Fig. 6h,i and Supplementary Tables 23 and 24); (2) a larger cohort from a randomized, adaptive, phase 2 trial (CombiVacS)16 (Extended Data Fig. 5b,c and Supplementary Tables 25 and 26). The trial sought to determine the reactogenicity and immunogenicity of a second dose of BNT162b2 (Comirnaty, BioNTech) in individuals who had received a first dose of ChAdOx1s (Vaxzevria, Astra Zeneca). Our assay reliably detected CXCL10 expression in 141 vaccinated individuals enrolled in this study. In line with previously published data from the CombiVacS trial where upregulation of IFN-γ was observed after heterologous vaccination16, we also observed a significant increase in CXCL10 expression in individuals who received a second dose (of BNT162b2) compared to those that received the placebo (only a first dose of ChAdOx1s, but no second dose) (Extended Data Fig. 5b,c) and (3) a small population of vaccinated participants stimulated with either a small subset of peptides covering the hotspots in the wildtype (Wuhan) or the delta variant (Fig. 6d and Supplementary Table 27). The latter experiment was performed to assess whether our dqTACT assay could be rapidly deployed to assess the protection of individual T cell repertoires to emerging variants. These data demonstrate a slight, but not statistically significant decrease of T cell activation toward delta in participants vaccinated with the wildtype (hotspot pool) strain consistently across the different platforms dqTACT (Fig. 6e), ELLA (Fig. 6f) and ELISpot (Fig. 6g). This is consistent with the substantial preservation of Spike-specific T cells response against mutated Spike protein present in different variants of concern (VOC) observed in most vaccinated and convalescent individuals36,37.
Discussion
A lack of rapid, user-friendly, accessible, scalable and accurate diagnostic methods to quantify cellular immunity prevents large population studies, affecting long-term vaccination strategies and public health responses to global pandemics, such as the one being caused by SARS-CoV-2. Considering that diagnostic centers around the world have ramped up the setup of qPCR with reverse transcription (RT–qPCR)-based facilities, we developed a qPCR-based dqTACT assay, which is amenable to periodic and repeated testing of patient samples, as it requires only 1 ml of blood and a 24-hour turnaround time. Clinical validation of this assay in response to recent draft guidance from the US Food and Drug Administration and European Medicines Agency is ongoing in a Clinical Laboratory Improvement Amendments (CLIA)-certified microbiology laboratory.
We here describe three approaches, with different advantages and limitations, to detect SARS-CoV-2-specific cellular immunity following overnight incubation of whole blood with viral peptides.
First, we implemented a next-generation sequencing approach (TACTseq). The advantage of this approach, which could be further implemented by a targeted amplification panel of 15–20 genes, is the possibility of capturing the variability of the response and measuring cytokines produced by both T cells and other myeloid cells in the blood. The limitations are a longer turnaround time, high cost and the need for skilled technical personnel.
Second, we developed a qPCR-based method (qTACT assay) on a 96- or 384-well platform (BioRad CFX). The advantages of this approach are the accuracy and sensitivity of qPCR probes, the opportunity to combine more than two fluorophores to measure the expression of 2–4 genes and the scalability and potential automation of the process. The limitations include a relatively longer processing time (roughly 24 h per 200 samples), the need to purify RNA (by standard RNA-purification kits/columns), a moderate price and a medium level of technical skill.
Third, we optimized a direct qPCR-based method (dqTACT assay) on multiple RT–qPCR platforms. The advantages of this approach are the accuracy of qPCR probes and the reduced processing time, cost and/or skill required. Overall, this is an easy-to-implement protocol that requires minimal training of the operator, thus reducing technical errors.
In qTACT and dqTACT assays, we use CXCL10 mRNA levels, after antigen-specific activation with peptide pools as a proxy for IFN-γ secreted by antigen-specific T cells, in vitro. CXCL10 protein levels have been recently proposed as a biomarker of impaired T cell responses in elderly people suffering from severe COVID-19 (ref. 38). However, these data are not related to an antigen-specific analysis. Serum CXCL10 levels are higher because these patients have inflammation, not because the production of CXCL10 was induced after an antigen-specific T cell activation done in vitro, such as in the qTACT and dqTACT assays. The latter are designed to test T cells of vaccinated or convalescent participants when they are in a state of ‘memory’, not during a generic activation.
We conclude that the derived profile of SARS-CoV-2-specific T cell activation by qTACT and dqTACT assays in different cohorts of naïve, COVID-19 recovered and vaccinated individuals, provides robust information about their level of SARS-CoV-2-specific cellular immunity. A diagnostic method that can be easily adapted to detect the degree of cellular immunity is an urgently needed complement to the currently available tests measuring viral presence or antibody titers, and design future vaccination strategies according to the levels of immune protection in the population. A recent publication describing an alternative test, which uses T cell receptor next-generation sequencing to determine cellular immunity to SARS-CoV-2, shows that there is interest in such a test in the wider scientific community39.
In conclusion, the assays presented here are based on the ability of SARS-CoV-2 T cells to respond to different peptides covering different proteins of the virus. With the possibility to use different peptide pools, our approach represents a flexible strategy that can be easily used to detect the presence of T cells responding to (1) different viral proteins (that is nucleoprotein, polymerase and so on), that have an important role in protection8,40, and (2) emerging mutant strains, thus immediately gauging the impact that viral mutations might have on cellular immunity36,37. This could be relevant for immunocompromised populations41,42, and to evaluate vaccine effectiveness in a time-sensitive manner.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-022-01347-6.
Methods
SARS-CoV-2 peptide pools.
The spike protein (1,276 amino acids long) requires a total of 253 15-mer peptides overlapping by 10 amino acids to cover the entire protein. We refined the pool (SpG) encompassing 40.5% of the spike protein (55 peptides)14. SpG encompasses most of the SARS-CoV2 spike epitope published so far. Pool hotspot (HS) contains peptides covering the nonconserved Spike-Wuhan regions affected by mutations present in the delta variant (24 peptides). Pool delta MT contains peptides from pool delta hotspot with the amino acid mutations present in the Spike protein of the delta variant. A similar strategy (15-mer peptides) was also used to cover nucleoprotein (NP2) as previously described14,17.
Whole-blood culture with SARS-CoV-2 peptide pools.
Here, 320 μl of whole blood drawn on the same day were mixed with 80 μl of Roswell Park Memorial Institute medium and stimulated with pools of SARS-CoV-2 peptides (S or nucleoprotein, 2 μg ml−1) or DMSO control at 37 °C. After 15–17 h of stimulation, the supernatant (plasma) was collected and stored at −80 °C until quantification of cytokines.
RNA-seq.
Whole blood was treated with ACK lysis buffer for 15 min at room temperature to remove red blood cells. Cells were resuspended in Trizol (300 µl) and RNA was extracted using Zymo’s Direct-zol extraction kit as per the manufacturer’s instructions. Sequencing libraries were prepared from the eluted RNA using the Ovation Ultralow V2 DNA-seq Library Preparation Kit following the manufacturer’s instruction (NuGEN) at the Mount Sinai Oncological Sciences Sequencing Facility. Sequencing was performed on an Illumina NextSeq 500 instrument to produce 75-bp single-end reads. Demultiplexed FASTQ files were subsequently returned for analysis.
RNA-seq data analysis.
Transcript expression was quantified from RNA-seq data using Salmon (v.1.2.1) (ref. 43) against an index built from the Ensembl44 GRCh38 v.99 transcriptome model with default parameters. Pseudocounts were imported into an R v.4.0.3 environment45 and summarized to the gene level using tximeta v.1.8.2. Differential expression analysis was conducted separately for naïve and convalescent individuals using DESeq2 (v.1.3.0) (ref. 46) with default parameters. Only protein-coding genes with at least ten total counts were included for each analysis, and donor information was included in the design. P values were corrected using the Benjamini–Hochberg method (two-sided). Significantly upregulated genes were defined as those with an adjusted P ≤ 0.05 and log2FC > 1. Gene set enrichment analysis (GSEA) was performed using fgsea (v.1.16.0) on differential gene expression signatures generated using DESeq2 and ranked according to the resulting test statistic (‘stat’ column). Gene sets were obtained from msigdbr (v.7.2.1). The results were further validated using gprofiler2 (v.0.2.0) using default parameters47.
qTACT assay.
Samples used for RNA extraction were diluted 1:1 with RNA/DNA shield (Zymo) and incubated at room temperature with proteinase K at a 1:100 dilution (20 mg ml−1 stock). Samples were then frozen at −80 °C until RNA extraction could be performed. Samples stored in RNA/DNA shield were thawed at room temperature before RNA extraction. Samples were vortexed and mixed with Trizol reagent (Life Technologies) at a 1:1 dilution. After vortexing, samples were processed using Zymo’s Direct-zol 96-well extraction kit as per the manufacturer’s instructions. Eluted RNA was diluted in TE buffer, aliquoted and stored at −80 °C or used immediately for qPCR analysis. Real-time quantification was performed on a BioRad CFX96/CFX384 or Hyris bCUBE v.2.0. Then 5 μl of diluted RNA was used with the TaqPath 1-Step Multiplex MasterMix (Applied Biosystems) and primers/probes targeting ACTIN (internal control) and other target genes, as described (sequences available in Supplementary Table 29).
dqTACT assay.
Samples used for direct amplification from whole blood were diluted 1:4 and stored at −80 °C or used immediately for qPCR analysis. Then 2 μl of diluted whole blood was mixed with SCRIPT Direct RT–qPCR ProbesMaster (Jena Bioscience) and primers/probes targeting ACTIN (internal control) and other target genes, as described. Quantification was performed using the Hyris bCUBE 2.0 (data acquired using bAPP (v.1.5.6) online software from Hyris), CFX96, CFX384 as specified. Sequences of primers and probes used are available in Supplementary Table 29.
Cytokine quantification and analysis.
Cytokine concentrations in the plasma were quantified using ELLA with microfluidic multiplex cartridges measuring IFN-γ and IL-2 following the manufacturer’s instructions (ProteinSimple). The level of cytokines present in the plasma of DMSO controls was subtracted from the corresponding peptide pool stimulated samples.
Spike-specific IgG quantification.
The ACCESS SARS-CoV-2 CLIA (Beckman Coulter Inc.) was used for semiquantitative detection of IgG directed against S protein receptor-binding domain using serum obtained from venipuncture blood. Samples were tested on a UniCel Dxl 800 high-performance analyzer.
Olink immunoassay.
Cytokine concentrations in the plasma were analyzed using Olink multiplex assay platform with inflammatory panel (Olink Bioscience), according to the manufacturer’s instructions. The inflammatory panel includes 92 proteins associated with human inflammatory conditions. Briefly, an incubation master mix containing pairs of oligonucleotide-labeled antibodies to each protein was added to the samples and incubated for 16 h at 4 °C. Each protein was targeted with two different epitope-specific antibodies increasing the specificity of the assay. Presence of the target protein in the sample brought the partner probes in close proximity, allowing the formation of a double strand oligonucleotide PCR target. On the following day, the extension master mix in the sample initiated the specific target sequences to be detected and generated amplicons using PCR in 96-well plates. For the detection of the specific protein, Dynamic array integrated fluidic circuit 48 × 48 chip was primed, loaded with 45 protein specific primers and mixed with sample amplicons including three inter-plate controls and three negative controls. Real-time microfluidic qPCR was performed in Biomark (Fluidigm) for the target protein quantification.
Data were analyzed using real-time PCR analysis software via ΔΔCt method and normalized protein expression manager. Data were normalized using internal controls in every single sample, inter-plate control and negative controls and correction factor and expressed as log2 scale, which is proportional to the protein concentration. One normalized protein expression difference equals to the doubling of the protein concentration.
ELISpot.
Enzyme-linked immunospot (ELISpot) flat-bottomed, 96-well nitrocellulose plates (MAHA S4510, Millipore) were coated with IFN-γ mAb (2 μg ml−1, 1-D1K; MABTECH) and incubated for 2 h at 37 °C. After washing with PBS, plates were blocked with 10% human AB serum for 2 h at 37 °C. Cells were washed, concentrated, plated into each well titrating down in twofold serial dilutions starting from 400,000 PBMCs, in the presence of peptides, negative control (DMSO) and positive control (phorbol 12-myristate 13-acetate and ionomycin (PMA)/ionomycin). Plates were incubated for 24 h in a CO2 incubator. After incubation, the plates were semi-automatically washed with PBS and then IFN-γ mAb (0.2 μg ml−1, 7-B6–1-biotin; MABTECH) was added to each well. After incubation for 2 h at 37 °C, plates were washed and incubated with streptavidin-alkaline phosphatase (1 μg ml−1, Roche) for 1 h at room temperature. After washing unbound streptavidin-alkaline phosphatase, substrate (5-bromo-4-chloro-3-indolyl phosphate/NBT; Sigma-Aldrich) was added and incubated for 12–15 min for spots to develop. After incubation, plates were washed to remove substrate and dark-violet spots were evaluated using the CTL Immunospot analyzer and software (Cellular Technology Limited) as routinely performed in the laboratory38.
Flow cytometry.
Flow cytometry analyses were carried out in whole-blood samples labeled for viability (Blue Live/dead fixable dye, BioLegend), washed, and stained for extracellular markers for 45 min at 4 °C. For intracellular staining, samples were fixed in 4% paraformaldehyde (Electron Microscopy Services) and treated with permeabilization buffer (eBioscience) before staining with labeled antibodies to detect intracellular cytokines, CD154 and CXCL10. The list of antibodies used for surface and cytoplasmic staining are shown in the Supplementary Table 28. Cells were subsequently acquired on a five-laser Cytek Aurora device (Cytek Biosciences) and data analyzed on Cytobank (https://cytobank.org/). For sorting experiments, whole-blood samples were exclusively stained for the surface markers CD16, CD3, HLA-DR, CD66b, CD15, CD8, CD4 and CD14. Four different populations (neutrophils, monocytes, CD4+ and CD8+ T cells) were sorted with purities >96% by using a fluorescence-activated cell sorting Aria II Cell Sorter device (BD Biosciences).
Statistical analysis.
All illustrations were prepared using ggplot2 and ggpubr in an R v.4.0.3. Statistical significance in figures is reported as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. For RNA-seq data (Fig. 1b and Extended Data Fig. 1), displayed P values were generated using DESeq2 as described in the RNA-seq data analysis section. For all plots, displayed P values were generated using a two-sided Wilcoxon rank sum test in R and corrected using the Benjamini–Hochberg method. ROC curves and associated 95% CIs for each assay were computed in R using the pROC package (v.1.18.0)48. Fisher exact tests and associated odds ratios were computer in R using the fisher.test function using a two-sided approach.
Ethics statement.
The trial complies with the principles of the Declaration of Helsinki and Good Clinical Practice. This study was approved by the Spanish Agency of Medicines and Healthcare Products and by the Ethics Committee at University Hospital La Paz (CombiVacS data). The study protocols for the collection of clinical specimens from individuals with and without SARS-CoV-2 infection were reviewed and approved by Hospital La Paz, Hospital 12 de Octubre, Hospital Gregorio Marañón, IIS-Fundación Jimenez Díaz, Hospital Universitario Marqués de Valdecilla-IDIVAL and Hospital Puerta de Hierro Clinical Research Ethics Committee (CEIm), the SingHealth Centralised Institutional Review Board (reference CIRB/F/2018/2387) and Mount Sinai Hospital Institutional Review Board (no. 11–00866). All study participants provided their written informed consent for the collection of samples and their subsequent analysis.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. CXCL10 mRNA levels can correlate with IFNG mRNA levels and can be reliably used as a proxy for T cell activation.

A. Normalized counts of candidate genes selected for downstream validation based on differential expression versus DMSO. Comparisons show significance calculated using DESeq2 (two-sided) and corrected using the Benjamini-Hochberg method. Naive, N = 11; convalescent N = 8; vaccinated, N = 16 biologically independent samples. B. Correlation between candidate gene and IFNG expression (normalized gene counts). Treatment and group is indicated by color/symbol (see legend). Correlation R and p-values were calculated using Spearman’s index. Grey bands represent the 95% confidence interval of the linear model. C. Concordance between ELLA and TACTseq NGS assays. For 24 subjects, IFN-γ protein secretion was quantified by ELLA (y-axis) and CXCL10 mRNA by Illumina NGS sequencing TACTseq (x-axis). D. Validation of candidate genes in naive (black) and convalescent (pink) subjects stimulated with NP2 or spike SpG peptide pools (qTACT assay). Naive, N = 13; convalescent, N = 16 biologically independent samples. P-values were calculated using a two-sided Wilcoxon rank sum method E. Average normalized variance in relative CXCL10 expression minus DMSO across replicate qTACT runs on the same samples. For A and D, the box bounds represent the first quartile (bottom), median (center) and the third quartile (top). The whiskers represent the range of samples up to 1.5 times the interquartile range. Beyond this point, the samples are shown as outliers.
Extended Data Fig. 2 |. Gating strategies for flow cytometry experiments.

a. Gating strategy for main Fig. 2 panel B. b. Gating strategy for main Fig. 2 panel D.
Extended Data Fig. 3 |. Comparing TACTseq assay to other T cell assays and dqTACT on different PCR machines.

A. Concordance between ELLA and TACTseq assays. For 24 subjects, IFN-γ secretion was quantified by ELLA (y-axis) and CXCL10 mRNA by TACTseq (x-axis). The quantification shown is with the DMSO control subtracted from the spike peptide stimulated sample. Each dot represents a unique subject color coded based on their COVID-19 and vaccination statuses. The dashed line represents thresholds for each assay. B. Concordance between ELISpot and TACTseq assays. For 24 subjects, IFN-γ producing cells were quantified by ELISpot (y-axis) and CXCL10 mRNA by TACTseq (x-axis). The quantification shown is with the DMSO control subtracted from the spike peptide stimulated sample. Each dot represents a unique subject color coded based on their COVID-19 and vaccination statuses. The dashed line represents thresholds for each assay. C. Concordance between ELISpot and ELLA assays. For 85 samples, IFN-γ protein secretion was quantified by ELLA (y-axis) and IFN-γ producing cells were quantified by ELISpot (y-axis). The quantification shown is with the DMSO control subtracted from the spike peptide stimulated sample. Each dot represents a unique subject color coded based on their COVID-19 and vaccination statuses. The dashed line represents thresholds for each assay. D. Nonparametric Spearman’s correlation between dqTACT samples run on Bio-Rad CFX384 (y-axis) and Hyris bCUBE 2.0 (x-axis). Bands represent 95% confidence interval. E. Nonparametric Spearman’s correlation between dqTACT samples run on Bio-Rad CFX96 (y-axis) and Applied Biosystems AB 7500 (x-axis). Bands represent 95% confidence interval.
Extended Data Fig. 4 |. Comparing dqTACT and Olink assays.
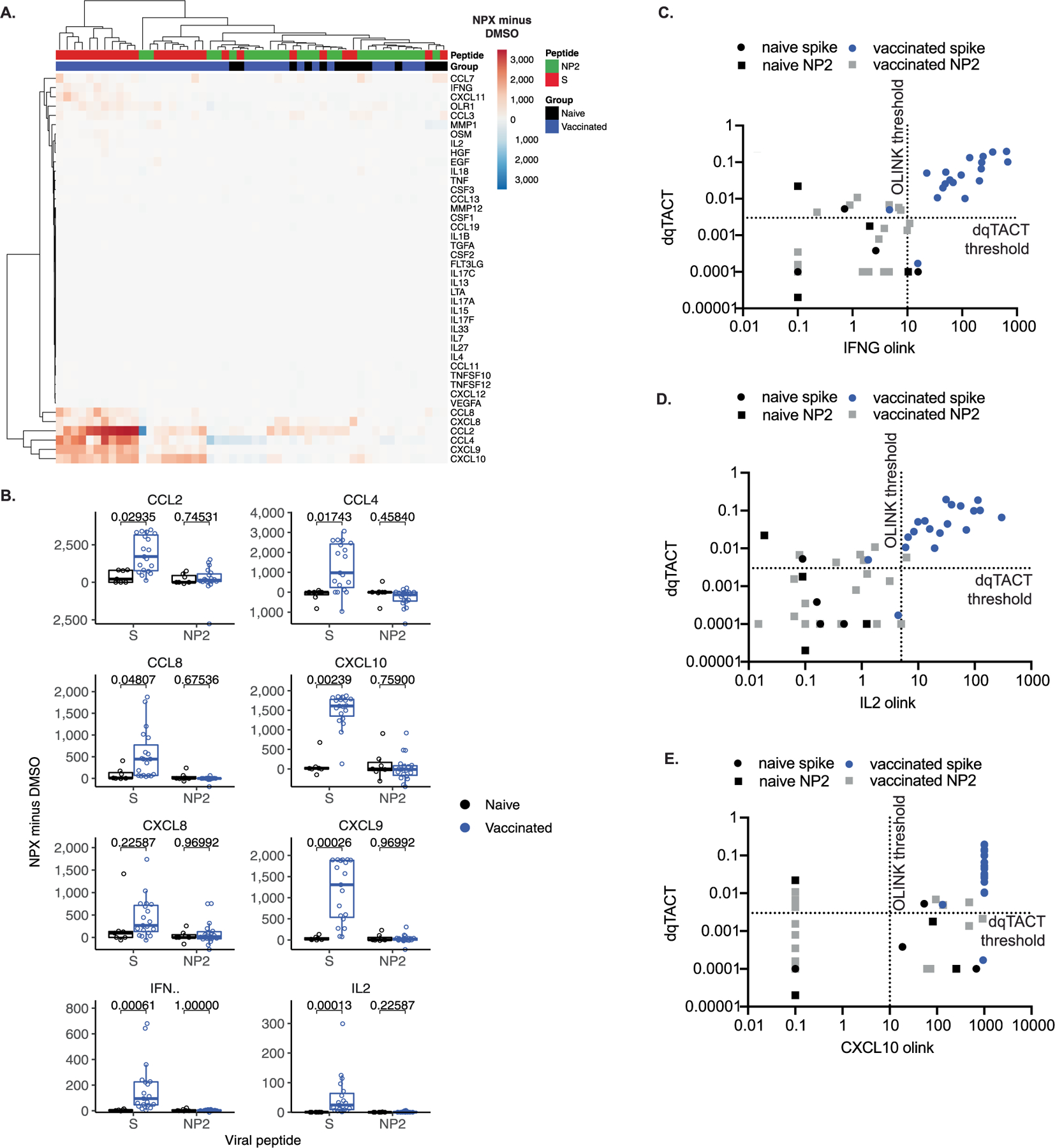
A. Heatmap displaying normalized protein expression (NPX) values minus DMSO across 42 peptides measured by Olink. B. Boxplot displaying normalized protein expression (NPX) values minus DMSO. Comparisons show significance for the Wilcoxon Rank Sum two-sided test, corrected using the Benjamini-Hochberg method. Naive, N = 7; vaccinated, N = 19 biologically independent samples. The box bounds represent the first quartile (bottom), median (center), and the third quartile (top). The whiskers represent the range of samples up to 1.5 times the interquartile range. Beyond this point, samples are shown as outliers. C-E. Concordance between Olink and dqTACT assays. For 23 samples, IFN-γ (C), IL2 (D), and CXCL10 (E) protein secretion was quantified by O-link (x-axis) and CXCL10 mRNA by qTACT (y-axis). The quantification shown is with the DMSO control subtracted from the spike peptide stimulated sample. Each dot represents a unique subject color coded based on their COVID-19 and vaccination statuses. The dashed line represents thresholds for each assay.
Extended Data Fig. 5 |. Using the dqTACT assay for a large clinical trial cohort (CombiVacS).

A. Concordance between ELLA and qTACT assays for IFN-γ/IFNG. For n = 91 subjects, IFN-γ secretion was quantified by ELLA (x-axis) and IFNG mRNA by qTACT (y-axis). Colors on their COVID-19 and vaccination statuses (see legend). The dashed lines represent thresholds for each assay. B-C. Quantification CXCL10 mRNA using dqTACT (B) or IFN-γ protein section using ELLA (C) in subjects enrolled in the CombiVacS trial. All subjects received a first dose of ChAdOx1s from AstraZeneca (Dose 1 (AZ)). The patients were then divided into two groups and received either a second placebo dose (Dose 1 (AZ) + Dose 2 (placebo)) or a second dose of BNT162b2 from Pfizer (Dose 1 (AZ) + Dose 2 (Pfizer)). The dqTACT assay was completed as shown in Fig. 1A (bottom). Comparisons show significance for the Wilcoxon rank sum two-sided test. For (B), naive, N = 13; dose 1 (AZ), N = 142, dose 1 (AZ) + dose 2 (placebo), N = 49; dose 1 (AZ) + dose 2 (Pfizer), N = 92 biologically independent samples. For (C), naive, N = 43; dose 1 (AZ), N = 155, dose 1 (AZ) + dose 2 (placebo), N = 52; dose 1 (AZ) + dose 2 (Pfizer), N = 99 biologically independent samples.
Supplementary Material
Acknowledgments
Research reported in this publication was supported in part by an ISMMS seed fund to E.G. and a Dean’s office grant to E.G. and I.M. We gratefully acknowledge use of the services and facilities of the Tisch Cancer Institute supported by the National Cancer Institute (NCI) Cancer Center Support grant (no. P30 CA196521), in particular the Hess sequencing core and the BiNGS shared facility. M.S. was supported by an NCI training grant (no. T32CA078207). J.S.Y.H. is supported by the Charles H. Revson Foundation. We acknowledge the technical contribution of D.A. Sánchez, J. Baranda, S. Baztan-Morales, M. Castillo de la Osa, A. Comins-Boo, C. del Álamo Mayo, S. Gil-Manso, B. Gonzalez, S. Hatem, J. Irure-Ventura, I. Miguens, S. Muñoz Martinez, M. Pereira, C. Rodrigues-Guerreiro, M. Rodriguez-Garcia, M.P. Rojo-Portolés and D. San Segundo. We also acknowledge Beckman Coulter for donating the equipment required for the determination of spike-specific IgG antibodies. W.M. was supported by grant no. NCI K00CA212474. This work was supported by ISMMS seed fund to J.O.; Instituto de Salud Carlos III, grant no. COV20–00668 to R.C.R.; Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation (COVID-19 Research Call grant no. COV20/00181) cofinanced by European Development Regional Fund ‘A way to achieve Europe’ to E.P.-A.; Instituto de Salud Carlos III, Spain (grant no. COV20/00170); Government of Cantabria, Spain (grant no. 2020UIC22-PUB-0019) to M.L.H.; Instituto de Salud Carlos III (grant no. PI16CIII/00012) to P.P.; Fondo Social Europeo e Iniciativa de Empleo Juvenil YEI (grant no. PEJ2018–004557-A) to M.P.E. and grant no. REDInREN 016/009/009 ISCIII. This project has received funding from the European Union’s Horizon 2020 research and innovation program VACCELERATE under grant agreement no. 101037867 to J.O. S.G. is supported by grant nos. U24CA224319, U01DK124165 and P30 CA196521. We acknowledge F. Buongiorno and S. Romano for technical help and B. Corneo, T. Blenkinsop, C. Schaniel, R. Kumar and M. Cerrone for help in coordinating recruitment.
Footnotes
Competing interests
A.B., A.T. and N.L.B. declare the filling of a patent application relating to the use of peptide pools in whole blood for detection of SARS-CoV-2 T cells (pending). E.G., J.O., M.S., D.T. and D.L.O. declare the filling of a patent application relating to the qTACT and dqTACT assays (pending). S.G. reports consultancy and/or advisory roles for Merck and OncoMed, and research funding from Bristol-Myers Squibb, Celgene, Genentech, Immune Design, Janssen R&D, Pfizer, Regeneron and Takeda. G.N., I.D.N. and L.C. are employees of Hyris Ltd, manufacturer of the bCUBE machine described in this article. C.L., G.S. and M.F. are employees of Synlab Italy. The other authors declare no competing interests.
Additional information
Extended data are available for this paper at https://doi.org/10.1038/s41587-022-01347-6.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-022-01347-6.
Peer review information Nature Biotechnology thanks Yu Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All data generated during this study are included in this published article (and its supplementary information files). RNA-seq source data are available as gene counts on the Gene Expression Omnibus at accession GSE178757.
References
- 1.Bhatraju PK et al. COVID-19 in critically ill patients in the Seattle Region—case series. N. Engl. J. Med 382, 2012–2022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med 180, 934–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagan N et al. BNT162b2 mRNA COVID-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med 384, 1412–1423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahan K et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wajnberg A et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370, 1227–1230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoletti A, Tan AT & Le Bert N The T-cell response to SARS-CoV-2: kinetic and quantitative aspects and the case for their protective role. Oxford Open Immunol 2, iqab006 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sette A & Crotty S Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184, 861–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swadling L et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 601, 110–117 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dan JM et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cucchiari D et al. Cellular and humoral response after MRNA-1273 SARS-CoV-2 vaccine in kidney transplant recipients. Am. J. Transplant 21, 2727–2739 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan AT et al. Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. J. Clin. Invest 131, e152379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilpeläinen AE A. Highly functional cellular immunity in SARS-CoV-2 non-seroconvertors is associated with immune protection. Preprint at bioRxiv (2021).
- 13.Sekine T et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183, 158–168 e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bert N et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med 218, e20202617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chia WN et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe 2, e240–e249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borobia A et al. Reactogenicity and immunogenicity of BNT162b2 in subjects having received a first dose of ChAdOx1S: initial results of a randomised, adaptive, phase 2 trial (CombiVacS). Lancet 398, 121–130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bert N et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Hillus D et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost ÿimmunisation with ChAdOx1-nCoV19 and BNT162b2: a prospective cohort study. Lancet Respir. Med 9, 1255–1265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozano-Ojalvo D et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep 36, 109570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrone L et al. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin Microbiol. Infect 27, 286 e287–286 e213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murugesan K et al. Interferon-gamma release assay for accurate detection of severe acute respiratory syndrome coronavirus 2 T-cell response. Clin. Infect. Dis 73, e3130–e3132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu FC et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395, 1845–1854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folegatti PM et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu FC et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 396, 479–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroemer M et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J. Infect 82, 282–327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramasamy MN et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet 396, 1979–1993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prendecki M et al. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. Lancet 397, 1178–1181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan AT et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 34, 108728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford T et al. Cryopreservation-related loss of antigen-specific IFNgamma producing CD4(+) T-cells can skew immunogenicity data in vaccine trials: lessons from a malaria vaccine trial substudy. Vaccine 35, 1898–1906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalimuddin S et al. Early T cell and binding antibody responses are associated with Covid-19 RNA vaccine efficacy onset. Med 2, 682–688 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichikawa A et al. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am. J. Respir. Crit. Care Med 187, 65–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luster AD, Unkeless JC & Ravetch JV Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315, 672–676 (1985). [DOI] [PubMed] [Google Scholar]
- 33.Koh CM et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Hsu AY, Peng Z, Luo H & Loison F Isolation of human neutrophils from whole blood and buffy coats. J. Vis. Exp 175 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Zhu W, Zeng NF & Wang N Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS Implementations. In NESUG Proceedings: Health Care and Life Sciences 19, 67 (2010). [Google Scholar]
- 36.Gao Y et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med 28, 472–476 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarke A et al. Alessandro Sette SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185, 847–859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moderbacher CR et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalai S Clinical validation of a novel T-cell receptor sequencing assay for identification of recent or prior SARS-CoV-2 infection. Preprint at medRxiv (2021). [DOI] [PMC free article] [PubMed]
- 40.Peng Y An immunodominant NP105–113-B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID-19 disease. Nat. Immunol 23, 50–61 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malard F et al. Weak immunogenicity of SARS-CoV-2 vaccine in patients with hematologic malignancies. Blood Cancer J 11, 142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aleman A et al. Variable cellular responses to SARS-CoV-2 in fully vaccinated patients with multiple myeloma. Cancer Cell 39, 1442–1444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howe KL et al. Ensembl 2021. Nucleic Acids Res 49, D884–D891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI et al. Tximeta: reference sequence checksums for provenance identification in RNA-seq. PLoS Comput. Biol 16, e1007664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raudvere U et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robin X et al. pROC: an open-source package for R and S+. to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article (and its supplementary information files). RNA-seq source data are available as gene counts on the Gene Expression Omnibus at accession GSE178757.


