Abstract
Routine laboratory surveillance has identified an unprecedented and ongoing exceedance of Cryptosporidium spp. across the United Kingdom, notably driven by C. hominis transmission, since 14 August 2023. Information from 477 reported cases in England and Wales, followed up with a standardised exposure questionnaire as of 25 September 2023, identified foreign travel in 250 (54%) of 463 respondents and swimming in 234 (66%) of 353 cases. A significant, common exposure has not yet been identified in first analyses.
Keywords: Cryptosporidium, cryptosporidiosis, United Kingdom, swimming pools, travel
The majority of disease caused by infection with Cryptosporidium in the United Kingdom (UK) is due to two species, Cryptosporidium hominis and Cryptosporidium parvum. National and international outbreaks can occur, but most outbreaks are localised and associated with private or public water supplies, swimming pools, animal contact, person-to-person spread or food consumption [1]. Here we describe the initial investigation into an unprecedented and ongoing nation-wide increase in cases of cryptosporidiosis, first noted in August 2023, and present findings from early descriptive analyses.
Exceedance detection and enhanced surveillance
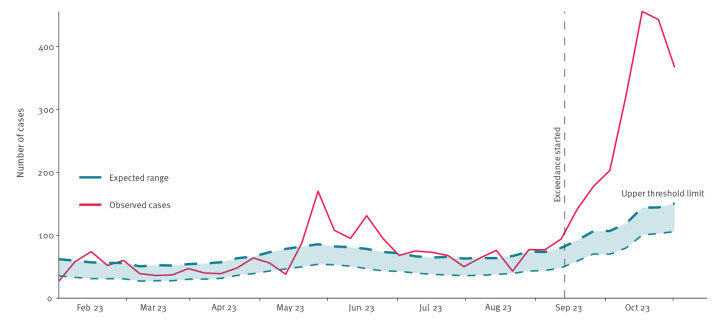
Across the UK, routine surveillance of cryptosporidiosis relies on mandatory laboratory notification. Since the International Organization for Standardization (ISO) week 33 2023 (starting on 14 August), and as at week 39 2023 (ending on 1 October), the combined weekly number of laboratory notifications of Cryptosporidium spp. detections in England, Wales and Northern Ireland has exceeded the expected upper threshold (Figure); an exceedance in Scotland has only been present since ISO week 39.
Figure.
Laboratory notifications of Cryptosporidium species in England, Wales and Northern Ireland, by week of specimen, 2023
Observed case numbers are compared to an exceedance threshold as derived from the Farrington Flexible algorithm, using published methodology [10]; the model can account for seasonal effects and changing trends. The upper threshold limit reflects the 99.5% percentile of a negative binomial distribution for the expected case frequency.
Given the scale and geographical spread of the exceedance across regions and nations of the UK, a single local exposure is an unlikely cause.
In England, local practice varies in the routine collection of exposure information in cryptosporidiosis cases. To allow for consistent capture of information as part of this investigation, a standardised electronic questionnaire was deployed. This includes questions on foreign travel, food and water exposures and interaction with animals. Data collection through this method has been ongoing since ISO week 38 (and in retrospect), for cases meeting the following definition: any person resident in England, with a clinical specimen dated 14 August 2023 or later, positive for Cryptosporidium (any species). In Wales, a standard questionnaire for gastrointestinal disease has been in place since before the exceedance.
Reference laboratory genotyping
Across England and Wales, standard practice is that Cryptosporidium spp. positive stool samples from local laboratories are referred to the Cryptosporidium Reference Unit (CRU, Swansea, UK) for genotyping. In 2022 [2], ca 70% of specimens were referred (2,913 in total, of which 2,698 were successfully genotyped); 63% (n = 1,694) were C. parvum and 34% (n = 919) C. hominis. Among genotyped specimens, 16% (n = 440) reported international travel, 65% (n = 284) of which were C. hominis.
Subtyping beyond species level is done to investigate clusters or outbreaks by sequencing the gp60 gene [3] using real-time PCR amplicons; this is underway as part of this investigation for cases who have completed a questionnaire.
Descriptive analysis of laboratory-reported cases
Between week 33 and 39, there have been 2,411 laboratory confirmed cases of cryptosporidiosis in the UK (2,032 in England, 163 in Wales, 127 in Scotland and 89 in Northern Ireland). Cases have been seen across almost all regions of the four UK nations.
Reference laboratory data suggest the exceedance is particularly due to C. hominis transmission (Table 1). Investigation by gp60 sequencing of an initial 76 specimens from C. hominis cases shows that five are family Ia, 21 are family Ib, 16 are family Id, six are Ie and 28 are If. Of the most prevalent gp60 families, only a single If subtype was found, IfA12G1R5, whereas there were three Ib subtypes, IbA9G3, IbA10G2 and IbA13G3, and three Id subtypes, IdA14, IdA16 and IdA17G1. The high prevalence of IfA12G1R5 is unusual in the UK [1,4].
Table 1. Notified cases of cryptosporidiosis, United Kingdom, ISO weeks 33–39 2023a (n = 2,411).
| Characteristics | England | Wales | Scotland | Northern Ireland |
|---|---|---|---|---|
| Local laboratory | ||||
| All Cryptosporidium species | 2,032 | 163 | 127 | 89 |
| Exceedance threshold first reached | Week 33 | Week 35 | Week 39 | NA |
| Reference laboratoryb | ||||
| C. parvum | 431 | 10 | 0 | |
| C. hominis | 929 | 25 | 0 | |
| Other Cryptosporidium species | 19 | 0 | 0 | |
| Not typable | 32 | 0 | 0 | |
| Not yet processed | 784 | 92 | 89 | |
ISO: International Organization for Standardization; NA: not available.
a At time of publication and subject to change.
b In England and Wales, local laboratories are requested to refer all Cryptosporidium positive stool samples for genotyping to the same reference laboratory. Scotland has a separate reference laboratory, and Northern Ireland specimens are not routinely genotyped. There is a lag between initial laboratory notification and genotyping data. Where specimen date is not available, date received by the reference laboratory is used.
Descriptive analysis of questionnaire data
In England, as of 2 October, 406 standardised exposure questionnaires have been completed for individuals reporting a symptom onset date of 1 August (ISO week 31) or later, with species data available for 235 (58%). Comparable questionnaire data from Wales are available for an additional 71 cases reporting symptom onset from 14 August (ISO Week 33).
Foreign travel was reported by 250 (54%) of 463 Cryptosporidium spp. cases and by 139 (65%) of 213 C. hominis cases (Table 2). Of the 394 cryptosporidiosis cases resident in England who provided information on travel, 215 (55%) reported foreign travel in the 14 days preceding their illness, of which 96 (45%) noted travel to Spain (Spanish mainland and/or the Balearic Islands).
Table 2. Characteristics and exposures of cryptosporidiosis cases in weeks 31–39 in England and weeks 33-39 in Wales, United Kingdom, 2022 (n = 209) and 2023 (n = 477).
| Characteristics | Cryptosporidium spp. |
C. hominis
W31–39 2023b n = 215 |
C. parvum
W31–39 2023b n = 85 |
|||||
|---|---|---|---|---|---|---|---|---|
| W31–39 2022a
n = 209 |
W31–39 2023b
n = 477 |
|||||||
| n | % | n | % | n | % | n | % | |
| Age (years) | n = 208 | n = 475 | n = 215 | n = 85 | ||||
| 0–4 | 35 | 17 | 68 | 14 | 34 | 16 | 11 | 13 |
| 5–9 | 46 | 22 | 91 | 19 | 44 | 20 | 13 | 15 |
| 10–19 | 30 | 14 | 77 | 16 | 36 | 17 | 16 | 19 |
| 20–39 | 61 | 29 | 141 | 30 | 60 | 28 | 26 | 31 |
| 40–59 | 26 | 13 | 67 | 14 | 36 | 17 | 7 | 8 |
| 60–79 | 8 | 4 | 30 | 6 | 5 | 2 | 12 | 14 |
| > 80 | 2 | 1 | 1 | < 1 | 0 | 0 | 0 | 0 |
| Sex | n = 209 | n = 476 | n = 215 | n = 85 | ||||
| Male | 88 | 42 | 218 | 46 | 101 | 47 | 36 | 42 |
| Female | 121 | 58 | 258 | 54 | 114 | 53 | 49 | 58 |
| Symptom duration (days) | n = 95 | n = 224 | n = 91 | n = 46 | ||||
| 0–4 | 6 | 6 | 26 | 12 | 11 | 12 | 6 | |
| 5–9 | 33 | 35 | 73 | 33 | 30 | 33 | 14 | |
| 10–14 | 42 | 44 | 85 | 38 | 31 | 34 | 21 | |
| 15–19 | 12 | 13 | 21 | 9 | 11 | 12 | 3 | |
| ≥ 20 | 2 | 2 | 19 | 8 | 8 | 9 | 2 | |
| Foreign travelc | n = 208 | n = 463 | n = 213 | n = 85 | ||||
| Yes | 118 | 57 | 250 | 54 | 139 | 65 | 24 | 28 |
| No | 90 | 43 | 213 | 46 | 74 | 35 | 61 | 72 |
| Swimmingc | n = 111 | n = 353 | n = 155 | n = 70 | ||||
| Yes | 61 | 55 | 234 | 66 | 124 | 80 | 26 | 37 |
| No | 50 | 45 | 119 | 34 | 31 | 20 | 44 | 63 |
| Farm animal exposurec | n = 110 | n = 384 | n = 164 | n = 73 | ||||
| Yes | 20 | 18 | 71 | 18 | 34 | 21 | 14 | 19 |
| No | 90 | 82 | 313 | 82 | 130 | 79 | 59 | 81 |
| Food exposuresc | ||||||||
| Salad at home | n = 164 | n = 49 | n = 28 | |||||
| Yes | NA | 77 | 47 | 17 | 19 | |||
| No | 87 | 53 | 32 | 9 | ||||
| Pasteurised cow’s milk | n = 182 | n = 57 | n = 27 | |||||
| Yes | NA | 120 | 66 | 37 | 21 | |||
| No | 62 | 34 | 20 | 6 | ||||
| Tap water | n = 219 | n = 71 | n = 37 | |||||
| Yes | NA | 163 | 74 | 53 | 75 | 30 | ||
| No | 56 | 26 | 18 | 25 | 7 | |||
| Bottled water | n = 231 | n = 77 | n = 41 | |||||
| Yes | NA | 137 | 59 | 55 | 71 | 17 | ||
| No | 94 | 41 | 22 | 29 | 24 | |||
| Ate at a restaurant | n = 346 | n = 136 | n = 58 | |||||
| Yes | NA | 210 | 61 | 89 | 65 | 36 | ||
| No | 136 | 39 | 47 | 35 | 22 | |||
C.: Cryptosporidium; NA: not available; spp.: multiple species; W: ISO week.
a Cases from four regions in England and Wales where questionnaire data were available and symptom onset was between ISO week 31 and 39 2022. Data may be incomplete, especially for ISO weeks 31 and 32. Heterogeneity in questionnaires used in 2022 means that comparisons to 2023 data should be done with caution.
b Cases from all regions in England (between ISO week 31 and 39 2023) and Wales (between ISO week 33 and 39 2023), where questionnaire and genotyping data were available, based on symptom onset. Percentages not included where n < 60.
c In 14 days before onset of illness.
Comparisons to available questionnaire data from four regions in England and Wales from a similar period in 2022 (Table 2), which is done cautiously given the heterogeneity in surveys used in that year and the availability of data, suggest the 2023 exceedance may be associated with greater exposure to swimming in the 14 days preceding illness (odds ratio (OR) = 1.61; 95% confidence interval (CI): 1.04–2.48), despite minimal difference in the proportion reporting foreign travel.
Analysis of 2023 questionnaires for potential setting exposures (e.g. swimming pools or water parks) has not detected significant case clusters that could alone explain the large exceedance. However, to date, two local exposures mutual to a small number of cases have been identified and have been actively investigated by environmental health teams in local authorities.
Public health measures
Routine public health measures to prevent transmission of common gastrointestinal infections are in place. As yet, survey responses have not identified common specific exposures or settings that may explain large numbers of cases. Public health action within the UK has included stakeholder communications to raise awareness and highlight the importance of avoiding use of swimming pools while symptomatic, and in the 14 days after resolution of symptoms. We are working with the National Travel Health Network and Centre (NaTHNaC) to ensure appropriate travel advice is available. We are liaising with international partners, including European Centre for Disease Prevention and Control (ECDC), to inform further investigations.
Identification and investigation of any local common exposures will be ongoing, within the remit of local health protection teams. Data collection, and analysis at a national level, will continue until the exceedance resolves.
Discussion
We have described an ongoing and unusual national exceedance in disease caused by Cryptosporidium spp., most notably driven by C. hominis incidence, for which no single exposure has been identified. While C. hominis infections (dominantly transmitted person-to-person via the faecal-oral route) are expected to increase at this time of year, the magnitude of the increase warrants further investigation.
Responses to surveillance questionnaires suggest many cases may relate to international travel, most notably to Spain and other Mediterranean countries. The exceedance may in part reflect increasing summer travel to the European continent in 2023, although travel trend data are not yet available for this year (historical data suggests Spain accounted for 40–50% of UK foreign travel in the years before the COVID-19 pandemic, and 33% of travel in 2022 [5]). Investigations in previous similar exceedances found international travel to be a common exposure [6].
Our initial findings would suggest that swimming (either in the UK or abroad), including the use of pools, and foreign travel to a variety of destinations may underlie the current increase. However, at this stage other sources, for example contaminated food, cannot be excluded as contributing to the exceedance. Ongoing gp60 subtyping may yield large unexplained clusters warranting further investigation. Furthermore, continuation of a sentinel scheme to detect genetic clusters of C. parvum by multilocus variable number tandem repeat analysis (MLVA) [7] initiated as a pilot in Wales and the north-west of England [8] has yet to identify any notable clusters during this exceedance.
This exceedance has reinforced the considerable disease burden that can result from cryptosporidiosis (56% of the 224 cases responding to the questionnaire and reporting an illness duration of more than 10 days), as well as the typical age-sex distribution of Cryptosporidium spp. infections. Previous studies have shown that Cryptosporidium spp. are highly transmissible within households, particularly those with children, and that a notable number of cases are under-reported [9]. The importance and utility of standard surveillance approaches within countries has also been demonstrated; rapid roll out of a single questionnaire has allowed for hypothesis generation and analysis at a national level.
Ethical statement
Ethical approval was not sought for this work as data were collected as part of routine or enhanced surveillance activities. The analysis was completed to inform public health practice.
Funding statement
RE, RV and NL receive support from the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections; RV receives support from the NIHR HPRU in Emerging and Zoonotic Infections; and RE receives support from the NIHR HPRU in Emergency Preparedness and Response. The views expressed are those of the author(s) and not necessarily those of the NIHR, UK Health Security Agency, or the Department of Health and Social Care. Funding was not applicable to this study.
Data availability
Data are available on reasonable request to the authors. Restrictions apply to the availability of data linked to patient information.
Acknowledgements
Thanks is given to all regional Field Service, Health Protection, Travel Health and International Health Regulations (IHR) teams within UKHSA, Public Health Wales, Public Health Scotland and the HSC Public Health Agency (Northern Ireland); Environmental Health colleagues within local government; the national Cryptosporidium Reference Unit team; and all who have supported this surveillance activity.
Conflict of interest: None declared.
Authors’ contributions: Data generation, processing and/or analysis by LP, TI, CJ, GK, HR, JH, AW, RJ, CR, CW, OO, LB, MN, PM, KE, GR and RC. Drafting of the paper led by LP and supported by TI, CJ, NY, RE and RV. Management of the exceedance was led by RV, RE and TM with 4-UK national contributions from CW, LB, MN and PM.
References
- 1. Chalmers RM, Robinson G, Elwin K, Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors. 2019;12(1):95. 10.1186/s13071-019-3354-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Public Health Wales Microbiology. Annual report of referrals and Cryptosporidium genotyping; England and Wales, 2022. Swansea: Public Health Wales; 2023. Available from: https://phw.nhs.wales/services-and-teams/cryptosporidium-reference-unit/annual-report-of-referrals-and-cryptosporidium-genotyping-england-and-wales-2022
- 3. Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68(7):4117-34. 10.1128/IAI.68.7.4117-4134.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bacchetti R, Connelly L, Browning L, Alexander CL. Changing molecular profiles of human cryptosporidiosis cases in Scotland as a result of the coronavirus disease, COVID-19 pandemic. Br J Biomed Sci. 2023;80:11462. 10.3389/bjbs.2023.11462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Office for National Statistics (ONS). Travel trends: 2022. Newport: ONS; 26 May 2023. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/leisureandtourism/articles/traveltrends/2022
- 6. Fournet N, Deege MP, Urbanus AT, Nichols G, Rosner BM, Chalmers RM, et al. Simultaneous increase of Cryptosporidium infections in the Netherlands, the United Kingdom and Germany in late summer season, 2012. Euro Surveill. 2013;18(2):20348. 10.2807/ese.18.02.20348-en [DOI] [PubMed] [Google Scholar]
- 7. Robinson G, Pérez-Cordón G, Hamilton C, Katzer F, Connelly L, Alexander CL, et al. Validation of a multilocus genotyping scheme for subtyping Cryptosporidium parvum for epidemiological purposes. Food Waterborne Parasitol. 2022;27:e00151. 10.1016/j.fawpar.2022.e00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Risby, Robinson G, Chandra N, King G, Vivancos R, Smith R, et al. Application of a new multilocus variable number tandem repeat analysis (MLVA) scheme for the seasonal investigation of Cryptosporidium parvum cases in Wales and the north west of England, spring 2022. Curr Res Parasitol Vector Borne Dis. 2023;100151. 10.1016/j.crpvbd.2023.100151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKerr C, Chalmers RM, Elwin K, Ayres H, Vivancos R, O’Brien SJ, et al. Cross-sectional household transmission study of Cryptosporidium shows that C. hominis infections are a key risk factor for spread. BMC Infect Dis. 2022;22(1):114. 10.1186/s12879-022-07086-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noufaily A, Enki DG, Farrington P, Garthwaite P, Andrews N, Charlett A. An improved algorithm for outbreak detection in multiple surveillance systems. Stat Med. 2013;32(7):1206-22. 10.1002/sim.5595 [DOI] [PubMed] [Google Scholar]