Abstract
Simple Summary
Loss of the CDKN2A gene and its protein p16 is a common event in pleural mesothelioma, which is frequently investigated to confirm the diagnosis. The loss of CDKN2A is often accompanied by simultaneous loss of the genes located in the vicinity, including the MTAP gene and protein. Detection of CDKN2A loss is usually achieved by relatively expensive fluorescent in situ hybridization (FISH) analysis. Here, we investigated if inexpensive immunohistochemistry (IHC) for p16 and MTAP could be used as an alternative detection method. Comparing CDKN2A and MTAP gene status, analyzed by the copy-number variation array, with p16/MTAP IHC revealed high sensitivity and specificity of the IHC for detecting gene loss. Our data show that p16/MTAP IHC can be used as an alternative to p16 FISH for detection of CDKN2A/MTAP loss. We recommend combined MTAP and p16 immunohistochemistry to confirm the diagnosis of PM.
Abstract
CDKN2A deletion is a common alteration in pleural mesothelioma (PM) and frequently associated with co-deletion of MTAP. Since the standard detection method for CDKN2A deletion and FISH analysis is relatively expensive, we here investigated the suitability of inexpensive p16 and MTAP IHC by comparing concordance between IHC and OncoScan CNV arrays on samples from 52 PM patients. Concordance was determined using Cohen’s kappa statistics. Loss of CDKN2A was associated with co-deletion of MTAP in 71% of cases. CDKN2A-MTAP copy-number normal cases were also IHC positive in 93% of cases for p16 and 100% for MTAP, while homozygous deletion of CDKN2A-MTAP was always associated with negative IHC for both proteins. In cases with heterozygous CDKN2A-MTAP loss, IHC expression of p16 and MTAP was negative in 100% and 71%, respectively. MTAP and p16 IHC showed high sensitivity (MTAP 86.5%, p16 100%) and specificity (MTAP 100%, p16 93.3%) for the detection of any gene loss. Loss of MTAP expression occurred exclusively in conjunction with loss of p16 labeling. Both p16 and MTAP IHC showed high concordance with Oncoscan CNV arrays (kappa = 0.952, p < 0.0001, and kappa = 0.787, p < 0.0001 respectively). We recommend combined MTAP and p16 immunohistochemistry to confirm the diagnosis of PM.
Keywords: pleural mesothelioma, p16, MTAP, molecular pathology, diagnostics, immunohistochemistry, copy-number array, CDKN2A
1. Introduction
Pleural mesothelioma (PM) is an aggressive neoplasm arising from the mesothelial cells lining the pleural cavity and is strongly associated with previous exposure to asbestos [1]. The algorithm for diagnosis of malignant PM requires histomorphologic evidence of invasion and immunohistochemical evidence of the mesothelial nature of the tumor [2]. However, in cases of small biopsy specimens, in situ mesothelial proliferations, and cell blocks, this approach is insufficient to make a clinically meaningful diagnosis. In such cases, additional studies such as fluorescence in situ hybridization (FISH) of cyclin-dependent kinase inhibitor 2A (CDKN2A) and BRCA-1-associated protein-1 (BAP1) immunohistochemistry are useful to confirm the malignant nature of the lesion [3,4,5,6,7,8,9,10]. Despite optimal specificity, CDKN2A FISH has variable sensitivity in PM with preserved BAP1 expression, ranging from 58% to 80% depending on the study and histological subtype of PM [7,8,9,10].
The CDKN2A locus encodes the p16 protein and resides on chromosome 9p21 in proximity to a cluster of genes harboring CDKN2B and methylthioadenosine phosphorylase (MTAP). MTAP is a key enzyme that cleaves methylthioadenosine, resulting in precursor substrates required for adenosine and methionine salvage pathways [11]. CDKN2A deletion is a frequent alteration in PM, occurring in up to 74% of cases [12,13,14,15,16,17]. Due to its proximity to CDKN2A, homozygous deletion of MTAP occurs frequently in various tumors, including mesotheliomas. This is associated with low levels of mRNA of both genes, which may explain the concurrent loss of p16 and MTAP protein expression and has opened the possibility of IHC instead of FISH to determine CDKN2A gene status [14]. Disadvantages of CDKN2A FISH are technical complexity, longer turnaround times, and higher costs. In contrast to FISH and other molecular tests, IHC is a technically feasible and cost-effective substitute that can be applied in any routine lab. Some early studies showed poor agreement between p16 and MTAP protein expression and CDKN2A status determined by FISH, suggesting that p16 and MTAP IHC are not reliable biomarkers for detecting CDKN2A loss [6,15]. Recently, however, evidence has accumulated that loss of MTAP protein expression is associated with a homozygous deletion of CDKN2A determined by FISH [5,16,17,18,19]. The aim of this study is to evaluate the diagnostic utility of MTAP and p16 immunohistochemistry in the simultaneous detection of CDKN2A and MTAP alterations using the copy-number variation (CNV) assay.
2. Materials and Methods
2.1. Tissue Samples and Tissue Microarray Construction
All patients gave written informed consent, the study was approved by the cantonal Ethics Committee Zurich (KEK-ZH-Nr. 2012-0094), and the study was conducted in accordance with the Declaration of Helsinki. Formalin-fixed paraffin-embedded tissue (FFPE) blocks containing diagnostic biopsies of 52 PMs between 1999 and 2015 were retrieved from the archives of our pathology department. Tissue processing was performed overnight using 4% neutral buffered formalin. Corresponding slides of all cases were independently reviewed by two experienced pathologists (B.V. and A.S.) and histologically categorized into epithelioid, biphasic, and sarcomatoid PM according to the latest WHO guidelines [20]. Additional staining, including mesothelial (CK5/6, calretinin) and epithelial markers (Ber-Ep4, Claudin-4), was performed to confirm the mesothelial nature of the tumor. In cases with a biphasic or sarcomatoid PM, additional pancytokeratin and podoplanin (D2-40) stains were applied [2].
Tissue blocks were used for array-based genome-wide copy-number analysis and construction of a tissue microarray (TMA). TMA construction was accomplished with a custom-made, semiautomatic tissue arrayer (Beecher Instruments, Sun Prairie, WI, USA) as previously described [21]. Only representative tumor blocks containing sufficient tumor tissue to perform CNV array and construction of TMA were selected, and four tissue cores, 0.6 mm in diameter, were taken from each case and transferred into a recipient paraffin block. In order to capture the diversity of PM, cores were taken from separately marked areas.
2.2. Copy-Number Variation Arrays (Oncoscan)
Array-based genome-wide copy-number analysis was conducted using OncoScan FFPE microarrays (Affymetrix, Santa Clara, CA, USA) as previously described [3]. Considering potential spatial heterogeneity cores were taken from separately marked areas within the tumor. From each patient, samples of normal tissue were collected and analyzed in parallel with tumor samples.
2.3. Immunohistochemistry for p16 and MTAP
IHC was performed using an automated single-staining procedure (Benchmark Ultra; Ventana Medical Systems, Oro Valley, AZ, USA). Antigen retrieval for p16 and MTAP was performed using Ventana CC1/EDTA buffer for 48 and 32 min, respectively. Using negative and positive on-slide-controls, including reactive mesothelial proliferations, 4 µm sections of the TMA were stained with monoclonal antibodies against p16 (clone E6H4, ready-to-use, Ventana Medical Systems-Tucson, Oro ValleyAZ, USA; Roche) incubated for 4 min at 37 °C and MTAP (clone EPR6893, Abcam Limited, Hangzhou, China) at 1:500 dilution, incubated for 32 min at room temperature.
Any p16 nuclear staining with or without cytoplasmatic staining was regarded as positive, while loss of p16 nuclear immunoreactivity was considered negative. Strong nuclear and/or cytoplasmatic staining for MTAP was interpreted as positive, whereas complete loss of cytoplasmic labeling was considered negative. Because of the variable expression of MTAP, staining was regarded as negative when staining intensity was weaker than the internal positive control, such as stromal or inflammatory cells. Heterogenous expression of p16 or MTAP was regarded as positive, and the percentage of positively stained cells was recorded. To validate the immunohistochemical results CDKN2A FISH analysis on gross slides was performed in selected cases, two with homozygous, two with heterozygous deletion, and two with retained CDKN2A as determined by CNV. Homozygous deletion by FISH was defined as 20% or more of 100 nuclei showing a loss of both CDKN2A signals, Loss of one CDKN2A signal in more than 50% together with less than 20% loss of both signals was regarded as heterozygous deletion.
2.4. Statistical Analysis
In order to determine the suitability of p16 and MTAP IHC for detecting loss of p16 and MTAP protein expression in samples with known homozygous or heterozygous deletion determined by Oncoscan CNV arrays, sensitivity and specificity were calculated. Furthermore, for assessment of the concordance between IHC and CNV, Cohen’s kappa was calculated. All calculations were performed using IBM SPSS Statistics 26. p < 0.05 was considered as statistically significant. Receiver operating characteristic (ROC) curves were generated using GraphPad Prism v.9 (GraphPad Software, Boston, MA, USA)
3. Results
Histological examination of the diagnostic biopsies revealed 43 epithelioid (82.7%), 6 biphasic (11.5%), and 3 sarcomatoid PM (5.8%). Basic patient demographics are summarized in Table 1, while the results of the IHC and CNV arrays are summarized in Figure 1.
Table 1.
Basic patient demographics.
| Median Age (Range) | 60 (35–73) |
|---|---|
| Gender | |
| Male | 45 (86.5%) |
| Female | 7 (13.5%) |
| Histotype | |
| Epithelioid | 43 (82.7%) |
| Biphasic | 6 (11.5%) |
| Sarcomatoid | 3 (5.8%) |
| Pathological Stage (IMIG, 8th edition) | |
| IA | 2 (3.8%) |
| IB | 23 (44.2%) |
| II | 0 (0.0%) |
| IIIA | 8 (15.4%) |
| IIIB | 15 (28.8%) |
| unknown | 4 (7.7%) |
| Median OS from diagnosis (range) | 14.95 months (3.94–79.86) |
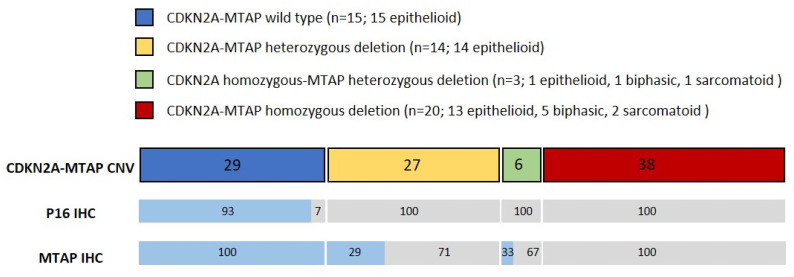
Figure 1.
Comparison of p16/MTAP immunohistochemistry (IHC) and CDKN2A-MTAP copy-number loss determined by CNV array. Numbers indicate percentage of each group compared to total number of patients (n = 52) for CNV, and to number of patients from each group for IHC, respectively. Blue represents IHC positive, and gray represents IHC negative cases.
3.1. Copy-Number Variation Array (Oncoscan)
CNV analysis identified homozygous deletion of both CDKN2A and MTAP in 20 cases (38%). In 14 cases (27%), heterozygous deletion of both CDKN2A and MTAP was found. In three cases (6%), homozygous deletion of CDKN2A was accompanied by a heterozygous loss of the MTAP gene. Fifteen cases (29%) revealed normal CDKN2A and MTAP copy numbers. Copy-number loss of MTAP in the absence of CDKN2A deletion was not detected.
Heterozygous loss of both CDKN2A and MTAP was identified in 15 and 14 cases of epithelioid PM. In one case of epithelioid PM, a homozygous loss of CDKN2A combined with a heterozygous loss of MTAP was detected. Fifteen cases with epithelioid PM revealed normal copy numbers of both genes were found. In five biphasic and two sarcomatoid PM, homozygous deletion of CDKN2A and MTAP was noted. One biphasic and one sarcomatoid PM showed homozygous CDKN2A loss combined with a heterozygous deletion of MTAP. None of the cases with a biphasic or sarcomatoid PM showed a WT pattern.
3.2. Concordance of Immunohistochemistry and Copy-Number Variation Array
Consistent with the immunoreactivity in the external controls, p16 positivity in the PM cases was characterized by a diffuse nuclear and cytoplasmatic staining pattern. Strong expression of MTAP was observed in 20 cases. In comparison to stromal cells, MTAP-negative cases showed a faint to moderately cytoplasmic staining (example shown in (Figure 2B)).
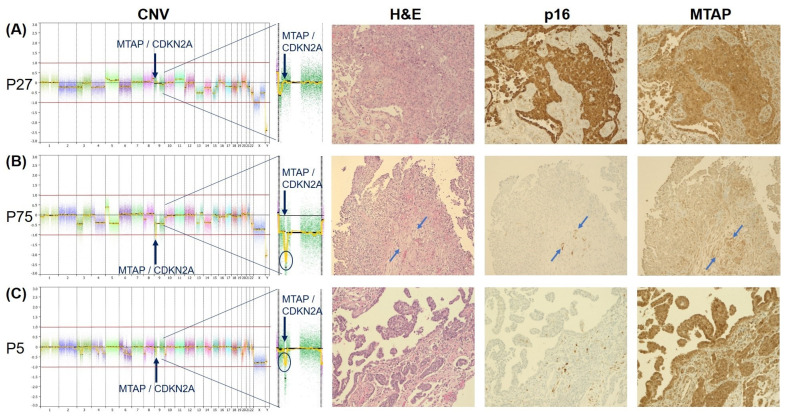
Figure 2.
CNV profiles and IHC staining patterns of p16 and MTAP in a case with WT ((A), P27), a case with homozygous CDKN2A-MTAP deletion ((B), P75), and a case with heterozygous CDKN2A-MTAP deletion ((C), P5), respectively. While the case with homozygous loss (B) shows consistent loss of MTAP staining, MTAP immunoreactivity is retained in some cases with heterozygous loss, as shown here in (C). Note the positive internal control for p16 and MTAP of stromal cells between the negative tumor cell infiltrates as marked by blue arrows in (B). CNV results are shown with a zoom-in into chromosome 9, which harbors the CDKN2A and MTAP loci on q21.3. H&E, p16, and MTAP staining are shown in 20× magnification. (Figure S1 provides additional information).
PM with a wild-type (WT) CDKN2A-MTAP showed retained strong expression of MTAP in all cases, whereas p16 positivity was observed in all but one WT case. None of the WT cases showed negativity of both p16 and MTAP (Figure 2A). Conversely, homozygous loss of both CDKN2A and MTAP was associated with loss of p16 and MTAP expression in all cases (Figure 2B). Heterozygous loss of CDKN2A and MTAP was observed in 14 cases, which was accompanied by negative staining of p16 and MTAP in 100% and 71%, respectively (Figure 2C). In three cases, homozygous loss of CDKN2A and heterozygous loss of MTAP were associated with loss of p16 in all and MTAP expression in two cases. Similar to CDKN2A-MTAP by CNV, retained expression of p16 in the absence of MTAP was not observed in any of the cases. CDKN2A FISH in the six selected cases corresponded with the results of the CNV array.
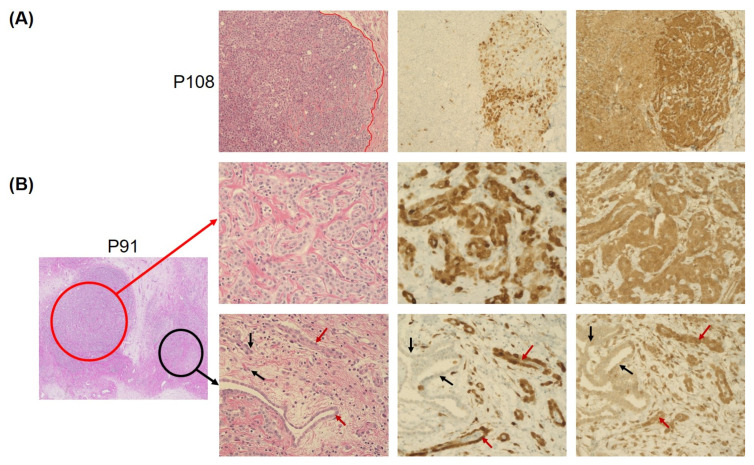
Nine mesotheliomas (17%) showed a heterogeneous p16 and MTAP expression, with labeling of both markers in a proportion of cores or tumor cells ranging from 10 to 90% of cells. Six of these cases were WT, as determined by the CNV array. Gross slides of these cases showed spatial loss of both p16 and MTAP (Figure 3), ranging from a focal loss in a smaller cell cluster (Figure 3B) to up to 50% loss in some tumor areas (Figure 3A). In one case with a homozygous deletion of both CDKN2A and MTAP and one case with a homozygous CDKN2A deletion and heterozygous MTAP deletion, strong MTAP expression was seen in 10% of the tumor cells, whereas p16 was negative in both cases. In one mesothelioma with heterozygous CDKN2A-MTAP deletion, positive MTAP expression was seen in 60% of the tumor cells, whereas p16 was completely negative.
Figure 3.
Heterogeneity of p16 and MTAP protein expression in PM observed in 17% of cases. (A) shows an example of a tumor sample with wild-type CDKN2A/MTAP and retained protein expression with very high spatial heterogeneity, and p16 and MTAP negativity in up to 50% of tumor cells (tumor area left of red line). The wild-type case presented in (B), however, showed the expected 100% p16 and MTAP positivity in one area of the obtained section (red circle, middle row), combined with only focal loss of both p16 and MTAP in around 10% of the tumor cells in a different area (black circle, bottom row). Staining positive tumor cells are marked with red arrow, while staining negative tumor cells are marked with black arrows. (Magnification: 20×).
3.3. Sensitivity and Specificity of p16 and MTAP Immunohistochemistry
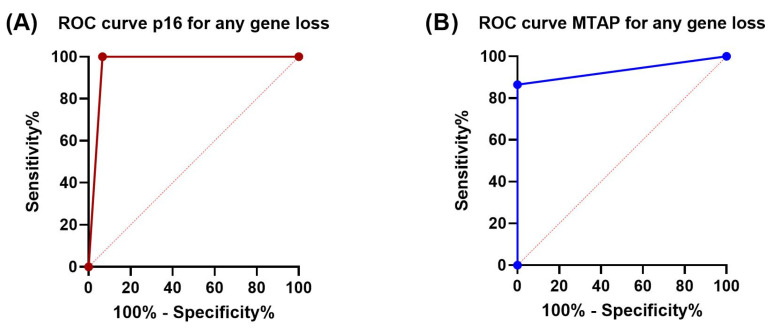
P16 and MTAP immunohistochemistry showed a specificity of 100% and 93.3% for detection of CDKN2A and MTAP copy-number alterations detected by CNV array, respectively. Compared to the results of the CNV array, sensitivity of p16 (100%) was better than that of MTAP (86.5%) in detecting any type of loss (homozygous or heterozygous). Concordance (Cohen’s kappa) for detection of any loss between Oncoscan CNV array and IHC was 0.952 for p16 and 0.787 for MTAP (p < 0.0001 for both). P16 IHC showed the same sensitivity for detection of both homozygous and heterozygous loss as determined by CNV. In the case of MTAP, however, sensitivity for detection of a homozygous loss was 100%, while a heterozygous loss was only detected by IHC with a sensitivity of 70.6%. Accordingly, Cohen’s kappa for detection of a heterozygous MTAP loss was reduced to 0.692 (p > 0.0001), while kappa for identifying homozygous MTAP deletion reached a perfect value of 1.0 (p > 0.0001). Sensitivity, specificity, and Cohen’s kappa are summarized in Table 2. ROC curves and the corresponding AUCs are depicted in Figure 4.
Table 2.
Overview of concordance, sensitivity, and specificity of IHC.
| Any Loss | Homozygous Loss | Heterozygous Loss | ||||
|---|---|---|---|---|---|---|
| p16 | MTAP | p16 | MTAP | p16 | MTAP | |
| Cohen’s kappa | 0.952 | 0.787 | 0.944 | 1.0 | 0.931 | 0.692 |
| Sensitivity | 100% | 86.5% | 100% | 100% | 100% | 70.6% |
| Specificity | 93.3% | 100% | 93.3% | 100% | 93.3% | 100% |
Figure 4.
ROC curves showing diagnostic sensitivity and specificity of p16 and MTAP IHC for homozygous and heterozygous loss of CDKN2A and MTAP genes: (A) P16 sensitivity 100%, specificity 93%, AUC 0.97 (95% CI: 0.89–1.0); (B) MTAP sensitivity 86.5%, specificity 100%, AUC 0.93 (95% CI: 0.86–1.0). AUC is annotaed with 95% CI using the Wilson/Brown method.
4. Discussion
Using a CNV array for genome-wide detection of copy-number alterations, we have shown for the first time that loss of CDKN2A in PM is consistently associated with co-deletion of the MTAP gene. The genomic loss was also associated with loss of both p16 and MTAP protein expression in all cases with homozygous deletion of CDKN2A-MTAP and in 100% and 71% with heterozygous deletion of CDKN2A and MTAP, respectively. Compared with CNV results, p16 IHC showed a sensitivity of 100% and a specificity of 93.3%, while MTAP IHC showed a sensitivity of 86.5% and a specificity of 100% for detecting loss. These data support the concept that combined p16 and MTAP immunohistochemistry parallels CDKN2A-MTAP gene status.
In this study, CDKN2A-MTAP co-deletion was associated with loss of both p16 and MTAP protein expression in all cases with homozygous deletion and in the majority of cases with heterozygous deletion of CDKN2A-MTAP. Consistent with our findings, various studies using technologies, including FISH, NGS, and single-nucleotide polymorphism microarray, have shown that homozygous CDKN2A deletion is associated with MTAP co-deletion and that isolated MTAP deletion does not occur without concurrent CDKN2A loss [12,13,22]. Using FISH, Illei et al. [12] observed discordant deletion of CDKN2A without MTAP loss in 9% of their cases, a finding that possibly reflects the heterozygous MTAP deletion combined with homozygous CDKN2A loss found in three of our cases (6%). This supports the concept that loss of MTAP deficiency is only present in the context of a homozygous loss of CDKN2A, specifically when the 9p21.3 deletion is large enough to encompass the MTAP gene.
Homozygous deletion of CDKN2A determined by FISH is formally considered to be diagnostic of malignancy if more than 20% of the tumor nuclei show loss of both 9p21 signals [6]. However, FISH results are highly variable ranging from no detectable loss and heterozygous deletion to homozygous deletion of CDKN2A both in malignant as well in reactive mesothelial proliferations, which is mainly attributable to artifactual loss of signals because of nuclear sectioning [23,24,25]. Previous published work investigating p16 protein expression as a substitute for CDKN2A gene status has shown a poor concordance between p16 immunohistochemistry and homozygous CDKN2A deletion by FISH in PM indicating that p16 labeling cannot serve as a surrogate for CDKN2A genomic loss [6]. In contrast to these results, but consistent with recent observations in peritoneal mesotheliomas [13], we found loss of p16 expression in all cases with homozygous CDKN2A deletion determined by CNV analysis. Similar to FISH, we cannot rule out that loss of p16 protein in the 14 cases with heterozygous deletion of CDKN2A is the result of spatial intra-tumoral heterogeneity [24]. Other mechanisms, such as mini deletions not detected by the CNV array, point mutations, or DNA methylation of the p16 promoter gene may also be attributable to loss of p16 labeling [26,27].
The poor concordance between p16 expression and CDKN2A loss has shifted attention to MTAP IHC as a surrogate marker for CDKN2A gene status. In an initial study, Zimling et al. [15] observed loss of MTAP expression in 65% and 23% of the malignant and benign mesothelial proliferations, suggesting that MTAP IHC is not useful for PM diagnostics. However, in recent years, there has been a growing body of evidence for high specificity of MTAP protein expression for detection of homozygous CDKN2A deletion by FISH [5,16,17,18,19]. MTAP turned out to be 100% specific for diagnosing PM, with sensitivity ranging from 43 to 65%. Consistent with these observations, MTAP immunohistochemistry in our study was 86.5% sensitive and 100% specific for CDKN2A-MTAP gene status. As mentioned before, MTAP deletion without CDKN2A co-deletion did not occur in any of the PM tumor samples, and loss of MTAP expression occurred exclusively in conjunction with loss of p16 labeling. Positive staining for p16 thus excludes loss of MTAP. Because the interpretation of MTAP immunohistochemistry can be challenging [18], the addition of p16 staining helps to better distinguish MTAP-positive from -negative cases. According to the literature, MTAP deficiency is due to either deletion or epigenetic silencing of the MTAP gene [28,29]. Therefore, tests for detecting genomic deletion, including FISH and CNV, may underestimate MTAP loss in PM. Therefore, the most appropriate test for demonstrating MTAP deficiency would be detection of loss of protein expression by immunohistochemistry. In our cases, different patterns of combined CDKN2A-MTAP deletion were associated with complete loss of p16 IHC independent of whether homozygous or heterozygous loss was present. In the case of MTAP, however, loss of protein expression was observed in 100% of cases with homozygous loss, but also in 71% (12/17) cases with a heterozygous MTAP deletion. Accordingly, the concordance between IHC and CNV only reached a kappa of 0.692 for detection of a heterozygous loss. Of the 17 heterozygous MTAP cases, 3 were associated with a homozygous deletion and 14 with a single-copy loss of CDKN2A. Recently, Chapel et al. [22] presented similar data with loss of MTAP expression in 10 of 15 (67%) tumors with heterozygous MTAP deletion. Based on the heterogenous MTAP staining in a subset of cases with MTAP deletion, they hypothesized the presence of topographical subclones with MTAP deletion. Another explanation for the loss of MTAP expression in those cases with heterozygous MTAP deletion could be MTAP silencing, as has been described in melanoma, where epigenetic dysregulation of MTAP and other genes contribute to tumor progression and invasion. We did not find any reports on MTAP mutations or mini deletions, which could explain loss of MTAP protein expression. However, in knockout mice heterozygous for MTAP appear to be indistinguishable from WT mice, but died because of T-cell lymphoma. In comparison with normal controls, levels of MTAP, RNA, and MTAP expression by IHC were significantly reduced in the tissues in these mice infiltrated with lymphoma [29]. Whether these observations can be extended to heterozygous MTAP deletions in PM remains to be elucidated.
In the present study, topographical loss of both p16 and MTAP protein in part of the tumor tissue was observed in 17% of our cases, the majority of which were WT (66%) according to CNV analysis (Figure 2). Also, in gross slides of WT cases, small groups of p16-MTAP-negative tumor cells could be identified between aggregates expressing both p16 and MTAP. Consistent with our findings heterogeneity of MTAP staining intensity in PM has been reported in several studies [5,18,22]. Berg and co-workers [5] described two cases with negative MTAP expression in 50% and 75% of the tumor cells (similar to the case described in Figure 2), whereas Chapel et al. [18] found a heterogeneous MTAP retention in four cases with homozygous CDKN2A deletion determined by FISH. Using combined p16 and MTAP IHC, we were able to discriminate tumor areas with sustained p16-MTAP expression from tumor cell aggregates that were negative for both markers. In three cases (6%), a heterozygous MTAP deletion combined with homozygous CDKN2A deletion was found, which was associated with loss of both p16 and MTAP expression in two cases. The concurrent biallelic loss of CDKN2A next to heterozygous MTAP gene deletion probably reflects genetic tumor progression and characterizes the spatial heterogeneity of PM [24].
This study has several limitations. First, it is difficult to compare FISH and CNV in assessing CDKN2A and MTAP copy-number status in PM. The FISH assay is still regarded as the gold standard for detection of copy-number loss in PM, because it allows visual identification of homozygous and heterozygous deletions. Nevertheless, molecular assays, such as CNV, enable the simultaneous analysis of both CDKN2A and MTAP of more cells, including the distinction of single- and two-copy-number loss, and are not compromised by artificial loss of signals due to cutting of sections. Second, The number of female patients was relatively low (16%) but comparable with those found in the general population (17%) [20]. In a larger cohort of 222 patients, who had been treated in our hospital, 23 (10%) were females. Loss of MTAP protein was found in 47% of the female cases, which is comparable with the findings in the present cohort (43%). Third, several studies have used a three-tiered system to assess the H-score of MTAP expression or, for statistical reasons, applied different cut-offs for positive or negative staining [15,16]. In this study, labeling in a proportion of tumor cells in the TMA was interpreted as positive. Although in the vast majority of cases p16 and MTAP were either positive or negative, the diagnostic significance of heterogeneous p16 and MTAP staining in small biopsies or cytological specimens remains unclear. In such cases, the detection of copy-number loss by p16 FISH, especially of those tumor aggregates showing loss of both p16 and MTAP, may help to determine the malignant nature of the mesothelial proliferation.
5. Conclusions
In conclusion, combined p16 and MTAP immunohistochemistry is a cost-effective surrogate for CDKN2A-MTAP genetic alterations that can be reliably used to detect homozygous deletions and visualize the spatial heterogeneity of the CDKN2A-MTAP gene complex within the same tumor sample. Topographic labeling of both MTAP and p16 in the same tumor areas of cases with a heterogeneous staining pattern indicates a common mechanism of inactivation of CDKN2A and MTAP genes. To obtain a reliable diagnosis of PM, combined MTAP and p16 immunohistochemistry is recommended, especially in BAP-1-positive cases, small biopsies, and cell blocks.
Acknowledgments
We would like to thank Susanne Dettwiler and Fabiola Prutek for their excellent technical assistance with TMA construction.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15204978/s1, Figure S1. Negative and positive on-slide control for p16 (A,C) and MTAP (B,D) in a mammacarcinoma (A,B) and ovarian carcinoma (C,D), respectively (Magnification 20×).
Author Contributions
Conceptualization, B.V. and P.J.W.; methodology, B.V. and K.O.; validation, B.V., K.O. and A.S.; formal analysis, B.V., M.B.K., K.O. and A.S.; investigation, B.V. and K.O.; resources, B.V., I.O. and P.J.W.; data curation, B.V., M.B.K., M.M. and K.O.; writing—original draft preparation, B.V., M.B.K. and P.J.W.; writing—review and editing, M.M., K.O., U.W., A.S., H.M. and I.O.; visualization, B.V., M.B.K. and M.M.; supervision, B.V., I.O. and P.J.W.; project administration, B.V., I.O. and P.J.W.; funding acquisition, P.J.W. and I.O. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the cantonal Ethics Committee of the canton Zurich (KEK-ZH-Nr. 2012-0094).
Informed Consent Statement
All patients gave written informed consent.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
P.J.W. has received consulting fees and honoraria for lectures by Bayer, Janssen-Cilag, Novartis, Roche, MSD, Astellas Pharma, Bristol-Myers Squibb, Thermo Fisher Scientific, Molecular Health, Guardant Health, Sophia Genetics, Qiagen, Eli Lilly, Myriad, Hedera Dx, and AstraZeneca. I.O. has received honoraria for lectures by Roche and AstraZeneca; honoraria for participation in Advisory Boards by AstraZeneca, MSD, and BMS; institutional grants by Roche and Medtronic; honoraria for proctorship by Intuitive. All other authors have nothing to disclose.
Funding Statement
Parts of this project (Oncoscan arrays) were funded by an Innovation Pool Grant from the University of Zurich (to P.J.W.), and a grant by the Polianthes Foundation (to I.O.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Spirtas R., Heineman E.F., Bernstein L., Beebe G.W., Keehn R.J., Stark A., Harlow B.L., Benichou J. Malignant mesothelioma: Attributable risk of asbestos exposure. Occup. Environ. Med. 1994;51:804–811. doi: 10.1136/oem.51.12.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Husain A.N., Colby T.V., Ordóñez N.G., Allen T.C., Attanoos R.L., Beasley M.B., Butnor K.J., Chirieac L.R., Churg A.M., Dacic S., et al. Guidelines for Pathologic Diagnosis of Malignant Mesothelioma 2017 Update of the Consensus Statement From the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2018;142:89–108. doi: 10.5858/arpa.2017-0124-RA. [DOI] [PubMed] [Google Scholar]
- 3.Oehl K., Vrugt B., Wagner U., Kirschner M.B., Meerang M., Weder W., Felley-Bosco E., Wollscheid B., Bankov K., Demes M.C., et al. Alterations in BAP1 Are Associated with Cisplatin Resistance through Inhibition of Apoptosis in Malignant Pleural Mesothelioma. Clin. Cancer Res. 2021;27:2277–2291. doi: 10.1158/1078-0432.CCR-20-4037. [DOI] [PubMed] [Google Scholar]
- 4.Nasu M., Emi M., Pastorino S., Tanji M., Powers A., Luk H., Baumann F., Zhang Y.A., Gazdar A., Kanodia S., et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J. Thorac. Oncol. 2015;10:565–576. doi: 10.1097/jto.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg K.B., Dacic S., Miller C., Cheung S., Churg A. Utility of Methylthioadenosine Phosphorylase Compared With BAP1 Immunohistochemistry, and CDKN2A and NF2 Fluorescence In Situ Hybridization in Separating Reactive Mesothelial Proliferations From Epithelioid Malignant Mesotheliomas. Arch. Pathol. Lab. Med. 2018;142:1549–1553. doi: 10.5858/arpa.2018-0273-OA. [DOI] [PubMed] [Google Scholar]
- 6.Chiosea S., Krasinskas A., Cagle P.T., Mitchell K.A., Zander D.S., Dacic S. Diagnostic importance of 9p21 homozygous deletion in malignant mesotheliomas. Mod. Pathol. 2008;21:742–747. doi: 10.1038/modpathol.2008.45. [DOI] [PubMed] [Google Scholar]
- 7.Hida T., Hamasaki M., Matsumoto S., Sato A., Tsujimura T., Kawahara K., Iwasaki A., Okamoto T., Oda Y., Honda H., et al. BAP1 immunohistochemistry and p16 FISH results in combination provide higher confidence in malignant pleural mesothelioma diagnosis: ROC analysis of the two tests. Pathol. Int. 2016;66:563–570. doi: 10.1111/pin.12453. [DOI] [PubMed] [Google Scholar]
- 8.Hwang H.C., Pyott S., Rodriguez S., Cindric A., Carr A., Michelsen C., Thompson K., Tse C.H., Gown A.M., Churg A. BAP1 Immunohistochemistry and p16 FISH in the Diagnosis of Sarcomatous and Desmoplastic Mesotheliomas. Am. J. Surg. Pathol. 2016;40:714–718. doi: 10.1097/pas.0000000000000616. [DOI] [PubMed] [Google Scholar]
- 9.Sheffield B.S., Hwang H.C., Lee A.F., Thompson K., Rodriguez S., Tse C.H., Gown A.M., Churg A. BAP1 immunohistochemistry and p16 FISH to separate benign from malignant mesothelial proliferations. Am. J. Surg. Pathol. 2015;39:977–982. doi: 10.1097/PAS.0000000000000394. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Liao X., Gu Y., Fu L., Zhao J., Li L., Chen Z., Jiang J. Role of p16 deletion and BAP1 loss in the diagnosis of malignant mesothelioma. J. Thorac. Dis. 2018;10:5522–5530. doi: 10.21037/jtd.2018.08.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryukov G.V., Wilson F.H., Ruth J.R., Paulk J., Tsherniak A., Marlow S.E., Vazquez F., Weir B.A., Fitzgerald M.E., Tanaka M., et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illei P.B., Rusch V.W., Zakowski M.F., Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin. Cancer Res. 2003;9:2108–2113. [PubMed] [Google Scholar]
- 13.Krasinskas A.M., Bartlett D.L., Cieply K., Dacic S. CDKN2A and MTAP deletions in peritoneal mesotheliomas are correlated with loss of p16 protein expression and poor survival. Mod. Pathol. 2010;23:531–538. doi: 10.1038/modpathol.2009.186. [DOI] [PubMed] [Google Scholar]
- 14.Hustinx S.R., Leoni L.M., Yeo C.J., Brown P.N., Goggins M., Kern S.E., Hruban R.H., Maitra A. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: Evidence of homozygous deletion in a noninvasive precursor lesion. Mod. Pathol. 2005;18:959–963. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 15.Zimling Z.G., Jørgensen A., Santoni-Rugiu E. The diagnostic value of immunohistochemically detected methylthioadenosine phosphorylase deficiency in malignant pleural mesotheliomas. Histopathology. 2012;60:E96–E105. doi: 10.1111/j.1365-2559.2012.04196.x. [DOI] [PubMed] [Google Scholar]
- 16.Hida T., Hamasaki M., Matsumoto S., Sato A., Tsujimura T., Kawahara K., Iwasaki A., Okamoto T., Oda Y., Honda H., et al. Immunohistochemical detection of MTAP and BAP1 protein loss for mesothelioma diagnosis: Comparison with 9p21 FISH and BAP1 immunohistochemistry. Lung Cancer. 2017;104:98–105. doi: 10.1016/j.lungcan.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Churg A., Nabeshima K., Ali G., Bruno R., Fernandez-Cuesta L., Galateau-Salle F. Highlights of the 14th international mesothelioma interest group meeting: Pathologic separation of benign from malignant mesothelial proliferations and histologic/molecular analysis of malignant mesothelioma subtypes. Lung Cancer. 2018;124:95–101. doi: 10.1016/j.lungcan.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 18.Chapel D.B., Schulte J.J., Berg K., Churg A., Dacic S., Fitzpatrick C., Galateau-Salle F., Hiroshima K., Krausz T., Le Stang N., et al. MTAP immunohistochemistry is an accurate and reproducible surrogate for CDKN2A fluorescence in situ hybridization in diagnosis of malignant pleural mesothelioma. Mod. Pathol. 2020;33:245–254. doi: 10.1038/s41379-019-0310-0. [DOI] [PubMed] [Google Scholar]
- 19.Brcic L., Le Stang N., Gallob F., Pissaloux D., Sequeiros R., Paindavoine S., Pairon J.C., Karanian M., Dacic S., Girard N., et al. A Combination of MTAP and p16 Immunohistochemistry Can Substitute for CDKN2A Fluorescence In Situ Hybridization in Diagnosis and Prognosis of Pleural Mesotheliomas. Arch. Pathol. Lab. Med. 2023;147:313–322. doi: 10.5858/arpa.2021-0331-OA. [DOI] [PubMed] [Google Scholar]
- 20.Sauter J.L., Dacic S., Galateau-Salle F., Attanoos R.L., Butnor K.J., Churg A., Husain A.N., Kadota K., Khoor A., Nicholson A.G., et al. The 2021 WHO Classification of Tumors of the Pleura: Advances Since the 2015 Classification. J. Thorac. Oncol. 2022;17:608–622. doi: 10.1016/j.jtho.2021.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Hinterberger M., Reineke T., Storz M., Weder W., Vogt P., Moch H. D2-40 and calretinin—A tissue microarray analysis of 341 malignant mesotheliomas with emphasis on sarcomatoid differentiation. Mod. Pathol. 2007;20:248–255. doi: 10.1038/modpathol.3800736. [DOI] [PubMed] [Google Scholar]
- 22.Chapel D.B., Dubuc A.M., Hornick J.L., Sholl L.M. Correlation of methylthioadenosine phosphorylase (MTAP) protein expression with MTAP and CDKN2A copy number in malignant pleural mesothelioma. Histopathology. 2021;78:1032–1042. doi: 10.1111/his.14324. [DOI] [PubMed] [Google Scholar]
- 23.Chung C.T., Gda C.S., Hwang D.M., Ludkovski O., Pintilie M., Squire J.A., Tsao M.S. FISH assay development for the detection of p16/CDKN2A deletion in malignant pleural mesothelioma. J. Clin. Pathol. 2010;63:630–634. doi: 10.1136/jcp.2010.076794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oehl K., Vrugt B., Opitz I., Meerang M. Heterogeneity in Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2018;19:1603. doi: 10.3390/ijms19061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D., Hiroshima K., Matsumoto S., Nabeshima K., Yusa T., Ozaki D., Fujino M., Yamakawa H., Nakatani Y., Tada Y., et al. Diagnostic usefullness of p16/CDKN2A FISH in distinguishing sarcomatoid mesotheliomaand fibrous pleuritis. Am. J. Clin. Pathol. 2013;139:39–46. doi: 10.1309/AJCPT94JVWIHBKRD. [DOI] [PubMed] [Google Scholar]
- 26.Destro A., Ceresoli G.L., Baryshnikova E., Garassino I., Zucali P.A., De Vincenzo F., Bianchi P., Morenghi E., Testori A., Alloisio M., et al. Gene methylation in pleural mesothelioma: Correlations with clinico-pathological features and patient’s follow-up. Lung Cancer. 2008;59:369–376. doi: 10.1016/j.lungcan.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi N., Toyooka S., Yanai H., Soh J., Fujimoto N., Yamamoto H., Ichihara S., Kimura K., Ichimura K., Sano Y., et al. Frequent p16 inactivation by homozygous deletion or methylation is associated with a poor prognosis in Japanese patients with pleural mesothelioma. Lung Cancer. 2008;62:120–125. doi: 10.1016/j.lungcan.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Behrmann I., Wallner S., Komyod W., Heinrich P.C., Schuierer M., Buettner R., Bosserhoff A.K. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am. J. Pathol. 2003;163:683–690. doi: 10.1016/S0002-9440(10)63695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadariya Y., Yin B., Tang B., Shinton S.A., Quinlivan E.P., Hua X., Klein-Szanto A., Al-Saleem T.I., Bassing C.H., Hardy R.R., et al. Mice heterozygous for germ-line mutations in methylthioadenosine phosphorylase (MTAP) die prematurely of T-cell lymphoma. Cancer Res. 2009;69:5961–5969. doi: 10.1158/0008-5472.CAN-09-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.