Abstract
A bacterial mixed culture reductively dechlorinating trichlorobenzenes was established in a defined, synthetic mineral medium without any complex additions and with pyruvate as the carbon and energy source. The culture was maintained over 39 consecutive transfers of small inocula into fresh media, enriching the dechlorinating activity. In situ probing with fluorescence-labeled rRNA-targeted oligonucleotide probes revealed that two major subpopulations within the microbial consortium were phylogenetically affiliated with a sublineage within the Desulfovibrionaceae and the gamma subclass of Proteobacteria. The bacterial consortium grew by fermentation of pyruvate, forming acetate, propionate, CO2, formate, and hydrogen. Acetate and propionate supported neither the reduction of trichlorobenzenes nor the reduction of sulfate when sulfate was present. Hydrogen and formate were used for sulfate reduction to sulfide. Sulfate strongly inhibited the reductive dechlorination of trichlorobenzenes. However, when sulfate was depleted in the medium due to sulfate reduction, dechlorination of trichlorobenzenes started. Similar results were obtained when sulfite was present in the cultures. Molybdate at a concentration of 1 mM strongly inhibited the dechlorination of trichlorobenzenes. Cultures supplied with molybdate plus sulfate did not reduce sulfate, but dechlorination of trichlorobenzenes occurred. Supplementation of electron-depleted cultures with various electron sources demonstrated that formate was used as a direct electron donor for reductive dechlorination, whereas hydrogen was not.
Chlorobenzenes are widespread pollutants and accumulate in the food chain due to their hydrophobicity and strong persistence against chemical and microbial degradation (34). Anaerobic reductive dechlorination of chlorinated benzenes was demonstrated for enrichment cultures from biofilm reactors, sewage sludge, river sediment, and soil (3, 4, 15, 16, 22, 31, 37). Dechlorination pathways for all multiply chlorinated benzenes were elucidated (4, 15). Some dechlorination patterns can be rationalized by thermodynamic considerations (3, 13), but little is known about the microorganisms participating in chlorobenzene dechlorination.
Anaerobic bacteria transforming chlorobenzoates and/or chlorophenols have been isolated in pure cultures (5, 7, 18, 27, 39, 40, 45, 48). Desulfomonile tiedjei (12), strain 2CP-1 (7), Desulfitobacterium chlororespirans (39), and Desulfitobacterium sp. strain PCE1 (18) grow anaerobically by chlororespiration. So far, it has not been possible to evaluate whether the anaerobic dechlorination of chlorobenzenes proceeds via a similar mechanism, since pure cultures are not available.
While the effect of oxygen and nitrate on the dechlorination of chloroaromatics is reported to be negative for most cultures (32), the effect of sulfur oxyanions is controversial. Some reports stated an inhibitory role of sulfate in the reductive dehalogenation of various chlorinated or fluorinated aromatics (17, 19, 25, 26); other studies found only slight inhibition (24), no inhibition (14), or even a stimulated rate of dechlorination (17, 23). For one mixed culture, the mineralization of chlorophenols was concomitantly coupled to the reduction of sulfur oxyanions (20, 21). With pure cultures of D. tiedjei, it could be shown that sulfite and thiosulfate inhibited the dechlorination of 3-chlorobenzoate in growing cells, nongrowing cells, and cell extracts, while sulfate inhibited dechlorination only in growing cells (46).
The high toxicity (22) and the low solubility of chlorobenzenes in water prevented the successful isolation of bacteria with chlorobenzenes as electron acceptors. It is therefore essential to study alternative electron acceptors that could be used by chlorobenzene-dechlorinating bacteria and that could substitute for chlorobenzenes during enrichment and isolation. Information about reductive dechlorination of chlorobenzenes in the presence of other electron acceptors is also needed for the evaluation of dechlorination processes at natural sites and for in situ remediation projects. To our knowledge, detailed studies of the effects of alternative electron acceptors on the dechlorination of chlorobenzenes have not been reported so far.
The aim of the present study was to describe the physiological properties of a mixed culture effectively dechlorinating trichlorobenzenes and to determine the effects of various specific inhibitors and alternative electron acceptors. For these experiments, we used a stable, sediment-free mixed consortium growing in a defined, synthetic mineral medium. This consortium has been established in our laboratory from a fluidized bed bioreactor (1, 33) and reductively dechlorinates 1,2,3-trichlorobenzene to 1,3-dichlorobenzene and 1,2,4-trichlorobenzene to 1,4- and 1,3-dichlorobenzene. By inhibiting the activity of methanogenic bacteria using the specific inhibitor bromoethanesulfonate (BES), we showed that dechlorination occurs independently from methanogenic bacteria (1), as has also been shown for other enrichment cultures dechlorinating chlorobenzenes (22, 31).
MATERIALS AND METHODS
Chemicals.
1,2,3- and 1,2,4-trichlorobenzene were obtained from E. Merck AG (Darmstadt, Federal Republic of Germany [FRG]), and 2,4-dichlorotoluene was obtained from Aldrich (Steinhofen, FRG). Titanium(III) chloride (synthesis-grade solution) was obtained from Merck-Schuchard (Hohenbrunn, FRG). All other chemicals used were at least of analytical grade and were purchased from Sigma (Deisenhofen, FRG) or Merck. Gases were obtained in 99.999% (vol/vol) (N2 and H2) or 99.8% (vol/vol) (CO2) quality from Linde (Berlin, FRG); traces of oxygen were removed by use of a reduction column (Ochs, Göttingen, FRG).
Inoculum and culture conditions.
An inoculum obtained from a fluidized bed bioreactor (33) was used to establish a dechlorinating mixed culture in a sulfide-reduced, synthetic medium (1). The medium used for all experiments in the present study was a sulfur-limited, synthetic, bicarbonate-buffered mineral medium that was reduced with 0.8 mM Ti(III) citrate (49) unless stated otherwise. In some experiments, 1 mM Na2S was used as the reducing agent instead of Ti(III) citrate. Stock solutions of sulfide, sulfite, Ti(III) citrate, and pyruvate were made with anoxic water, sterilized, and stored under a nitrogen atmosphere. The basal medium contained (in grams per liter of deionized water): NaCl, 1; KH2PO4, 0.2; NH4Cl, 0.27; MgCl2 · 6H2O, 0.41; KCl, 0.52; and CaCl2 · 2H2O, 0.15. After autoclaving, the medium was cooled under an N2–CO2 (4:1, vol/vol) atmosphere to 60°C. 1,2,3-Trichlorobenzene was dissolved in 1,2,4-trichlorobenzene to obtain an equimolar solution, and 16.3 μl of this mixture was added per liter of medium. The medium was subsequently stirred for at least 24 h, resulting in a final concentration of 15 to 20 μM for each trichlorobenzene. To minimize chlorobenzene losses, all tubes, sealings, and valves were made of glass or Teflon. After cooling, NaHCO3 was added to a final concentration of 2.5 g/liter, and the medium was supplemented with 0.1% (vol/vol) trace element solution SL9 (47), 0.05% (vol/vol) vitamin solution, 0.1% (vol/vol) selenite-tungstate solution (47), and substrate solutions. The vitamin solution (36) was modified to contain 40 mg of p-aminobenzoate, 10 mg of biotin, 100 mg of nicotinic acid, 50 mg of pantothenic acid, 150 mg of pyridoxine, 100 mg of thiamine, and 100 mg of cobalamin per liter. Sulfur was present in the medium as a contaminant and was calculated from protein yields to be present at a concentration of about 3 μM (9). All additions were prepared aseptically under anoxic conditions. The pH was adjusted to 7.0 to 7.2 with sterile, anoxic HCl. Resazurin (0.5 mg/liter) was used as a redox indicator. Fifty or 30 ml of medium were placed in 100- or 60-ml serum bottles, respectively, and the reducing agent was added [0.8 mM Ti(III) or 1 mM Na2S]. The headspace was flushed with N2–CO2 (4:1, vol/vol), and the bottles were sealed with Teflon-lined butyl rubber septa and aluminum crimp caps. Prior to inoculation, the actual trichlorobenzene concentrations were determined. Pyruvate was added as a carbon and energy source at a final concentration of 10 mM. Hydrogen was added by injecting 5 ml of hydrogen with a sterile syringe, equivalent to 7.5 mM in 30 ml of liquid. When needed, the specific inhibitors BES and molybdate (35) were added at final concentrations of 4 and 2 mM, respectively. The inoculation was done anoxically after the medium was equilibrated with the reducing agent [Ti(III) citrate for 2 h or Na2S for at least 24 h] by use of glass syringes that had been flushed with sterile water to seal the syringes and to remove air bubbles. The inoculation volumes were 1 ml for 50-ml cultures and 0.5 ml for 30-ml cultures. Subcultures were established in fresh medium every 14 days. The first 9 transfers were done in sulfide-reduced medium (1), and 30 further transfers were done in Ti(III) citrate-reduced medium. Cultures were incubated statically at 28°C in the dark. Pasteurization was done at 80°C for 30 min. For exposure to a positive redox potential, the inoculum was taken up with a syringe, and air was sucked through the liquid until the redox indicator turned pink. After 60 s, the inoculum was injected into a culture vessel containing reduced medium.
Analytical procedures.
Chlorobenzene concentrations were determined by removing 1-ml aliquots of bacterial suspension from a culture bottle with a glass syringe, followed by extraction with 1 ml of hexane. Analysis of the extracts was done by gas chromatography (GC)-flame ionization detection with 2,4-dichlorotoluene as an internal standard (44). Dechlorination was expressed as the percentage of dichlorobenzenes formed relative to the sum of all chlorobenzenes within a culture vessel. For quantification of bacterial growth, cells were harvested by centrifugation (15 min, 10,000 × g), washed with phosphate-buffered saline (130 mM NaCl, 12 mM Na2HPO4/NaH2PO4 [pH 7.4]), and resuspended in sterile water. Protein concentrations were determined by the bicinchoninic acid procedure (41) with bovine serum albumin as the standard. Pyruvate, lactate, and formate concentrations were determined enzymatically (2). Hydrogen, methane, and hydrogen sulfide in the gas phase were analyzed by GC and thermal conductivity detection with a packed column (12 by 0.125 in. [ca. 30.5 by 0.32 cm]; inside diameter, 2 mm; Chromosorb 102; 60/80 mesh; Macherey & Nagel, Düren, FRG). Operating conditions were as follows: 50°C isotherm; carrier gas, nitrogen; inlet pressure, 200 kPa; detector current, 85 mA; injection volume, 10 μl; and split, none. The lower limit of detection of hydrogen was 0.05% (vol/vol), which corresponds to a nominal concentration of 27 μM. Nominal concentrations of methane and hydrogen were calculated by assuming that the amount of gases present within the gas phase was completely dissolved in the volume of the liquid phase. Gas pressure was measured by use of a piezoelectric sensor (Konrad, Berlin, FRG) with a sensitivity of 1 kPa. Acetate, propionate, and butyrate were quantified by GC with a Shimadzu model 14B apparatus equipped with a Permabond-FFAP column (25 m by 0.25 μm; inside diameter, 0.25 mm; Macherey & Nagel) and flame ionization detection. Operating conditions were as follows: 150°C for 5 min, increasing by 5°C/min to 170°C for 3 min; carrier gas, nitrogen; inlet pressure, 60 kPa; split, 1:50. Sulfate was quantified after precipitation with BaCl2 according to Madsen and Aamand (26). Sulfide was quantified as H2S in the gas phase (see above) and as free sulfide in the liquid phase by CuS precipitation (8). Standards were prepared by anoxically distributing standard amounts of Na2S into 60-ml serum bottles containing 30 ml of medium. Measurements of the standards were taken after 24 h at 28°C to allow equilibration between the gas and liquid phases.
In situ hybridization.
For in situ characterization of the bacterial consortium, 16S and 23S rRNA-targeted, fluorescence-labeled oligonucleotides were applied according to Manz et al. (28). The oligonucleotides used in this study were (i) EUB338, specific for the domain Bacteria (43); (ii) ARCH915, complementary to a region of the 16S rRNA conserved in the domain Archaea (42); (iii) ALF1b, BET42a, and GAM42a, specific for the alpha, beta, and gamma subclasses of Proteobacteria, respectively (28); (iv) CF319a/b, specific for the flavobacter-cytophaga group (29); (v) HGC, specific for gram-positive bacteria with a high G+C content of DNA (38); (vi) a comprehensive set of probes specific for the different lineages of mesophilic sulfate-reducing bacteria affiliated with the delta subclass of Proteobacteria, including probes specific for phylogenetic groups within the Desulfovibrionaceae (DSV698 and DSV1292) and a probe specific for a branch consisting of Desulfoarculus baarsii and D. tiedjei (DSMA488) (30); (vii) DMT273, a species-specific probe designed for in situ detection of D. tiedjei (5′-GCT AAC CAT CTC GGC CTT-3′; Escherichia coli positions 273 to 290); and (viii) non-EUB338, complementary to EUB338, serving as a negative control for nonspecific binding.
All probes were purchased 5′ labeled with the indocarbocyanine dye CY3 (Biometra, Göttingen, FRG). Fluorescence was detected by epifluorescence microscopy with a Zeiss (Oberkochen, FRG) Axioskop equipped with light filter 41007 (AF Analysentechnik, Tübingen, FRG) for CY3-labeled probes (excitation, 535 to 550 nm; dichroic mirror, 565 nm; emission, 610 to 675 nm). Epifluorescence microscopy was also used for direct detection of living methanogens containing cofactor F420 (Zeiss light filter set 05; excitation, 395 to 440 nm; dichroic mirror, 460 nm; emission, 470 nm) and for direct cell counting after staining with the fluorochrome 4′,6-diamidino-2-phenyl-indole-dihydrochloride (Zeiss light filter set 01; excitation, 365 nm; dichroic mirror, 395 nm; emission, 397 nm).
RESULTS
Dechlorination in a defined medium reduced by Ti(III).
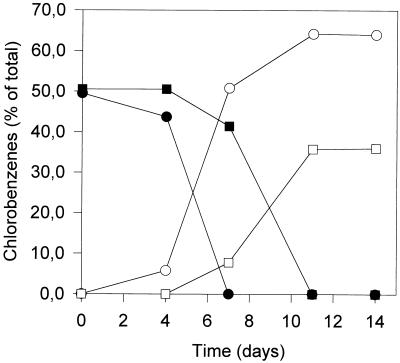
The defined synthetic medium used for the dechlorinating mixed culture described previously (1) was modified by use of Ti(III) as a reducing agent instead of sulfide. When this sulfur-limited medium was used, the number of subcultures which lost their dechlorinating activity was reduced considerably. In addition, the dechlorinating activity increased. A mixture of 20 μM 1,2,3-trichlorobenzene and 20 μM 1,2,4-trichlorobenzene was dechlorinated within 10 to 14 days after inoculation (Fig. 1), while in sulfide-reduced medium, dechlorination was complete only after 21 days. Abiotic reduction of trichlorobenzenes by Ti(III) was excluded by preparing Ti(III)-reduced, noninoculated controls. In these assays, no dechlorination products were detected. Inocula autoclaved or exposed to a positive redox potential for 60 s lost their dechlorinating activity completely.
FIG. 1.
Dechlorination of trichlorobenzenes by a culture growing from a small inoculum with 10 mM pyruvate as the carbon and energy source in Ti(III)-reduced medium. Symbols: •, 1,2,3-trichlorobenzene; ▪, 1,2,4-trichlorobenzene; ○, 1,3-dichlorobenzene; □, 1,4-dichlorobenzene.
Physiological activities.
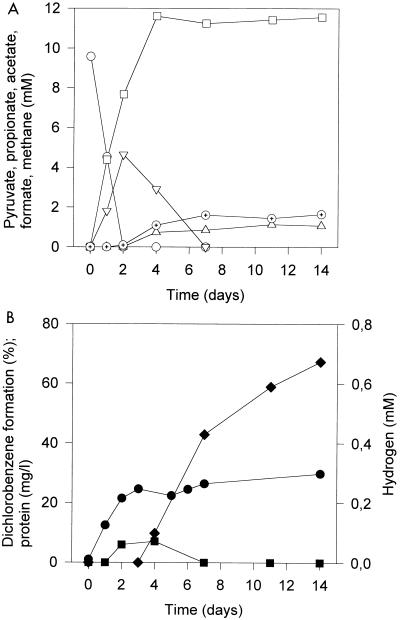
In Ti(III)-reduced medium, the pyruvate concentration decreased rapidly during the first 48 h of incubation (Fig. 2). Concurrently, acetate, formate, and hydrogen were formed, whereas lactate and butyrate were not detected. The main increase in bacterial biomass, measured as the amount of cell protein, also occurred during the first 48 h of incubation. Thereafter, the formate concentration decreased, methane and propionate were formed, and the dechlorination of trichlorobenzenes to dichlorobenzenes started. It is noteworthy that more acetate was produced than pyruvate was added. Acetate was not further oxidized. The hydrogen partial pressure did not reach values above 0.15% the gas phase (nominal concentration of about 60 μM), while formate concentrations reached 4.5 mM. The incubation time after which the dechlorination started varied with the batch of medium used. Therefore, all cultures within one experiment were set up from the same batch of medium, and the results were evaluated with respect to the cultures under standard conditions.
FIG. 2.
Concentrations of metabolites, growth, and dichlorobenzene formation in cultures initially supplied with 10 mM pyruvate in Ti(III)-reduced medium containing 20 μM sulfide as a source of sulfur. Symbols: (A) ○, pyruvate; □, acetate; ▿, formate; ▵, propionate; ⊕, methane; (B) ⧫, formation of dichlorobenzenes; •, protein; ▪, hydrogen. Methane and hydrogen data are given as the nominal concentrations. Data are means of triplicate cultures; standard deviations were below 10%.
Specific inhibition.
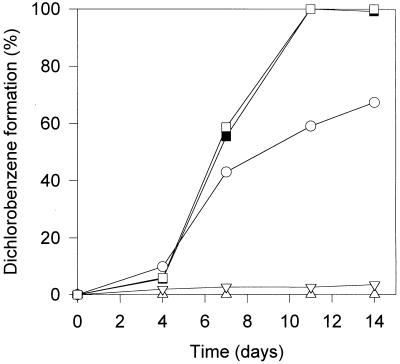
The addition of 4 mM BES to the culture medium resulted in a significant increase in the extent of trichlorobenzene dechlorination after 14 days of incubation (Fig. 3). Supplementation with BES and hydrogen did not lead to faster dechlorination than that with BES alone. The addition of 2 mM molybdate resulted in a drastic drop in dechlorinating activity to less than 5% after 14 days of incubation (Fig. 3). This strong inhibition by 2 mM molybdate was also observed in cultures also containing 1 mM sulfide as a source of sulfur.
FIG. 3.
Effect of specific inhibitors on the dechlorination of trichlorobenzenes in Ti(III)-reduced medium containing 10 mM pyruvate. Symbols: ○, control; □, 4 mM BES; ▪, 4 mM BES plus 15% (vol/vol) hydrogen; ▿, 2 mM molybdate; ▵, negative control, not inoculated. Data are means of triplicate cultures; standard deviations were below 10%.
When BES was added, the rate of pyruvate fermentation remained unchanged. However, no methane was produced, and the hydrogen partial pressure increased up to 1% the gas phase (nominal concentration of about 0.4 mM) at day 7 after inoculation. Formate production was comparable to that in cultures not amended with BES, but consumption of formate was slower. The same pattern was observed in molybdate-supplemented cultures: pyruvate fermentation was similar to that in cultures not inhibited by molybdate, but formate and hydrogen were only slowly consumed.
To investigate the effects of BES, hydrogen, and low sulfate concentrations on the dechlorination of trichlorobenzenes, cultures were set up with combinations of 4 mM BES, 2 mM sulfate, and hydrogen (nominal concentration of 7.5 mM). These cultures were analyzed for dechlorination products and gas composition after 7 days of incubation (Table 1). An analysis of gas composition confirmed the complete inhibition of methanogenesis in the presence of BES. When sulfate was added, sulfide was produced. When methanogenesis was not inhibited and sulfate was present, no hydrogen could be detected at day 7. However, dichlorobenzenes were formed. In none of the possible combinations did the addition of hydrogen lead to a significant increase in dechlorinating activity. BES (4 mM) or 2 mM sulfate increased the extent of dechlorination, but the simultaneous addition of 4 mM BES and 2 mM sulfate did not lead to a higher extent of dechlorination than that in cultures without supplements.
TABLE 1.
Trichlorobenzene dechlorination and gas production in cultures supplied with 10 mM pyruvate and different combinations of 4 mM BES, 2 mM sulfate, and 7.5 mM hydrogena
| Additionb
|
Analysis after 7 days of incubationc
|
|||||
|---|---|---|---|---|---|---|
| BES | SO4 | H2 | Dechlorinationd | CH4 (mM)a | H2 (mM)a | Sulfide (mM)e |
| − | − | − | 1.1 ± 0.25 | 0.06 ± 0.02 | 0.03 ± 0.01 | 0.0 ± 0 |
| − | − | + | 0.3 ± 0.46 | 0.03 ± 0.01 | 4.31 ± 0.25 | 0.0 ± 0 |
| − | + | − | 10.4 ± 1.61 | 0.49 ± 0.08 | 0.0 ± 0 | 1.7 ± 0.31 |
| − | + | + | 8.5 ± 3.04 | 1.00 ± 0.07 | 2.35 ± 0.14 | 1.9 ± 0.13 |
| + | − | − | 41.4 ± 15.9 | 0.0 ± 0 | 0.23 ± 0.02 | 0.0 ± 0 |
| + | − | + | 50.1 ± 0.96 | 0.0 ± 0 | 3.37 ± 0.21 | 0.0 ± 0 |
| + | + | − | 1.2 ± 0.12 | 0.0 ± 0 | 0.59 ± 0.03 | 1.5 ± 0.25 |
| + | + | + | 1.3 ± 0.49 | 0.0 ± 0 | 3.53 ± 0.17 | 1.5 ± 0.28 |
Nominal concentration.
+, present; −, absent.
Means of triplicate cultures ± standard deviations.
Dichlorobenzenes as a percentage of the total amount of chlorobenzenes.
Sum of sulfides in the liquid phase and H2S in the gas phase.
The addition to the medium of penicillin G at concentrations of up to 10 μg/ml did not influence the dechlorination of trichlorobenzenes. Pasteurization of cultures or culture inocula resulted in a complete loss of dechlorinating activity.
Bacterial composition.
Cultures grown in Ti(III)-reduced medium without inhibitors were composed of three dominant morphologically different bacterial subpopulations and several other specimens, which contributed in minor amounts to the bacterial consortium (Table 2). Bacteria of all three dominant morphologies grew rapidly during the first 2 days of cultivation. Between days 2 and 21 after inoculation, neither the total cell counts nor the proportions of the major subpopulations changed significantly. It was not possible to detect population changes that were linked with the dechlorination of trichlorobenzenes in the medium. One of the major subpopulations was formed by small, motile vibrios. Cells of this morphotype were not inhibited by BES, but molybdate had a strong inhibitory effect. The percentage of this population dropped from about 25% to below 2% of the total cell counts in cultures containing 2 mM molybdate. In situ hybridization with the probes DSV1292 and DSV698 resulted in the emission of strong probe-conferred fluorescence from these cells. The two other main subpopulations in the consortium were characterized microscopically as coccoid and small, rod-shaped morphotypes, neither of which was inhibited by the addition of BES or molybdate. Hybridization with probes encompassing all gram-negative, mesophilic sulfate-reducing bacteria (30) did not result in positively stained cells of the coccoid and small, rod-shaped morphotypes. The small, rod-shaped bacteria were shown to be affiliated with the gamma subclass of Proteobacteria by use of the fluorescent probe GAM42a.
TABLE 2.
Characterization of dominant members of the stable mixed culture by light microscopy, in situ hybridization, and inhibitor studiesa
| Morphotype | Relative abundance (%) | Growth in the presence of:
|
Gram stain reaction | Hybridization positive with probe(s) | |
|---|---|---|---|---|---|
| BES | Molybdate | ||||
| Vibrio | 25 | Yes | No | Negative | EUB338, DSV1292, and DSV698 |
| Coccus | 40 | Yes | Yes | Positive | EUB338 |
| Rod | 30 | Yes | Yes | Negative | EUB338 and GAM42a |
| Long rod | <5 | No | Yes | Negative | ARCH915 |
Data shown are for cultures after 7 days of incubation.
Long rods, forming a minor subpopulation in cultures not amended with BES, were identified as methanogenic members of the domain Archaea by detection of autofluorescence after excitation at 420 nm and strong epifluorescence signals after in situ hybridization with probe ARCH915; these cells were not present in cultures containing BES.
No epifluorescence signals were obtained after hybridization of the mixed culture with the 16S rRNA-targeted probes DSMA488 and DMT273 or other probes designed for the detection of different bacterial lineages within the Desulfobacteriaceae (30).
Elimination of methanogenic bacteria from the culture.
Since the dechlorinating activity increased considerably in the presence of BES, the consortium was transferred successively for three times in medium containing 4 mM BES. As a result, none of the succeeding cultures, used for all of the following experiments, showed methanogenesis or contained subpopulations of methanogenic bacteria.
Effect of sulfur oxyanions on trichlorobenzene dechlorination.
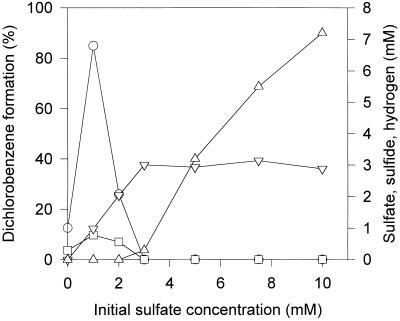
Ti(III)-reduced medium containing 10 mM pyruvate was supplemented with different concentrations of sulfur oxyanions and inoculated with an actively dechlorinating mixed culture. Figure 4 shows the extent of dechlorination as well as hydrogen, sulfate, and sulfide concentrations after 7 days of incubation for cultures supplied with sulfate. Initial sulfate concentrations of 1 or 2 mM increased the dechlorinating activity over that in cultures without sulfate, whereas initial sulfate concentrations above 2 mM inhibited the dechlorination of trichlorobenzenes completely. In cultures that were initially supplied with 1 or 2 mM sulfate, all sulfate was reduced to sulfide after 7 days of incubation. Sulfate was still present in cultures supplied with 3 to 10 mM initial sulfate concentrations. A parallel experiment was performed with various sulfite concentrations. Low concentrations (1 or 2 mM) increased while higher amounts (4 mM or more) inhibited the dechlorinating activity. In further experiments, the sulfate concentration was varied in the presence of 1 mM sulfide to exclude effects due to sulfur limitation at low sulfate concentrations. In these experiments, dechlorination was much faster with 1 mM sulfate than in cultures without sulfate, and high sulfate concentrations inhibited the dechlorination of trichlorobenzenes completely.
FIG. 4.
Dechlorination of trichlorobenzenes depends on the initial sulfate concentration. All cultures were Ti(III) reduced and contained 10 mM pyruvate. Symbols: ○, dichlorobenzene formation; □, hydrogen; ▿, sulfide; ▵, sulfate. Hydrogen data are given as nominal concentrations. Data represent values 7 days after inoculation and are means of triplicate cultures; standard deviations were below 10%.
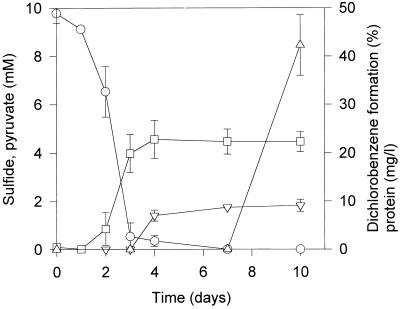
The physiological activities of cultures supplemented with 2 mM sulfate were monitored over 10 days (Fig. 5). The formation of sulfide started when pyruvate fermentation was almost finished, and reductive dechlorination occurred only after the depletion of sulfate in the medium. An increase in biomass was detected during pyruvate fermentation but not during sulfate reduction. An increase in biomass due to the reduction of trichlorobenzenes could not be detected.
FIG. 5.
Metabolic activities in a Ti(III)-reduced culture containing 10 mM pyruvate and 2 mM sulfate. Symbols: ○, pyruvate; □, protein; ▿, sulfide; ▵, formation of dichlorobenzenes. Data are means of triplicate cultures ± standard deviations.
Both molybdate and sulfate had strong effects on the dechlorination of trichlorobenzenes. A series of cultures with various concentrations of both effectors was analyzed for sulfate and trichlorobenzene reduction (Table 3). The production of sulfide confirmed the potential of sulfate reduction when no molybdate was present. The reduction of sulfate was completely inhibited by 1 or 3 mM molybdate. Trichlorobenzene reduction was inhibited by 3 mM molybdate in the presence of 0 to 3 mM sulfate or by 1 mM molybdate when no sulfate was present. Sulfate at a concentration of 1 or 3 mM neutralized the inhibition of dechlorination by 1 mM molybdate. As stated before, high sulfate concentrations prevented dechlorination in the absence of molybdate. However, the inhibition of chlorobenzene dechlorination by 3 mM sulfate was abolished by the addition of 1 mM molybdate.
TABLE 3.
Reductive dechlorination of trichlorobenzenes to dichlorobenzenes (DCB) and reduction of sulfate to sulfide after 14 days of incubation in cultures supplied with different concentrations of sulfate and molybdatea
| Initial sulfate concn (mM) | Analysis with initial molybdate concn (mM) of:
|
|||||
|---|---|---|---|---|---|---|
| 0
|
1
|
3
|
||||
| DCB (%) | Sulfide (mM) | DCB (%) | Sulfide (mM) | DCB (%) | Sulfide (mM) | |
| 0 | 99.5 | 0.0 | 0.6 | 0.0 | 0.1 | 0.0 |
| 1 | 100.0 | 1.0 | 99.1 | 0.0 | 0.4 | 0.0 |
| 3 | 5.2 | 2.5 | 96.0 | 0.0 | 0.4 | 0.0 |
Means of triplicate cultures; standard deviations were below 10%.
Electron donors.
To determine which of the different metabolic products from pyruvate fermentation was used as an electron donor for reductive dechlorination, cultures were set up with 10 mM pyruvate as the electron and carbon source. The addition of 3 mM sulfate resulted in the depletion of electron equivalents and consequently prevented reductive dechlorination (Table 4). After 2 weeks of incubation, it was confirmed that no dichlorobenzenes had been formed, and no hydrogen, formate, or pyruvate was available to the bacteria. At this time, one of the following additional electron donors was injected into the cultures: none; hydrogen (nominal concentration of 7.5 mM); formate, acetate, or pyruvate (10 mM each). The concentrations were high enough to allow complete reduction of the remaining sulfate ions and trichlorobenzenes. Finally, after a further 3 weeks of incubation, the cultures were analyzed again for dichlorobenzene formation. This experiment revealed that formate was readily used as an electron donor for reductive dechlorination of trichlorobenzenes, whereas hydrogen or acetate was not.
TABLE 4.
Dechlorination of trichlorobenzenes before and after supplementation with additional electron donorsa
| Dichlorobenzene formationb 2 wk after inoculation | Additional electron donor injected 2 wk after inoculation | Dichlorobenzene formationb 5 wk after inoculation |
|---|---|---|
| 0 | None | 0 |
| 0 | Hydrogen | 0 |
| 0 | Formate | 100 ± 0 |
| 0 | Acetate | 0 |
| 0 | Pyruvate | 61 ± 4.0 |
Cultures were initially supplied with 10 mM pyruvate and 3 mM sulfate.
Means of at least duplicate cultures ± standard deviations, reported as a percentage of the total amount of chlorobenzenes.
DISCUSSION
The successful isolation of a chlorobenzene-dechlorinating anaerobic bacterium in a pure culture has not been reported so far. A major problem in the enrichment and isolation of chlorobenzene-dechlorinating bacteria is to provide enough chlorobenzene in a water phase to sustain growth based on reductive dechlorination without reaching toxic levels. Holliger et al. (22) observed no dechlorination with concentrations higher than 40 μM 1,2,3-trichlorobenzene or 70 μM 1,3-dichlorobenzene. In our experiments, no dechlorination was found with 1,2,3-, or 1,2,4-trichlorobenzene concentrations exceeding 30 μM. The present study was therefore directed to determine the physiological activities of a dechlorinating culture and to evaluate selective enrichment conditions for chlorobenzene-dechlorinating bacteria. Further intentions of the study were the determination of the conditions under which dechlorination occurs, the identification of the actual electron donor, and the determination of the effects of specific bacterial inhibitors on the dechlorination process. A prerequisite to addressing these questions was the establishment of a stable, rapidly growing, and reproducibly dechlorinating culture in a defined medium. This kind of culture was obtained by use of a medium with a low sulfur concentration, with Ti(III) citrate as a reductant, and with pyruvate as a fermentable substrate.
The use of BES for several transfers was successful in eliminating methanogenesis from the culture. The stimulating effect of BES on trichlorobenzene dechlorination may be due to the elimination of methanogenic bacteria, which compete with dechlorinating bacteria for electron donors, or to a release of sulfur limitation caused by the use of BES as a source of sulfur.
The effects of sulfur oxyanions on the reductive dechlorination of chloroaromatic compounds in mixed cultures and in pure cultures of D. tiedjei are complex (see the introduction). Also, the capabilities of bacteria dechlorinating chloroaromatics to grow by use of sulfur oxyanions as terminal electron acceptors differ strongly. While D. tiedjei can grow by the reduction of sulfate, sulfite, or thiosulfate (11), dechlorinating Desulfitobacterium spp. use sulfite and thiosulfate but not sulfate as a terminal electron acceptor (5, 6, 18, 39, 48). This physiological difference corresponds to the phylogenetic distance between the two taxa. The myxobacterial isolate 2CP-1 does not use any of the sulfur oxyanions as a terminal electron acceptor (7). Since many of the dechlorinating bacteria use sulfur oxyanions as alternative electron acceptors, we investigated the effect of sulfur oxyanions on our trichlorobenzene-dechlorinating mixed culture in detail.
Within our consortium, the fermentation of pyruvate, sulfate reduction, and trichlorobenzene dechlorination occur strictly in succession. Dechlorination starts only after all of the sulfate is reduced to sulfide, and only one pair of electrons per molecule of pyruvate is used for sulfate reduction. The lack of trichlorobenzene dechlorination in the presence of sulfate may be due to interspecies competition for electrons between sulfate-reducing and dechlorinating bacteria or to intracellular channeling of electrons from the reductive dechlorinating to the sulfate-reducing enzyme systems within one organism (32). In the first case, the presence of sulfate should result in a growth-inhibiting effect on the dechlorinating bacteria. In the second case, sulfate should prevent the dechlorination reaction. The stimulating effect of sulfate and sulfite at low concentrations may be explained by stimulated growth of the dechlorinating bacteria due to sulfate or sulfite reduction. The effect cannot be explained solely by the release of sulfur limitation, since sulfate at a concentration of 1 mM stimulated dechlorination even when 1 mM sulfide was present. Our study further shows that the chlorobenzene-dechlorinating bacteria are not irreversibly inactivated by the presence of sulfate.
An inhibitory effect of molybdate on the dechlorination of chloroaromatic compounds was previously reported for mixed cultures (20, 23) and for D. tiedjei (10). Other reports stated that molybdate did not inhibit dechlorinating activity in mixed cultures and even neutralized the inhibitory effect of sulfate (19, 26). This neutralization of sulfate inhibition by molybdate was explained by interspecies competition for hydrogen between dechlorinating and sulfate-reducing bacteria that was shifted in favor of the dechlorinating bacteria by the addition of molybdate (26). The isolation from the latter culture of Desulfitobacterium hafniense (6), which is a sulfite- but not sulfate-reducing, spore-forming, pentachlorophenol-dechlorinating bacterium, is in accordance with this explanation. Within our consortium, low concentrations of molybdate also neutralized inhibition by sulfate. However, competition for electron donors cannot explain this result, because 1 mM molybdate in the absence of sulfate completely inhibited reductive dechlorination, even in the presence of 1 mM sulfide as a source of sulfur. A possible explanation including many of the observed effects is that the dechlorinating organism is a sulfate-reducing bacterium that does not perform sulfate reduction at a molybdate concentration of 1 mM. The chlorobenzene-dechlorinating enzyme system may be separated spatially from the sulfate-reducing enzyme system and may be inhibited by a high ratio of molybdate to sulfate. Nevertheless, the possibility that sulfate reduction and trichlorobenzene dechlorination are performed by separate species cannot be excluded completely. The differentiation of sulfate-reducing and dechlorinating bacteria by use of molybdate during enrichment was not possible with the microbial consortium.
Most known bacteria dechlorinating chloroaromatic compounds are phylogenetically affiliated with the genus Desulfitobacterium. All of them are gram-positive rods; with one exception (18), they form heat resistant endospores (5, 27, 39, 48); but none of them uses sulfate as a terminal electron acceptor. Within our consortium, only a coccus stains gram positive, the dechlorinating activity is not sensitive to penicillin G, the dechlorinating activity is irreversibly inactivated by pasteurization, and no dechlorination occurs as long as sulfate is present in the medium. Therefore, there is no indication that the dechlorinating bacteria in our consortium are related to the genus Desulfitobacterium.
Other dechlorinating bacteria described in the literature are members of the delta subclass of Proteobacteria (7, 11). By use of in situ hybridization techniques to monitor the presence of Proteobacteria within the culture, bacteria of the gamma and delta subclasses of Proteobacteria were found in major portions. No cells were detected by use of probes targeted at different specificity levels to the phylogenetic position of D. tiedjei. The detection limit of the in situ hybridization technique at 0.1% of the population corresponded to 104 cells/ml. However, because the concentrations of trichlorobenzenes in the medium were low, even small subpopulations could take part in reductive dechlorination, and these might not have been detectable by in situ hybridization.
With mixed bacterial cultures, a number of different substrates have been reported to promote reductive dechlorination of chlorobenzenes (1, 16, 22, 31). Our stable consortium uses pyruvate for growth; however, pyruvate cannot be the actual electron donor, since it was depleted when dechlorination started. The strict sequential use of sulfate and trichlorobenzenes as electron acceptors by our consortium allowed all available electrons to be scavenged when 3 mM sulfate was added. After fermentation of pyruvate, sulfate reduction continued with formate and hydrogen until those electron donors were depleted. Under these conditions, no trichlorobenzenes were dechlorinated. Since the addition of hydrogen or acetate did not result in the formation of dichlorobenzenes, the possibility that these compounds served as electron donors in the dechlorination process can be excluded. The conclusion that hydrogen is not involved in dechlorination is supported by a number of other experiments in which the addition of hydrogen did not increase the extent of dechlorination. In contrast, the addition of formate led to the formation of dichlorobenzenes, indicating that in our consortium formate is used as a direct electron donor for the reductive dechlorination of trichlorobenzenes.
Attempts to isolate a pure culture reductively dechlorinating trichlorobenzenes make use of formate as an effective electron donor and low concentrations of sulfate as a possible alternative electron acceptor for dechlorinating bacteria. Since low concentrations of molybdate in the presence of sulfate inhibited the reduction of sulfate but not trichlorobenzene dechlorination, we now use these characteristics as selective isolation conditions. We also hope that the results presented help in the evaluation of anaerobic chlorobenzene-dechlorinating processes at natural sites and remediation plants.
ACKNOWLEDGMENTS
We thank A. Zapf, Institut für Lebensmittelchemie, Technische Universität Berlin, for analysis of chlorobenzenes and P. Wendler for expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 193, Biological Treatment of Industrial Wastewaters.
REFERENCES
- 1.Adrian L, Manz W, Szewzyk U, Görisch H. Etablierung einer stabilen, Trichlorbenzol dechlorierenden Mischkultur und deren partielle Populationsbeschreibung mit Hilfe rRNA-gerichteter Oligonukleotidsonden. GWF Wasser-Abwasser. 1996;137:612–618. [Google Scholar]
- 2.Bergmeyer H U. Methods of enzymatic analysis. 3rd ed. Weinheim, Germany: Verlag Chemie; 1984. [Google Scholar]
- 3.Beurskens J E M, Dekker C G C, van den Heuvel H, Swart M, de Wolf J, Dolfing J. Dechlorination of chlorinated benzenes by an anaerobic microbial consortium that selectively mediates the thermodynamic most favorable reactions. Environ Sci Technol. 1994;28:701–706. doi: 10.1021/es00053a026. [DOI] [PubMed] [Google Scholar]
- 4.Bosma T N P, van der Meer J R, Schraa G, Tros M E, Zehnder A J B. Reductive dechlorination of all trichloro- and dichlorobenzene isomers. FEMS Microbiol Ecol. 1988;53:223–229. [Google Scholar]
- 5.Bouchard B, Beaudet R, Villemur R, McSween G, Lépine F, Bisaillon J-G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. doi: 10.1099/00207713-46-4-1010. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen N, Ahring B K. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int J Syst Bacteriol. 1996;46:442–448. [Google Scholar]
- 7.Cole J R, Cascarelli A L, Mohn W W, Tiedje J M. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl Environ Microbiol. 1994;60:3536–3542. doi: 10.1128/aem.60.10.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cord-Ruwisch R. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods. 1985;4:33–36. [Google Scholar]
- 9.Denger K, Kertesz M A, Vock E H, Schön R, Mägli A, Cook A M. Anaerobic desulfonation of 4-tolylsulfonate and 2-(4-sulfophenyl)butyrate by a Clostridium sp. Appl Environ Microbiol. 1996;62:1526–1530. doi: 10.1128/aem.62.5.1526-1530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeWeerd K A, Concannon F, Suflita J M. Relationship between hydrogen consumption, dehalogenation, and the reduction of sulfur oxyanions by Desulfomonile tiedjei. Appl Environ Microbiol. 1991;57:1929–1934. doi: 10.1128/aem.57.7.1929-1934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeWeerd K A, Mandelco L, Tanner R S, Woese C R, Suflita J M. Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic dehalogenating, sulfate-reducing bacterium. Arch Microbiol. 1990;154:23–30. [Google Scholar]
- 12.Dolfing J. Reductive dechlorination of 3-chlorobenzoate is coupled to ATP production and growth in an anaerobic bacterium, strain DCB-1. Arch Microbiol. 1990;153:264–266. doi: 10.1007/BF00249079. [DOI] [PubMed] [Google Scholar]
- 13.Dolfing J, Harrison B K. Redox and reduction potentials as parameters to predict the degradation pathway of chlorinated benzenes in anaerobic environments. FEMS Microbiol Ecol. 1993;13:23–29. [Google Scholar]
- 14.Drzyzga O, Jansen S, Blotevogel K-H. Mineralization of monofluorobenzoate by a diculture under sulfate-reducing conditions. FEMS Microbiol Lett. 1994;116:215–219. doi: 10.1111/j.1574-6968.1994.tb06703.x. [DOI] [PubMed] [Google Scholar]
- 15.Fathepure B Z, Tiedje J M, Boyd S A. Reductive dechlorination of hexachlorobenzene to tri- and dichlorobenzenes in anaerobic sewage sludge. Appl Environ Microbiol. 1988;54:327–330. doi: 10.1128/aem.54.2.327-330.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fathepure B Z, Vogel T M. Complete degradation of polychlorinated hydrocarbons by a two-stage biofilm reactor. Appl Environ Microbiol. 1991;57:3418–3422. doi: 10.1128/aem.57.12.3418-3422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genthner B, Sharak R, Price II W A, Pritchard P H. Anaerobic degradation of chloroaromatic compounds in aquatic sediments under a variety of enrichment conditions. Appl Environ Microbiol. 1989;55:1466–1471. doi: 10.1128/aem.55.6.1466-1471.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerritse J, Renard V, Pedro Gomes T M, Lawson P A, Collins M D, Gottschal J C. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch Microbiol. 1996;165:132–140. doi: 10.1007/s002030050308. [DOI] [PubMed] [Google Scholar]
- 19.Gibson S A, Suflita J M. Anaerobic biodegradation of 2,4,5-trichlorophenoxyacetic acid in samples from a methanogenic aquifer: stimulation by short-chain organic acids and alcohols. Appl Environ Microbiol. 1990;56:1825–1832. doi: 10.1128/aem.56.6.1825-1832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Häggblom M M, Young L Y. Chlorophenol degradation coupled to sulfate reduction. Appl Environ Microbiol. 1990;56:3255–3260. doi: 10.1128/aem.56.11.3255-3260.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Häggblom M M, Young L Y. Anaerobic degradation of halogenated phenols by sulfate-reducing consortia. Appl Environ Microbiol. 1995;61:1546–1550. doi: 10.1128/aem.61.4.1546-1550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holliger C, Schraa G, Stams A J M, Zehnder A J B. Enrichment and properties of an anaerobic mixed culture reductively dechlorinating 1,2,3-trichlorobenzene to 1,3-dichlorobenzene. Appl Environ Microbiol. 1992;58:1636–1644. doi: 10.1128/aem.58.5.1636-1644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennes C, Wu W-M, Bhatnagar L, Zeikus J G. Anaerobic dechlorination and mineralization of pentachlorophenol and 2,4,6-trichlorophenol by methanogenic pentachlorophenol-degrading granules. Appl Microbiol Biotechnol. 1996;44:801–806. doi: 10.1007/BF00178622. [DOI] [PubMed] [Google Scholar]
- 24.Kohring G-W, Zhang X, Wiegel J. Anaerobic dechlorination of 2,4-dichlorophenol in freshwater sediments in the presence of sulfate. Appl Environ Microbiol. 1989;55:2735–2737. doi: 10.1128/aem.55.10.2735-2737.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn E P, Townsend G T, Suflita J M. Effect of sulfate and organic carbon supplements on reductive dehalogenation of chloroanilines in anaerobic aquifer slurries. Appl Environ Microbiol. 1990;56:2630–2637. doi: 10.1128/aem.56.9.2630-2637.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madsen T, Aamand J. Effects of sulfuroxy anions on degradation of pentachlorophenol by a methanogenic enrichment culture. Appl Environ Microbiol. 1991;57:2453–2458. doi: 10.1128/aem.57.9.2453-2458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madsen T, Licht D. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl Environ Microbiol. 1992;58:2874–2878. doi: 10.1128/aem.58.9.2874-2878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 30.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. Abundance and spatial organization of gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol., in press.
- 31.Middeldorp P J M, de Wolf J, Zehnder A J B, Schraa G. Enrichment and properties of a 1,2,4-trichlorobenzene-dechlorinating methanogenic microbial consortium. Appl Environ Microbiol. 1997;63:1225–1229. doi: 10.1128/aem.63.4.1225-1229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohn W W, Tiedje J M. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowak J, Kirsch N-H, Hegemann W, Stan H-J. Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture isolated from Saale river sediment. Appl Microbiol Biotechnol. 1996;45:700–709. [Google Scholar]
- 34.Oliver B G, Nicol K D. Chlorobenzenes in sediments, water, and selected fish from lakes Superior, Huron, Erie, and Ontario. Environ Sci Technol. 1982;16:532–536. doi: 10.1021/es00114a014. [DOI] [PubMed] [Google Scholar]
- 35.Oremland R S, Capone D G. Use of specific inhibitors in biogeochemistry and microbial ecology. Adv Microb Ecol. 1988;10:285–383. [Google Scholar]
- 36.Pfennig N. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int J Syst Bacteriol. 1978;28:283–288. [Google Scholar]
- 37.Ramanand K, Balba M T, Duffy J. Reductive dehalogenation of chlorinated benzenes and toluenes under methanogenic conditions. Appl Environ Microbiol. 1993;59:3266–3272. doi: 10.1128/aem.59.10.3266-3272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 39.Sanford R A, Cole J R, Löffler F E, Tiedje J M. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling the oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl Environ Microbiol. 1996;62:3800–3808. doi: 10.1128/aem.62.10.3800-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelton D R, Tiedje J M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 42.Stahl D A, Amann R I. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Sequencing and hybridization techniques in bacterial systematics. Chichester, England: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 43.Stahl D A, Devereux R, Amann R I, Flesher B, Lin C, Stromley J. Ribosomal RNA based studies of natural microbial diversity and ecology. In: Hattori T, Ishida Y, Maruyama Y, Morita R, Uchida A, editors. Recent advances in microbial ecology. Tokyo, Japan: Japan Scientific Societies Press; 1989. pp. 669–673. [Google Scholar]
- 44.Stan H-J, Kirsch N H. GC-FID determination of chlorobenzene isomers in methanogenic batch-cultures from river sediments. Int J Environ Anal Chem. 1995;60:33–40. [Google Scholar]
- 45.Steward C C, Dixon T C, Chen Y P, Lovell C R. Enrichment and isolation of a reductively debrominating bacterium from the burrow of a bromometabolite-producing marine hemichordate. Can J Microbiol. 1995;41:637–642. [Google Scholar]
- 46.Townsend G T, Suflita J M. Influence of sulfur oxyanions on reductive dehalogenation activitis in Desulfomonile tiedjei. Appl Environ Microbiol. 1997;63:3594–3599. doi: 10.1128/aem.63.9.3594-3599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tschech A, Pfennig N. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch Microbiol. 1984;137:163–167. [Google Scholar]
- 48.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 49.Zehnder A J B, Wuhrmann K. Titanium(III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]