Abstract
Mixed-linked glucanases (MLGases), which are extracellular enzymes able to hydrolyze β1,3-1,4-glucans (also known as mixed-linked glucans or cereal β-glucans), were identified in culture filtrates of the plant-pathogenic fungus Cochliobolus carbonum. Three peaks of MLGase activity, designated Mlg1a, Mlg1b, and Mlg2, were resolved by cation-exchange and hydrophobic-interaction high-performance liquid chromatography (HPLC). Mlg1a and Mlg1b also hydrolyze β1,3-glucan (laminarin), whereas Mlg2 does not degrade β1,3-glucan but does degrade β1,4-glucan to a slight extent. Mlg1a, Mlg1b, and Mlg2 have monomer molecular masses of 33.5, 31, and 29.5 kDa, respectively. The N-terminal amino acid sequences of Mlg1a and Mlg1b are identical (AAYNLI). Mlg1a is glycosylated, whereas Mlg1b is not. The gene encoding Mlg1b, MLG1, was isolated by using PCR primers based on amino acid sequences of Mlg1b. The product of MLG1 has no close similarity to any known protein but does contain a motif (EIDI) that occurs at the active site of MLGases from several prokaryotes. An internal fragment of MLG1 was used to create mlg1 mutants by transformation-mediated gene disruption. The total MLGase and β1,3-glucanase activities in culture filtrates of the mutants were reduced by approximately 50 and 40%, respectively. When analyzed by cation-exchange HPLC, the mutants were missing the two peaks of MLGase activity corresponding to Mlg1a and Mlg1b. Together, the data indicate that Mlg1a and Mlg1b are products of the same gene, MLG1. The growth of mlg1 mutants in culture medium supplemented with macerated maize cell walls or maize bran and the disease symptoms on maize were identical to the growth and disease symptoms of the wild type.
The cell walls of monocotyledonous plants are composed of a variety of macromolecules, including cellulose, arabinoxylan, xyloglucan, pectin, and proteins. One of the major hemicelluloses of the walls of plants in the Poaceae is mixed-linked glucan (also called β1,3-1,4-glucan or β-glucan), in which unbranched chains of β1,4-glucose are disrupted by periodic β1,3 linkages at a ratio of about 2:1. Several lines of evidence suggest that this polysaccharide is important for the control of plant cell expansion (4), and therefore it might have a critical role in maintaining the structural integrity of the wall during pathogen attack. Furthermore, the enzymes that degrade cereal mixed-linked glucans have potential medical and industrial significance because such glucans are an important component of soluble fiber in human diets and a major factor affecting the quality of fermented beverages (41).
Enzymes that can degrade mixed-linked glucan are called mixed-linked glucanases (MLGases), β1,3-1,4-glucanases, β-glucanases, or lichenases. Some MLGases can also degrade other glucans (for example, β1,3-glucans and β1,4-glucans) (16, 28, 31, 35). Genes encoding MLGases have been cloned from a number of bacteria (31, 35, 38) and higher plants (34, 42). An enzyme with MLGase activity has been purified from the fungus Rhizopus arrhizus (6), but to the best of our knowledge no genes encoding MLGases have previously been isolated from fungi.
Cochliobolus carbonum, an ascomycetous pathogen of maize, penetrates into and ramifies through intact leaves and in the process obtains nutrients for growth from the plant cell cytoplasm and walls. For penetration, ramification, and nutrient assimilation, both as a pathogen and during the saprophytic phase of its life cycle, C. carbonum produces a variety of extracellular enzymes that can degrade the polymers of the plant cell wall, including pectinases, xylanases, β1,3-glucanases, cellulases, β-xylosidase, α-arabinosidase, and proteases (40).
A common feature of microbial extracellular degradative enzymes is redundancy; that is, most microorganisms make two or more chromatographically separable proteins that have the same or similar enzymatic activities. C. carbonum is no exception to this rule, because it secretes, for example, at least four endo-β1,4-xylanases (2) and three proteases (21). Enzymatic redundancy can be due to multiple genes encoding proteins with similar or overlapping enzymatic activities, to alternative RNA processing (3), and/or to different posttranslational modifications. Apparent redundancy can also be caused by artifactual processes, such as partial proteolysis during fermentation or purification. Although difficult to establish by purely biochemical methods, the biogenic relationships between redundant enzymes can be substantially clarified by analyzing microbial strains specifically mutated in one or more of the encoding genes (1, 2, 21, 32).
In this study, we identified and characterized three extracellular enzymes (Mlg1a, Mlg1b, and Mlg2) that degrade β-glucan from the filamentous fungus C. carbonum and demonstrated by performing cloning and targeted gene disruption experiments that one gene encodes two of the three MLGases.
MATERIALS AND METHODS
Fungal culture and maintenance.
C. carbonum race 1 strain 367-2A, which is a progeny of strain SB111 (= ATCC 90305), was grown on V8 juice agar plates. For MLGase production, two fungal plugs (5 mm2) were inoculated into a 1,000-ml Erlenmeyer flask containing 125 ml of mineral salts, 0.2% yeast extract, and trace elements (39) and grown in still culture for 9 days at 21 to 23°C. The supplemental carbon sources tested were Country Life maize bran (Country Life Natural Foods, Pullman, Mich.), Mother’s Oat Bran cereal, and Quaker Oat Bran cereal (the latter two were obtained from The Quaker Oats Company, Chicago, Ill.). For routine enzyme production, cultures were grown on 1% maize bran plus 0.2% sucrose.
Enzyme assays.
Routine glucanase assays were performed by using a reducing sugar assay (18) with barley β-glucan (catalog no. G6513; Sigma) as the substrate. Laminarin (catalog no. L9634; Sigma) was used to test for β1,3-glucanase activity, and Avicel PH-101 (catalog no. 11365; Fluka), high-viscosity carboxymethyl cellulose (catalog no. C5013; Sigma), low-viscosity carboxymethyl cellulose (catalog no. C5678; Sigma), α-cellulose (catalog no. C8002; Sigma), and microgranular Whatman cellulose were used to examine β1,4-glucanase activities. Most assays were performed by using substrate at a concentration of 0.2%; laminarin was used at a concentration of 0.1%. The substrates were dissolved or suspended in 50 mM sodium acetate buffer (pH 5.0), and most assays were performed at 37°C for 30 min with 10 to 20 μl of enzyme. When cellulosic substrates were used, the assay was performed for 17 h at 37°C. After the reaction mixtures were heated at 100°C for 10 min, 200 μl of each reaction mixture was placed in a 96-well microtiter plate and cooled to 22°C, and the A410 was determined with an enzyme-linked immunosorbent assay plate reader (Bio-Tek). One unit of activity was defined as 1 nmol of glucose released per μl of enzyme per min at 37°C.
Viscometric assays were performed with a no. 200 tube viscometer and 0.5% barley β-glucan in 50 mM sodium acetate (pH 5.0) at 37°C. Viscometry readings were taken every 3 min for 20 min.
Protein purification.
Concentration and purification of MLGase activities from culture filtrates by using low-pressure DEAE-cellulose chromatography and dialysis were performed by the method of Murphy and Walton (21), except that the 25 mM sodium acetate buffer was adjusted to pH 4.0. Fractionation on a polysulfylethyl aspartamide cation-exchange high-performance liquid chromatography (HPLC) column (The Nest Group, Southboro, Mass.) was accomplished by using a 30-min linear gradient from buffer A (25 mM sodium acetate [pH 4.0]) to buffer B (25 mM sodium acetate [pH 4.0] plus 0.4 M KCl) at a flow rate of 1 ml/min. The peak of UV absorption (at 280 nm) containing Mlg1a and Mlg1b was collected, adjusted to 1.7 M ammonium sulfate, and applied to a hydrophobic-interaction HPLC (HI-HPLC) column (Biogel TSK-Phenyl-5PW; Bio-Rad, Richmond, Calif.) (21). Fractions containing Mlg1a and Mlg1b activities were then individually passed over a gel filtration HPLC column (7.5 by 300 mm; Ultraspherogel SEC3000; Beckman). Purified Mlg1a and Mlg1b were lyophilized and were sequenced directly from the N terminus, as well as after digestion with trypsin and separation of peptides by microbore HPLC, by automated Edman degradation. Mlg2 was purified by using the same methods that were used for Mlg1a and Mlg1b through the HI-HPLC step. The fractions containing Mlg2 activity were then fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% acrylamide), transferred to a ProBlot membrane (Applied Biosystems, Foster City, Calif.) (19), stained with 0.1% Coomassie brilliant blue R-250 in 40% methanol, and destained with 50% methanol. Mlg2 was excised from the blot and digested with trypsin. The resulting peptides were separated by HPLC and sequenced by automated Edman degradation.
The methods used for SDS-PAGE and glycoprotein detection by periodic acid-Schiff staining have been described previously (14, 37). The pH optima for the three enzymes were determined as described previously (21).
Nucleic acid manipulations.
DNA and RNA were isolated as described by Pitkin et al. (25) and Chomczynski and Sacchi (5), respectively. The methods used for genomic and cDNA library screening, probe labeling, DNA blotting, and hybridization have been described previously (21, 32). Sequencing with gene-specific primers was performed by automated fluorescent sequencing at the Michigan State University Department of Energy Plant Research Laboratory Plant Biochemistry Facility by using an Applied Biosystems model 373A sequencer for analysis of the products. The transcription start site of MLG1 was determined by using an Amplifinder RACE kit (Clonetech, Palo Alto, Calif.) (10). First-strand cDNA synthesis was primed with the reverse complement oligonucleotide GAAGGCGGGCCAAGAGCC (starting at nucleotide 727). PCR primer CGTGCGTGGGATCAGCGATATCTTC (reverse complement) (starting at nucleotide 439) and the anchor primer provided with the RACE kit were used to amplify the 5′ end of the MLG1 transcript.
Cloning of MLG1.
The PCR conditions used to amplify MLG1 and the protocol used to clone the PCR fragments have been described previously (21). Template DNA that generated the 340-bp product was isolated from phage lysate of a cDNA library prepared from mRNA from C. carbonum grown on maize cell walls (25). Total genomic DNA was used as a template for a reaction that yielded a 460-bp product. PCR primers ATHGAYACNTAYGAYGC and ATRTCDATYTCNCCYTG (H = A, C, or T; Y = C or T; N = any nucleotide; R = A or G; D = A, G, or T), corresponding to the sequences IDTYDA and QGEIDI, respectively, were used at an annealing temperature of 55°C for PCR amplification of a fragment of MLG1. Oligonucleotide sequence AARTTYAAYTTYGARGA, corresponding to amino acid sequence KFNFED, was end labeled (29) and hybridized at 45°C to the PCR products to confirm that a fragment of MLG1 had been amplified. The MLG1 PCR products were cloned into pBluescript II SK+ at the SmaI restriction site and sequenced. A 7.0-kb BamHI MLG1 genomic fragment from a λEMBL3 phage that hybridized to the MLG1 cDNA was subcloned into pBluescript II SK+.
Targeted gene disruption of MLG1.
The transformation vector was made by digesting an MLG1 cDNA clone, pC4-2.1, with XhoI and SalI to liberate a 345-bp fragment internal to the MLG1 locus, treating the fragment with T4 DNA polymerase, and ligating it into the SmaI restriction site of pHYG1 (36). The resulting vector, pJM5, was linearized at the unique SmaI restriction site and used to transform wild-type C. carbonum 367-2A.
Protoplast isolation and transformation have been described previously (1, 32). Transformants were selected for their ability to grow in the presence of 100 U of hygromycin B (Calbiochem, La Jolla, Calif.) per ml. Single spores were isolated twice to ensure nuclear homogeneity.
For pathogenicity tests on germinating young seedlings, 10 seeds of susceptible maize cultivar Pr (genotype hm/hm) and 10 seeds of resistant cultivar Pr1 (genotype Hm/Hm) were surface sterilized for 10 min in 10% (vol/vol) commercial sodium hypochlorite (household bleach), washed five times with water, soaked in water for 17 h, and planted at a depth of 2 cm in soil in 13-cm-diameter clay pots. The pots were watered with 100 ml of a preparation containing 105 fresh conidia per ml. Germination and growth were monitored daily for 10 days. Pathogenicity tests on 14-day-old maize seedlings were performed by inoculating leaves of susceptible hybrid Pr X K61 (genotype hm/hm) and resistant cultivar Great Lakes (genotype Hm/Hm) with a fine mist of a preparation containing 104 conidia per ml suspended in 0.1% Tween 20. Disease symptoms were observed daily until the plants were dead.
Nucleotide sequence accession number.
The nucleotide sequence of MLG1 has been deposited in the GenBank database under accession no. U81606.
RESULTS AND DISCUSSION
Characterization and purification of Mlg1a, Mlg1b, and Mlg2.
Several carbon sources were tested for optimal production of extracellular MLGase by C. carbonum. Maize bran was a better inducer than two commercial oat bran products (data not shown). Like endopolygalacturanase, exo-β1,3-glucanase, xylanase, β-xylosidase, α-arabinosidase, and protease production, adding 0.2% sucrose enhanced production of MLGase. Unlike xylanase, exo-β-1,3-glucanase, or endopolygalacturanase activities, however, some MLGase activity was still produced when C. carbonum was grown on 2% (wt/vol) sucrose as the sole carbon source (data not shown) (21, 27).
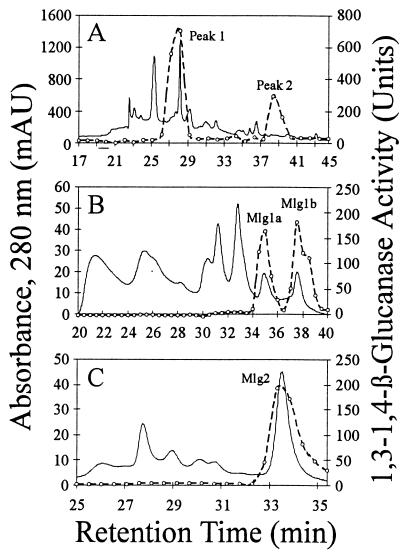
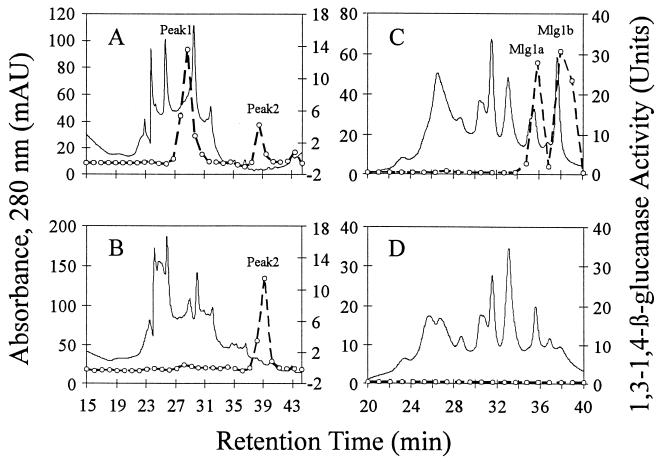
After concentration by rotary evaporation, dialysis, and passage through an anion-exchange column to remove acidic proteins and pigments, MLGase activities were fractionated by cation-exchange HPLC. One major peak of activity (peak 1) and one minor peak of activity (peak 2) were resolved (Fig. 1A). The two peaks were then separately applied to HI-HPLC columns for further purification (Fig. 1B and C). Cation-exchange HPLC peak 1 (Fig. 1A) was resolved into two peaks of activity, designated Mlg1a and Mlg1b (Fig. 1B). Peak 2 (Fig. 1A) remained a single peak of MLGase activity and was designated Mlg2 (Fig. 1C). Mlg1a and Mlg1b were subsequently chromatographed by using the gel filtration method to purify them to electrophoretic homogeneity.
FIG. 1.
Purification of Mlg1a, Mlg1b, and Mlg2. (A) Cation-exchange chromatography of culture filtrates. (B) HI-HPLC of peak 1 from panel A. (C) HI-HPLC of peak 2 from panel A. Solid lines, A280; dashed lines, MLGase activity.
The molecular masses of Mlg1a, Mlg1b, and Mlg2, as determined by SDS-PAGE, are 33.5, 31, and 29.5 kDa, respectively. Mlg1a and Mlg1b are endo-acting enzymes, as determined by their ability to rapidly reduce the viscosity of a β-glucan solution relative to the simultaneous appearance of reducing sugars (data not shown). The temperature and pH optima for all three enzymes are approximately 55°C and 5.0, respectively. The activity of Mlg1a and Mlg1b against β1,3-glucan is comparable to the activity against β-glucan. Neither Mlg1a nor Mlg1b exhibited activity in long-term assays (17 h) against any of the β1,4-glucan substrates tested. Based on their HI-HPLC retention times and their ability to degrade β1,3-glucan, Mlg1a and Mlg1b are probably responsible for the two peaks of β1,3-glucanase activity remaining in culture filtrates of exg1 (encoding exo-β1,3-glucanase) mutants of C. carbonum (30). Mlg2 has no detectable activity against β1,3-glucan, but in long (17-h) incubations shows some ability to degrade several β1,4-glucans (low-viscosity carboxymethyl cellulose, Whatman cellulose, and Avicel). Mlg2 is 180 to 340 times less active against these β1,4-glucans than it is against β-glucan. In a 17-h incubation, Mlg2 showed no ability to degrade two other β1,4-glucan substrates, high-viscosity carboxymethyl cellulose and α-cellulose. Therefore, on the basis of their substrate preferences, Mlg1a and Mlg1b can be considered bifunctional β1,3-1,4/β1,3-glucanases and Mlg2 can be considered a β1,3-1,4-glucanase.
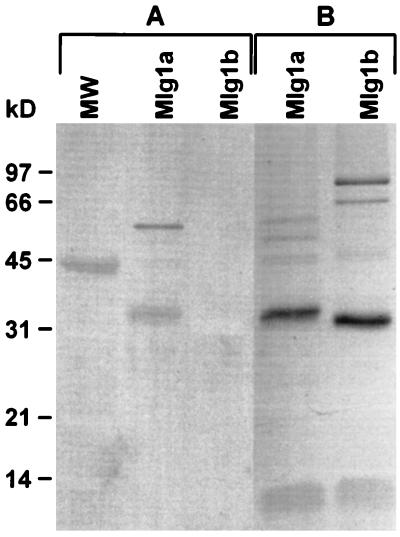
Mlg1a is glycosylated, whereas Mlg1b is not (Fig. 2). In this experiment, partially purified preparations were used so that the other proteins could serve as positive and negative glycosylation controls. Although glycosylation can alter the pH optima, temperature optima, or thermostabilities of enzymes (7, 20), Mlg1a and Mlg1b have similar temperature optima and thermostabilities (data not shown).
FIG. 2.
Glycosylation of Mlg1a and Mlg1b. (A) Protein blot from SDS-PAGE gel of partially purified Mlg1a and Mlg1b stained with periodic acid-Schiff reagent (37). (B) SDS-PAGE of the samples in panel A stained with Coomassie brilliant blue R-250. Mlg1a and Mlg1b are the bands at 33.5 and 31 kDa, respectively. The positions of the molecular size standards are shown on the left; ovalbumin (45 kDa) is a glycoprotein.
At least the first five amino acids of the mature Mlg1a and Mlg1b proteins are identical (Table 1). In an analysis of the N-terminal sequence of Mlg1b (22 amino acids) by BLASTP (12) we found no strong similarity to any sequence in the nonredundant databases. Two internal tryptic peptides of Mlg1b were sequenced, and one of these peptides (peptide 3) overlaps the N-terminal peptide (Table 1). Peptide 2 from Mlg1b has 77% identity to an MLGase from the prokaryote Rhodothermus marinus (PIR S48201).
TABLE 1.
Experimentally determined amino acid sequences of Mlgla, Mlglb, and Mlg2 and comparison to the sequences deduced from the nucleotide sequence of MLG1
| Protein | Peptide | Peptide sequence
|
|
|---|---|---|---|
| Exptla | Deduced from DNA sequenceb | ||
| Mlgla | 1 | AAYNLI | AAYNLI |
| Mlglb | 1 | AAYNLIDTYDA(A)N(W)AAKFNFED | AAYNLIDTYDAsNWAsKFNFED |
| 2 | GPNWP(A)QGE(I)D(I) | GPNWPnQGEIDI | |
| 3 | FNFEDIADPDT | FNFEDIADPDT | |
| Mlg2 | 1 | FTVNQ(C)SANAY | |
| 2 | YDVYPIGSSQGMVNVAGR | ||
| 3 | GFPINSQNLITYQFGTEAFTGGP | ||
Purified proteins were sequenced at the N terminus (Mlgla peptide 1 and Mlglb peptide 1) or by using tryptic peptides (all of the other peptides). Parentheses indicate uncertainty.
Lowercase letters in the deduced sequence indicate discrepancies between the experimental and deduced sequences.
For final purification of Mlg2, proteins in the HI-HPLC fractions containing MLGase activity were separated by SDS-PAGE and blotted, and Mlg2 was excised from the blot for sequencing. As the N terminus was blocked, internal amino acid sequences were obtained from three tryptic peptides (Table 1). BLASTP analysis indicated that peptide 2 of Mlg2 is 56% identical to a cellulase, EglS, from the prokaryote Streptomyces rochei (GenBank accession no. X73953) and that peptide 3 is 78% identical to the F1 carboxymethyl cellulase of Aspergillus aculeatus (PIR S12610). EglS (24) and F1 carboxymethyl cellulase (23) are members of cellulase family H (11) or the glycosyl hydrolase family 12 (15). To the best of our knowledge, neither EglS nor F1 carboxymethyl cellulase has been tested with mixed-linked glucan as the substrate. Although some cellulases also degrade mixed-linked glucans (16), Mlg2 is clearly distinct from the cellulases of C. carbonum, which do not degrade mixed-linked glucan (36). Cloning and sequencing of the gene for Mlg2 are in progress.
Isolation and characterization of MLG1.
Two 96-fold degenerate oligonucleotides based on the amino acid sequences IDTYDA and QGEIDI (Table 1) were used in a PCR to amplify a fragment of the encoding gene. This primer combination yielded a single, 340-bp PCR product when DNA from a C. carbonum cDNA library was used as a template and a single, 460-bp PCR product when C. carbonum genomic DNA was used as a template. To confirm that these PCR products encoded Mlg1b, they were blotted and probed with a third 36-fold degenerate oligonucleotide based on the amino acid sequence KFNFED (Table 1). Both products hybridized to the third oligonucleotide and were therefore cloned and sequenced. Sequencing indicated that the PCR products are identical except for the presence of two introns of 57 and 64 bp in the PCR product amplified from genomic DNA.
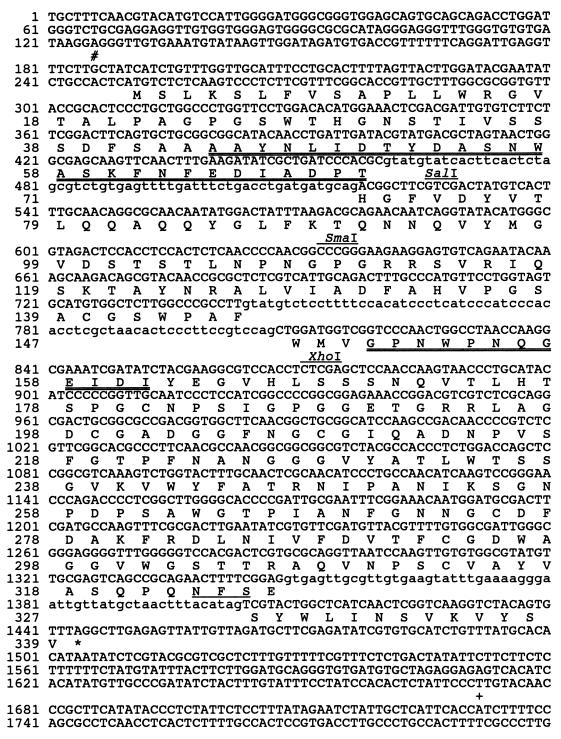
C. carbonum cDNA and genomic libraries were screened by using the cDNA-derived PCR fragment as a probe. A 1.34-kb MLG1 cDNA (C4-2.1) was isolated from the cDNA library. and sequenced. A 7.0-kb BamHI fragment of DNA (MLG1-2B) containing the MLG1 genomic locus was subcloned, and both strands were sequenced. Figure 3 shows the sequence of MLG1 and the deduced amino acid sequence. The transcription start site of MLG1 was determined by analyzing the sequences of three independent RACE products (10). The MLG1 transcript has a 64-bp 5′ untranslated region. The deduced translation start site (CACTCATGTCT) (Fig. 3) conforms with the consensus sequence for Neurospora crassa translation initiation (CAMMATGGCT, where M = C or A) (8). Three introns (Fig. 3) were identified by comparing the sequences of the cDNA and the RACE products with the genomic clone sequence. The 5′ and 3′ splice sites, the splice branch sites, and the lengths of the introns are consistent with introns in N. crassa and in other genes of C. carbonum (1, 2, 8, 21, 25, 32, 36). Like most other fungal genes, including those of C. carbonum, MLG1 has no obvious AATAAA polyadenylation signal sequence preceding the polyadenylation site (13).
FIG. 3.
Nucleotide sequence and deduced amino acid sequence of MLG1. Amino acids are indicated below the corresponding codons. The amino acid sequences of the mature N terminus and the internal tryptic peptide are indicated by double underlining. The three introns are indicated by lowercase letters. The SalI, SmaI, and XhoI restriction sites indicated were used to construct and linearize pJM5 for the gene disruption experiments. The transcription start site is indicated by a pound sign, and the polyadenylation site is indicated by a plus sign (the pound and plus signs refer to the nucleotides below them). The predicted N-glycosylation site (amino acid 323) is underlined.
The open reading frame of cDNA C4-2.1 is predicted to encode a mature protein of 31.8 kDa, which is in good agreement with the size of Mlg1b, 31 kDa, as determined by SDS-PAGE. The predicted pI of the mature protein is 4.99. The program SignalP version 1.1 (22) predicts that there is a signal peptide cleavage site between amino acids 19 and 20 (VTA/LPA); if this is the true cleavage site, then Mlg1b must undergo additional processing to generate the mature protein. There is a unique predicted N-glycosylation site (NFS) near the C-terminal end (Fig. 3). There are a few discrepancies between the experimentally determined sequence of Mlg1b and the deduced sequence of MLG1 (Table 1). However, in our experience (33), these discrepancies are within the range of experimental error in protein sequencing, and the gene disruption experiments (see below) established that MLG1 does encode the activities catalyzed by Mlg1a and Mlg1b.
The amino acid sequence of Mlg1b has little overall similarity to the amino acid sequence of any other protein in the nonredundant databases. The best matches are with two prokaryotic glucanases, a β1,3-glucanase from Oerskovia xanthineolytica (BLAST score, 51; P = 0.84) and an MLGase from Rhodothermus marinus (BLAST score, 50; P = 0.92) (9, 35). The overall amino acid similarity and identity of Mlg1b to the MLGase of Rhodothermus marinus are 50 and 22%, respectively. The longest stretch of identity to the MLGase of Rhodothermus marinus is a five-amino-acid motif (GEIDI) surrounding an active site Glu residue; this motif is also at the active site of most MLGases from Bacillus spp. and other prokaryotes (17, 26, 35). This similarity to the bacterial MLGases identifies Mlg1b as a member of the family 16 glycosyl hydrolases (15). The sequence of Mlg1b has no detectable similarity to the sequence of any known plant MLGase or any β1,3-glucanase, including Exg1 from C. carbonum (30).
Transformation-mediated gene disruption of MLG1.
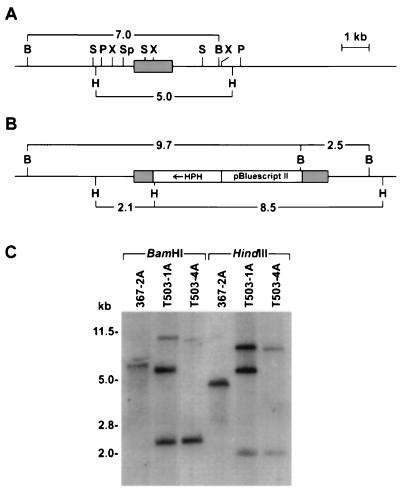
A 345-bp SalI-XhoI fragment, which is within the open reading frame of MLG1 (Fig. 3), was subcloned into Cochliobolus transformation vector pHYG1. The resulting plasmid, pJM5, was linearized with SmaI and introduced into wild-type C. carbonum 367-2A by transformation of protoplasts. Hygromycin B-resistant transformants were characterized by DNA gel blot analysis. Figure 4A shows the wild-type MLG1 locus, whereas Fig. 4B shows the predicted map for single integration of pJM5 into MLG1. The results of a DNA gel blot analysis of strain T503-4A (Fig. 4C, lanes 3 and 6) are consistent with the pattern predicted for a single insertion event, as shown in Fig. 4B. The pattern of hybridization seen for strain T503-1A (Fig. 4C, lanes 2 and 5) is consistent with tandem integration of the 6-kb pJM5 vector.
FIG. 4.
Analysis of the MLG1 locus in the wild type and mlg1 mutants. (A) Restriction map of the wild-type MLG1 locus, showing the location of the MLG1 transcript (shaded box). (B) Predicted restriction map of the MLG1 locus with a single insertion of transforming plasmid pJM5. (C) DNA blot of the wild type (367-2A) and two transformants (T503-1A and T503-4A). Total genomic DNA was digested with BamHI (lanes 1 to 3) or HindIII (lanes 4 to 6), fractionated by agarose gel electrophoresis, blotted, and probed with the MLG1 cDNA C4-2.1. The disappearance of a 7.0-kb BamHI band and a 5.0-kb HindIII band and the appearance of 9.7- and 2.5-kb BamHI bands and 2.1- and 8.5-kb HindIII bands are as predicted from homologous integration of pJM5. The additional bands in digests of T503-1A are as predicted for homologous integration of more than one copy of pJM5. The shaded areas in panels A and B indicate MLG1 sequences. B, BamHI, S, SalI; P, PstI; X, XhoI; Sp, SphI; H, HindIII. HPH indicates the hygromycin resistance gene.
The total MLGase and β1,3-glucanase activities in culture filtrates of Mlg1b mutants T503-1A and T503-4A grown on maize bran were reduced by approximately 50 and 40%, respectively. However, the growth (mycelial mat dry weight) of T503-1A or T503-4A was similar to the growth of the wild type (data not shown). Apparently, the residual MLGase activity and other enzymes capable of degrading the other components of maize bran are sufficient to support normal growth of the mlg1 mutant.
MLGase activities from the wild type and mlg1 mutant T503-1A were purified in parallel through the HI-HPLC step. When analyzed by cation-exchange HPLC, the mlg1 mutant was missing peak 1; peak 2, corresponding to Mlg2, was still present (Fig. 5A and B). Thus, Mlg2 is not encoded by MLG1. HI-HPLC analysis (Fig. 5C and D) indicated that both Mlg1a and Mlg1b are missing in the mlg1 mutant (Fig. 5D).
FIG. 5.
Analysis of MLGase in the mlg1 mutant. (A and B) Cation-exchange HPLC analysis of MLGase from the wild type (A) and mlg1 mutant T503-1A (B). (C and D) HI-HPLC analysis of peak 1 from cation-exchange HPLC of the wild type (C) and from cation-exchange HPLC of the mlg1 mutant (D). Solid lines, A280; dashed lines, MLGase activity.
Because Mlg1a and Mlg1b have the same substrate specificities and the same N-terminal amino acid sequences and because mlg1 mutants have neither Mlg1a nor Mlg1b activity, we conclude that MLG1 encodes both Mlg1a and Mlg1b. The different chromatographic behaviors of Mlg1a and Mlg1b are probably due to different glycosylation, because Mlg1a is glycosylated whereas Mlg1b is not, Mlg1a is 2.5 kDa larger than Mlg1b, and the MLG1 gene product has one predicted N-glycosylation site. The two products of MLG1 are probably not due to different intron splicing of the MLG1 transcript (3) because all three introns of MLG1 contain either stop codons or frameshifts (Fig. 3).
Pathogenicity of mlg1 mutants.
As the highest amounts of β-glucan are in young maize seedlings (4), we tested whether infection of young seedlings by C. carbonum was impeded by mutating MLG1. There were no measurable differences in lesion morphology and development or the percentage of seedlings that germinated when the plants were inoculated with either wild-type strain 367-2A or mlg1 mutant strain T503-1A or T503-4A. The wild type and the mlg1 mutants were also indistinguishable with regard to lesion size, color, and rate of lesion formation when they were spray inoculated onto leaves of 14-day-old maize seedlings. Thus, MLG1 does not by itself make a significant contribution to the virulence of C. carbonum.
ACKNOWLEDGMENTS
We thank Joe Leykam of the Macromolecular Facility, Michigan State University, for sequencing the peptides of Mlg1a, Mlg1b, and Mlg2 and for synthesizing the oligonucleotides used for DNA sequencing and PCR, Tom Newman of the Department of Energy Plant Research Laboratory Biochemistry Facility for automated DNA sequencing, and Fabienne Hamburger for technical assistance.
This work was supported by grant DEFG02-91ER20021 from the United States Department of Energy Division of Energy Biosciences and by grant 96-35303-3241 from the United States Department of Agriculture National Research Initiative Competitive Grants Program. J.M.G. was the recipient of a fellowship from the Michigan State University Biotechnology Training Program (National Institutes of Health grant T32-GM08350).
REFERENCES
- 1.Apel P C, Panaccione D G, Holden F R, Walton J D. Cloning and targeted gene disruption of XYL1, a β1,4-xylanase gene from the maize pathogen Cochliobolus carbonum. Mol Plant Microbe Interact. 1993;6:467–473. doi: 10.1094/mpmi-6-467. [DOI] [PubMed] [Google Scholar]
- 2.Apel-Birkhold P C, Walton J D. Cloning, disruption, and expression of two endo-β1,4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl Environ Microbiol. 1996;62:4129–4135. doi: 10.1128/aem.62.11.4129-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boel E, Hansen M T, Hjort I, Høegh I, Fiil N P. Two different types of intervening sequences in the glucoamylase gene from Aspergillus niger. EMBO J. 1984;3:1581–1585. doi: 10.1002/j.1460-2075.1984.tb02014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpita N C. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single step method of RNA extraction by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Clark D R, Johnson J, Jr, Chung K H, Kirkwood S. Purification, characterization, and action-pattern studies on the endo-(1→3)-β-d-glucanase from Rhizopus arrhizus QM1032. Carbohydr Res. 1978;61:457–477. doi: 10.1016/s0008-6215(00)84505-x. [DOI] [PubMed] [Google Scholar]
- 7.Doan D N P, Fincher G B. Differences in the thermostability of barley (1→3, 1→4)-beta-glucanases are only partly determined by N-glycosylation. FEBS Lett. 1992;309:265–271. doi: 10.1016/0014-5793(92)80786-g. [DOI] [PubMed] [Google Scholar]
- 8.Edelmann S E, Staben C. A statistical analysis of sequence features within genes from Neurospora crassa. Exp Mycol. 1994;18:70–81. [Google Scholar]
- 9.Ferrer P, Halkier T, Hedegaard L, Savva D, Diers I, Asenjo J A. Nucleotide sequence of a β-1,3-glucanase isoenzyme IIA gene of Oersokovia xanthineolytica LL G109 (Cellulomonas cellulans) and initial characterization of the recombinant enzyme expressed in Bacillus subtilis. J Bacteriol. 1996;178:4751–4757. doi: 10.1128/jb.178.15.4751-4757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gish W, States D J. Identification of protein coding regions by database similarity search. Nature Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 13.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J, editor. Gene structure in eukaryotic microbes. London, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 14.Hames B D, Rickwood D. Gel electrophoresis, a practical approach. Oxford, United Kingdom: IRL Press; 1981. [Google Scholar]
- 15.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Høj P B, Rodriguez E B, Stick R V, Stone B A. Differences in active site structure in a family of β-glucan endohydrolases deduced from the kinetics of inactivation by epoxyalkyl β-oligoglucosides. J Biol Chem. 1989;264:4939–4947. [PubMed] [Google Scholar]
- 17.Keitel T, Ortwin S, Borriss R, Heinemann U. Molecular and active-site structure of a Bacillus β-1,3-1,4-glucanase. Proc Natl Acad Sci USA. 1993;90:5287–5291. doi: 10.1073/pnas.90.11.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 19.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 20.Meldgaard M, Svendsen L. Different effects of N-glycosylation on the thermostability of highly homologous bacterial (1,3-1,4)-beta-glucanases secreted from yeast. Microbiology. 1994;140:159–166. doi: 10.1099/13500872-140-1-159. [DOI] [PubMed] [Google Scholar]
- 21.Murphy J M, Walton J D. Three extracellular proteases from Cochliobolus carbonum: cloning and targeted disruption of ALP1. Mol Plant Microbe Interact. 1996;9:290–297. doi: 10.1094/mpmi-9-0290. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Ooi T, Shinmyo A, Okada H, Hara S, Ikenaka T, Murao S, Arai M. Cloning and sequence analysis of a cDNA for cellulase (FI-CMCase) from Aspergillus aculeatus. Curr Genet. 1990;18:217–222. doi: 10.1007/BF00318384. [DOI] [PubMed] [Google Scholar]
- 24.Perito B, Hanhart E, Irdani T, Iqbal M, McCarthy A J, Mastromei G. Characterization and sequence analysis of a Streptomyces rochei A2 endoglucanase-encoding gene. Gene. 1994;148:119–124. doi: 10.1016/0378-1119(94)90244-5. [DOI] [PubMed] [Google Scholar]
- 25.Pitkin J W, Panaccione D G, Walton J D. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 26.Planas A, Juncosa M, Lloberas J, Querol E. Essential catalytic role of Glu134 in endo-β-1,3-1,4-d-glucan 4-glucanohydrolase from B. licheniformis as determined by site-directed mutagenesis. FEBS Lett. 1992;308:141–145. doi: 10.1016/0014-5793(92)81262-k. [DOI] [PubMed] [Google Scholar]
- 27.Ransom R F, Walton J D. Purification and characterization of extracellular β-xylosidase and α-arabinosidase from the plant pathogenic fungus Cochliobolus carbonum. Carbohydr Res. 1997;297:357–364. [Google Scholar]
- 28.Sakellaris H, Pemberton J M, Manners J M. Characterization of an endo-1,3(4)-β-d-glucanase gene from Cellvibrio mixtus. FEMS Microbiol Lett. 1993;109:269–272. doi: 10.1016/0378-1097(93)90031-v. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schaeffer H J, Leykam J, Walton J D. Cloning and targeted gene disruption of EXG1, encoding exo-β1,3-glucanase, in the phytopathogenic fungus Cochliobolus carbonum. Appl Environ Microbiol. 1994;60:594–598. doi: 10.1128/aem.60.2.594-598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schimming S, Schwarz W H, Staudenbauer W L. Structure of the Clostridium thermocellum gene licB and the encoded β-1,3-1,4-glucanase. Eur J Biochem. 1992;204:13–19. doi: 10.1111/j.1432-1033.1992.tb16600.x. [DOI] [PubMed] [Google Scholar]
- 32.Scott-Craig J S, Panaccione D G, Cervone F, Walton J D. Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell. 1990;2:1191–1200. doi: 10.1105/tpc.2.12.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott-Craig J S, Panaccione D G, Pocard J-A, Walton J D. The cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7-kilobase open reading frame. J Biol Chem. 1992;67:26044–26049. [PubMed] [Google Scholar]
- 34.Slakeski N, Baulcombe D C, Devos K M, Doan D N P, Fincher G B. Structure and tissue-specific regulation of genes encoding barley (1→3,1→4)-β-glucan endohydrolases. Mol Gen Genet. 1990;224:437–449. doi: 10.1007/BF00262439. [DOI] [PubMed] [Google Scholar]
- 35.Spilliaert R, Hreggvidsson G O, Kristjansson J K, Eggertsson G, Palsdottir A. Cloning and sequencing of a Rhodothermus marinus gene, bglA, coding for a thermostable β-glucanase and its expression in Escherichia coli. Eur J Biochem. 1994;224:923–930. doi: 10.1111/j.1432-1033.1994.00923.x. [DOI] [PubMed] [Google Scholar]
- 36.Sposato P, Ahn J-H, Walton J D. Characterization and disruption of a gene in the maize pathogen Cochliobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol Plant Microbe Interact. 1995;8:602–609. doi: 10.1094/mpmi-8-0602. [DOI] [PubMed] [Google Scholar]
- 37.Strömqvist M, Guffman H. Periodic acid/Schiff staining of glycoproteins immobilized on a blotting matrix. BioTechniques. 1992;13:744–746. [PubMed] [Google Scholar]
- 38.Teather R, Erfle J D. DNA sequence of a Fibrobacter succinogenes mixed-linkage β-glucanase (1,3-1,4-d-glucan 4-glucanohydrolase) gene. J Bacteriol. 1990;172:3837–3841. doi: 10.1128/jb.172.7.3837-3841.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hoof A, Leykam J, Schaeffer H J, Walton J D. A single β1,3-glucanase secreted by the maize pathogen Cochliobolus carbonum acts by an exolytic mechanism. Physiol Mol Plant Pathol. 1991;39:259–267. [Google Scholar]
- 40.Walton J D. Deconstructing the cell wall. Plant Physiol. 1994;104:1113–1118. doi: 10.1104/pp.104.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood P J. Oat beta-glucan: structure, location, and properties. In: Webster F H, editor. Oats: chemistry and technology. St. Paul, Minn: American Association of Cereal Chemists; 1986. pp. 121–152. [Google Scholar]
- 42.Yun S J, Martin D J, Gengenbach B G, Rines H W, Somers D A. Sequence of a (1-3,1-4)-β-glucanase cDNA from oat. Plant Physiol. 1993;103:295–296. doi: 10.1104/pp.103.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]